Abstract
Crimean-Congo hemorrhagic fever (CCHF) virus is the cause of an important tick-borne disease of humans throughout regions of Africa, Europe, and Asia. Like other members of the genus Nairovirus, family Bunyaviridae, the CCHF virus M genome RNA segment encodes the virus glycoproteins. Sequence analysis of the CCHF virus (Matin strain) M RNA segment revealed one major open reading frame that potentially encodes a precursor polyprotein 1,689 amino acids (aa) in length. Comparison of the deduced amino acid sequences of the M-encoded polyproteins of Nigerian, Pakistani, and Chinese CCHF virus strains revealed two distinct protein regions. The carboxyl-terminal 1,441 aa are relatively highly conserved (up to 8.4% identity difference), whereas the amino-terminal 243 to 248 aa are highly variable (up to 56.4% identity difference) and have mucin-like features, including a high serine, threonine, and proline content (up to 47.3%) and a potential for extensive O-glycosylation. Analysis of released virus revealed two major structural glycoproteins, G2 (37 kDa) and G1 (75 kDa). Virus protein analysis by various techniques, including pulse-chase analysis and/or reactivity with CCHF virus-specific polyclonal and antipeptide antibodies, demonstrated that the 140-kDa (which contains the mucin-like region) and 85-kDa nonstructural proteins are the precursors of the mature G2 and G1 proteins, respectively. The amino termini of the CCHF virus (Matin strain) G2 and G1 proteins were established by microsequencing to be equivalent to aa 525 and 1046, respectively, of the encoded polyprotein precursor. The tetrapeptides RRLL and RKPL are immediately upstream of the cleavage site for mature G2 and G1, respectively. These are completely conserved among the predicted polyprotein sequences of all the CCHF virus strains and closely resemble the tetrapeptides that represent the major cleavage recognition sites present in the glycoprotein precursors of arenaviruses, such as Lassa fever virus (RRLL) and Pichinde virus (RKLL). These results strongly suggest that CCHF viruses (and other members of the genus Nairovirus) likely utilize the subtilase SKI-1/S1P-like cellular proteases for the major glycoprotein precursor cleavage events, as has recently been demonstrated for the arenaviruses.
Crimean-Congo hemorrhagic fever (CCHF) viruses are members of the genus Nairovirus of the family Bunyaviridae. The nairoviruses are predominantly tick-borne viruses. The two most important serogroups are the CCHF virus group, which includes CCHF and Hazara viruses, and the Nairobi sheep disease group, which includes Nairobi sheep disease and Dugbe viruses (21). CCHF virus was first described in the 1940s, when more than 200 cases of severe hemorrhagic fever occurred among agricultural workers in the Crimean peninsula (6). CCHF virus is now known to be present throughout much of sub-Saharan Africa (33, 43), Bulgaria, the former Yugoslavia, northern Greece, the former Soviet Union (particularly the former Central Asian republics), the Arabian peninsula, Iraq, Pakistan, and Xinjiang Province in northwest China.
CCHF case fatality is approximately 30%, with most deaths occurring 5 to 14 days after onset of illness (37). Humans commonly become infected by a bite from or contact with infected ticks or by contact with blood or tissues of infected livestock. Most humans who become infected live or work in close contact with livestock (sheep, goats, cattle, or ostriches) in areas where CCHF virus is endemic. Medical personnel can also become infected following treatment of or surgery on a patient with unsuspected CCHF virus.
The geographic distribution of CCHF virus cases corresponds most closely with the distribution of Hyalomma ticks, suggesting their principal vector role. Although other ixodid ticks can be infected, only some tick species of the Hyalomma, Dermacentor, and Rhipicephalus genera have been shown to be capable of transstadial transmission (i.e., passing the virus from larva to nymph to adult) of CCHF virus after feeding on a viremic host. Transovarial transmission (i.e., passage of virus to offspring) of CCHF virus has also been shown to occur with some of the tick species in these genera. Although virus can persist in ticks, vertebrates are needed to provide blood meals for the ticks, and they can become infected and develop viremias capable of supporting virus transmission to uninfected ticks (12, 19, 32). A variety of livestock (sheep, goats, cattle, and ostriches), large wild herbivores, hares, and hedgehogs can become infected with CCHF virus, and in contrast to human infections, these infections generally result in inapparent or subclinical disease (14, 30, 31, 36).
The RNA genome of members of the family Bunyaviridae consists of three negative-sense segments, S, M, and L, which minimally encode the virus nucleocapsid, glycoproteins, and L polymerase proteins, respectively (28). The M segment generally encodes two structural glycoproteins, G1 and G2. In addition, members of the genera Bunyavirus, Phlebovirus, and Tospovirus also encode a nonstructural glycoprotein referred to as NSM. The glycoproteins of viruses of the genus Nairovirus have so far been poorly characterized. Hazara virus, a member of the CCHF virus serogroup, was shown to possess three structural glycoproteins of 84, 45, and 30 kDa (11). Similarly, Clo Mor virus (Sakhalin serogroup) appeared to contain three virion-associated glycoproteins that were 90, 80, and 45 kDa (41, 42). However, like most members of the family Bunyaviridae, Qalyub virus (Qalyub serogroup) appeared to possess only two structural glycoproteins, 75 and 40 kDa (7). The Clo Mor and Qalyub viruses have also been shown to contain additional glycoproteins in cell lysates which appear, by use of pulse-chase analysis, to be precursors of the mature viral glycoproteins.
The most extensively studied glycoproteins of the Nairovirus genus are those of Dugbe virus (Nairobi sheep disease serogroup), in which two structural glycoproteins (73 and 35 kDa) were observed (5, 8, 20). Results obtained from N-terminal amino acid sequence analysis of the slower-migrating protein (G1) and experiments with antibodies targeting amino acids within the N-terminal portion of the predicted M segment open reading frame (ORF) polyprotein showed the order of the coding sequence to be G2 followed by G1. In addition, it was noted that the N terminus of G1 was about 50 amino acids (aa) downstream of the nearest predicted signal sequence, a feature not seen in glycoproteins of viruses of other genera of the family Bunyaviridae. Like the Clo Mor and Qalyub viruses, the Dugbe virus M segment appears to generate at least one precursor protein. In virus-infected cells, 110- and 85-kDa proteins were shown to share epitopes with G1, but only the 85-kDa protein demonstrated a precursor relationship to G1. In addition, a 70-kDa nonstructural protein was shown to possess amino acids in common with G2 and may also be a possible precursor protein. Within the family Bunyaviridae, such glycoprotein precursor-product relationships appear to be unique to viruses of the Nairovirus genus.
Of the members of the Nairovirus genus, CCHF virus is the most pathogenic for humans. The virus glycoproteins likely influence the vertebrate and tick host usage and cell tropism of the virus and the high pathogenicity of the virus during human infections. The purpose of this study was to identify the glycoproteins encoded by the CCHF virus genome and understand their processing patterns. We have deduced the nucleotide sequence of the entire M genome RNA segment of the Matin strain of CCHF virus (isolated from a patient in Pakistan), compared this sequence with those available in GenBank for all other viruses of the genus Nairovirus, and identified various conserved motifs and features. These include CCHF virus strain IbAr10200 (M. D. Parker, P. J. Glass, G. B. Jennings, R. Lofts, J. F. Smith, M. M. Miller, K. W. Spik, and R. Schoepp, unpublished data) and the more recent Chinese strains BA8402 and BA66019 (23) and Dugbe virus (20). At the amino terminus of the CCHF virus-encoded polyprotein, a highly variable mucin-like region was identified, a feature of nairoviruses that appears to be unique among other members of the family Bunyaviridae. The biosynthesis of glycoproteins in cells infected with CCHF virus strains Matin and IbAr10200 was also analyzed. In parallel, the complete M segment ORF of CCHF virus strain IbAr10200 was cloned into an expression vector, and the expression and processing of the glycoproteins were analyzed and compared with those of CCHF virus-infected cells. In addition, the exact N termini of both the G1 and G2 proteins were identified, and the experimentally determined cleavage sites as well as predicted cleavage recognition sites are discussed.
MATERIALS AND METHODS
Viruses, cell lines, and antibodies.
CCHF virus strains IbAr10200 (originally isolated from Hyalomma excavatum ticks from Sokoto, Nigeria, in 1966) and Matin (originally isolated from a patient in Pakistan in 1976) were used in this study. Viruses were grown in Vero E6 or SW-13 cells. Cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 2% fetal bovine serum and antimicrobial agents. All work with CCHF virus was conducted in the biosafety level 4 laboratory at the Centers for Disease Control and Prevention (CDC). SW-13 cells were used in transfection experiments for expression of CCHF virus strain IbAr10200 glycoproteins. These cells were maintained in DMEM supplemented with 10% fetal bovine serum. Vaccinia virus expressing bacteriophage T7 RNA polymerase was grown in HeLa cells, and titers were determined in CV-1 cells.
Hyperimmune mouse ascitic fluid (HMAF) raised against the CCHF virus IbAr10200 strain was provided by Thomas Ksiazek (CDC). Anthony Sanchez (CDC) provided antibodies to the mucin region of CCHF virus IbAr10200 strain. These antibodies were produced by replacing the mucin region of Ebola virus GP (inserted in an expression vector) with that of CCHF virus IbAr10200 strain and injecting the plasmid DNA into mice. The exact region of the IbAr10200 strain glycoprotein ORF used in these vaccinations included aa 17 to 243, which eliminated most of the predicted signal peptide at the N terminus and stopped immediately preceding the potential furin cleavage site RSKR (see Results). Monoclonal antibody against the nucleocapsid (N) protein of CCHF virus IbAr10200 strain was kindly provided by Jonathan Smith (U.S. Army Medical Research Institute of Infectious Diseases). Rabbit antipeptide antibodies were designed based on deduced amino acid sequences of various regions of the IbAr10200 strain polyprotein sequence and were generated under contract with Research Genetics. These included antibodies to aa 540 to 551 in the G2 region (EIHGDNYGGPGD) and aa 1388 to 1399 in the G1 region (ETDYTKNFHFHS) (see Results).
Analysis of the nucleotide sequence of CCHF virus Matin strain M segment and comparison with that of other CCHF virus strains.
In order to target the M segment RNA termini of viruses belonging to the genus Nairovirus, initial primers were designed on the basis of M segment sequences available in GenBank for other viruses in the genus Nairovirus. At this phase of the project, these included CCHF virus IbAr10200 strain (U39455) and Dugbe virus (M94133). The primer designed to amplify the 5′ (cDNA sense) termini was named NAIRO5 (5′-TCTCAAAGAIATACITGCGGCAC-3′), and the primer for the 3′ termini was termed NAIRO3 (5′-TCTCAAAGAIAIIGTGGCGGCA-3′), with I representing inosine. RNA was isolated from Vero E6 cells infected with CCHF virus Matin strain with TriPure reagent (Boehringer Mannheim), purified with chloroform-isoamyl alcohol (24:1), and precipitated with isopropyl alcohol. Reverse transcription of the entire M RNA segment was performed with 5 μl of RNA, Nairovirus terminus primers (300 ng each), SuperScript II (Gibco-BRL) reverse transcriptase (RT; 200 U), 2 μl of 0.1 M dithiothreitol [DTT], 1 μl of a mix of the four deoxynucleoside triphosphates (10 mM each ), 4 μl of 5× RT buffer, and RNasin (40 U) in a 20-μl reaction mixture. The reaction mixture was incubated at 42°C for 2 h and then boiled for 3 min. PCR was done with 2 μl of the cDNA, 2 μl of the mix of the four deoxynucleoside triphosphates (10 mM each), 10 μl of primers (10 pmol/μl), 10 μl of dimethyl sulfoxide, 10 μl of 10× buffer with 15 mM MgCl2, and 2.6 U of Expand High-Fidelity polymerase (Boehringer Mannheim) in a 100-μl reaction mixture.
PCR thermocycler conditions were used as recommended by the manufacturer, with an annealing temperature of 55°C. PCR primers included the Nairovirus terminus primers and internal primers designed from the CCHF virus IbAr10200 strain sequence. As more CCHF virus Matin strain sequence became available, additional primers were designed and utilized. Nucleotide sequences were generated, aligned, and compared as described previously (27). Available virus M segment sequences used in comparisons included those for Dugbe virus (M94133), CCHF virus Nigerian strain IbAr10200, isolated from a tick (U39455), and CCHF virus Chinese strains BA8402, isolated from a tick (AF350449), and BA66019, isolated from a CCHF virus patient (AF350448).
Prediction servers provided by the Center for Biological Sequence Analysis, BioCentrum-DTU, Technical University of Denmark, were used to further analyze amino acid sequences. Prediction of transmembrane helices in proteins was done with TMHMM, version 2.0 (16). SignalP (version 1.1) allowed prediction of signal sequence cleavage sites (22). Predictions of mucin-type GalNAc O-glycosylation sites were conducted with the NetOGlyc 2.0 prediction server (13).
Preparation and analysis of CCHF virus particles and infected cell proteins.
Vero E6 or SW-13 cells were infected with CCHF virus IbAr10200 or Matin strain at a multiplicity of infection of approximately 0.3 or less. The virus proteins were radiolabeled approximately 17 to 24 h postinfection with 100 to 200 μCi of [35S]cysteine or 100 μCi of [3H]leucine (NEN) per ml for various labeling and chase periods, which are indicated in the figure legends. Cell monolayers, supernatants, and viral pellets were then harvested. Cell monolayers were lysed with 1% Triton X-100 in TNE buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 3 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). Supernatant was harvested and analyzed directly or used for virus pelleting. Virus was pelleted through a 20% sucrose cushion in SW41 or SW40 tubes at 30,000 rpm for 4 to 5 h at 4°C. Pellet was resuspended in 1% sodium dodecyl sulfate (SDS) and either left untreated or digested with endoglycosidase H (50 mU in sodium acetate buffer, pH 5.5) at 37°C overnight (Boehringer Mannheim). Samples were loaded onto an SDS-10% polyacrylamide gel electrophoresis (PAGE) system as previously described (10).
Radiolabeled cell lysates and supernatants were used for radioimmunoprecipitation analysis. Briefly, protein A-Sepharose CL-4B (Amersham Pharmacia Biotech) was incubated with antibody in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Triton X-100, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate) for 2 h at 4°C and then radiolabeled virus samples were added and incubated for another 2 h. Protein A-Sepharose beads were washed first in RIPA buffer, followed by buffer A (50 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 0.5% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS, 0.5% bovine serum albumin) and then buffer B (50 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 0.5% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS). Samples were loaded onto 10% NuPAGE Bis-Tris or 3 to 8% Tris-acetate precast gel systems (Invitrogen).
For Western blot analysis, virus infections and harvesting of cell monolayers and supernatants were conducted in SW-13 cells as stated above except without the addition of radiolabel. Supernatants were concentrated by vacuum-dry centrifugation. Samples were electrophoresed on 3 to 8% NuPAGE Tris-acetate gels. Proteins were blotted onto nitrocellulose membranes with a semidry blotting system (Sigma). Membranes were probed with polyclonal antibodies specific for the mucin region of CCHF virus IbAr10200 strain glycoprotein and antipeptide antibodies generated against distinct peptide sequences within the deduced G1 and G2 protein sequences of the CCHF virus IbAr10200 strain. Peroxidase-labeled goat anti-rabbit and anti-mouse immunoglobulin G (Kirkegaard and Perry Laboratories), and 3,3′-diaminobenzidine were used to visualize the reactive proteins.
N-terminal sequencing of CCHF virus (Matin strain) proteins.
The specific identity of individual virus glycoprotein bands was analyzed by N-terminal sequencing. Briefly, unlabeled virus pelleted from the supernatants of CCHF (Matin strain) virus-infected Vero E6 cells were subjected to SDS-10% PAGE, and the gel was then soaked in 3-cyclohexylamino-1-propanesulfonic acid (CAPS) transfer buffer (10 mM CAPS, 0.5 mM DTT, 10% methanol [pH 11.0]). A polyvinylidene difluoride membrane was rinsed in methanol and then soaked with filter paper in CAPS buffer. Transfer of proteins to the membrane was done with a semidry blotting apparatus (Sigma). The membrane was then soaked in 1 mM DTT and stained with 0.1% Ponceau S containing 1 mM DTT in order to visualize the protein bands. Excised protein bands were subjected to N-terminal sequencing, which was performed on a model 492 Procise protein sequencing system (Applied Biosystems/Perkin-Elmer Corporation) by using the Edman degradation technique.
Cloning of CCHF virus IbAr10200 strain M segment ORF.
Primers to amplify the virus M segment ORF were designed on the basis of available CCHF virus IbAr10200 strain M segment sequence (U39455). In addition, SalI and KpnI restriction sites were incorporated on the primers that would amplify the region encoding the protein's N terminus and C terminus, respectively. PCR products representing the two halves of the ORF were generated. The N-terminal and C-terminal PCR products were cloned into a pCR2.1 vector (Invitrogen), and three independent clones of each were sequenced to verify correct insertion. For each half, all three clones were identical, but a small number of sequence differences relative to the reported CCHF virus IbAr10200 strain M segment ORF (U39455) were noted. The differences amounted to 10 nucleotides (nt) and 8 aa substitutions in the compared ORF sequences, excluding the primer-derived sequences, which contributed to the 23 nt and 25 nt at the beginning and end of the ORF, respectively.
For protein expression studies, the two cDNA halves were joined with a shared NdeI restriction site present within the ORF sequence, and the full-length ORF insert was subcloned into a pBluescript II KS vector (Stratagene) with the incorporated SalI and KpnI sites flanking the glycoprotein ORF. The ligated plasmid was then transformed into Escherichia coli DH5α competent cells (Life Technologies). While growing the bacteria under normal conditions at 37°C, it was found that the inserted gene appeared to be unstable, resulting in expression of proteins of aberrant molecular masses. In order to maintain the stability of the plasmid, original plasmid DNA (pCR2.1) with the correct sequence was transformed into E. coli STBL2 competent cells (Life Technologies), following the manufacturer's recommendations. Bacteria were grown at 30°C, and DNA was isolated with DNA purification columns (Qiagen).
In order to characterize the features of the G1 protein, the region from nt 2880 to the end of the glycoprotein ORF were amplified by primers G1F (5′-CACCATGGAAGTAAGTAAC-3′) and G1R (5′-GGTACCCTAGCCAATGTGTGTTTTTGT-3′). The resulting PCR products were cloned into a pCDNA3.1 Directional Topo expression vector (Invitrogen). This construct utilized an ATG start codon immediately preceding the closest upstream hydrophobic region from the N terminus of G1 (see Results). This hydrophobic region was included to function as the signal peptide for the G1 construct. STBL2 competent cells were used for the transformation experiments and were grown at 30°C. A G1 clone was obtained, and the nucleotide sequence was verified.
Transfection, radioimmunoprecipitation, and protein analyses.
Transfection and analyses of virus protein expression were performed as previously described (39). Briefly, SW-13 cells (5 × 105) were infected with vaccinia virus vTF7-3 (multiplicity of infection = 5) for 60 min. Upon removal of the virus inoculum, DNA (5 μg) complexed with Lipofectamine Plus reagent (5 μg) (Life Technologies) was added to the cells and incubated at 37°C in a CO2 incubator for 15 h. The cells were then placed in cysteine-free medium for 60 min, followed by labeling with 100 μCi of [35S]cysteine (New England Nuclear) for different periods. The label was removed, and the cells were further chased in DMEM containing 10% fetal bovine serum for the indicated time periods. The labeled cells were then lysed with RIPA buffer with 1 mM phenylmethylsulfonyl fluoride, and the clarified supernatants were immunoprecipitated with CCHF virus-specific polyclonal or antipeptide antibodies. Protein A-Sepharose beads were added to the immune complexes and incubated for 3 to 15 h. After extensive washing, the proteins were resolved with a 10% NuPAGE gel system (Invitrogen) and analyzed by autoradiography.
Nucleotide sequence accession number.
The CCHF virus IbAr10200 strain ORF sequence represented in our clone has been deposited in GenBank under accession number AF467768.
RESULTS
General features of the CCHF virus Matin strain M segment sequence and comparison with other strains.
The complete nucleotide sequence of the M segment of CCHF virus Matin strain was determined (GenBank accession no. AF467769) with the exception of the terminal primer regions (the first 23 nt at the 5′ end [cDNA sense] and the last 22 nt at the 3′ end). The M segment of the Matin strain is 5,367 nt in length (assuming no deletions or insertions in the primer regions) and encodes a polyprotein 1,689 aa in length. The CCHF virus Matin strain M segment nucleotide and deduced amino acid sequences were compared with those available for other CCHF virus strains (Table 1). The Chinese strain BA88166 sequence (AF338470; B.-J. Ma, C.-S. Hang, and Q. Tang, unpublished data) was not included because it was virtually identical to that of strain BA8402.
TABLE 1.
Comparison of CCHF virus Matin strain glycoprotein sequence with that of other CCHF virus strainsa
| Sequence | Strain | % Differences
|
|||
|---|---|---|---|---|---|
| Matin | 10200 | BA8402 | BA66019 | ||
| M segment ORF | Matin | 18.9 | 7.0 | 15.5 | |
| (5,070 nt, 1,689 aa) | 10200 | 14.0 | 19.3 | 18.5 | |
| BA8402 | 6.2 | 14.8 | 15.7 | ||
| BA66019 | 11.7 | 14.8 | 12.1 | ||
| Variable region (N- | Matin | 35.3 | 14.3 | 26.9 | |
| terminal 248 aa) | 10200 | 53.5 | 37.2 | 34.2 | |
| BA8402 | 23.8 | 56.4 | 28.4 | ||
| BA66019 | 38.7 | 53.1 | 42.3 | ||
| Conserved region | Matin | 16.1 | 5.7 | 13.5 | |
| (C-terminal 1,441 aa) | 10200 | 7.3 | 16.3 | 15.9 | |
| BA8402 | 3.2 | 7.8 | 13.5 | ||
| BA66019 | 7.0 | 8.4 | 6.9 | ||
Sequence lengths are given relative to the Matin strain sequence. Values above the diagonal in each group are percent nucleotide (nt) differences in identity; values below the diagonals are percent amino acid (aa) differences in identity.
The IbAr10200 strain M segment sequence was found to be the most divergent of the CCHF virus strain sequences, with the virus exhibiting sequence identity differences of 18.5 to 19.3% within the M segment ORF nucleotide sequence and 14.0 to 14.8% within the encoded amino acid sequence (Table 1). The closest sequence relationship was seen between CCHF virus Matin and BA8402 strains, with identity differences of 7.0% in the M segment ORF nucleotide sequence and 6.2% in the full-length encoded glycoprotein amino acid sequence. In contrast, 62.9 to 63.4% difference in amino acid identity was seen on comparison of the M segment-encoded CCHF virus polyprotein with that of Dugbe virus.
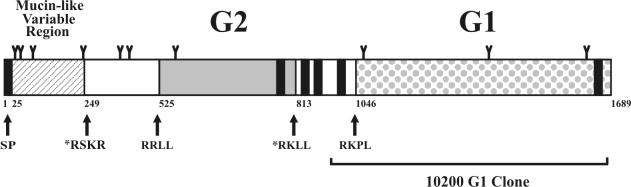
The encoded polyprotein sequence appears to possess a signal peptide at the amino terminus, with signalase cleavage predicted to occur between aa 24 and 25 (SEG-IH) for the CCHF virus Matin strain. The predicted cleavage sites for the other CCHF viruses are between aa 27 and 28 in BA8402 (SHG-LS), aa 21 and 22 in BA66019 (LWS-LE), and aa 22 and 23 in IbAr10200 (THG-SH). Five additional membrane-spanning regions are predicted for the CCHF virus polyprotein (Fig. 1). The deduced Dugbe virus M segment-encoded polyprotein is predicted to have a similar pattern of transmembrane regions.
FIG. 1.
Schematic representation of the glycoprotein ORF of CCHF virus Matin strain. SP represents the signal peptide predicted by the prediction server SignalP V1.1 (22). Cleavage of the signal peptide for the Matin strain was predicted to occur between aa 24 and 25 (SEG-IH) based on neural networks trained on eukaryotic data. The remaining five black bars represent transmembrane helices within the glycoprotein ORF with the prediction server TMHMM 2.0 (16). With outside meaning the lumenal side, TMhelix meaning transmembrane helix, and inside meaning the cytoplasmic side, the precise locations of the various regions of the Matin strain glycoprotein ORF predicted by the TMHMM server are as follows: aa 1 to 701 (outside), aa 702 to 724 (TMhelix), aa 725 to 825 (inside), aa 826 to 848 (TMhelix), aa 849 to 857 (outside), aa 858 to 880 (TMhelix), aa 881 to 973 (inside), aa 974 to 996 (TMhelix), aa 997 to 1599 (outside), aa 1600 to 1622 (TMhelix), and aa 1623 to 1689 (inside). The tetrapeptide RRLL (aa 521 to 524) precedes the confirmed N terminus of G2, and RKPL (aa 1042 to 1045) precedes the confirmed N terminus of G1, and they are presumed to be cleavage recognition sites possibly used by the pyrolysin-like subtilase SKI-1/S1P. The asterisks denote potential cleavage recognition sites: RSKR, aa 249 to 252 (furin) and RKLL, aa 809 to 812 (SKI-1). Note, however, that RKLL resides on the cytoplasmic side of the membrane. Y-shaped projections represent predicted N-linked glycosylation sites. The region of amino acid sequence included in an expression construct of the G1 region of CCHF virus IbAr10200 strain is indicated (G1 plus the 79 aa upstream, which includes the nearest predicted hydrophobic transmembrane domain to act as a signal peptide). Numbers below the bar graph are amino acid positions. Schematic diagram not drawn to scale.
Further analysis of the CCHF virus Matin strain polyprotein predicted 10 potential N-linked glycosylation sites, not including those present in predicted hydrophobic membrane-spanning domains or those predicted to be on the cytoplasmic side of the membrane (Fig. 1). One of these sites contains an aspartic acid (NDT), which may reduce the likelihood of this site being utilized. Six of the 10 sites are conserved among all the CCHF virus strains. The entire CCHF virus Matin strain-encoded polyprotein contains a remarkable 79 cysteines, all of which are conserved among all the CCHF viruses, suggesting extensive disulfide linkages.
More detailed comparison of the CCHF virus amino acid sequences revealed the N-terminal 243 to 248 aa to be highly variable, a feature also recently noted by Papa et al. (23). This variability is quite striking when this region alone is compared with the remainder of the M segment-encoded polyprotein. For the variable region, the nucleotide differences observed among the CCHF virus strains were 14.3 to 37.2% (Table 1). This translated to a surprisingly high (23.8 to 56.4%) difference in amino acid identity for the encoded protein region. In contrast, nucleotide differences for the remainder of the ORF were 5.7 to 16.3%, with the encoded protein region displaying only 3.2 to 8.4% difference in amino acid identity (Table 1). Analysis of the Dugbe virus M segment-encoded polyprotein relative to that of the CCHF viruses indicated that Dugbe virus possesses an equivalent highly variable region, although it is approximately 155 aa shorter.
More detailed analysis of the variable region showed that the serine, threonine, and proline (S, T, and P) content in this region is especially high, being 43.1 to 47.3%, compared with 18.7 to 19.2% in the remainder of the polyprotein (Table 2). Comparison of the ST content alone shows 29.0 to 34.7% for the variable domain and 15.2 to 15.5% in the more conserved remainder of the protein. For the variable region, 52.3 to 62.5% of the total ST content was predicted to possess O-linked glycosylation, in contrast to only 2.7 to 4.0% in the conserved region (Table 2). A graphic representation of the extent of O-glycosylation predicted for the CCHF virus Matin strain is shown in Fig. 2. A heavy concentration of O-glycosylation within the variable domain, particularly aa 54 to 247, can be seen. A small area of the more conserved region (aa 331 to 344) also displays a high potential for O-linked glycosylation. These features suggest that this N-terminal region of the polyprotein may represent a mucin-like domain. A similar mucin-like domain predicted to be rich in O-linked glycosylation can be seen in the Dugbe virus M segment-encoded polyprotein, but it is significantly smaller (only 47 aa) relative to that of the CCHF viruses (194 aa in the Matin strain).
TABLE 2.
O-glycosylation predictions of CCHF virus glycoproteina
| Region | Strain | Composition (% of total aa)
|
O-glycosylated S,T (% of total S,T) | ||
|---|---|---|---|---|---|
| S,T,P | S,T | O-glycosylated S,T | |||
| Variable | Matin | 47.2 | 33.5 | 17.7 | 53.0 |
| 10200 | 47.3 | 32.9 | 20.6 | 62.5 | |
| BA8402 | 46.4 | 34.7 | 18.1 | 52.3 | |
| BA66019 | 43.1 | 29.0 | 17.7 | 61.1 | |
| Conserved | Matin | 19.2 | 15.5 | 0.4 | 2.7 |
| 10200 | 18.7 | 15.4 | 0.4 | 2.7 | |
| BA8402 | 19.2 | 15.5 | 0.6 | 4.0 | |
| BA66019 | 18.7 | 15.2 | 0.6 | 3.7 | |
aa, amino acids; S, serine; T, threonine; P, proline; O-glycosylated S,T, percentage of serines and threonines predicted to be O-glycosylated.
FIG. 2.
Predicted O-glycosylation sites in CCHF virus Matin strain glycoprotein ORF. Prediction results were generated with the prediction server NetOGlyc 2.0, which produces neural network predictions of mucin type GalNAc O-glycosylation sites in mammalian proteins (13). Where the potential is greater than the threshold, O-glycosylation is predicted for that site. The higher the potential, the higher the confidence of the prediction.
CCHF virus encodes two major structural glycoproteins.
To identify the structural glycoproteins encoded by the CCHF virus M RNA segment, CCHF virus (Matin strain) virus-infected Vero E6 cell cultures were labeled with [35S]cysteine, and virus was pelleted from the infected cell supernatant. The viral pellets were either untreated or digested with endoglycosidase H and analyzed by SDS-10% PAGE (Fig. 3A). Three prominent bands were visible for the untreated virions (Fig. 3A, lane 1). The mobilities of these bands correspond to proteins of approximately 37, 55, and 75 kDa. Immunoprecipitation experiments with a monoclonal antibody specific for the CCHF virus N protein identified the 55-kDa protein as N (data not shown). Consistent with this, the 55-kDa band did not shift in mobility on treatment with endoglycosidase H (Fig. 3A, lane 2). The two other prominent protein bands (37 and 75 kDa) showed a shift in migration when treated with endoglycosidase H, suggesting that these were glycoproteins possessing N-linked glycosylation (Fig. 3A, lane 2). On digestion with endoglycosidase H, the approximately 37-kDa protein band shifted to a faster-migrating, more diffuse band which appeared to contain two major forms. While the protein with the faster mobility obviously possesses glycans sensitive to endoglycosidase H digestion, the slower-migrating form may possess complex endoglycosidase H-resistant modifications (such as those occurring in the medial Golgi). The 75-kDa protein shifted to a faster-migrating form following digestion with endoglycosidase H, again indicating the presence of high-mannose or hybrid structures. In keeping with the nomenclature used for the glycoproteins of other members of the family Bunyaviridae, the 37-kDa protein (faster migrating on PAGE analysis) will be referred to as G2, and the 75-kDa protein (slower migrating) will be referred to as G1.
FIG. 3.
(A) Autoradiograph of CCHF virus (Matin strain) pellets grown in Vero E6 cells labeled overnight with [35S]cysteine and either untreated or digested with endoglycosidase H. Virus samples were resolved on SDS-10% PAGE. (B) Lanes 1 and 2, autoradiograph of CCHF virus (IbAr10200 strain) virus-infected SW-13 cells and supernatant (Sup) labeled overnight with [35S]cysteine, immunoprecipitated with polyclonal HMAF against CCHF virus IbAr10200 strain, and resolved on a 10% NuPAGE gel system. Lanes 3 and 4, same as lanes 1 and 2 except samples were resolved on 3 to 8% NuPAGE. Lane 5, autoradiograph of supernatant from CCHF (Matin strain) virus-infected SW-13 cells labeled overnight with [3H]leucine, immunoprecipitated as in lanes 1 to 4, and resolved on 10% NuPAGE.
CCHF virus mature G2 and G1 glycoproteins are produced by proteolytic processing of the M segment-encoded polyprotein.
In order to identify the N terminus of the G2 and G1 glycoproteins expressed by CCHF virus Matin strain, unlabeled virus pellets were prepared and proteins were separated on SDS-10% PAGE and blotted onto a polyvinylidene difluoride membrane. The G2 and G1 proteins were visualized with Ponceau S and excised for N-terminal sequencing analysis. The N termini of the G2 and G1 proteins were determined to begin at aa 525 and 1046, respectively, relative to the sequence of the Matin strain M segment encoded polyprotein (shown diagrammatically in Fig. 1). Since the signal peptide for the polyprotein is predicted to be aa 1 through 24 and the N terminus of G2 is 500 aa downstream of the predicted signalase cleavage site, additional protease processing must be occurring to generate the mature G2 protein (Fig. 1). Also, no other predicted membrane-spanning hydrophobic domains are present in this 500-aa region of the polyprotein, indicating that the N terminus is not generated by an additional signalase cleavage event. Similarly, the N terminus of the mature G1 is located approximately 50 aa downstream of the nearest predicted membrane-spanning hydrophobic domain, suggesting that it is also not generated from a precursor protein by signalase cleavage (Fig. 1). Analysis of the Dugbe virus M segment-encoded polyprotein suggests a similar processing pattern for the Dugbe virus mature G1 and G2 proteins.
On further analysis of the protease cleavage sites that must be utilized to generate the N termini of the CCHF virus mature glycoproteins, it was noted that the tetrapeptides RRLL and RKPL immediately precede the G2 and G1 cleavage sites, respectively. In addition, these tetrapeptides are conserved in the glycoprotein sequences of all the CCHF virus strains examined to date. Interestingly, although the N terminus of the Dugbe virus G2 has not been directly determined, by alignment with the CCHF virus sequences, the Dugbe virus G2 N terminus would be predicted to be at aa 375 and be immediately preceded by the tetrapeptide RKLL. In addition, the Dugbe virus G1 N terminus has been determined previously (20). It aligns well with the G1 N terminus of the CCHF virus and is also immediately preceded by the tetrapeptide RKLL. These data suggest that proteolytic cleavage events involved in the generation of the N termini of the G2 and G1 proteins may be common for viruses of the genus Nairovirus in general. More surprisingly, similar tetrapeptides have recently been implicated in the major protease cleavage events involved in the processing of the arenavirus GPC glycoprotein precursor into the mature GP1 and GP2 proteins. The tetrapeptides RRLL and RKLL immediately precede the known GPC cleavage sites of Lassa and Pichinde viruses, respectively (2, 18), and the cellular protease subtilase SKI-1/S1P carries out the cleavage, at least in the case of the RRLL site in Lassa virus (17, 38). In addition, Guanarito virus (another arenavirus), the cause of Venezuelan hemorrhagic fever, possesses the tetrapeptide RKPL (identical to that seen in the CCHF virus glycoprotein precursor) immediately prior to the predicted GPC cleavage site (34). These conserved features suggest similarities in glycoprotein processing between viruses of the genus Nairovirus, family Bunyaviridae, and viruses of the family Arenaviridae and implicate SKI-1 or related proteases in the major glycoprotein cleavage events for CCHF and Dugbe viruses.
The CCHF virus M segment-encoded polyprotein sequences were analyzed further for any other potential cleavage sites. The sequence RKLL was found in all of the CCHF virus sequences analyzed (aa 809 to 812 for the Matin strain) at a position in the polyprotein that could potentially be the C terminus of G2 and result in a predicted G2 protein comparable in size to the 37-kDa G2 protein observed on PAGE analysis (Fig. 1). However, no similar sequence was observed in the comparable region of the Dugbe virus polyprotein sequence. One additional potential cleavage site observed in all CCHF virus sequences was the sequence RSKR (aa 249 to 252 for the Matin strain), a previously described furin protease cleavage site (reviewed in reference 15). However, a similar sequence was absent in the comparable region of the Dugbe virus polyprotein, and no data are available to suggest that this site is utilized.
CCHF virus glycoproteins in infected cells and supernatants.
To identify potential CCHF virus glycoprotein precursor proteins present within infected cell cytoplasm as well as virus proteins that may be secreted from virus-infected cells, SW-13 cells were infected with CCHF virus IbAr10200 strain and labeled with [35S]cysteine. The viral proteins were then immunoprecipitated with polyclonal HMAF raised against CCHF virus IbAr10200 strain and visualized on 10% and 3 to 8% NuPAGE gel systems (Fig. 3B). As seen previously in [35S]cysteine-labeled virions, the G1, N, and G2 proteins were clearly seen in the infected cell supernatants (Fig. 3B, lanes 2 and 4). In addition, a 160-kDa protein was also seen (Fig. 3B, lane 4).
Two additional virus proteins of approximately 85 and 140 kDa (referred to as P85 and P140, respectively) were observed within infected cells (Fig. 3B, lanes 1 and 3). While the 140-kDa and 160-kDa proteins have similar mobilities on a 10% gel (Fig. 3B, lanes 1 and 2), they can be distinguished as individual proteins on a 3 to 8% gel (Fig. 3B, lanes 3 and 4). The observation of P85 and P140 in infected cells but not in virions or cell supernatants suggests that these proteins may be precursor proteins. Analysis of the CCHF virus glycoprotein amino acid sequences revealed that the variable mucin-like region (minus the predicted signal peptide and assuming the furin recognition site RSKR is cleaved) (Fig. 1) contains no cysteines. Based on this observation, the CCHF virus Matin strain was labeled with [3H]leucine instead of [35S]cysteine, and infected supernatants were immunoprecipitated with polyclonal HMAF to look for any unique protein (structural or secreted) that was not observed previously. However, no additional proteins were observed, with only the P160, G1, N, and G2 proteins being visible (Fig. 3B, lane 5).
Processing of virus glycoproteins.
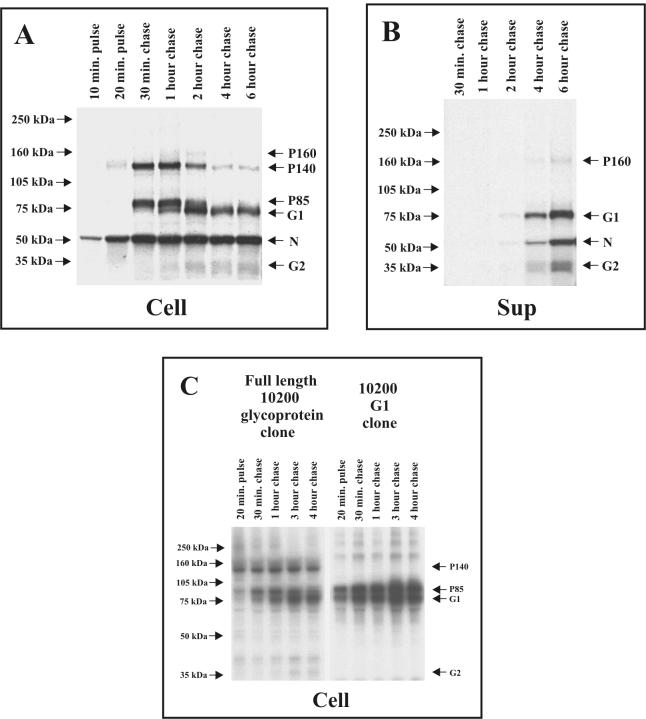
To determine if the proteins present only in infected cells undergo any kind of processing, pulse-chase experiments were conducted. SW-13 cells were infected with IbAr10200 strain virus, pulse-labeled with [35S]cysteine for 10 or 20 min, and then chased for 0.5, 1, 2, 4, or 6 h (Fig. 4A and B). The virus N protein was already detectable in infected cells following the 10-min pulse. Both N and P140 were detected by the 20-min pulse. The maximum amount of P140 was detectable during the 30-min and 1-h chases and then began to decrease. Similarly, P85 was detectable as early as the 30-min chase, peaked at the 1-h chase time point, and then decreased to undetectable levels by the 4-h chase time point. Neither of these proteins was detectable in the supernatant. These data are consistent with P140 and P85 representing precursor proteins.
FIG. 4.
(A and B) Autoradiographs of pulse-chase analyses of CCHF virus-infected cell cultures. SW-13 cells were infected with CCHF virus IbAr10200 strain, pulse-labeled with [35S]cysteine, and chased with unlabeled medium for the indicated time periods. Cell lysates (Cell) and supernatants (Sup) were harvested, immunoprecipitated with polyclonal HMAF against CCHF virus IbAr10200 strain, and then resolved on a 3 to 8% NuPAGE gel system. (C) Autoradiograph of pulse-chase analyses of cell lysates from transfected cell cultures. SW-13 cells were transfected with the CCHF virus IbAr10200 strain full-length glycoprotein ORF expression construct and an expression plasmid encoding for G1 plus the upstream hydrophobic region to function as a signal peptide. Cells were pulse-labeled with [35S]cysteine and chased for the indicated time periods. Cell lysates were harvested, immunoprecipitated with polyclonal HMAF against CCHF virus IbAr10200 strain, and resolved on a 10% NuPAGE system.
As P140 and P85 levels decreased, G2 and G1 protein levels increased, suggesting a precursor-product relationship. G2 and G1 were detectable in the infected cells at the 1-h chase time point and began to appear in the cell supernatant by 2 h of chase. The P160 protein was detectable in the cells only at the 2-h chase time point. P160 began to appear in the supernatant at the 4-h chase time point and increased by the 6-h chase time point. This suggests that the disappearance of P160 from the infected cell and its subsequent appearance in the supernatant were due to its release into the medium either in virions or as a secreted protein.
To further characterize the CCHF virus glycoproteins and their processing, an expression plasmid was constructed which contained the entire CCHF virus IbAr10200 strain M segment ORF inserted within pBluescript II KS. SW-13 cells were transfected with the CCHF virus ORF clone and pulse-labeled with [35S]cysteine for 20 min, followed by various lengths of unlabeled chase periods. Following immunoprecipitation with polyclonal HMAF raised against CCHF virus IbAr10200 strain, the expressed virus proteins were analyzed on 3 to 8% gels (Fig. 4C). Results similar to those obtained with virus-infected cells were observed. P140 was detected as early as the 20-min pulse time point, with amounts beginning to decrease following the 4-h chase. Also, P85 was detectable at the 20-min pulse time point, with amounts decreasing by 4 h of chase. These results are consistent with those obtained with CCHF virus-infected cell lysates and supernatants and suggest that P140 and P85 are glycoprotein precursors. G1 could be readily detected in the 30-min chase and continued to increase through subsequent chase periods. G2 was more difficult to visualize but could be detected after 3 and 4 h of chase.
To more precisely examine the relationship of P140 and P85 to G1 and G2, a plasmid was constructed that would express only the G1 coding region of the IbAr10200 strain plus the first hydrophobic region upstream of the N-terminal cleavage site for G1 (Fig. 1). This region (approximately 50 aa upstream of G1) was included to act as a signal peptide for the G1 protein. None of the ORF region that would encode G2 or any polypeptides N terminal to G2 was included. When this G1 clone was expressed in SW-13 cells, the P140 and G2 proteins were no longer detected. However, both P85 and G1 proteins were detected, suggesting that P140 is likely a precursor to G2, whereas P85 is a precursor of G1 (Fig. 4C). These results indicate that P85 may be processed from the larger M ORF polyprotein by signalase cleavage following the hydrophobic domain immediately upstream of G1, with subsequent cleavage by a subtilase SKI-1-like protease to produce the mature G1.
Analysis with individual glycoprotein-specific antibodies.
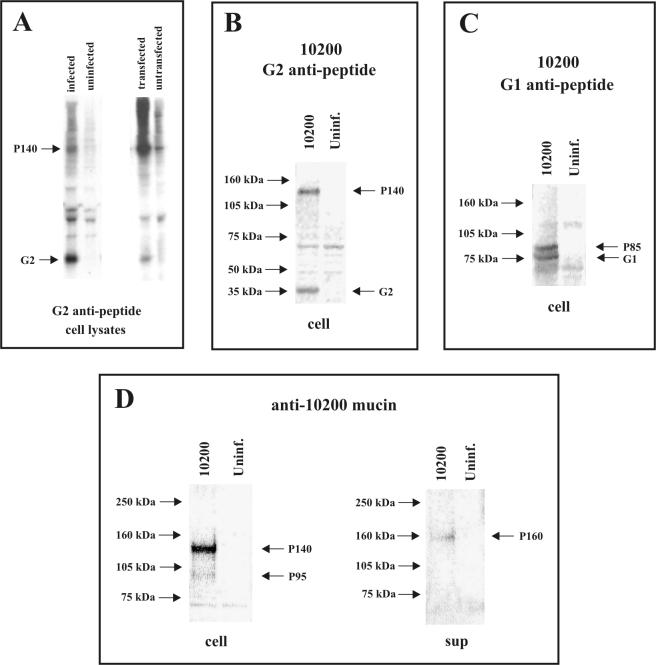
To verify the identities of the various individual glycoproteins seen in CCHF virus-infected cell and supernatant preparations, we used a variety of specific antibodies targeting various regions of the virus M segment-encoded polyprotein. SW-13 cells were either infected with the CCHF virus IbAr10200 strain or transfected with the full-length IbAr10200 strain glycoprotein clone and radiolabeled with [35S]cysteine. A G2-specific antipeptide antibody (generated against aa 540 to 551 of the IbAr10200 strain ORF sequence) was used in immunoprecipitation assays of cell lysates. Autoradiographs of the precipitated proteins resolved on a 10% NuPAGE gel showed reactivity of the G2 antipeptide to the 37-kDa G2 protein as expected (Fig. 5A). In addition, the P140 protein was also precipitated by the G2 antipeptide antibody.
FIG. 5.
(A) Autoradiograph of [35S]cysteine-labeled SW-13 cell lysates which were infected with CCHF virus IbAr10200 strain or transfected with the IbAr10200 full-length glycoprotein ORF construct and immunoprecipitated with G2 antipeptide. Antipeptide antibody was generated against aa 540 to 551 of the IbAr10200 strain glycoprotein sequence and was therefore G2 specific. Immunoprecipitated proteins were resolved on 10% NuPAGE. (B and C) Western blot analysis of cell lysates harvested from CCHF (IbAr10200 strain) virus-infected SW-13 cell cultures, resolved on 3 to 8% NuPAGE and reacted with antipeptide antibodies. Panel B shows cell lysates reacted with the same G2-specific antipeptide antibody used in panel A. Panel C shows cell lysates reacted with a G1-specific antipeptide antibody raised against aa 1388 to 1399 of the IbAr10200 strain sequence. (D) Western blot analysis of cell lysates (cell) and supernatants (sup) harvested from SW-13 cells infected with CCHF virus IbAr10200 strain, resolved on 3 to 8% NuPAGE and reacted with a mucin-specific antibody. The mucin-specific antibody was generated by DNA immunization of mice with a chimeric expression construct containing sequence from the variable mucin-like region of the IbAr10200 strain (without most of the predicted signal peptide and preceding the RSKR tetrapeptide).
To determine whether coprecipitation of G2 and P140 was due to the presence of G2 peptides within the P140 protein or if it was due simply to close interaction of P140 and G2, Western blot analysis was done, allowing separation of the proteins before reaction to the G2 antipeptide antibody. Unlabeled virus proteins for this experiment were generated by infecting SW-13 cells with CCHF virus IbAr10200 strain and harvesting cell lysates. Results of this analysis on a 3 to 8% NuPAGE gel showed G2 antipeptide antibody reactivity to both G2 and P140, confirming the presence of G2 peptides in both G2 and P140 (Fig. 5B). These results, in combination with the results obtained by pulse-chase analyses, strongly implicate P140 as a precursor protein to G2.
The same unlabeled CCHF virus (IbAr10200 strain) infected cell lysates were also analyzed by Western blot analysis on a 3 to 8% gel with an antipeptide antibody generated against aa 1388 to 1399 of the CCHF virus IbAr10200 strain glycoprotein sequence, which specifically targets G1. Results showed reactivity to 75- and 85-kDa proteins similar to the 75-kDa G1 and P85 observed in radioimmunoprecipitation assays of cell lysates, indicating that P85 contains G1 peptides (Fig. 5C). The pulse-chase results showing P85 processing into G1, in addition to these Western blot results indicating G1 sequence within P85, provide compelling evidence that P85 is a precursor protein of G1.
Polyclonal antibodies to the variable mucin-like region of the IbAr10200 strain of CCHF virus were generated in mice (see Materials and Methods). These antibodies were also used in our Western blot studies of CCHF virus-infected cultures. Following infection of SW-13 cells with the IbAr10200 strain virus, cell lysates and supernatants were harvested and analyzed on a 3 to 8% gel with the antibody targeting the CCHF virus IbAr10200 strain mucin region. Results showed reactivity of the antibody with the P140 protein in infected cells and the P160 protein in the supernatant, indicating the presence of the mucin-like domain in both proteins (Fig. 5D). Our results also revealed a possible second mucin-associated protein with mobility corresponding to approximately 95 kDa (P95) in addition to P140 in infected cells. P95 may represent a cleavage product of P140, but further studies are needed to examine this possibility more thoroughly.
DISCUSSION
While the M segments and their encoded glycoproteins have been well characterized for viruses of other genera of the family Bunyaviridae, much less is known for the viruses of the genus Nairovirus. In the case of CCHF virus, the difficulty of working with the virus and the containment conditions (biosafety level 4) required have resulted in few studies being carried out even though the virus is an important cause of hemorrhagic fever in humans throughout broad areas of Africa, Europe, and Asia. Here we present the complete nucleotide sequence of the M segment of the Matin strain of CCHF virus, which was originally isolated from a patient in Pakistan in 1976. Together with the recently available M segment sequences for Nigerian and Chinese CCHF virus strains (23), this now provides insight into the extent of M segment and glycoprotein diversity of CCHF viruses over much of the geographic range of the virus and the time span from 1965 to 1984. High diversity is evident, with up to 19.3% nucleotide sequence variation and 14.8% amino acid differences observable among the virus M segment and glycoprotein precursor proteins, respectively. The presence of distinct virus genotypes in different geographic areas correlates well with observations based on comparison of partial CCHF virus S segment nucleotide sequences (25), although greater sequence diversity is observed in the M RNA segment and encoded proteins than in the S segment and nucleocapsid protein. This diversity, particularly in the glycoprotein sequences, likely reflects the use of different principal vector tick hosts and virus-amplifying vertebrate species in the virus life cycle in the different geographic regions. For instance, Hyalomma marginatum ticks are thought to play an important role in West Africa, whereas Hyalomma asiaticum is the implicated tick species in China (3, 37, 44).
The published data concerning the synthesis and processing of the glycoproteins of the viruses of the genus Nairovirus provide a rather complex and inconsistent picture. Hazara and Clo Mor viruses appear to possess three structural glycoproteins encoded by their M genome RNA segments, while only two glycoproteins have been described for Qalyub and Dugbe viruses, all with various electrophoretic mobilities (5, 7, 8, 11, 20, 41, 42). One of the striking features revealed on analysis of the M segment-encoded polyprotein of the CCHF viruses was the presence of two distinct regions: a highly variable region, which includes the N-terminal 243 to 248 aa of the protein, and a relatively conserved region, which encompasses the remainder of the protein. The variable N-terminal domain varied by up to approximately 37% at the nucleotide level and 56% at the amino acid level. As indicated by the excess of amino acid substitutions relative to nucleotide substitutions, most of the nucleotide changes are nonsynonymous, resulting in a rapidly evolving variable region compared with the more conserved region. The driving force behind this rapid evolution of CCHF virus glycoproteins may be positive selection for the virus to evade the host immune system (9).
The similarities of this N-terminal variable region with host cell mucins were also quite unexpected. The high percentage of serines, threonines, and prolines (up to 47.3%), many of which were conserved among the CCHF virus strains, and the predicted extensive O-glycosylation would indicate that the mucin-like feature must play some important structural or functional role in the CCHF virus life cycle. The presence of a similar mucin-like domain near the N terminus of the Dugbe virus M segment-encoded polyprotein, albeit much shorter (47 aa) than that seen in CCHF viruses, suggests that this property may be conserved broadly among viruses of the genus Nairovirus. No evidence of any similar mucin-like regions could be identified on analysis of glycoprotein sequences of representative members of the other four genera of the Bunyaviridae family, suggesting that this feature is unique to members of the genus Nairovirus.
The function of this variable mucin-like region is currently unknown. There are no cysteine residues within this region (the N-terminal 243 to 248 aa, excluding the predicted signal peptide), suggesting the lack any disulfide linkages. Mucins are described as large glycoproteins with a high degree of O-glycosylation on serines and threonines (4, 35). This abundance of O-glycosylation may offer carbohydrate heterogeneity, which could enable viruses to bind to a variety of molecules (1). The presence of many prolines within mucin-like sequences, which can induce β-turn conformations, probably allows close packing of O-linked oligosaccharides. A large abundance of these closely packed O-linked sugars may offer protection from proteases. Curiously, the glycoprotein of Ebola virus, another human pathogen causing hemorrhagic fever, also possesses a variable and highly O-glycosylated mucin-like region within the middle third of its glycoprotein sequence (26). The mucin-like domain of Ebola virus glycoprotein has been shown to be a major determinant of pathogenicity, as expression of Ebola virus glycoprotein in explanted human and porcine blood vessels caused massive endothelial cell loss and subsequent substantial increase in vascular permeability, while deletion of the mucin-like domain prevented these effects (45). It remains to be seen if the mucin-like domain of CCHF virus has similar pathological functions.
The cumulative data presented here demonstrate that CCHF virus possesses two major virion-associated glycoproteins, G1 and G2, and that these are encoded by the virus M RNA segment in the order 5′-G2-G1-3′. This is consistent with earlier findings for Dugbe virus (20). The data for CCHF virus, together with that available for Dugbe virus, demonstrate that glycoprotein synthesis and processing for the members of the genus Nairovirus are quite distinct from those of members of the other family Bunyaviridae genera. For the viruses of the other genera, the mature G1 and G2 proteins are immediately preceded by a signal peptide sequence, and there is no evidence of a polyprotein precursor, suggesting that cotranslational cleavage of the G1 and G2 proteins of these viruses occurs via a cellular signalase (28). In contrast, within the Dugbe virus polyprotein precursor sequence, the only potential signal sequence for G1 is located about 50 aa upstream of the amino terminus of the mature G1 protein (20). The existence of a precursor protein for G1 was supported by the identification of an 85-kDa protein that shared epitopes with G1 and was found to be processed to G1 during pulse-chase studies of Dugbe virus-infected cells. In our study, we show that the N terminus of the mature G1 protein of CCHF virus is also located approximately 50 aa downstream of the closest potential signal peptide hydrophobic region. Similarly, we were unable to identify any obvious signal peptide immediately upstream of the confirmed N terminus of the CCHF virus mature G2. Obviously, translocation of G2 into the endoplasmic reticulum lumen must be affected by a hydrophobic region that lies further upstream.
Virus protein analysis by various techniques, including pulse-chase analysis and reactivity with CCHF virus-specific polyclonal and antipeptide antibodies, demonstrated that the P140 (which contains the mucin-like region) and P85 nonstructural proteins are the precursors of the mature G2 and G1 proteins, respectively. P140 appears to represent the protein region from the mucin-like domain through the G2 region and possibly to the cleavage site generating the N terminus of the P85 precursor of G1 (Fig. 1). A variety of lines of evidence, particularly the synthesis of P85 by the G1 expression plasmid, suggest that P85 extends from about 50 aa upstream of the mature G1 N terminus to the C terminus of G1 (presumably aa 1689 relative to the Matin strain). The N terminus of P85 is likely generated from the polyprotein by cotranslational signalase cleavage prior to subsequent processing into the mature G1.
The N termini of the CCHF virus Matin strain G2 and G1 proteins were determined by microsequencing to be equivalent to aa 525 and 1046 of the encoded polyprotein precursor, respectively. The tetrapeptides RRLL and RKPL are immediately upstream of the N termini of mature G2 and G1, respectively, and were shown to be completely conserved among the predicted polyprotein sequences of all the CCHF virus strains. In addition, Dugbe virus possesses the tetrapeptide RKLL immediately upstream of the confirmed G1 cleavage site (20) and the predicted G2 cleavage site (aa 375). These tetrapeptides are identical to or closely resemble the tetrapeptides that represent the major cleavage recognition sites present in the glycoprotein precursors of arenaviruses, such as Lassa fever virus (RRLL) and Pichinde virus (RKLL). The glycoprotein precursor GPC of Lassa virus, another hemorrhagic fever virus, is posttranslationally processed into GP1 and GP2 by proteolytic cleavage at a site preceded by the cleavage motif RRLL (18). Mutations in this tetrapeptide sequence inhibited the processing.
In addition, recent studies have shown that Lassa virus GPC cleavage occurs in the endoplasmic reticulum and is mediated by SKI-1/S1P, a protease belonging to the pyrolysin branch of subtilases, following the RRLL sequence (17). This unique protease plays a distinct role in cholesterol metabolism (24) and is involved in the generation of neuroactive peptides (29). The presence of the sequence RKLL immediately preceding the Pichinde GPC cleavage site suggests that SKI-1 or a related protease is also involved in this cleavage. In addition, the RKPL tetrapeptide motif seen in the CCHF virus glycoprotein precursor also precedes the predicted GPC cleavage site in Guanarito virus, an arenavirus responsible for hemorrhagic fever in Venezuela (34). These results strongly suggest that CCHF viruses (and other members of the genus Nairovirus) likely utilize the subtilase SKI-1/S1P-like cellular proteases for the major glycoprotein precursor cleavage events that generate the N termini of the G2 and G1 mature glycoproteins.
Since G2 is estimated to be approximately 37 kDa, based on PAGE mobility, and the N terminus is known to be located at aa 525 for CCHF virus Matin strain, the RKLL tetrapeptide located at aa 809 to 812 may represent the G2 protein C terminus, which may also be generated by an SKI-1-like protease cleavage event (Fig. 1). This tetrapeptide is conserved in all the CCHF virus strains, although not in the equivalent region of the Dugbe virus sequence. However, if one examines the six potential membrane-spanning hydrophobic domains, beginning with the N-terminal signal peptide, the RKLL tetrapeptide at aa 809 to 812 would be predicted to lie on the cytoplasmic side of the membrane, assuming the protein transits the membrane at each of the hydrophobic domains (Fig. 1). If this model is correct, it is difficult to see how this tetrapeptide could be a protease cleavage site. Further studies will be needed to more precisely determine the C terminus of the CCHF virus G2 protein and the mechanism of its generation.
CCHF virus G1 protein probably extends from aa 1046 (for Matin strain) to the end of the glycoprotein ORF (Fig. 1), since a predicted transmembrane domain is present at the C terminus to act as a membrane anchor, followed by an approximately 65-aa cytoplasmic tail. Such a protein would be compatible with the 75-kDa estimate that was determined on the basis of the mobility of G1 on PAGE analysis.
Multiple sequence alignments of CCHF viruses also revealed the presence of a conserved tetrapeptide motif, RSKR, at the junction of the variable mucin-like region and the remainder of the glycoprotein. This motif conforms to the tetrapeptide consensus sequence R-X-K/R-R recognized by furin, an enzyme belonging to the kexin family of subtilases and involved in the cleavage of a number of glycoproteins in the Golgi apparatus (reviewed in reference 15). Although furin activation has been shown to occur for several viral glycoproteins, including human immunodeficiency virus type 1, influenza virus, and Ebola virus (40), there is currently no evidence that furin cleavage occurs at the RSKR site in the CCHF virus glycoprotein precursor. If furin cleavage did occur, the resulting products (aa 25 to 252 [assuming the predicted signal peptide is cleaved off] and aa 253 to 524, relative to the Matin strain sequence) would likely be soluble proteins due to the lack of identifiable transmembrane domains. These may be secreted or bound to a membrane structural protein. However, additional experiments are in progress to further examine potential furin cleavage.
An additional protein, P160, was observed in virus-infected cells and supernatants and found to react with CCHF virus (IbAr10200 strain) virus-specific polyclonal HMAF in immunoprecipitation experiments and with mucin-specific antibodies in Western blot analyses. This may represent a more glycosylated form of the precursor protein P140, as additional O-linked glycans are added late in the secretory pathway. However, P160 may represent a distinct protein, since it would be unexpected to find a precursor protein in the supernatant. Further studies are under way to attempt to determine more precisely the origin of this protein.
The data presented here for CCHF virus together with earlier studies on other viruses of the genus Nairovirus show that their M segment polyprotein processing and mature G1 and G2 synthesis are much more complex than those for viruses of the other genera of the family Bunyaviridae. Our study also uncovered unsuspected similarities between the glycoproteins of CCHF virus and those of other hemorrhagic fever viruses. These included the identification of a glycoprotein mucin-like domain similar to that present in Ebola virus and the likely use of SKI-1-like proteases for the major glycoprotein proteolytic processing events, similar to Lassa fever and Guanarito viruses. With the foundation provided by this study, further studies to more precisely define and understand the complexity and significance of these elements of CCHF virus glycoproteins should be possible.
Acknowledgments
We thank Anthony Sanchez for sharing the chimeric IbAr10200 mucin antibody with us and for invaluable input at the initial stages of this project. We also thank Jonathan Smith for sharing his knowledge and graciously providing us with monoclonal antibody and Tamara Crews of the Scientific Resources Program for protein microsequencing.
REFERENCES
- 1.Brockhausen, I. 1995. Biosynthesis of O-glycans of the N-acetylgalactosamine-α-ser/thr linkage type, p. 201-259. In J. Montreuil, J. F. G. Vliegenthart, and H. Schachter (ed.), Glycoproteins. Elsevier Science, New York, N.Y.
- 2.Burns, J. W., and M. J. Buchmeier. 1993. Glycoproteins of the arenaviruses, p. 17-35. In M. S. Salvato (ed.), The arenaviridae. Plenum Press, New York, N.Y.
- 3.Camicas, J. L., J. P. Cornet, J. P. Gonzalez, M. L. Wilson, F. Adam, and H. G. Zeller. 1994. Crimean-Congo hemorrhagic fever in Senegal — present status of the knowledge on the ecology of the CCHF virus. Bull. Soc. Pathol. Exot. 87:11-16. [PubMed] [Google Scholar]
- 4.Carraway, K. L., and S. R. Hull. 1991. Cell surface mucin-type glycoproteins and mucin-like domains. Glycobiology 1:131-138. [DOI] [PubMed] [Google Scholar]
- 5.Cash, P. 1985. Polypeptide synthesis of Dugbe virus, a member of the Nairovirus genus of the Bunyaviridae. J. Gen. Virol. 66:141-148. [DOI] [PubMed] [Google Scholar]
- 6.Chumakov, M. 1963. Study of viral haemorrhagic fevers. J. Hyg. Epidemiol. 7:125-140. [PubMed] [Google Scholar]
- 7.Clerx, J. P. M., and D. H. L. Bishop. 1981. Qalyub virus, a member of the newly proposed Nairovirus genus (Bunyaviridae). Virology 108:361-372. [DOI] [PubMed] [Google Scholar]
- 8.El-Ghorr, A. A., A. C. Marriott, V. K. Ward, T. F. Booth, S. Higgs, E. A. Gould, and P. A. Nuttall. 1990. Characterization of Dugbe virus by biochemical and immunochemical procedures with monoclonal antibodies. Arch. Virol. 1990(Suppl. 1):169-179. [Google Scholar]
- 9.Endo, T., K. Ikeo, and T. Gojobori. 1996. Large-scale search for genes on which positive selection may operate. Mol. Biol. Evol. 13:685-690. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann, H., S. T. Nichol, H.-D. Klenk, C. J. Peters, and A. Sanchez. 1994. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology 199:469-473. [DOI] [PubMed] [Google Scholar]
- 11.Foulke, R. S., R. R. Rosato, and G. R. French. 1981. Structural polypeptides of Hazara virus. J. Gen. Virol. 53:169-172. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, J. P., J. L. Camicas, J. P. Cornet, O. Faye, and M. L. Wilson. 1992. Sexual and transovarian transmission of Crimean-Congo haemorrhagic fever virus in Hyalomma truncatum ticks. Res. Virol. 143:23-28. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, J. E., O. Lund, N. Tolstrup, A. A. Gooley, K. L. Williams, and S. Brunak. 1998. NetOGlyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 15:115-130. [DOI] [PubMed] [Google Scholar]
- 14.Hoogstraal, H. 1979. The epidemiology of tick-borne Crimean-Congo hemorrhagic fever in Asia, Europe, and Africa. J. Med. Entomol. 15:307-417. [DOI] [PubMed] [Google Scholar]
- 15.Klenk, H.-D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 16.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 17.Lenz, O., J. ter Meulen, H.-D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenz, O., J. ter Meulen, H. Feldmann, H.-D. Klenk, and W. Garten. 2000. Identification of a novel consensus sequence at the cleavage site of the Lassa virus glycoprotein. J. Virol. 74:11418-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logan, T. M., K. J. Linthicum, C. L. Bailey, D. M. Watts, D. J. Dohm, and J. R. Moulton. 1990. Replication of Crimean-Congo hemorrhagic fever virus in four species of ixodid ticks (Acari) infected experimentally. J. Med. Entomol. 27:537-542. [DOI] [PubMed] [Google Scholar]
- 20.Marriott, A. C., A. A. El-Ghorr, and P. A. Nuttall. 1992. Dugbe Nairovirus M RNA: nucleotide sequence and coding strategy. Virology 190:606-615. [DOI] [PubMed] [Google Scholar]
- 21.Nichol, S. T. 2001. Bunyaviruses, p. 1603-1633. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 22.Nielsen, H., S. Brunak, and G. von Heijne. 1999. Machine learning approaches to the prediction of signal peptides and other protein sorting signals. Protein Eng. 12:3-9. [DOI] [PubMed] [Google Scholar]
- 23.Papa, A., B. Ma, S. Kouidou, Q. Tang, C. Hang, and A. Antoniadis. 2002. Genetic characterization of the M RNA segment of Crimean-Congo hemorrhagic fever virus strains, China. Emerg. Infect. Dis. 8:50-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawson, R. B., D. Cheng, M. S. Brown, and J. L. Goldstein. 1998. Isolation of cholesterol-requiring mutant Chinese hamster ovary cells with defects in cleavage of sterol regulatory element-binding proteins at site 1. J. Biol. Chem. 273:28261-28269. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez, L. L., G. O. Maupin, T. G. Ksiazek, P. Rollin, A. S. Khan, T. F. Schwarz, R. S. Lofts, J. F. Smith, A. M. Noor, C. J. Peters, and S. T. Nichol. 1997. Molecular investigation of a multisource outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates. Am. J. Trop. Med. Hyg. 57:512-518. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez, A., A. S. Khan, S. R. Zaki, G. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae: Marburg and Ebola viruses, p. 1279-1304. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 27.Sanchez, A. J., K. D. Abbott, and S. T. Nichol. 2001. Genetic identification and characterization of Limestone Canyon virus, a unique Peromyscus-borne hantavirus. Virology 286:345-353. [DOI] [PubMed] [Google Scholar]
- 28.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 29.Seidah, N. G., S. J. Mowla, J. Hamelin, A. M. Mamarbachi, S. Benjannet, B. B. Touré, A. Basak, J. S. Munzer, J. Marcinkiewicz, M. Zhong, J. C. Barale, C. Lazure, R. A. Murphy, M. Chretien, and M. Marcinkiewicz. 1999. Mammalian subtilase/kexin isozyme SKI-1: a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc. Natl. Acad. Sci. USA 96:1321-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shepherd, A. J., R. Swanepoel, P. A. Leman, and S. P. Shepherd. 1987. Field and laboratory investigation of Crimean-Congo haemorrhagic fever virus (Nairovirus, family Bunyaviridae) infection in birds. Trans. R. Soc. Trop. Med. Hyg. 81:1004-1007. [DOI] [PubMed] [Google Scholar]
- 31.Shepherd, A. J., R. Swanepoel, S. P. Shepherd, G. M. McGillivray, and L. A. Searle. 1987. Antibody to Crimean-Congo hemorrhagic fever virus in wild mammals from southern Africa. Am. J. Trop. Med. Hyg. 36:133-142. [DOI] [PubMed] [Google Scholar]
- 32.Shepherd, A. J., R. Swanepoel, S. P. Shepherd, P. A. Leman, and O. Mathee. 1991. Viraemic transmission of Crimean-Congo haemorrhagic fever virus to ticks. Epidemiol. Infect. 106:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson, D. I., E. M. Knight, G. Courtois, M. C. Williams, M. P. Weinbren, and J. W. Kibukamusoke. 1967. Congo virus: a hitherto undescribed virus occurring in Africa. I. Human isolations—clinical notes. East Afr. Med. J. 44:86-92. [PubMed] [Google Scholar]
- 34.Spiropoulou, C. F., S. Kunz, P. E. Rollin, K. P. Campbell, and M. B. A. Oldstone. 2002. New World arenavirus clade C, but not clade A and B, viruses utilizes alpha-dystroglycan as its major receptor. J. Virol. 76:5140-5146. [DOI] [PMC free article] [PubMed]
- 35.Strous, G. J., and J. Dekker. 1992. Mucin-type glycoproteins. Crit. Rev. Biochem. Mol. Biol. 27:57-92. [DOI] [PubMed] [Google Scholar]
- 36.Swanepoel, R., A. J. Shepherd, P. A. Leman, S. P. Shepherd, G. M. McGillivray, M. J. Erasmus, L. A. Searle, and D. E. Gill. 1987. Epidemiologic and clinical features of Crimean-Congo hemorrhagic fever in southern Africa. Am. J. Trop. Med. Hyg. 36:120-132. [DOI] [PubMed] [Google Scholar]
- 37.Swanepoel, R. 1994. Crimean-Congo haemorrhagic fever, p. 723-729. In J. A. W. Coetzer, G. R. Thomson, and R. C. Tustin (ed.), Infectious diseases of livestock, with special reference to Southern Africa. Oxford University Press, Cape Town, South Africa.
- 38.Touré, B. B., J. S. Munzer, A. Basak, S. Benjannet, J. Rochemont, C. Lazure, M. Chrétien, and N. G. Seidah. 2000. Biosynthesis and enzymatic characterization of human SKI-1/S1P and the processing of its inhibitory prosegment. J. Biol. Chem. 275:2349-2358. [DOI] [PubMed] [Google Scholar]
- 39.Vincent, M. J., N. U. Raja, and M. A. Jabbar. 1993. Human immunodeficiency virus type 1 Vpu protein induces degradation of chimeric envelope glycoproteins bearing the cytoplasmic and anchor domains of CD4: role of the cytoplasmic domain in Vpu-induced degradation in the endoplasmic reticulum. J. Virol. 67:5538-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volchkov, V. E., H. Feldmann, V. A. Volchkova, and H.-D. Klenk. 1998. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. USA 95:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watret, G. E., C. R. Pringle, and R. M. Elliott. 1985. Synthesis of bunyavirus-specific proteins in a continuous cell line (XTC-2) derived from Xenopus laevis. J. Gen. Virol. 66:473-482. [DOI] [PubMed] [Google Scholar]
- 42.Watret, G. E., and R. M. Elliott. 1985. The proteins and RNAs specified by Clo Mor virus, a Scottish Nairovirus. J. Gen. Virol. 66:2513-2516. [DOI] [PubMed] [Google Scholar]
- 43.Woodall, J. P., M. C. Williams, and D. I. Simpson. 1967. Congo virus: a hitherto undescribed virus occurring in Africa. II. Identification studies. East Afr. Med. J. 44:93-98. [PubMed] [Google Scholar]
- 44.Yan, Y. C., L. X. Kong, L. Lee, Y. Q. Zhang, F. Li, B. J. Cai, and S. Y. Gao. 1985. Characteristics of Crimean-Congo hemorrhagic fever virus (Xinjiang strain) in China. Am. J. Trop. Med. Hyg. 34:1179-1182. [PubMed] [Google Scholar]
- 45.Yang, Z.-Y., H. J. Duckers, N. J. Sullivan, A. Sanchez, E. G. Nabel, and G. J. Nabel. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6:886-889. [DOI] [PubMed] [Google Scholar]