Abstract
The emergence of antiretroviral (ARV) drug-resistant human immunodeficiency virus type 1 (HIV-1) quasispecies is a major cause of treatment failure. These variants are usually replaced by drug-sensitive ones when the selective pressure of the drugs is removed, as the former have reduced fitness in a drug-free environment. This was the rationale for the design of structured ARV treatment interruption (STI) studies for the management of HIV-1 patients with treatment failure. We have studied the origin of drug-sensitive HIV-1 quasispecies emerging after STI in patients with treatment failure due to ARV drug resistance. Plasma and peripheral blood mononuclear cell samples were obtained the day of treatment interruption (day 0) and 30 and 60 days afterwards. HIV-1 pol and env were partially amplified, cloned, and sequenced. At day 60 drug-resistant variants were replaced by completely or partially sensitive quasispecies. Phylogenetic analyses of pol revealed that drug-sensitive variants emerging after STI were not related to their immediate temporal ancestors but formed a separate cluster, demonstrating that STI leads to the recrudescence and reemergence of a sequestrated viral population rather than leading to the back mutation of drug-resistant forms. No evidence for concomitant changes in viral tropism was seen, as deduced from env sequences. This study demonstrates the important role that the reemergence of quasispecies plays in HIV-1 population dynamics and points out the difficulties that may be found when recycling ARV therapies with patients with treatment failure.
The different variants of human immunodeficiency virus type 1 (HIV-1) present in infected individuals have been described as quasispecies of related but distinct viruses; this plasticity of phenotype allows the virus to occupy a large adaptive landscape from which novel phenotypes may readily emerge (7, 17, 23). These variants are generated continuously due to the high replication rate of HIV-1 (50), the high frequency of recombination among viral genomes (4), and the lack of proofreading activity of the viral reverse transcriptase (RT) (41). When the selective pressure of antiretroviral (ARV) therapy is exerted on such a population, drug-resistant mutants may emerge and consequently lead to treatment failure (7, 37). The emergence of such viral variants has been extensively described and is found even in the setting of highly active ARV therapies (11, 39, 46). When this selective pressure is removed, the emergence of drug-sensitive quasispecies may be expected, as they would be predicted to have a higher fitness in a drug-free environment (9, 15, 19, 20, 22, 25, 34, 40, 42, 43). This rationale led to the many structured treatment interruption (STI) studies carried out on patients with treatment failure and multidrug-resistant viruses (10, 12, 16, 26, 45, 59). In those cohorts, a rise in plasma viral load and a concomitant fall in CD4+ cell count was observed after treatment interruption; in approximately half of the patients, drug susceptibility shifted from resistant to sensitive. However, the origin of these drug-sensitive quasispecies which emerged after STI has not been clearly defined. Understanding the mechanisms responsible for the observed changes in HIV-1 drug resistance and in viral replication after STI will provide a greater insight into HIV-1 evolution and will help define future therapeutic strategies for patients suffering treatment failure.
The objective of this work was to determine the origin of drug-sensitive quasispecies arising after STI. New drug-sensitive HIV variants may be the result of point mutations (“back mutations”) in the drug-resistant quasispecies circulating immediately before STI, or they may be reemerging ancestral viral variants that had circulated before the drug-resistant viruses associated with treatment failure arose (and which had been sequestrated or suppressed during therapy). In addition we assessed whether the increase in viral load and decrease in CD4+ cell count observed after STI was associated with a change in the biological phenotype of the virus from non-syncytium-inducing/CCR5-tropic to syncytium-inducing/CXCR4-tropic. Like STI, this latter phenotype has been associated with faster replication, more rapid decline in CD4 numbers, and disease progression (6, 8, 21, 28, 56). We were able to show that viral quasispecies replicating after treatment interruption were not related to its immediate temporal ancestor but formed a separate cluster, demonstrating that STI leads to the recrudescence and reemergence of a sequestrated viral population rather than the back mutation of drug-resistant forms. No evidence for concomitant changes in viral tropism was seen.
(The present study constitutes a part of Gustavo H. Kijak's doctoral work at the University of Buenos Aires.)
MATERIALS AND METHODS
Population under study.
Four patients chronically infected with HIV-1 who had treatment failure and were participating in an STI protocol for 60 days were studied. Treatment failure was defined as the presence of two consecutive samples with a viral load of more than 1,000 HIV-1 RNA copies/ml obtained 14 days apart after being on ARV therapy for more than 6 months. All patients gave informed consent before undertaking STI. Demographic, epidemiological, and clinical characteristics of the studied individuals are shown in Table 1.
TABLE 1.
Demographic, epidemiological, and clinical characteristics of four individuals chronically infected with HIV-1 with treatment failure undergoing structured treatment interruption
| Patient | Sexa | Age (yr) | Length (yr) of infectionb | Route of infectionc | Clinical statusd | Previous treatment (time on treatment) | Last treatment (duration) |
|---|---|---|---|---|---|---|---|
| 1 | M | 33 | 7 | IDU | C3 | SQV + RTV, 3TC, d4T (4 mo) | AZT, ddI, ABC (12 mo) |
| 2 | M | 33 | 6 | Sexual | C2 | AZT, ddC, ddI, 3TC, d4T, HU, IDV, NVP NFV, SQV + RTV (6 y) | 3TC, d4T, IDV + RTV (9 mo) |
| 3 | M | 32 | 8 | IDU | B3 | AZT, ddC, ddI, 3TC, d4T, HU, IDV, EFV, NFV, RTV, SQV (8 y) | ABC, d4T, EFV (12 mo) |
| 4 | F | 32 | 6 | Sexual | A1 | AZT, ddC (4 mo) | d4T, ddI, NVP (3 y) |
F, female; M, male.
Time elapsed since infection until STI.
IDU, intravenous drug use.
Clinical status according to the Centers for Disease Control and Prevention classification system (5a).
Plasma viral load was assessed by the b-DNA method with a detection limit of 50 copies/ml (Bayer HIV RNA 3.0 Assay; Bayer Co., Tarrytown, N.Y.). Cell populations from peripheral blood were studied by flow cytometry (Coulter XL; Coulter Co., Hialeah, Fla.).
Samples.
Whole blood was drawn with EDTA as an anticoagulant the day of (day 0) and 30 and 60 days after ARV treatment interruption. Plasma was separated and stored at −70°C until use. After concentration of plasma viral particles by centrifugation at 23,500 × g at 4°C for 1 h, plasma HIV-1 RNA was extracted by using a QIAamp Viral RNA Mini Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer's instructions. Peripheral blood mononuclear cells (PBMC) were separated in a Ficoll-Hypaque gradient (54), and two million PBMC were stored at −70°C until use. Genomic DNA was extracted by using a QIAamp DNA Mini Kit (Qiagen GmbH) according to the manufacturer's recommendations.
pol and env amplification and cloning.
cDNA of the protease (PR) and RT coding regions of the HIV-1 pol gene was synthesized with a solution containing 10 μl of extracted viral RNA, 10 pmol of primer fph4 (5′TGAATGATTCCYAATGCATATTGTGAGTCTGT 3′, positions 4038 to 4069), and 1 μl of 10 mM deoxynucleoside triphosphate mix at 65°C for 5 min. The mixture was cooled on ice for 2 min and a solution containing 200 U of SuperScript II Rnase H− Reverse Transciptase (Life Technololgies, Gaithersburg, Md.), 5× first-strand buffer, 2 μl of 0.1 M dithiothreitol, 20 U of RNasin RNase Inhibitor (Promega, Madison, Wis.), and 120 ng of random hexamers (Promega) in a final volume of 20 μl was added. The reaction was allowed to proceed at 42°C for 1 h. A fragment of the pol gene (positions 2145 to 3408) was amplified in a nested PCR. In the first PCR round, a solution containing 2.5 U of Taq DNA polymerase (Qiagen GmbH), 10× PCR buffer, 1 μl of 10 mM deoxynucleoside triphosphate mix, and 20 pmol of outer primers PU1 (5′TAAGTGTTTCAAYTGTGGCAAAGAAGGRCA 3′, positions 1959 to 1988) and PL1 (5′CYTGYTTCTGTATTTCTGCTAYTAAGTCTTTGT3′, positions 3514 to 3546) was added to 5 μl of cDNA or proviral DNA extracted from PBMC in a final volume of 50 μl. In the second-round PCR, 3 μl of first-round PCR product and inner primers PT1 (2) and RTD (5′TGGTTCCCCTAAGGAGTTACATA3′, positions 3385 to 3409) were used. Reactions were allowed to proceed according to the following program: 3 min at 94°C, then 30 cycles of denaturing at 94°C for 15 s, annealing at 60°C for 30 s, primer extension at 72°C for 90 s, and a final extension at 72°C for 10 min.
cDNA of the third hypervariable loop of gp120 (V3 loop) was prepared as described above, except for the use of primer V3F (5′ATGGGAGGGGCATACATTGC3′, positions 7521 to 7540). A fragment of the env gene (positions 6957 to 7506) was amplified in a nested PCR as described above, except for the use of outer primers 325H− (44) and V3A (62), inner primers V3G (5′GCCACATGTTTATAATTTG3′, positions 7488 to 7506) and V3E (62), and an annealing temperature of 52°C. PCR products were purified with a QIAquick PCR Purification Kit (Qiagen GmbH). Primers V3F and V3G were kindly provided by Linqi Zhang (Aaron Diamond AIDS Research Center, Rockefeller University, New York, N.Y.).
The obtained pol and V3 loop fragments were cloned into pCR2.1 TOPO vector (Invitrogen, Carlsbad, Calif.) and used to transfect chemically competent TOP10 One Shot Escherichia coli according to the manufacturers' instructions. Colonies were grown on Luria-Bertani plates containing 50 μg of kanamycin/ml, and the presence of the expected insert was assessed by colony-PCR with the same primers utilized in the second-round PCR followed by electrophoresis into an ethidium bromide-stained 1.2% agarose gel. Selected colonies were transferred to 0.5 ml of Luria-Bertani medium with 50 μg of kanamycin/ml and was incubated overnight at 37°C, and the insert-containing plasmid was purified with a QIAprep Spin Miniprep Kit (Qiagen GmbH).
pol and env sequencing.
The complete HIV-1 PR and RT codons 1 to 261 were sequenced directly from purified PCR products (population-based sequencing) and from purified cloned fragments by using primers PT2, PT3, R-3, R-4, R-6, and R-7 (2). The HIV-1 gp120 V3 loop (positions 7110 to 7217) was sequenced by using the second-round PCR primers. The Protein/DNA Technology Center and Howard Hughes Medical Institute Biopolymer Facility at The Rockefeller University performed DNA sequencing as follows. Sequencing reaction mixtures were assembled with BigDye Terminator v3.0 Cycle Sequencing Ready Reaction kits (Applied Biosystems, Foster City, Calif.) containing AmpliTaq DNA polymerase, FS. Extension products were purified by using Edge Biosystems gel filtration plates and were run on an ABI 3700 DNA capillary-gel sequencer. Sequence data were generated from raw data files by using Sequencing Analysis v.3.4.1 (Applied Biosystems).
The obtained sequences were visualized with Chromas 2.01 (Technelysium, Helensvale, Queensland, Australia) and were assembled by using SeqMan II, which was included in the DNASTAR software package (Madison, Wis.) (5).
Phylogenetic analysis.
pol and env sequences were trimmed to equivalent lengths and sequence alignments were generated by using Clustal X (58). Phylogenetic relatedness was established by using the PHYLIP software package version 3.5c (distributed by Joseph Felsenstein, Department of Genetics, University of Washington, Seattle [ftp://evolution.genetics.washington.edu/pub/phylip/]). Evolutionary distances were estimated by using DNADIST (Kimura two-parameter method [31]; transition/transversion ratio, 2), and phylogenetic relationships were determined by using NEIGHBOR (neighbor-joining method) (53). Reproducibility of branching patterns (18) was done with SEQBOOT (bootstrap method; 1,000 replicates), and the consensus tree was generated with CONSENSE. The obtained trees were visualized by using TREEVIEW (48).
In order to rule out the effect of homoplasies (47), pol phylogenetic analysis was also performed without taking into account the drug-resistance-associated codons (49) that corresponded in each case.
Sequence quality control was performed according to Los Alamos National Laboratory recommendations (Los Alamos National Laboratory HIV Sequence Database, Los Alamos, N.M. [http:hiv-web.lanl.gov]), and it indicated that no contaminations had occurred.
MEGA version 2.1 (36) (distributed by Sudhir Kumar, Arizona State University, Tempe [http://www.megasoftware.net]) was used to estimate within- and between-sample uncorrected nucleotide distances.
The base numbering stated throughout the text follows that for the HIV-1 strain HXB2 (GenBank accession number K03455) (33).
Drug resistance genotyping.
Previously described drug-resistance-associated mutations in the PR and RT were sought (49), and genotyping results were interpreted for abacavir (ABC), didanosine (ddI), lamivudine (3TC), stavudine (d4T), zalcitabine (ddC), zidovudine (AZT), delavirdine (DLV), efavirenz (EFV), nevirapine (NVP), amprenavir (APV), indinavir (IDV), nelfinavir (NFV), ritonavir (RTV), and saquinavir (SQV) by using the drug resistance interpretation beta test (HIV RT and Protease Sequence Database; Stanford University [http://hiv-4.stanford.edu/cgi-bin/hivtestweb.pl]) (27).
HIV-1 coreceptor usage.
HIV-1 envelope sequences corresponding to the gp120 V3 loop were used to predict coreceptor usage on the basis of the presence of basic or acidic residues at positions 275 and 287 of the env gene, corresponding to positions 11 and 25 of the V3 loop (1, 24).
Statistical analyses.
The Mann-Whitney U test (Tadpole III program; Cambridge, United Kingdom) was employed for statistical analyses.
Nucleotide sequence accession numbers.
All the HIV-1 sequences related to this work have been submitted to GenBank and were given accession numbers AF470720 through AF471027.
RESULTS
Virological and immunological evolution during STI.
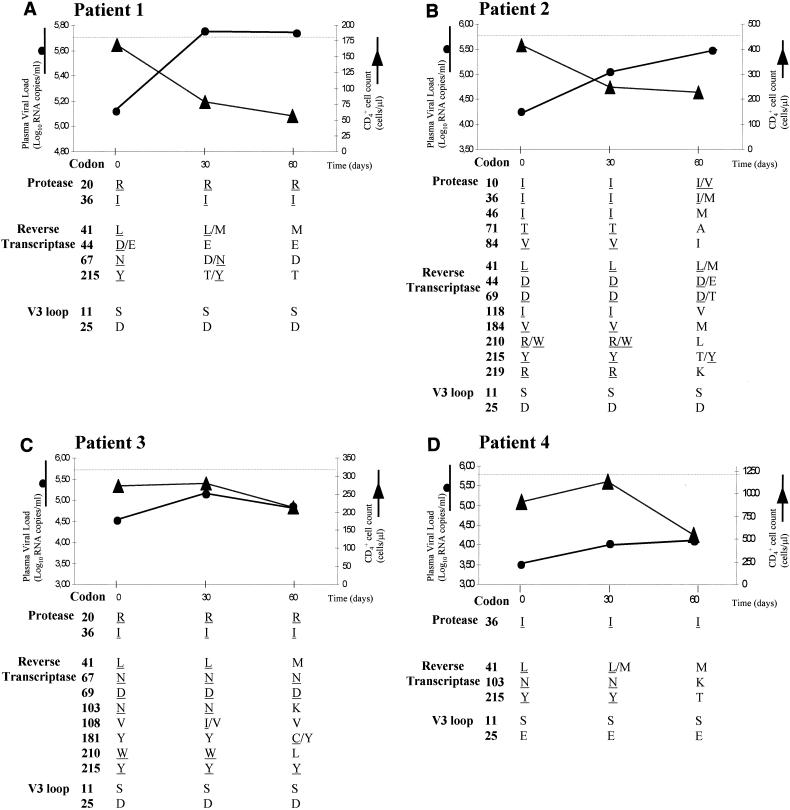
Four chronically HIV-1-infected patients who had detectable plasma HIV-1 RNA in spite of being on highly active ARV therapies had their treatments interrupted for 60 days. The evolution of CD4+ cell count and plasma viral load during STI is shown in Fig. 1. In all the cases, an increase of plasma viremia and a decrease of CD4+ cell count was observed after 60 days of treatment interruption. In two patients the increase of plasma viral load was detected simultaneously with the drop in the CD4+ cell count, whereas in the other two patients this increase preceded the CD4+ cell decline.
FIG. 1.
Changes in plasma viral load, CD4+ cell count, ARV drug resistance, and HIV-1 V3 loop population-based genotypes of four chronically HIV-1-infected patients with therapeutic failure during structured treatment interruption. Plasma viral load was assessed with the b-DNA assay (Bayer HIV RNA 3.0 Assay; Bayer Co.) with an upper detection limit of 500,000 copies/ml (represented as dotted lines). Drug-resistance-associated mutations, as defined by the Los Alamos National Laboratory (49), are underlined.
Drug resistance genotype during STI.
Consensus sequencing of the PR and RT coding regions of the pol gene of samples taken at day 0 showed the presence of mutations associated with resistance to the last ARV treatment received for all of the patients (Fig. 1). Patient 1 carried mutations that conferred high-level resistance to AZT and that are associated with low- or intermediate-level resistance to other components both of his last ARV treatment (ddI and ABC) and of the preceding treatment (d4T). Patient 2 had mutations associated with high- or intermediate-level resistance to all the drugs of his last ARV treatment (d4T, 3TC, and IDV + RTV) together with mutations associated with reduced susceptibility to ARV agents he had received in previous therapy schemes (ddI, ddC, d4T, NFV, and SQV). Similarly, patient 3 possessed mutations associated with high- or intermediate-level resistance to all the drugs of his last ARV treatment (ABC, d4T, and EFV) and mutations associated with resistance to some components of his previous ARV treatments (AZT, ddI, and ddC). Patient 4 carried mutations associated with high-level resistance to two of the components of her last ARV scheme (d4T and NVP), which also conferred resistance to an agent of an earlier ARV treatment (AZT).
Evolution of drug-resistance-associated mutations after treatment interruption is shown in Fig. 1. In patients 1 and 4 all the primary mutations found on day 0 were not detected by population-based sequencing by day 60. On the other hand, for patients 2 and 3 several drug-resistance-associated mutations persisted after STI, while a sensitive genotype was detected at other positions. Secondary mutations in the PR did not change over time for all but patient 2. For some codons in the PR and RT regions, wild-type-mutant mixtures were observed at days 30 and 60 after STI.
Of particular note in the sample obtained at day 60 from patient 3 was the presence of a new mutation, Y181C, associated with resistance to the nonnucleoside reverse transcriptase inhibitors (13, 52, 60). In the interviews the patient mentioned having completely interrupted ARV treatment, but ARV drug levels were not measured.
Phylogenetic analysis of quasispecies emerging during STI.
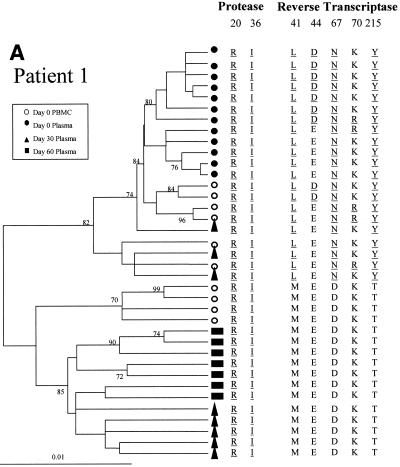
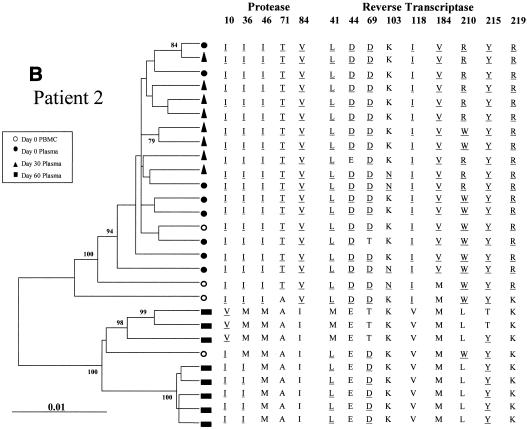
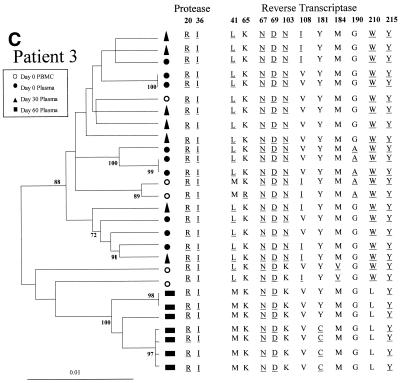
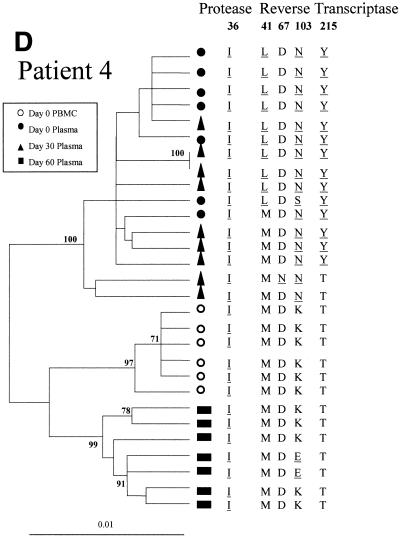
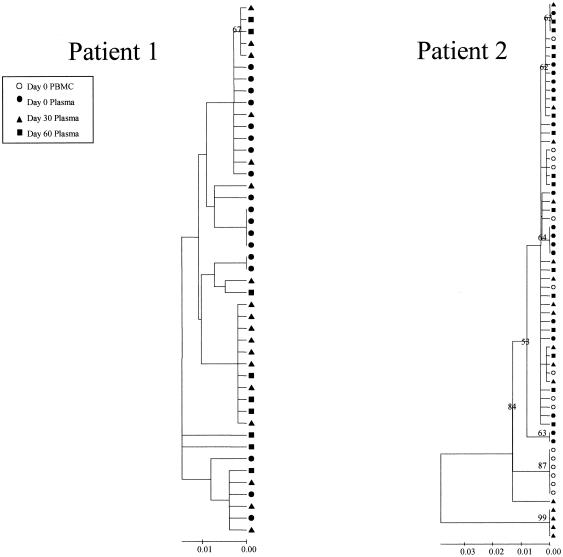
The changes in the sequence observed at days 30 and 60 after treatment interruption in all the patients may be due to back mutations on the viral quasispecies circulating at day 0 or to the reemergence of ancestral viral subpopulations that had a sensitive genotype. In order to discriminate between these alternative hypotheses, the entire PR coding region and a portion of the RT coding region of HIV-1 pol were amplified, cloned, and analyzed by phylogenetic methods (Fig. 2). For all four patients the clones from plasma samples taken at day 60 clustered in a separate group from those taken at day 0, with high bootstrap values (over 85%).
FIG. 2.
Phylogenetic trees of partial HIV-1 pol sequences (complete PR and RT codons 1 to 261) for patients 1 (A), 2 (B), 3 (C), and 4 (D) during antiretroviral treatment interruption. Trees were constructed as described in Materials and Methods. Filled symbols represent plasma-derived sequences, and open symbols represent PBMC-derived sequences. Samples were taken the day of therapy interruption (day 0) and 30 and 60 days later. Numbers at branch nodes refer to the number of bootstrap repetitions (out of 1,000) in which the sequences grouped together; only frequencies greater than 70% are shown. Sequence dissimilarity (distance) is shown on the horizontal axis. The genotypes at drug-resistance-associated codons in HIV-1 PR and RT are indicated next to the corresponding clone. Drug-resistance-associated mutations, as defined by the Los Alamos National Laboratory (49), are underlined.
Patient 1.
For patient 1, the clones derived from the day 0 plasma sample showed the drug resistance profile observed by the direct sequencing of bulk amplified material (Fig. 2A). At day 30 a mixture of quasispecies having wild-type and resistant genotypes was observed. The drug-resistant clones clustered in the same group as sequences from day 0. In contrast, the drug-sensitive clones from day 30 clustered into a separate group, which also included all the clones derived from the day 60 plasma sample. All the quasispecies observed at day 60 had a completely sensitive genotype in RT and conserved secondary mutations in PR. When clones from baseline PBMC were studied, some variants carried drug-resistance-associated mutations on RT, clustering in the same group as the quasispecies from day 0. The PBMC-derived clones carrying a sensitive genotype at the RT clustered into a group that was distinct from both of the two clades previously described.
Patient 2.
For patient 2, all the clones obtained from the day 0 and day 30 plasma samples clustered in a group supported by a 100% bootstrap value and shared a drug-resistant genotype (Fig. 2B). At day 60 all of the plasma sample clones had sensitive genotypes at most of the codons on PR and RT. Two separate clusters supported by high bootstrap values were found within the day 60 clade, differing in the genotype at codons 10 and 36 of PR and 41, 69, and 215 of RT. One out of 4 clones derived from baseline PBMC clustered within the group of day 60 clones, while the other three were more related to day 0 and day 30 plasma-derived clones.
Patient 3.
For patient 3, all the clones from plasma samples from days 0 and 30 had a resistant genotype and clustered in a single group with a bootstrap value of 88% (Fig. 2C). At day 60 all of the plasma-derived clones had a wild-type genotype at most of the codons of RT but had conserved mutations at codons 67, 69, and 215 of RT and secondary mutations on PR. All of these clones clustered as a distinct group (100% bootstrap). Within the day 60 clade a subgroup of highly related clones carried mutation Y181C in the RT, while the remaining had a Tyr residue at that position. Some of the clones from baseline PBMC clustered in the same group as the day 0 and day 30 plasma clones, while others were located away from the two main clusters of plasma-derived clones (bootstrap values of 100% and 88%).
Patient 4.
In the case of patient 4, all of the plasma-derived clones from days 0 and 30 clustered in a close group (100% bootstrap value) (Fig. 2D). At day 30 all these clones had conserved mutations M36I of PR and K103N of RT observed in baseline clones, while a mixture of wild-type and resistant genotypes was observed at positions 41 and 215 of RT. At day 60 all the clones grouped in a separate cluster, and all of them had a wild-type genotype at codons 41, 67, and 215 of RT while conserving the secondary mutation on PR. Mutation K103E, detected in 2 out of 7 clones, has not been associated with resistance to NVP (49). All of the PBMC-derived clones from day 0 had a completely wild-type genotype in the RT and clustered in a separate group from the plasma-derived ones.
Pair-wise genetic distances and assessment of homoplasies.
The separate clustering of baseline drug-resistant and day 60 drug-sensitive plasma-derived clones was supported by the observation that, for all of the patients, the between-sample pair-wise genetic distances for the clones obtained at days 0 and 60 (mean, 0.031; median, 0.029) were significantly higher (P < 0.001) than the corresponding within-sample pair-wise distances for either time point (day 0 mean and median, 0.014 and 0.016, respectively; day 60 mean and median, 0.015 and 0.016, respectively).
In order to assess the role of the codons associated with resistance in the analyses performed, the sequence alignments were reanalyzed after deleting the resistance-associated codons involved in the changes described above. The phylogenetic trees constructed on the basis of these reduced alignments showed that the drug-susceptibility- and time-point-based clustering was conserved, with some changes in the local topology within clades (data not shown). Moreover, the bootstrap values that supported each major clade were not substantially reduced.
V3 loop phylogenetic analysis and inferred coreceptor usage.
Quasispecies utilizing the CXCR-4 coreceptor have been previously described as being associated with faster viral replication and disease progression (6, 8, 21, 28, 56). In order to assess whether the rapid increase of plasma viral load and decrease in CD4+ cell counts observed after STI was associated with the emergence of such variants, the third variable loop (V3 loop) region of gp120 was sequenced and the amino acids previously described as being associated with CXCR-4 usage (1, 24) were sought (Fig. 1). In all patients a Ser residue was found at position 11 in all the plasma- and PBMC-derived sequences taken throughout the 60-day period of the study. For three of the patients position 25 was occupied by an Asp, and for the fourth patient this codon was a Glu throughout the period of study. The coreceptor usage inferred from these V3 loop sequences indicates that these quasispecies are most likely to have utilized the CCR5 coreceptor, and there is no evidence of change during therapy interruption. Hence, utilization of CXCR-4 is unlikely to have been responsible for the increased viral replication and depletion of CD4+ cells seen.
The clones from the V3 loop region of gp120 from samples taken at different time points were phylogenetically analyzed, and the quasispecies from days 0, 30, and 60 were intermingled with no evidence for cladistic structuring (exemplified for patients 1 and 2 in Fig. 3). This contrasts with the time-point-associated topologies observed in the pol region (see above).
FIG. 3.
Phylogenetic trees of HIV-1 V3 loop sequences (positions 7110 to 7217) of patients 1 (left) and 2 (right) during ARV treatment interruption. The trees were constructed as described in Materials and Methods. Symbols are as described in the legend to Fig. 2. Bootstrap repetitions greater than 50% are shown.
DISCUSSION
In the present study we analyzed the evolution of HIV-1 quasispecies during STI in four patients with treatment failure due to HIV-1 drug resistance. All of the mutations in the PR and RT detected at baseline conferred resistance to some or all of the components of the last ARV scheme. In some cases these mutations were also associated with decreased susceptibility to ARV drugs taken by the patients in former treatments, so the possibility of their selection during previous schemes and their conservation during subsequent treatments should not be ruled out (55). After 60 days off therapy, completely drug-sensitive viruses were detected in two patients. For one individual some drug-resistance-associated mutations were not detected 60 days after treatment interruption, while at other codons the coexistence of wild-type and mutated quasispecies was observed. On the other hand, for 1 out of 4 patients the genotype changed to a sensitive one in some codons, but other drug-resistance-associated mutations were conserved after STI.
Two major hypotheses may be proposed to explain the source of the drug-sensitive quasispecies found after STI: (i) they may have appeared as the major drug-resistant quasispecies replicating immediately before STI-experienced point mutations (back mutations) at drug-resistance-associated codons of PR and RT, or (ii) they may represent drug-sensitive ancestral viral variants that had circulated before treatment failure, were suppressed during ARV treatment, and reemerged when the selective pressure of the drugs disappeared. In a phylogenetic analysis, the representation of the first scenario would correspond to a phylogenetic tree in which drug-resistant and drug-sensitive quasispecies would be intermingled, as they would be genetically closely related, only differing at a few codons implied in drug resistance. In this case, the differences among quasispecies would not be enough for a drug-susceptibility-based topology to be strongly statistically supported. Furthermore, a phylogenetic analysis that excludes drug-resistance-associated codons would result in the same intermingled topology. By contrast, the latter hypothesis would be supported if viral variants would cluster separately according to their drug sensitivity, as they would not only differ in drug-resistance-associated mutations but also in other codons not subjected to the selective pressure of the drugs. Moreover, if the phylogenetic study was to be performed without taking into account drug-resistance-associated codons, the cluster structure should be conserved. It is important to note that the two exposed hypotheses are not to be considered mutually exclusive.
In the present work the source of the completely or partially drug-sensitive quasispecies that emerged after STI was studied by phylogenetic analyses, which strongly supported the hypothesis of the ancestral origin of viral variants circulating after cessation of ARV therapy. For all of the patients, drug-sensitive quasispecies detected after 60 days of STI clustered apart from the baseline drug-resistant ones, supported by bootstrap values of more than 85%. This drug-sensitivity-based topology was also seen when the corresponding drug-resistance-associated codons were not taken into account, ruling out homoplasies (i.e., characters identical in state due to parallel or convergent change) as the causes for the observed grouping (data not shown). The observation that the genetic distances between baseline and day 60 clones were significantly higher than the within-sample distances corresponding to each time point further supported the ancestral origin hypothesis.
The fact that a reversion to drug-resistance-associated genotype after STI was observed simultaneously at most of the involved codons also supports the ancestral origin of these quasispecies. Moreover, in those patients in which a reversion of genotype at codon 215 of RT was observed a fully wild-type genotype (i.e., threonine residue) was observed, but this was not observed for similarly drug-sensitive reversion intermediates (i.e., aspartic acid, cysteine, serine, and asparagine residues) reported for primary HIV-1-infected patients (14, 61).
Completely or partially drug-sensitive quasispecies emerging at day 60 showed in the phylogenetic analysis of pol that they did not constitute one homogeneous cluster but rather a subdivided one. This suggests that they were originated by the reemergence of several viral clones. Due to the fact that the patients resumed ARV treatments after the 60-day STI, it was not possible to determine if the viral populations had a tendency to homogeneity or if further quasispecies continued to emerge.
Viral variants with a complete or partial drug-sensitive genotype were found at day 0 in PBMC samples, while most of the plasma baseline quasispecies had a fully drug-resistant genotype. The discrepancy in the genotype of HIV-1 genomes detected in plasma and PBMC has been previously described for the pol region (29, 32, 35, 38, 57) and also for other HIV-1 genomic regions (63). These variants detected in PBMC samples could represent ancestral proviruses integrated before the emergence of drug resistance. In the present study, drug-sensitive plasma quasispecies from day 60 were not closely phylogenetically related to the drug-sensitive variants from PBMC (except for 1 out of 4 clones detected in patient 2), which indicates that the viruses emerging after STI may come from minor variants integrated in PBMC or replicating in peripheral blood cells at very low levels during ARV treatment, from variants replicating in other compartments, or from the reactivation of quiescent viral reservoirs. Price et al. reported that the PR and RT sequences from HIV-1 derived from plasma and cerebrospinal fluid before and during STI show no evidence of compartmentalization (51), thus suggesting that major quasispecies replicating in cerebrospinal fluid may not be the source of the drug-sensitive virus emerging after STI.
It was previously reported that not all the patients with treatment failure undergoing STI experienced a reversal in their drug-resistance-associated genotype (10, 12, 16, 26, 45, 59). Elucidating the factors that could predict which patients would revert from their drug-resistant genotype to a drug-sensitive one after STI would be of great use for the physicians managing patients with ARV treatment failure. Factors that were associated with a shift in drug susceptibility were the patient clinical status, the number of drugs to which the patients had been exposed, CD4+ cell count and plasma viral load at study entry (26), baseline CD4+ cell count and baseline viral load/CD4+ cell count ratio (45), viral suppression during ARV therapy (10), and duration of treatment interruption (12).
According to the present knowledge of HIV-1 replication kinetics (50), fully wild-type viruses might be nearly extinct in patients in which drug-resistant variants emerged many years ago and in the presence of a sustained ARV pressure for their selection, so their emergence after treatment interruption might not be detected. Nevertheless, variants that have emerged during the early stages of drug resistance selection may reemerge after STI if their fitness in a drug-free environment is (even slightly) higher than that of the drug-resistant quasispecies that replicated immediately before treatment interruption. The observation that only some patients showed a reversion in their genotype may thus be explained by the different availabilities of fully sensitive quasispecies in different individuals, associated with the time since the emergence of drug-resistant variants and the duration of the pressure that selected for them. Another point that should be taken into account is that if wild-type and resistant variants have equivalent replicative capacities in a drug-free environment, the resistant quasispecies may persist much longer than otherwise expected.
In patients 1 and 4 at day 30, mixtures of quasispecies with different drug susceptibilities were detected. In the case of patient 1, variants carrying the same drug-resistance-associated mutations as those found at baseline coexisted with quasispecies with a complete drug-sensitive genotype. The former clustered with the drug-resistant variants of day 0, while the latter were more closely related to the wild-type variants detected at day 60. The fact that the drug-resistant quasispecies were not detected at day 60 reflects that in this case, the fitness of the drug-sensitive quasispecies in an ARV drug-free environment was higher (9, 15, 19, 20, 22, 25, 34, 40, 42, 43). As for patient 4, some variants from day 30 conserved all the mutations found at day 0, whereas others carried only some of them. In this case, however, all the day 30 quasispecies clustered with the drug-resistant variants from day 0, regardless of their drug-resistance-associated genotype. The partially sensitive variants from day 30 could be drug-resistant quasispecies that were selected subsequently during ARV treatment and reemerged after treatment interruption or could reflect a back-mutation process from the completely drug-resistant variants from day 0. This could indicate that the back mutation and reemergence of ancestral variants are processes that are taking place simultaneously after treatment interruption, and if ancestral quasispecies had a higher replicative capacity they would outgrow the partially sensitive ones.
In all the studied patients, secondary mutations in the PR were observed at day 0. In the case of patients 1, 3, and 4, these mutations remained throughout the STI in spite of the reversion observed at other codons. These mutations could be polymorphisms present at the time of HIV-1 infection. The fact that none of these individuals had primary drug-resistance-associated mutations in the PR and previous reports showing that secondary mutations in the PR are highly prevalent in the local HIV-1 chronically infected drug-naïve population (30) support this hypothesis. On the other hand, secondary mutations in the PR found in patient 2 at day 0 were not observed in some of the quasispecies emerging after STI. In this case these mutations have probably been selected by ARV drugs along with the primary ones.
When the evolution of sequences of the V3 loop of gp120 during STI was investigated, viral quasispecies from different time points were intermingled compared to the time-point- and drug-susceptibility-based topology observed in the pol region, suggesting that these regions of the genome are evolving independently. These results are in concordance with the observations of Brown and Cleland (3), which showed that selection acting at the pol region has no significant effect on the evolution of the env gene due to the possibility of recombination between the two distant HIV-1 genomic regions.
After treatment interruption, a higher rate of viral replication and a rapid decline in CD4+ cells was observed in this study, in concordance with previous reports (10, 12, 16, 26, 45, 59). In order to determine if these changes in viral replication and cytopathogenicity were caused by the emergence of variants with a syncytium-inducing/CXCR4 phenotype, the coreceptor usage by HIV-1 was inferred from V3 loop sequences. Basic amino acidic residues at positions 11 and 25, previously associated with CXCR4 coreceptor usage (1, 24), were not found in any of the patients belonging to this study. These results indicate that the observed virologic and immunologic effects of STI were not related to a change of tropism but rather to an increase in viral fitness associated with changes in the PR and RT. Nevertheless, as changes in regions of the env gene other than the V3 loop have been reported to be related to the syncytium-inducing/CXCR4 phenotype (21), changes in tropism after STI cannot be ruled out.
In conclusion, the present study demonstrates that drug-susceptible viral variants emerging after ARV treatment interruption in patients with therapeutic failure are of ancestral origin and are not derived from drug-resistant quasispecies by back mutation. These results show the important role that reemergence of viral variants play in HIV-1 population dynamics. This work has major implications in the clinical setting, since it points out the difficulties that may be found when recycling ARV therapies in patients with treatment failure. The sources of the emerging ancestral viruses still need to be elucidated, and this knowledge will be helpful in understanding the mechanism of HIV-1 evolution and in the design of ARV treatment strategies.
REFERENCES
- 1.Bhattacharyya, D., B. R. Brooks, and L. Callahan. 1996. Positioning of positively charged residues in the V3 loop correlates with HIV type 1 syncytium-inducing phenotype. AIDS Res. Hum. Retrovir. 12:83-90. [DOI] [PubMed] [Google Scholar]
- 2.Boden, D., A. Hurley, L. Zhang, Y. Cao, Y. Guo, E. Jones, J. Tsay, J. Ip, C. Farthing, K. Limoli, N. Parkin, and M. Markowitz. 1999. HIV-1 drug resistance in newly infected individuals. JAMA 282:1135-1141. [DOI] [PubMed] [Google Scholar]
- 3.Brown, A. J., and A. Cleland. 1996. Independent evolution of the env and pol genes of HIV-1 during zidovudine therapy. AIDS 10:1067-1073. [PubMed] [Google Scholar]
- 4.Burke, D. S. 1997. Recombination in HIV: an important viral evolutionary strategy. Emerg. Infect. Dis. 3:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burland, T. G. 2000. DNASTAR's Lasergene sequence analysis software. Methods Mol. Biol. 132:71-91. [DOI] [PubMed] [Google Scholar]
- 5a.Centers for Disease Control. 1992. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. Morbid. Mortal. Wkly. Rep. 41:1-19. [PubMed] [Google Scholar]
- 6.Cheng-Mayer, C., D. Seto, M. Tateno, and J. A. Levy. 1998. Biologic features of HIV-1 that correlate with virulence in the host. Science 240:80-82. [DOI] [PubMed] [Google Scholar]
- 7.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 8.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in co-receptor use co-receptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 11.Deeks, S. G. 2001. International perspectives on antiretroviral resistance. Nonnucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S25-S33. [DOI] [PubMed] [Google Scholar]
- 12.Delaugerre, C., M. A. Valantin, M. Mouroux, M. Bonmarchand, G. Carcelain, C. Duvivier, R. Tubiana, A. Simon, F. Bricaire, H. Agut, B. Autran, C. Katlama, and V. Calvez. 2001. Re-occurrence of HIV-1 drug mutations after treatment re-initiation following interruption in patients with multiple treatment failure. AIDS 15:2189-2191. [DOI] [PubMed] [Google Scholar]
- 13.Demeter, L. M., R. W. Shafer, M. Para, G. Morse, W. Freimuth, T. C. Merigan, and R. C. Reichman. 1995. Delavirdine (DLV) susceptibility of HIV-1 isolates obtained from patients receiving DLV monotherapy (ACTG 260). J. Acquir. Immune Defic. Syndr. Hum. Retrovir. 10:23. [Google Scholar]
- 14.de Ronde, A., M. van Dooren, L. van Der Hoek, D. Bouwhuis, E. de Rooij, B. van Gemen, R. de Boer, and J. Goudsmit. 2001. Establishment of new transmissible and drug-sensitive human immunodeficiency virus type 1 wild types due to transmission of nucleoside analogue-resistant virus. J. Virol. 75:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devereux, H. L., V. C. Emery, M. A. Johnson, and C. Loveday. 2001. Replicative fitness in vivo of HIV-1 variants with multiple drug resistance-associated mutations. J. Med. Virol. 65:218-224. [DOI] [PubMed] [Google Scholar]
- 16.Devereux, H. L., M. Youle, M. A. Johnson, and C. Loveday. 1999. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS 13:F123-F127. [PubMed] [Google Scholar]
- 17.Eigen, M., and C. K. Biebricher. 1998. Sequence space and quasispecies distribution, p. 3-22. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics, vol. III. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 18.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 29:783-791. [DOI] [PubMed] [Google Scholar]
- 19.Goudsmit, J., A. de Ronde, E. de Rooij, and R. de Boer. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goudsmit, J., A. De Ronde, D. D. Ho, and A. S. Perelson. 1996. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J. Virol. 70:5662-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond, A. L., J. Lewis, J. May, J. Albert, P. Balfe, and J. A. McKeating. 2001. Antigenic variation within the CD4 binding site of human immunodeficiency virus type 1 gp120: effects on chemokine receptor utilization. J. Virol. 75:5593-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrigan, P. R., S. Bloor, and B. A. Larder. 1998. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J. Virol. 72:3773-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland, J. J., J. C. De La Torre, and D. A. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1-20. [DOI] [PubMed] [Google Scholar]
- 24.Hung, C. S., N. Vander Heyden, and L. Ratner. 1999. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J. Virol. 73:8216-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamichi, T., S. C. Berg, H. Imamichi, J. C. Lopez, J. A. Metcalf, J. Falloon, and H. C. Lane. 2000. Relative replication fitness of a high-level 3′-azido-3′-deoxythymidine-resistant variant of human immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr->Gly) at codon 69. J. Virol. 74:10958-10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izopet, J., P. Massip, C. Souyris, K. Sandres, B. Puissant, M. Obadia, C. Pasquier, E. Bonnet, B. Marchou, and J. Puel. 2000. Shift in HIV resistance genotype after treatment interruption and short-term antiviral effect following a new salvage regimen. AIDS 14:2247-2255. [DOI] [PubMed] [Google Scholar]
- 27.Kantor, R., R. Machekano, M. J. Gonzales, K. Dupnik, J. M. Shapiro, and R. W. Shafer. 2001. Human Immunodeficiency Virus Reverse Transcriptase and Protease Sequence Database: an expanded data model integrating natural language text and sequence analysis programs. Nucleic Acids Res. 29:296-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson, A., K. Parsmyr, E. Sandstrom, E. M. Fenyo, and J. Albert. 1994. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 32:364-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaye, S., E. Comber, M. Tenant-Flowers, and C. Loveday. 1995. The appearance of drug resistance-associated point mutations in HIV type 1 plasma RNA precedes their appearance in proviral DNA. AIDS Res. Hum. Retrovir. 11:1221-1225. [DOI] [PubMed] [Google Scholar]
- 30.Kijak, G. H., S. E. Pampuro, M. M. Avila, C. Zala, P. Cahn, M. A. Wainberg, and H. Salomon. 2001. Resistance profiles to antiretroviral drugs in HIV-1 drug-naive patients in Argentina. Antivir. Ther. 6:71-77. [PubMed] [Google Scholar]
- 31.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 32.Koch, N., N. Yahi, F. Ariasi, J. Fantini, and C. Tamalet. 1999. Comparison of human immunodeficiency virus type 1 (HIV-1) protease mutations in HIV-1 genomes detected in plasma and in peripheral blood mononuclear cells from patients receiving combination drug therapy. J. Clin. Microbiol. 37:1595-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korber, B. T., B. T. Foley, C. L. Kuiken, et al. 1998. Numbering positions in HIV relative to HXB2CG, p. III-102-III-111. In B. Korber, B. Hahn, B. Foley, F. McCutchan, C. Kuiken, J. W. Mellors, and J. Soroski (ed.), Human retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 34.Kosalaraksa, P., M. F. Kavlick, V. Maroun, R. Le, and H. Mitsuya. 1999. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an in vitro competitive HIV-1 replication assay. J. Virol. 73:5356-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozal, M. J., R. W. Shafer, M. A. Winters, D. A. Katzenstein, and T. C. Merigan. 1993. A mutation in human immunodeficiency virus reverse transcriptase and decline in CD4 lymphocyte numbers in long-term zidovudine recipients. J. Infect. Dis. 167:526-532. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 37.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 38.Loveday, C., S. Kaye, M. Tenant-Flowers, M. Semple, U. Ayliffe, I. V. Weller, and R. S. Tedder. 1995. HIV-1 RNA serum-load and resistant viral genotypes during early zidovudine therapy. Lancet 345:820-824. [DOI] [PubMed] [Google Scholar]
- 39.Loveday, C. 2001. International perspectives on antiretroviral resistance. Nucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S10-S24. [DOI] [PubMed] [Google Scholar]
- 40.Maeda, Y., D. J. Venzon, and H. Mitsuya. 1998. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J. Infect. Dis. 177:1207-1213. [DOI] [PubMed] [Google Scholar]
- 41.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Picado, J., A. V. Savara, L. Shi, L. Sutton, and R. T. D'Aquila. 2000. Fitness of human immunodeficiency virus type 1 protease inhibitor-selected single mutants. Virology 275:318-322. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKeating, J. A., C. Shotton, J. Cordell, S. Graham, P. Balfe, N. Sullivan, M. Charles, M. Page, A. Bolmstedt, S. Olofsson, et al. 1993. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J. Virol. 67:4932-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, V., C. Sabin, K. Hertogs, S. Bloor, J. Martinez-Picado, R. D'Aquila, B. Larder, T. Lutz, P. Gute, E. Weidmann, H. Rabenau, A. Phillips, and S. Staszewski. 2000. Virological and immunological effects of treatment interruptions in HIV-1 infected patients with treatment failure. AIDS 14:2857-2867. [DOI] [PubMed] [Google Scholar]
- 46.Miller, V. 2001. International perspectives on antiretroviral resistance. Resistance to protease inhibitors. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S34-S50. [DOI] [PubMed] [Google Scholar]
- 47.Page, R. D. M., and E. C. Holmes. 1998. Molecular evolution. A phylogenetic approach. Blackwell Science Ltd., Oxford, United Kingdom.
- 48.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:53-60. [DOI] [PubMed] [Google Scholar]
- 49.Parikh, U., J. Hammond, C. Calef, B. Larder, R. Schinazi, and J. Mellors. 2000. Mutations in retroviral genes associated with drug resistance, p. 106-161. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, J. Mullins, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium 2000. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 50.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 51.Price, R. W., E. E. Paxinos, R. M. Grant, et al. 2001. Cerebrospinal fluid response to structured treatment interruption after virological failure. AIDS 15:1251-1259. [DOI] [PubMed] [Google Scholar]
- 52.Richman, D., C. K. Shih, I. Lowy, J. Rose, P. Prodanovich, S. Goff, and J. Griffin. 1991. Human immunodeficiency virus type 1 mutants resistant to nonnucleoside inhibitors of reverse transcriptase arise in tissue culture. Proc. Natl. Acad. Sci. USA 88:11241-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 54.Salomon, H., A. Belmonte, K. Nguyen, Z. Gu, M. Gelfand, and M. A. Wainberg. 1994. Comparison of cord blood and peripheral blood mononuclear cells as targets for viral isolation and drug sensitivity studies involving human immunodeficiency virus type 1. J. Clin. Microbiol. 32:2000-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarmati, L., E. Nicastri, S. G. Parisi, G. D'Ettorre, P. Narciso, G. Mancino, I. Gallo, V. Abbadessa, N. E. Dalle, C. Traina, V. Vullo, and M. Andreoni. 2001. Failure of stavudine-lamivudine combination therapy in antiretroviral-naïve patients with AZT-like HIV-1 resistance mutations. J. Med. Virol. 65:631-636. [DOI] [PubMed] [Google Scholar]
- 56.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, M. S., K. L. Koerber, and J. S. Pagano. 1993. Zidovudine-resistant human immunodeficiency virus type 1 genomes detected in plasma distinct from viral genomes in peripheral blood mononuclear cells. J. Infect. Dis. 167:445-448. [DOI] [PubMed] [Google Scholar]
- 58.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verhofstede, C., F. V. Wanzeele, B. Van Der Gucht, N. DeCabooter, and J. Plum. 1999. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS 13:2541-2546. [DOI] [PubMed] [Google Scholar]
- 60.Winslow, D. L., S. Garber, C. Reid, H. Scarnati, D. Baker, M. M. Rayner, and E. D. Anton. 1996. Selection conditions affect the evolution of specific mutations in the reverse transcriptase gene associated with resistance to DMP 266. AIDS 10:1205-1209. [DOI] [PubMed] [Google Scholar]
- 61.Yerly, S., A. Rakik, S. K. De Loes, B. Hirschel, D. Descamps, F. Brun-Vezinet, and L. Perrin. 1998. Switch to unusual amino acids at codon 215 of the human immunodeficiency virus type 1 reverse transcriptase gene in seroconvertors infected with zidovudine-resistant variants. J. Virol. 72:3520-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, Y. M., S. C. Dawson, D. Landsman, H. C. Lane, and N. P. Salzman. 1994. Persistence of four related human immunodeficiency virus subtypes during the course of zidovudine therapy: relationship between virion RNA and proviral DNA. J. Virol. 68:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]