Abstract
Foamy viruses (FVs) are nonpathogenic, widely spread complex retroviruses which have been isolated in nonhuman primates, cattle, cats, and more recently in horses. The equine foamy virus (EFV) was isolated from healthy horses and was characterized by molecular cloning and nucleotide sequence analysis. Here, to further characterize this new FV isolate, the location of the transcriptional cap and poly(A) addition sites as well as the main splice donor and acceptor sites were determined, demonstrating the existence of the specific subgenomic pol mRNA, one specific feature of FVs. Moreover, similar to what has been described for the human foamy virus (HFV), the prototype of FVs, a replication-defective EFV genome was identified during persistent infection. At the protein level, the use of specific antibodies allowed us to determine the size and the subcellular localization of EFV Gag, Env, and Tas, the viral transactivators. While EFV Gag was detected in both the cytoplasm and the nucleus, EFV Env mainly localized in the Golgi complex, in contrast to HFV Env, which is sequestered in the endoplasmic reticulum. In addition, electron microscopy analysis demonstrated that EFV budding occurs at the plasma membrane and not intracellularly, as is the case for primate FVs. Interestingly, EFV Tas was detected both in the nucleus and the cytoplasm of Tas-transfected cells, in contrast to the strict nuclear localization of other FV Tas but similar to the equine infectious anemia virus Tat gene product. Taken together, our results reveal that this new FV isolate exhibits remarkable features among FVs, bringing new insights into the biology of these unconventional retroviruses.
Foamy viruses (FVs) are complex retroviruses with unique features among the retroviral family (15, 21). The most evident of them lies in their name, since they induce the formation of multinucleated giant cells which present numerous vacuoles, giving the monolayer culture a foam aspect. In contrast, FVs establish a persistent innocuous infection in their natural hosts (20). The mechanisms of viral persistence are not elucidated yet, but in contrast to the case for lentiviruses, genetic variability probably does not constitute the basis of this equilibrium (39). In the case of the so-called human foamy virus (HFV), the prototype of FVs, it has been suggested that a defective variant (named ΔHFV or HFVΔTas) negatively interferes with the replication of the parental counterpart (20, 34, 36). HFVΔTas is generated by alternative splicing of the wild-type pregenomic RNA and contains a 301-bp deletion in the gene of the viral transactivator, leading to the formation of an intronless auxiliary bet gene. Another feature of FVs is the synthesis of a specific subgenomic mRNA for the expression of viral enzymatic products. Indeed, in retroviruses, Pol is synthesized as a Gag-Pol fusion protein derived either from a frameshift event or through stop codon suppression. Instead, in FVs, Pol is expressed from a spliced mRNA by translational initiation at the first AUG in the pol gene independently of Gag expression (42). This particularity, together with the late-occurring reverse transcription which leads to incorporation of infectious viral DNA into virions, is more reminiscent of hepadnaviruses than of retroviruses (15, 42).
FVs are highly prevalent in nonhuman primates, and three nonprimate FVs have been isolated to date: the bovine foamy virus (BFV), the feline foamy virus (FFV), and the equine foamy virus (EFV). This latter was recently isolated from blood samples of naturally infected healthy horses after coculture of phytohemagglutinin (PHA)-activated lymphocytes derived from seropositive horses with permissive human U373-MG cells (40). Nucleotide sequence analysis reveals that EFV is phylogenetically the most distant FV compared to the HFV prototype, but it presents the classical FV genomic organization. In particular, EFV codes for auxiliary proteins located downstream of the env gene. FV Tas (for transactivator of spumaviruses, originally called Bel1) is encoded by ORF1 and transactivates viral gene expression by direct binding to the viral DNA on specific sequences in the long terminal repeat (LTR) and in an internal promoter (IP), another specific feature of FVs (13, 23). The transcriptional unit in the 5′ LTR directs the synthesis of mRNAs giving rise to structural (Gag and Env), enzymatic (Pol), and auxiliary (Tas, Bet) proteins, whereas the IP, located within the 3′ end of the env gene, allows the expression of auxiliary proteins. Transcriptional activity of both promoters depends on the presence of the homologous viral transactivator Tas, although the basal activity of the IP is constantly higher (22). These two promoters were previously identified in EFV and were found to be sensitive to the presence of the homologous Tas (40). Finally, sequence analysis revealed that the EFV envelope (Env) glycoprotein does not harbor the characteristic endoplasmic reticulum (ER) retrieval motif located in the C terminus of primate FV Env (12). Since this dilysine motif has been shown to be responsible for the budding of HFV from the membranes of the ER (11), its absence in EFV Env should lead to a change in the viral budding site.
Here, to further characterize this new isolate, the transcriptional cap sites in the 5′ LTR and the IP as well as the poly(A) addition site in the 3′ LTR were determined by primer extension and reverse transcription (RT)-PCR analysis. The main splice sites were mapped by RT-PCR, demonstrating the existence of the characteristic subgenomic pol mRNA. We have also identified a Tas-defective EFV genome which has been detected mainly during persistent infections. Subcloning of the EFV gag, env, and tas genes allowed us to determine the size and the subcellular localization of the corresponding viral products by radioimmunoprecipitation assay and confocal microscopy. While EFV Gag harbors a nucleocytoplasmic distribution, similar to other FVs, EFV Env is mainly detected in the Golgi complex but not in the ER. In line with these observations, electron microscopy analysis demonstrated that EFV buds at the plasma membrane. Interestingly, EFV Tas was detected both in the nucleus and the cytoplasm, in contrast to the strict nuclear localization of other known FV transactivators but similar to the subcellular localization of Tat from the equine infectious anemia virus (EIAV). Despite this similarity, EFV Tas was unable to transactivate the EIAV LTR.
MATERIALS AND METHODS
Cells and virus.
Mycoplasma-free EFV stocks were produced on ED cells, a horse fibroblast cell line, maintained in RPMI medium supplemented with l-glutamine (2 mM), penicillin (100 μg/ml), streptomycin (50 μg/ml), HEPES (240 mM), and 8% heat-inactivated fetal calf serum. Simian Cos6 and hamster BHK21 cells were maintained in Dulbecco's modified Eagle medium (Gibco-BRL) supplemented with l-glutamine (2 mM), penicillin (100 μg/ml), streptomycin (50 μg/ml), HEPES (240 mM), and 5% fetal calf serum. Following EFV-induced cell lysis, BHK21 cells were maintained in culture to establish a persistent infection, an approach already employed to generate cell lines persistently infected with HFV (34).
Nucleic acids analysis.
For Southern blotting, genomic DNA was extracted with the QIAamp Tissue Kit (Qiagen), digested by NheI, and separated in a 0.8% agarose gel. Hybridization was carried out overnight at 42°C with a radiolabeled (Prime-a-Gene labeling system; Promega) NheI probe (nucleotides 9445 to 11078) derived from an λEMBL3 clone harboring a full-length EFV genome (λrEFV) (40) in a solution containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 5× Denhardt's solution, and 50% formamide. Washings were performed in 0.1× SSC-0.1% SDS buffer at 50°C for 30 min.
For Northern blotting, total RNAs from EFV-infected cells were extracted by Trizol (Invitrogen) according to the manufacturer's instructions. About 10 μg of RNA was loaded on a 1% formaldehyde-agarose gel, blotted to a nitrocellulose filter according to the method of Fourney et al. (8), and hybridized as described above with a α-32P-labeled U3-R-specific probe, amplified from λrEFV (sense, 5′-CAGAAAGGGATTAAAGGA-3′; antisense, 5′-GCAGAGCAAATTAAGTGC-3′).
Splice and polyadenylation signal.
Viral RNAs were reverse transcribed with the OneStep RT-PCRv Kit (Qiagen) performed on total RNAs extracted from EFV-infected cells. Primers used for determination of the EFV splicing pattern are available on request. RT-PCR performed to determine the polyadenylation signal was performed with the primer 5′-CCAAGCGTGACGCATTCTGT-3′ (U3-specific oligonucleotide) and a 20-nucleotide poly(dT) primer. RT-PCR products were cloned into the pGEMT vector (Promega) and were sequenced.
Primer extension.
Primer extensions were carried out by using Superscript RT (Invitrogen) on total RNA extracted from EFV-infected ED cells. Minus-strand primers (LTR+1, GAGCAAATTAAGTGCATC; IP+1, CAGGCCAGATGTCTTCTA) were end-labeled in a reaction mixture containing 100 pmol of primer, 1× kinase buffer, 2.8 μM [γ-32P]ATP (NEG-035C; NEN), and 5 U of T4 polynucleotide kinase (New England Biolabs) for 45 min at 37°C. The reaction was stopped by incubation at 55°C for 5 min and then was purified on a G50 column. Sequences were determined with the ThermoSequenase kit (United States Biochemicals).
Recombinant DNA.
λrEFV was the source of viral DNA for the following constructs. The EFV Gag expression plasmid was generated by insertion of a 2.6-kb StuI-BglII fragment into the SmaI site of pSG5M (provided by Libin Ma, Leiden, The Netherlands). The EFV Env expression plasmid was PCR cloned from λrEFV (primers are available on request) into the pTarget vector (Promega). Note that the pSGEFV-Tas and pSGEFV-Bet vectors, expressing Tas and Bet, respectively, have been already described (16, 40). The vector pSGHFV-Env, coding for HFV Env, was described by Giron et al. (10) and was given the name p1EnvΔBel.
Transactivation experiments.
The EIAV LTR was extracted from pEIAV/eTAR/CAT (kindly provided by P. Bieniasz and B. Cullen) with BamHI and HindIII and was inserted in the promoterless pGL3basic upstream of the firefly luciferase gene (Promega). The EIAV Tat-expressing vector was provided by P. Bienasz and B. Cullen. Plasmids directing the expression of EFV Tas (pSVEFV-Tas) and of the firefly luciferase under the control of the EFV LTR (pEFV-LTR) were already described (40). The pRL-CMV vector, directing the expression of the renilla luciferase under the control of the cytomegalovirus immediate-early promoter, was used for normalization (Dual-Luciferase Reporter Assay System; Promega).
Antibodies.
Polyclonal antibodies against EFV were generated following infection of two male New Zealand White rabbits (R77 and R98, 8 months of age) with a single subcutaneous dose of EFV as already described (35). These antibodies, as well as polyclonal anti-HFV antibodies (35), were diluted at 1/400. To generate mouse polyclonal antibodies against EFV Gag and EFV Env, mice were DNA immunized. Briefly, mice were first treated with cardiotoxin (10 μM; Sigma), and 100 μg of Gag- or Env-expressing plasmids diluted in phosphate-buffered saline (PBS) was injected bilaterally into each quadriceps muscle 1 week later. Plasmid injection was repeated twice during the two following weeks, and sera were collected 20 days after the last DNA injection (a precise procedure is described at the web site http://www.dnavaccine.com/).
Antibodies used also included a monoclonal antibody against a centriolar protein, diluted at 1:50 (CTR910; provided by M. Bornens [1]), a monoclonal antibody against GM130, diluted at 1:100 (provided by F. Perez), and a goat anti-lactate dehydrogenase (LDH) antibody (used at a 1:2,000; Sigma). These antibodies were detected with anti-immunoglobulin G fluorescein isothiocyanate (FITC) or Texas Red-coupled antibodies (Biosys) or anti-mouse or anti-rabbit peroxidase-conjugated antibodies (diluted at 1:8,000; Sigma). When used alone, these secondary antibodies gave no staining on infected or transfected cells.
Protein analysis.
For transfection experiments, about 5 × 105 cells were transfected with 1 μg of recombinant plasmid and the Lipofectin reagent (Gibco-BRL) as specified by the manufacturer. All plasmids used for transfection were purified on anion-exchange resin columns (Qiagen).
For immunofluorescence, 48 h posttransfection cells were fixed with 4% paraformaldehyde at 4°C for 10 min, permeabilized with methanol at 4°C for 5 min, and then washed in PBS-0.1% Tween-0.2% bovine serum albumin. Nuclei were revealed with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) at 250 ng/ml. For confocal analysis cells were embedded in Mowiol (Calbiochem) and were examined with a Bio-Rad MRC-1024 confocal imaging system (Hertforshire, United Kingdom) and an inverted Diaphot 300 Nikon microscope. Images were collected with an oil immersion lens (60×, NA I0.4 plan Apochromat). For fluorescein, a krypton/argon ion laser (Ion Laser Technology Inc., Salt Lake City, Utah) operating with a 488-nm line was used. Texas Red-X was excited at 568 nm, and fluorescence above 590 nm was captured. For DAPI excitation, an Enterprise ion laser (Coherent Laser Group, Santa Clara, Calif.) operating with a 353-nm nonline was used. FITC, Texas Red, and DAPI images were pseudocolored green for FITC, blue for DAPI, and red for Texas Red.
For immunoprecipitation, 36 h posttransfection or 72 h postinfection cells were labeled with [35S]methionine-cysteine (75 μCi/ml, 1.175 Ci/mmol specific activity; Dupont NEN) for 7.5 h. Cells were harvested by being scraped into cold PBS and then lysed in a solution containing 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.05% SDS, and 1 mM phenylmethylsulfonyl fluoride for 30 min at 4°C. Nuclei were separated from the lysate by centrifugation at 12,000 × g for 5 min at 4°C and were lysed in the same buffer containing 1 M NaCl. Immune complexes with Protein A Sepharose were centrifuged and washed four times in lysis buffer and analyzed by SDS-5 to 15% polyacrylamide gel electrophoresis, followed by autoradiography.
For Western blot analysis, 48 h posttransfection cells were heat disrupted at 100°C for 5 min in Laemmli sample buffer. About 30 μg of protein extracts were resolved by SDS-polyacrylamide gel electrophoresis, transferred by electroblotting onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore), and revealed by enhanced chemiluminescence (Amersham).
Electron microscopy.
Monolayers were fixed in situ with 1.6% glutaraldehyde (Taab Laboratory Equipment Ltd., Reading, United Kingdom) in 0.1 M Sörensen phosphate buffer, pH 7.3 to 7.4, for 1 h at 4°C. Cells were scraped from their plastic substratum and were centrifuged. The resulting pellets were successively postfixed with 2% aqueous osmium tetroxide for 1 h at room temperature, dehydrated in ethanol, and embedded in Epon. Ultrathin sections were collected on 200-mesh copper grids coated with Formvar and carbon and stained with uranyl acetate and lead citrate prior to being observed with a Philips 400 transmission electron microscope at 80 kV and 2,800× to 36,000× magnification.
RESULTS
Characterization of the transcriptional cap and poly(A) addition sites.
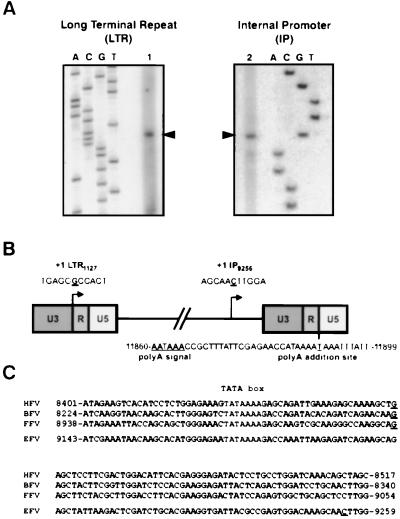
Besides the 5′ LTR, an internal promoter located at the 3′ end of the env gene was also described for EFV (40). To further characterize these transcriptional units as well as the 3′ LTR poly(A) addition site, total RNA extracted from EFV-infected horse ED cells was analyzed in primer extension reactions and by RT-PCR, respectively. Sequence analysis was performed by using radiolabeled specific antisense primers located downstream of the putative transcriptional cap sites predicted by DNA sequence alignment (Fig. 1A) (40). As shown in Fig. 1B, the transcriptional cap site in the LTR is located at position 1127. The 5′ end of EFV transcripts initiated at the IP is mapped to a cytosine at position 9256, which is 79 nucleotides 3′ of the TATA box, 53 bp downstream of known FV IP cap sites (Fig. 1C). Concomitantly, RT-PCR analysis using specific U3 and poly(dT) primers allowed us to locate the poly(A) addition site at position 11890 (Fig. 1B).
FIG. 1.
Characterization of the transcriptional cap and poly(A) addition sites of EFV. (A) Primer extension performed on total RNA from EFV-infected ED cells revealed that the LTR cap site (1) is located at position 1127 (left panel), whereas the IP cap site (2) is located at position 9256 (right panel). These sites are represented on a schematic diagram (B). The polyadenylation signal has been located at position 11860 on the basis of sequence analysis and the poly(A) addition site at position 11890 on the basis of RT-PCR results. (C) Sequence alignment reveals that the EFV IP cap site is more distant from the TATA box than other described FV cap sites (the TATA box is in bold lettering, and cap sites are underlined).
RT-PCR analysis reveals a complex EFV splicing pattern.
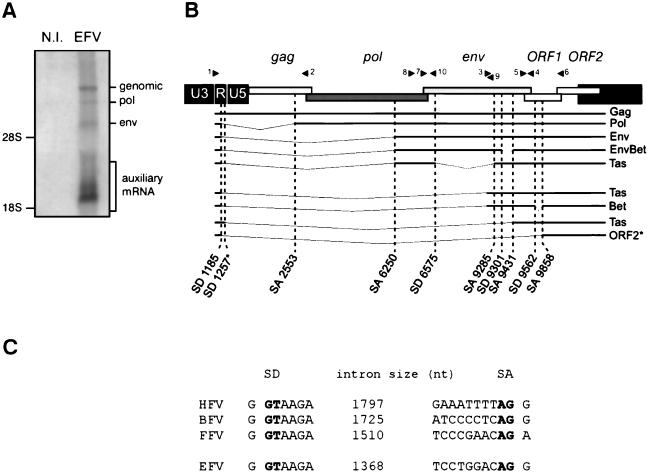
To assess the presence of the specific subgenomic pol transcript and to build a splicing map of EFV transcriptional expression, total RNA from EFV-infected horse ED cells were collected and first analyzed by Northern blot. As shown in Fig. 2A, the different viral mRNA species can be readily detected by using a radiolabeled U3-R probe. To identify the splice donor and acceptor sites, RT-PCR analysis was performed with distinct sets of primers, and the corresponding products were subcloned into the pGEM-T vector and sequenced. After this procedure the main splice donor and acceptor sites were identified, each splice being confirmed in three independent clones. The EFV splicing pattern was finally drawn (Fig. 2B), demonstrating the existence of a specific transcript coding for the recently described Env-Bet fusion protein (10, 19) and also the existence of a subgenomic pol mRNA, one characteristic of FVs. For the latter, comparison with other FVs confirmed the previously highlighted consensus flanking the splice sites (Fig. 2C). Note that the EFV leader sequence (LS) is 58 nucleotides long, the shortest LS discovered among retroviruses, in agreement with what has been reported for other FVs. Interestingly, we have isolated in one clone a splice donor site in the R region of the LTR which is located 72 bp from the consensual one (position 1257) and which fuses to the acceptor splice site at position 9431. Similarly, a single clone (SD1185-SA9858), allowing the synthesis of a putative ORF2 product, has been isolated. Whether this mRNA is efficiently translated into a viral polypeptide remains to be determined.
FIG. 2.
EFV splicing pattern. (A) The different viral mRNA species from EFV-infected ED cells are detected by Northern blot with a specific radiolabeled U3-R probe. N.I., not infected. (B) RT-PCR assays performed on total RNA from EFV-infected cells with distinct sets of primers (black arrowheads) reveal a complex pattern of viral mRNA expression. The splice donor (SD) and splice acceptor (SA) sites are indicated for each viral mRNA species. Note the presence of a second SD site at the R region of the 5′ LTR (position 1257). A putative ORF2-encoding mRNA was also isolated. These two mRNA species were found in a single clone (*). (C) Alignment of the pol mRNA splice junctions of HFV, BFV, FFV, and EFV. SD and SA are indicated in bold lettering. nt, nucleotides.
EFV generates a Tas-defective genome.
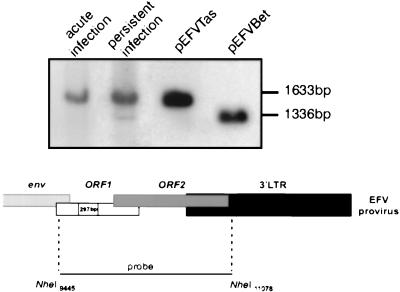
In the case of HFV, several lines of evidence suggest that a defective molecular form harboring a deletion in the tas gene (HFVΔTas) could participate in the establishment of viral persistence (2, 34, 35). Therefore, we sought to determine whether EFV generates such a defective viral genome. For that purpose genomic DNA was extracted from acutely or persistently infected BHK21 cells and was digested with the NheI enzyme; those sites flank the bel region (Fig. 3). The pSVEFV-Tas and pSGEFV-Bet plasmids, which were digested by the same enzyme, were used as size controls. Using the 1,633-bp NheI fragment as a probe, a single band can be detected in acutely infected cells. This band comigrates with the fragment derived from pSVEFV-Tas and thus likely represents the wild-type EFV DNA genome (40). Interestingly, in persistently infected BHK21 cells two signals were detected, one band corresponding to the wild-type virus and a second, shorter band of approximately 300 bp. The plasmid coding for EFV Bet, which harbors a 297-bp deletion in the tas gene (16), gave rise to a band of a similar size. Therefore, the shorter form detected during EFV persistent infections likely represents a defective EFV genome harboring a deletion in the tas gene (Fig. 3). This was further confirmed by the lack of hybridization of the shorter band when using the 297-bp Bet intron as a probe (data not shown). These results demonstrated that a nonprimate FV generates a tas-defective genome during persistent infection.
FIG. 3.
Characterization of a Tas-defective EFV DNA genome. Genomic DNA from BHK21 cells acutely or persistently infected with EFV was analyzed by Southern blot using a probe encompassing the bel region. Plasmids coding for EFV Tas (pSVEFV-Tas) or EFV Bet (pSGEFV-Bet, harboring a 297-bp deletion in the tas gene) were used as size controls. A Tas-defective DNA genome is detected in cells persistently infected with EFV.
Characterization of EFV proteins.
To characterize the EFV proteins, polyclonal antibodies against EFV antigens were generated following infection of two rabbits with one unique dose of EFV stock. As early as 5 weeks postinfection their sera were unambiguously found positive against the main EFV polypeptides (data not shown). This method was successfully employed to raise powerful rabbit polyclonal antibodies against HFV (35). Moreover, specific antisera against EFV Gag and Env were raised by DNA immunization of mice following injection of their respective expressing vectors.
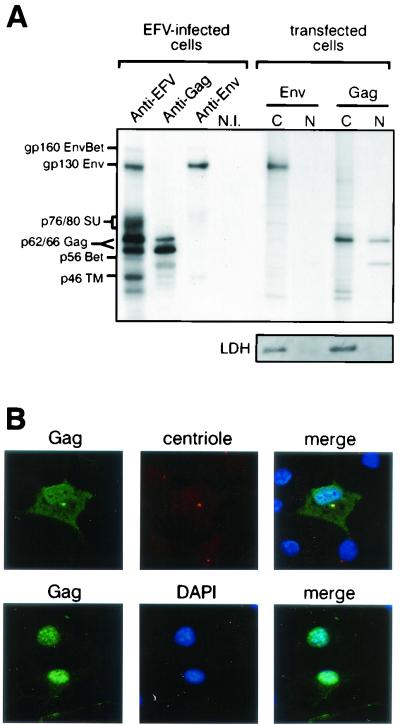
As shown in Fig. 4A, specific bands from EFV-infected ED cells were immunoprecipitated with the sera from EFV-infected rabbits, while no reactivity was observed on protein extracts from mock-infected cells or when using sera from uninfected rabbits (data not shown). The sizes of the main viral products, predicted on the basis of sequence analysis (40), have been confirmed here, as the characteristic Gag doublet was detected at 66 and 62 kDa, the Env precursor at 130 kDa, and the Bet protein at 56 kDa (16). The use of specific sera confirmed these observations and allowed the specific detection of the transmembrane (TM) and surface (SU) subunits of Env, detected at 46 kDa and as a smear between 76 and 80 kDa, respectively, (Fig. 4A). A band at 160 kDa can also be detected with anti-EFV antibodies and with anti-Env antiserum, which likely represents an Env-Bet fusion protein (10, 19) (Fig. 4A). To further characterize the EFV structural proteins, the subcellular distribution of Gag and Env proteins was studied by immunoprecipitation on nuclear and cytoplasmic fractions of Cos6 cells transfected with either pSGEFV-Gag or pSGEFV-Env. The Env glycoprotein was mainly detected as the 130-kDa precursor in the cytoplasm, while Gag was also detected in the nucleus, consistent with the data reported previously for HFV Gag (9, 38) (Fig. 4A). Note that only the Env precursor can be detected when Env is expressed alone. The efficiency of the subcellular fractionation was controlled by Western blot by using a monoclonal antibody directed against the LDH, a soluble protein, strictly confined into the cytoplasm.
FIG. 4.
Characterization of the EFV proteins. (A) Proteins extracted from EFV-infected ED cells were immunoprecipitated with a rabbit anti-EFV antiserum or mouse anti-Gag or anti-Env antibodies. The EFV Gag doublet is detected at 66/62 kDa, whereas the Env precursor is detected at 130 kDa and the TM and SU subunits are detected near 46 and 76 to 80 kDa, respectively. EFV Bet is detected at 56 kDa. A 160-kDa protein which is detected with both anti-EFV and anti-Env antisera likely represents an Env-Bet fusion glycoprotein. Immunoprecipitation of nuclear (N) and cytoplasmic (C) fractions of Env- or Gag-expressing BHK21 cells revealed that, while Env is mainly detected in the cytoplasm, Gag is distributed in both the cytoplasm and the nucleus. The efficiency of the subcellular fractionation is assessed by Western blot by using a monoclonal antibody directed against the LDH. (B) Confocal analysis of Gag-transfected BHK21 cells performed 20 h posttransfection (top) or 48 h posttransfection (bottom). Whereas Gag is detected at the vicinity of the centrosome (detected with an anti-centriole antibody, CTR910) early after transfection, its localization is mainly nuclear 48 h posttransfection. Nuclei were revealed with DAPI.
Finally, EFV Gag subcellular localization was further explored by immunofluorescence on pSGEFV-Gag-transfected Cos6 cells with rabbit anti-EFV sera. Confocal analysis revealed that, early after transfection (Fig. 4B, top), EFV Gag was detected at the vicinity of the centrosome, as described for HFV Gag (37; C. Petit, M. L. Giron, J. Tobaly-Tapiero, P. Bittoun, E. Réal, Y. Jacob, N. Tordo, and A. Saïb, unpublished data) and suggested for FFV (3), and later it was detected in the nucleus (Fig. 4B, bottom).
EFV buds at the plasma membrane.
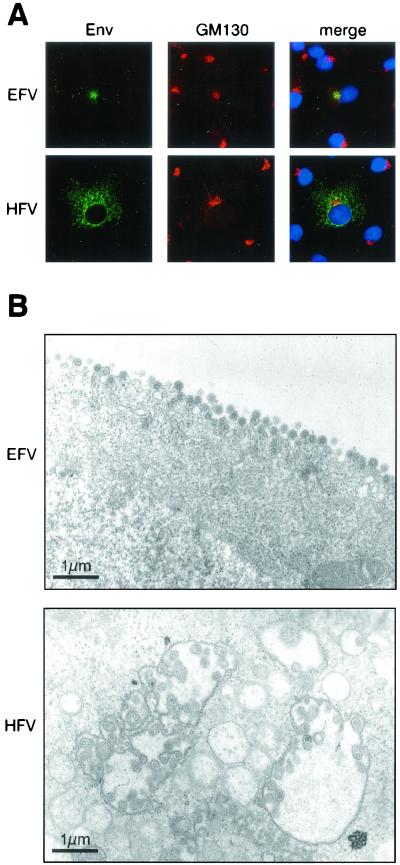
Since HFV Env harbors an ER retrieval motif responsible for its sequestration in the ER (11, 12), we wondered whether EFV Env, lacking this motif, could target the plasma membrane by following the classical route of a viral glycoprotein (28). Cos6 cells were transfected with either pSGEFV-Env or pSGHFV-Env, and 2 days later immunofluorescence analysis of fixed and permeabilized transfected cells was performed with rabbit anti-HFV or anti-EFV sera. Confocal microscopy showed that at the steady state EFV Env mainly colocalized with GM130, a cis-Golgi matrix protein (25), whereas HFV Env harbored a perinuclear staining corresponding to the ER (Fig. 5A). Given that the proteolytic maturation of retroviral glycoproteins occurs in the distal trans-Golgi network (28), this observation is in agreement with our biochemical data showing that EFV Env is not cleaved into SU and TM subunits when expressed alone. Moreover, in a fusion assay EFV Env does not yield enhanced cell fusion activity (data not shown). Finally, to determine whether the absence of the dilysine motif in EFV Env could direct viral budding to the plasma membrane, electron microscopy analysis was performed on EFV-infected BHK21 cells. Figure 5B reveals that EFV virions bud exclusively from the plasma membrane, whereas HFV budding mainly occurs from intracellular membranes, demonstrating that this new FV isolate follows a distinct pathway to exit the cell.
FIG. 5.
Subcellular distribution of the EFV glycoprotein. (A) Confocal microscopy of pSGEFVEnv- or pSGHFVEnv-transfected Cos6 cells reveals that EFV Env mainly localized in the cis-Golgi complex stained with anti-GM130 antibodies. Nuclei are stained with DAPI in merge images. (B) Electron micrographs of ultrathin sections of EFV- or HFV-infected BHK21 cells. Images represent budding of viral particles at the plasma membrane for EFV and at internal membranes for HFV. The bars correspond to 1 μm.
The EFV transactivator is not strictly confined to the nucleus.
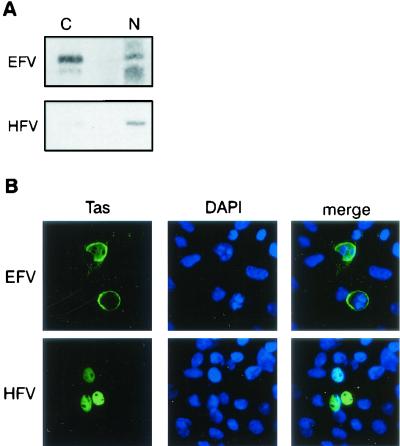
One common feature of all cloned FV Tas is their strict nuclear localization, which is required to achieve their transactivation function (27). To study the subcellular distribution of EFV Tas, Cos6 cells were transfected with pSVEFV-Tas and pSVHFV-Tas, which was used as a control. Forty-eight hours posttransfection, nuclear and cytoplasmic fractions were analyzed by Western blot with rabbit anti-EFV or anti-HFV sera. Figure 6A shows that whereas HFV Tas was mainly detected in the nucleus, EFV Tas was present in both the nucleus and the cytoplasm of transfected cells. The use of a monoclonal antibody directed against LDH assessed the efficiency of the subcellular fractionation (data not shown). This result was further confirmed by confocal microscopy using anti-EFV and anti-HFV polyclonal sera, revealing nuclear and cytoplasmic perinuclear stainings for EFV Tas, whereas HFV Tas is solely detected in the nucleus (Fig. 6B). Similar observations were obtained with hamster BHK21 or human U373-MG cells (data not shown). Interestingly, a similar subcellular distribution was also reported for Tat from EIAV, which also constitutes an exception from the lentiviral family (4, 33). Given that EFV Tas and EIAV Tat share a similar subcellular distribution and that HFV Tas was shown to transactivate the LTR of human immunodeficiency virus type 1 (14, 17), we sought to investigate a possible transcriptional interaction between EFV and EIAV. For that purpose, Cos6 cells were transfected with an EIAV Tat- or an EFV Tas-expressing vector together with a vector carrying a luciferase reporter gene under the control of either the EIAV or the EFV LTR. As shown in Table 1, whereas EIAV Tat and EFV Tas transactivated their homologous promoters, EFV Tas was unable to transactivate the LTR of EIAV. Similar results were obtained with hamster BHK21, canine Cf2Th, and horse ED cells (data not shown).
FIG. 6.
Subcellular distribution and transactivation properties of EFV Tas. (A) Western blot analysis of nuclear and cytoplasmic fractions of pSVEFV- and pSVHFV-Tas-transfected Cos6 cells. EFV Tas is distributed both in the nucleus and the cytoplasm, whereas HFV Tas is mainly nuclear. (B) Confocal microscopy of cells expressing EFV Tas or HFV Tas confirms the biochemical data and reveals that EFV Tas is mainly perinuclear. Nuclei are stained with DAPI.
TABLE 1.
Cross-transactivation experiments between EFV and EIAV fail to detect a transactivation of the EIAV LTR by EFV Tas
| Vector | Transactivation fold increase for:
|
|
|---|---|---|
| EFV LTR | EIAV LTR | |
| Mock transfection | 1 ± 0.4 | 1 ± 0.5 |
| EFV Tas | 218 ± 70 | 0.8 ± 0.3 |
| EIAV Tat | 5.8 ± 0.2 | 27.5 ± 8.6 |
DISCUSSION
EFV presents the main characteristics defining the FV retroviral genus, such as the induction of specific cytopathic effects in cell culture and major sequence homologies with other FVs (40). Here we demonstrated the existence of the specific spliced pol mRNA giving rise to Pol independently of Gag synthesis, one feature of FVs. The locations of other donor and acceptor splice sites described in this report are consistent with what has been defined for other FVs (29). Interestingly, we were able to identify a Tas-defective genome only in BHK21 cells persistently infected with EFV and not during acute infection, in agreement with previous reports on HFV (20, 26, 30, 34, 36). Further experiments are needed to determine whether EFVΔTas is free or integrated during chronic infection. Also, whether EFVΔTas is implicated in viral persistence, as has been suggested for HFVΔTas (34), remains to be elucidated.
Construction of expression vectors coding for the main viral products and the generation of specific antibodies against the main EFV proteins allowed us to determine the molecular sizes and to determine the subcellular localizations of EFV Gag, Env, and Tas. Note that the molecular characterization of EFV Bet has been described elsewhere (16). As for other FVs, EFV Gag is detected in both the nucleus and the cytoplasm of Gag-transfected cells but concentrates in the nucleus 48 h posttransfection, consistent with previous reports on HFV Gag (38). However, soon after transfection Gag exclusively localizes in the cytoplasm, close to the microtubule organizing center, as reported for HFV (37; C. Petit et al., unpublished data) and suggested for FFV (3). The situation is quite different concerning the Env glycoprotein. In the case of HFV Env, sequestration in the ER prevents its cleavage by a subtilisin-like endoprotease into mature TM and SU subunits which occurs in the distal trans-Golgi apparatus (12, 28). Strikingly, EFV Env, which lacks this retrieval motif, is only detected as an immature precursor when expressed alone and localized in the cis-Golgi complex. However, despite this uncommon Env subcellular distribution, electron microscopy studies have shown that EFV mainly buds at the plasma membrane. Therefore, interaction of Env with another viral component seems to be required for its progression through the secretory pathway. Several lines of evidence support the idea that Gag could play such a role for HFV. First, syncytia formation and therefore cell surface expression of HFV Env, requires coexpression of Gag (32). Moreover, an interaction between Gag and its homologous envelope is absolutely required to allow FV virus egress, which is in contrast to what has been described for other retroviruses (7, 18, 27, 31, 41). Our results support the idea that intracellular trapping of FV Env is not essentially due to the presence of an ER retrieval dilysine motif as previously suggested (12) but rather seems to require other Env domains. Finally, note that EFV Gag, like other FV Gag, harbors no obvious membrane targeting motif, such as a myristylation signal (40). Instead, a conserved Arg residue located in a region resembling the cytoplasmic targeting and retention signal of the Mason-Pfizer monkey virus (5) has been described in the N terminus of EFV Gag (7). Although this domain has been shown to be responsible for intracellular HFV capsid assembly, whether this motif plays a similar role for EFV remains to established.
The particular subcellular localization of EFV Tas constitutes another specific feature of this new FV isolate. Indeed, in contrast to all described FV Tas which are strictly nuclear (3, 21), EFV Tas also localizes in the cytoplasm of transfected cells. This situation is unique among FVs and is rather intriguing, since transactivation of the viral promoters should occur in the nucleus following proviral integration. Interestingly, the transactivator of the equine lentivirus EIAV shares a similar subcellular distribution (4, 33). In that case also, the situation appears to be unique among lentiviruses. The reason for this is not particularity well understood, but it has been suggested that cellular cofactors involved in Tat-mediated transactivation of the EIAV LTR are recruited by Tat in the cytoplasm prior to nuclear migration (4). In the case of EFV Tas and in regards to all FV Tas, it could be interesting to identify such cellular cofactors which might be involved in the regulation of their subcellular distribution and therefore in the modulation of LTR and/or IP transcription. Nucleocytoplasmic distribution of EFV Tas may constitute a new mechanism to regulate viral gene expression, as reported for several viral or cellular transcription factors (reviewed in reference 6). However, despite a similar subcellular distribution of their transactivator, no cross-transactivation has been pointed out between these two equine retroviruses, once again in contrast to what has been reported between HFV and human immunodeficiency virus (14, 17, 24).
Taken together, the results presented in this report show that EFV presents unusual properties among FVs. Whether theses characteristics are shared by all nonprimate FVs remains to be determined.
Acknowledgments
We thank the Labo Photo Hematologic for photographic work; Michel Schmidt for confocal analysis; M. C. Guillemin, B. Gicquel, and Stephan Zientara for the preparation of rabbit antisera; and P. Bienasz and B. Cullen for their kind gift of an EIAV Tat-expressing vector and pEIAV/eTAR/CAT. We thank Hugues de Thé for continuous support.
This work was financed by Association de Recherche sur le Cancer (grant no. 5981), F. Lacoste, and Conseil Général de la Manche.
REFERENCES
- 1.Bailly, E., M. Doree, P. Nurse, and M. Bornens. 1989. p34cdc2 is located in both nucleus and cytoplasm; part is centrosomally associated at G2/M and enters vesicles at anaphase. EMBO J. 8:3985-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock, M., M. Heinkelein, D. Lindemann, and A. Rethwilm. 1998. Cells expressing the human foamy virus (HFV) accessory Bet protein are resistant to productive HFV superinfection. Virology 250:194-204. [DOI] [PubMed] [Google Scholar]
- 3.Bodem, J., M. Zemba, and R. M. Flügel. 1998. Nuclear localization of the functional Bel 1 transactivator but not of the gag proteins of the feline foamy virus. Virology 251:22-27. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, R., B. M. Peterlin, and D. Derse. 1992. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous trans activators. J. Virol. 66:2000-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, G., S. Park, B. Choi, S. Hong, J. Lee, E. Hunter, and S. S. Rhee. 1999. Identification of a cytoplasmic targeting/retention signal in a retroviral Gag polyprotein. J. Virol. 73:5431-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cyert, M. S. 2001. Regulation of nuclear localization during signaling. J. Biol. Chem. 276:20805-20808. [DOI] [PubMed] [Google Scholar]
- 7.Eastman, S. W., and M. L. Linial. 2001. Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J. Virol. 75:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fourney, R. M., J. Miyakoshi, R. S. Day, and M. C. Paterson. 1988. Northern blotting: efficient RNA staining and transfer. Focus 10:5-7. [Google Scholar]
- 9.Giron, M. L., S. Colas, J. Wybier, F. Rozain, and R. Emanoil-Ravier. 1997. Expression and maturation of human foamy virus Gag precursor polypeptides. J. Virol. 71:1635-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giron, M. L., H. de Thé, and A. Saïb. 1998. An evolutionarily conserved splice generates a secreted env-Bet fusion protein during human foamy virus infection. J. Virol. 72:4906-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goepfert, P. A., K. Shaw, G. Wang, A. Bansal, B. H. Edwards, and M. J. Mulligan. 1999. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J. Virol. 73:7210-7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goepfert, P. A., K. L. Shaw, G. D. Ritter, Jr., and M. J. Mulligan. 1997. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J. Virol. 71:778-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, F., W. S. Blair, J. Fukushima, and B. R. Cullen. 1996. The human foamy virus Bel-1 transcription factor is a sequence-specific DNA binding protein. J. Virol. 70:3902-3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller, A., E. D. Garrett, and B. R. Cullen. 1992. The Bel-1 protein of human foamy virus activates human immunodeficiency virus type 1 gene expression via a novel DNA target site. J. Virol. 66:3946-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecellier, C. H., and A. Saïb. 2000. Foamy viruses: between retroviruses and pararetroviruses. Virology 271:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Lecellier, C. H., W. Vermeulen, F. Bachelerie, M. L. Giron, and A. Saïb. 2002. Intra- and intercellular trafficking of the foamy virus auxiliary Bet protein. J. Virol. 76:3388-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, A. H., K. J. Lee, S. Kim, and Y. C. Sung. 1992. Transactivation of human immunodeficiency virus type 1 long terminal repeat-directed gene expression by the human foamy virus bel1 protein requires a specific DNA sequence. J. Virol. 66:3236-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindemann, D., and A. Rethwilm. 1998. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J. Virol. 72:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linial, M. 2000. Why aren't foamy viruses pathogenic? Trends Microbiol. 8:284-289. [DOI] [PubMed] [Google Scholar]
- 21.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Löchelt, M., M. Aboud, and R. M. Flügel. 1993. Increase in the basal transcriptional activity of the human foamy virus internal promoter by the homologous long terminal repeat promoter in cis. Nucleic Acids Res. 21:4226-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Löchelt, M., W. Muranyi, and R. M. Flügel. 1993. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc. Natl. Acad. Sci. USA 90:7317-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marino, S., C. Kretschmer, S. Brandner, C. Cavard, A. Zider, P. Briand, S. Isenmann, E. F. Wagner, and A. Aguzzi. 1995. Activation of HIV transcription by human foamy virus in transgenic mice. Lab. Investig. 73:103-110. [PubMed] [Google Scholar]
- 25.Marra, P., T. Maffucci, T. Daniele, G. D. Tullio, Y. Ikehara, E. K. Chan, A. Luini, G. Beznoussenko, A. Mironov, and M. A. De Matteis. 2001. The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat. Cell Biol. 3:1101-1113. [DOI] [PubMed] [Google Scholar]
- 26.Meiering, C. D., K. E. Comstock, and M. L. Linial. 2000. Multiple integrations of human foamy virus in persistently infected human erythroleukemia cells. J. Virol. 74:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meiering, C. D., and M. L. Linial. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moulard, M., and E. Decroly. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469:121-132. [DOI] [PubMed] [Google Scholar]
- 29.Muranyi, W., and R. M. Flügel. 1991. Analysis of splicing patterns of human spumaretrovirus by polymerase chain reaction reveals complex RNA structures. J. Virol. 65:727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neves, M., J. Périès, and A. Saïb. 1998. Study of human foamy virus proviral integration in chronically infected murine cells. Res. Virol. 149:393-401. [DOI] [PubMed] [Google Scholar]
- 31.Pietschmann, T., M. Heinkelein, M. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietschmann, T., H. Zentgraf, A. Rethwilm, and D. Lindemann. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J. Virol. 74:4474-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosin-Arbesfeld, R., P. Mashiah, D. Willbold, P. Rosch, S. R. Tronick, A. Yaniv, and A. Gazit. 1994. Biological activity and intracellular location of the Tat protein of equine infectious anemia virus. Gene 150:307-311. [DOI] [PubMed] [Google Scholar]
- 34.Saïb, A., M. H. Koken, P. van der Spek, J. Périès, and H. de Thé. 1995. Involvement of a spliced and defective human foamy virus in the establishment of chronic infection. J. Virol. 69:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saïb, A., M. Neves, M. L. Giron, M. C. Guillemin, J. Valla, J. Périès, and M. Canivet. 1997. Long-term persistent infection of domestic rabbits by the human foamy virus. Virology 228:263-268. [DOI] [PubMed] [Google Scholar]
- 36.Saïb, A., J. Périès, and H. de Thé. 1993. A defective human foamy provirus generated by pregenome splicing. EMBO J. 12:4439-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saïb, A., F. Puvion-Dutilleul, M. Schmid, J. Périès, and H. de Thé. 1997. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J. Virol. 71:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schliephake, A. W., and A. Rethwilm. 1994. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 68:4946-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schweizer, M., H. Schleer, M. Pietrek, J. Liegibel, V. Falcone, and D. Neumann-Haefelin. 1999. Genetic stability of foamy viruses: long-term study in an African green monkey population. J. Virol. 73:9256-9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tobaly-Tapiero, J., P. Bittoun, M. Neves, M. C. Guillemin, C. H. Lecellier, F. Puvion-Dutilleul, B. Gicquel, S. Zientara, M. L. Giron, H. de Thé, and A. Saïb. 2000. Isolation and characterization of an equine foamy virus. J. Virol. 74:4064-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilk, T., V. Geiselhart, M. Frech, S. D. Fuller, R. M. Flügel, and M. Löchelt. 2001. Specific interaction of a novel foamy virus Env leader protein with the N-terminal Gag domain. J. Virol. 75:7995-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]