Abstract
Varicella-zoster virus (VZV) transcription is limited in latently infected human ganglia. Note that much of the transcriptional capacity of the virus genome has not been analyzed in detail; to date, only VZV genes mapping to open reading frames (ORFs) 4, 21, 29, 62, and 63 have been detected. ORF 62 encodes the major immediate-early virus transcription transactivator IE62, ORF 29 encodes the major virus DNA binding protein, and ORF 21 encodes a protein associated with the developing virus nucleocapsid. We analyzed the cellular location of proteins encoded by ORF 21 (21p) and ORF 29 (29p), their phosphorylation state during productive infection, and their ability form a protein-protein complex. The locations of both 21p and 29p within infected cells mimic those of their herpes simplex virus type 1 (HSV-1) homologues (UL37 and ICP8); however, unlike these homologues, 21p is not phosphorylated and neither 21p nor 29p exhibits a protein-protein interaction. Transient transfection assays to determine the effect of 21p and 29p on transcription from VZV gene 20, 21, 28, and 29 promoters revealed no significant activation of transcription by 21p or 29p from any of the VZV gene promoters tested, and 21p did not significantly modulate the ability of IE62 to activate gene transcription. A modest increase in IE62-induced activation of gene 28 and 29 promoters was seen in the presence of 29p; however, IE62-induced activation of gene 28 and 29 promoters was reduced in the presence of 21p. A Saccharomyces cerevisiae two-hybrid analysis of 21p indicated that the protein can activate transcription when tethered within a responsive promoter. Together, the data reveal that while VZV gene 21 and HSV-1 UL37 share homology at the nucleic acid level, these proteins differ functionally.
Varicella-zoster virus (VZV) is a ubiquitous human pathogen. Unlike other neurotropic alphaherpesviruses for which primary infection is often asymptomatic, VZV infection produces malaise, fever, and an extensive vesicular rash (1, 27). In immunocompetent individuals, varicella is limited due to the development of adaptive immunity. However, in immunocompromised individuals, primary varicella can result in life-threatening complications. Of concern for both immunocompetent and immunodeficient individuals is the seeding of sensory ganglia and establishment of latency during primary VZV infection. Reactivation of latent VZV in elderly individuals typically results in zoster. After the rash of zoster resolves, many elderly patients continue to experience intense pain (postherpetic neuralgia). In some cases, reactivated VZV spreads to cerebral arteries, resulting in stroke (41). The neurological complications associated with VZV reactivation underscore the need to understand the molecular biology of latent VZV infection (27).
Just as the severity of primary infection varies among humans, so does the VZV DNA burden in latently infected ganglia. In previous studies, from 37 to >3,500 copies of VZV DNA have been detected per 100 ng of total human ganglionic DNA (18, 55). PCR of latent virus DNA has demonstrated that the latent virus genome assumes a circular structure, and in situ hybridization as well as PCR evidence indicates that the neuron is the primary site of virus latency (12, 35, 36, 42). Analyses of RNA from latently infected human ganglia by Northern blotting, reverse Northern blotting, reverse transcription nested PCR, cDNA cloning followed by DNA sequencing, quantitative reverse transcription-PCR, and in situ hybridization have all revealed limited transcription of VZV genes. With the caveat that less than 20% of the VZV genome has been studied in detail, only transcripts mapping to open reading frames (ORFs) 4, 21, 29, 62, and 63 have been detected during latency (14-16, 18, 36, 53). The translation products of the latently transcribed VZV genes have been even more difficult to detect; IE63, the immediate-early protein encoded by ORF 63, was the first VZV translation product detected in latently infected human ganglia (47), and another study has since detected proteins encoded by ORFs 4, 21, 29, 62, and 63 during latency (46).
Propagation of VZV in tissue culture is difficult due to the highly cell-associated nature of the virus. Hence, the functional analysis of VZV proteins has been largely by analogy to herpes simplex virus type 1 (HSV-1), the prototype neurotropic human alphaherpesvirus. The HSV-1 homologues of VZV ORFs 21 and 29 are UL37 and ICP8, respectively. HSV-1 UL37 encodes an early tegument-associated 120-kDa phosphoprotein that functions in virus maturation (2, 22, 24, 30, 64, 65). The UL37 protein contains a putative nuclear export signal that may account for its cytoplasmic location (50, 51). However, in HSV-1-infected cells, UL37 interacts with ICP8 and this complex is transported into the nucleus (2). Within the nucleus, the UL37-ICP8 protein complex can be isolated by DNA affinity chromatography (66).
HSV-1 ICP8 encodes the major virus DNA binding protein and is synthesized before viral DNA replication. The 133-kDa zinc metalloprotein is initially diffusely located in the nucleus, where it associates with duplex chromatin (4, 29, 43, 59). As virus infection proceeds, ICP8 is localized only in areas containing the virus DNA polymerase and newly replicated virus DNA (9, 57, 63). ICP8 stimulates HSV-1 DNA polymerase activity and functions in the redistribution of cellular DNA replication complexes (10, 21, 45, 59, 67, 69). ICP8 binding to single-stranded DNA facilitates localized DNA melting, thus potentiating the helicase activity associated with the HSV-1 origin binding protein (6, 7, 23, 32-34, 44, 49, 56). In addition to its effect on DNA replication, ICP8 also stimulates the transcription of late virus genes (25). Expression of late HSV-1 transcripts is reduced in cells infected with HSV-1 virus lacking functional ICP8 (11). The decrease in gene transcription in ICP8-deficient HSV-1-infected cells is not solely due to reduced virus DNA replication, suggesting that ICP8 may form multifunctional complexes (52).
VZV ORF 29 protein (29p) is a nonstructural 130 kDa nuclear protein that can be extracted from infected cells by affinity chromatography on DNA-cellulose columns (39, 40). Site-specific binding of 29p to the late glycoprotein I (gI) promoter modulates the activation in trans of the gI gene by the major VZV immediate-early ORF 62 protein (IE62). In rat PC-12 neuronal cells, 29p reduces the ability of IE62 to activate gI transcription (8), whereas in permissive human T lymphocytes, 29p augments IE62-induced transcription from the gI promoter (31). VZV ORF 21 protein (21p) maps to a contiguous 3,113-bp ORF (17, 19, 68). In VZV-infected cells, the 115-kDa 21p is associated with maturing nucleocapsids (48). Both VZV gene 21 and 29 transcripts have been detected in latently infected human ganglia; however, the question regarding the nature of their function remains. The present study was undertaken to characterize two VZV proteins during productive infection whose genes are expressed during latency.
MATERIALS AND METHODS
Cells and virus and metabolic labeling.
VZV (strain Ellen) was propagated in a continuous human malignant melanoma cell line (MeWo) or in a continuous monkey kidney cell line (Vero). Virus was grown by cocultivation of infected cells with uninfected cells as described previously (26, 28). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL, Grand Island, N.Y.) supplemented with 10% heat-inactivated fetal calf serum. Proteins were metabolically labeled by first starving infected or control cell cultures of inorganic phosphate (Pi) or l-methionine in 2% fetal calf serum-supplemented DMEM for 20 min, followed by incubation in fresh medium containing either 1 mCi of [32P]Pi (carrier-free H332PO4) or l-[35S]methionine (specific activity, 1,175 Ci/mmole; ICN Pharmaceuticals, Inc., Irvine, Calif.)/ml. Where indicated, the labeling medium was removed and cultures were washed with DMEM and incubated for various times in fresh medium (chase).
Antiserum and immunoprecipitation.
Antibodies against VZV immediate-early gene 63 protein (IE63) or 21p were raised in rabbits as described previously (20, 48). Rabbit antibodies against a 12-amino acid peptide in the carboxy-terminal region of 29p were also raised (39). To resolve VZV proteins, cell extracts were prepared in radioimmunoprecipitation (RIPA) lysis buffer (20 mM Na2HPO4 [pH 7.6], 100 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). Where required, viscosity of the cell extract was reduced by centrifugation at 12,500 × g for 2 min through a shredding device (Qiashredder; Qiagen, Valencia, Calif.). Cell extracts were clarified by centrifugation (10,000 × g, 60 min, 4°C). Proteins were immunoprecipitated overnight at 4°C with the addition of the specific rabbit antiserum (1:200 final dilution) and collected on recombinant protein A-coated Sepharose beads (Sigma, St. Louis, Mo.). Beads were collected by low-speed centrifugation (2,000 × g for 2 min), washed twice in RIPA wash buffer I (20 mM Tris-HCl [pH 7.6], 400 mM NaCl, 0.5% NP-40, 1 mM EDTA), centrifuged through a 1 M sucrose cushion prepared in RIPA wash buffer II (20 mM Tris-HCl [pH 7.6], 40 mM NaCl, 0.1% SDS, 1 mM EDTA), and washed twice in RIPA wash buffer II. Immunoprecipitated proteins were released from beads by boiling in SDS sample buffer for 5 min and electrophoresed through denaturing polyacrylamide gels (13). The 35S signal was amplified by soaking for 60 min in Enlightning (NEN Life Science Products, Boston, Mass.) before drying. Radiolabeled proteins were located by exposure to a phosphorstorage screen, and intensities were quantitated with attendant ImageQuant software (Molecular Dynamics, San Francisco, Calif.).
In situ histochemistry.
VZV-infected Vero cells were grown on 6-mm-diameter glass disks. At daily intervals, cultures were processed for two-color analysis to detect 21p and 29p. Cells were fixed in ice-cold PLP (4% paraformaldehyde, 0.1 M l-lysine, 1.3 mM sodium m-periodate, 50 mM Na2HPO4 [pH 7.4]), permeabilized for 10 min in methanol-acetone (50:50), and blocked for 60 min in normal mouse serum diluted 1:20 in Tris-buffered saline (TBS) (20 mM Tris-HCl, 150 mM NaCl). Between all solution changes, cells were extensively washed in TBS. Cells were incubated for 60 min at room temperature with rabbit anti-29p serum (1:250), followed by incubation for 60 min with unconjugated goat anti-rabbit immunoglobulin (Ig) (Fab fragment) diluted 1:250 in TBS. Samples were incubated for 60 min with fluorescein isothiocyanate (FITC)-conjugated mouse anti-goat IgG diluted 1:250 in TBS, followed by incubation with rabbit anti-21p diluted 1:200 in TBS. After incubation for 60 min with Cy3-conjugated mouse anti-rabbit IgG diluted 1:500 in TBS, samples were mounted in 4′,6′-diamidine-2-phenylindole dihydrochloride (DAPI) containing mineral oil (Vectashield; Vector Laboratories, Burlingame, Calif.). All immunoreagents were obtained from Jackson ImmunoResearch Laboratories, West Grove, Pa., and prior to use, the rabbit anti-21p sera was adsorbed with acetone-fixed Vero cells. Transmission immunofluorescence microscopy was performed using a Nikon E-800 microscope equipped with equipped with epifluorescence, DAPI, fluorescein, and Texas Red barrier filters. Images were also collected using a Nikon Diaphot inverted confocal microscope with an attached argon ion laser. All images were captured using a Cooke SensiCam charge-coupled device camera at ×400 or ×630 magnification. Slide Book (Intelligent Imagining Innovations, Inc. Denver, Colo.) and Adobe PhotoShop version 6.0 (Mountain View, Calif.) software was used to deconvolute and overlay the digital images.
Chromatography of DNA binding proteins.
At 3 days postinfection, VZV-infected MeWo cells (5 × 108 cells) were labeled with [35S]methionine for 16 h. Cells were collected, washed in phosphate-buffered saline, resuspended in high-salt buffer (1.7 M KCl, 50 mM Tris-HCl [pH 7.6], 5 mM EDTA, 0.5 mM dithiothreitol, 0.12 mM phenylmethylsulfonyl fluoride), and lysed by sonication. Samples were clarified by centrifugation (20,000 × g, 30 min, 4°C), and the supernatant was brought to an 8% concentration in polyethylene glycol. After 60 min of incubation on ice and clarification, the supernatant was diluted with an equal volume of TEDPGK (50 mM Tris-HCl [pH 7.6], 5 mM EDTA, 0.5 mM dithiothreitol, 0.12 mM phenylmethylsulfonyl fluoride, 20% glycerol, 150 mM KCl) containing 0.4% NP-40 and 0.1% bovine serum albumin. After extensive dialysis against TEDPGK containing 0.2% NP-40, the sample was again clarified and applied to washed DNA-agarose beads (Amersham Pharmacia Biotech). Unbound proteins were removed by washing with 150 mM KCl in TEDPGK containing 0.2% NP-40. Proteins were eluted from the DNA-agarose beads by stepwise increase of the KCl concentration from 0.2 to 2.4 M. After elution, each fraction was brought to 0.1% with respect to bovine serum albumin, dialyzed against TBS (20 mM Tris-HCl [pH 7.6], 150 mM NaCl), and reduced to 1 ml by centrifugal concentration (Ultrafree-CL filters; Millipore Corp., Bedford, Mass.). Each fraction was divided into equal portions and precipitated with either anti-21p or anti-29p serum, and the immuncomplexes were collected on recombinant protein A-coated Sepharose beads, washed, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) as described above.
Saccharomyces cerevisiae two-hybrid analyses.
The Hybrid Hunter yeast two-hybrid system (Invitrogen, Carlsbad, Calif.) was used to detect in vivo protein-protein interactions. Initially, full-length ORF 21 and ORF 29 were shuttled from eukaryotic expression plasmids (21AM-pCIneo and 29 AM-pCIneo; for details, see below) into yeast pHybLex/Zeo (bait) and pYESTrp2 (prey) plasmids. The multiple cloning sites of both yeast plasmids were modified to ensure in-frame ORF insertion and multiple downstream translation termination signals. In subsequent experiments, various segments of ORF 21 and ORF 29 were amplified by PCR using high-fidelity DNA polymerase (Advantage-HF; Clontech, Palo Alto, Calif.) and subcloned in-frame into the yeast bait and prey plasmids. Oligonucleotide primers (Table 1) were selected from regions of 21p and 29p that lack consensus amino acid sequence homology to HSV-1 (ClustalW; European Bioinformatics Institute). The DNA sequences bracketing the cloning sites were determined for all constructs to confirm in-frame insertions.
TABLE 1.
Oligonucleotide primers used to construct yeast bait and prey plasmids
| 5′ to 3′ sequencea | ORFb | REc | Locationd | Directione | Clonef |
|---|---|---|---|---|---|
| CGTCGAGCTC ATG GAA GAA CCA ATT TGT | 21 | SacI | 30,759 | Forward | 21A, 21A1, 21A+B |
| CTCAGCGGCCGC CAT TCC AAT ACG AGC GTG AGT GAA TGG | 21 | NotI | 31,286 | Reverse | 21A1 |
| CGTCGAGCTC CCA TTC ACT CAC GCT CGT ATT GGT ATG | 21 | SacI | 31,260 | Forward | 21A2 |
| CTCAGCGGCCGC ACG GGT GTG TTC TGC TAC | 21 | NotI | 31,826 | Reverse | 21A, 21A2 |
| CGTCGAGCTC GTA GCA GAA CAC ACC CGT | 21 | SacI | 31,809 | Forward | 21B |
| CTCAGCGGCCGC GAG AGT TGC TAA AAA AAC | 21 | NotI | 32,909 | Reverse | 21B |
| CGTCGAGCTC GTT TTT TTA GCA ACT CTC | 21 | SacI | 32,892 | Forward | 21C |
| CTCAGCGGCCGC TTA AGG GTC ACT CCC ACT TGT | 21 | NotI | 33,875 | Reverse | 21C |
| CGTCGAGCTC ATG GAA ATT ACT CAG AAG | 29 | SacI | 50,857 | Forward | 29A |
| CTCAGCGGCCGC CGC CAG GCC TAT AAA CAT | 29 | NotI | 51,924 | Reverse | 29A |
| CGTCGAGCTC ATG TTT ATA GGC ATG GAG | 29 | SacI | 51,907 | Forward | 29B |
| ACTCGCGGCCGC CCT ATA AGT GTC CTG CAT | 29 | NotI | 52,581 | Reverse | 29B |
| CGTCGAGCTC ATG CAG GAC ACT TAT AGG | 29 | SacI | 52,564 | Forward | 29C |
| CTCAGCGGCCGC GGA TTT ACT GTT TGG TGG CAT | 29 | NotI | 53,361 | Reverse | 29C |
| CTGTCGAGCTC ATG CCA CCA AAC AGT AAA TCC | 29 | SacI | 53,341 | Forward | 29D |
| CTCAGCGGCCGC ATT TCC CGA CGA GGG ACC | 29 | NotI | 53,991 | Reverse | 29D |
| CGTCGAGCTC GGT CCC TCG TCG GGA AAT | 29 | SacI | 53,974 | Forward | 29E |
| CTCAGCGGCCGC TTA AAT CAT TTC CAT TGT AAT | 29 | NotI | 54,471 | Reverse | 29E |
Oligonucleotide sequences including leader (single underline) and restriction endonuclease (double underline) sites 5′ with respect to the VZV DNA sequence.
VZV open reading frames.
Restriction endonuclease (RE) sites used for cloning.
Locations on the VZV genome of the 5′ nucleotide.
Directions (5′ to 3′) with respect to ORF.
PCR fragment identifications (see also Fig. 6).
Saccharomyces cerevisiae (strain L40) was transformed with bait plasmids and selected for zeocin resistance. Bait-containing yeast were made competent and transformed with prey plasmids and were selected for tryptophan prototrophism. Protein-protein interaction was indicated by growth in minimal medium lacking tryptophan, histidine, uracil, and lysine and supplemented with zeocin. β-Galactosidase (β-Gal) filter assays were performed to confirm protein-protein interactions that induced expression of LacZ from the LexA promoter. In all transformation experiments, positive (pHybLex/Zeo-Fos and pYESTrp2-Jun) and negative (pHybLex/Zeo-lamin and pYESTrp2-Jun) control plasmid combinations were added to validate the transformation protocol and colony selection. All protocols were performed as suggested by the supplier (Invitrogen).
Eukaryotic expression and reporter plasmid construction.
Plasmids expressing 21p, 29p, and IE62 in eukaryotic cells were constructed by inserting the full-length ORFs 21 and 29 into pCI-neo (Invitrogen). Initially, plasmid DNA containing the selected ORFs was extracted (14). Restriction endonucleases were used to shuttle ORF 21 (Hsp921 and BanI), ORF 29 (HphI), and ORF 62 (TthIII1 and ScaI) into pAlter1 (Promega). Unique restriction endonuclease recognition sequences were placed outside ORF 21 (HindIII and SpeI), ORF 29 (HindIII and SpeI), and ORF 62 (EcoRI) by using oligonucleotide base site-directed mutagenesis of the pAlter1 inserts. The altered ORFs were then shuttled into pCIneo, yielding eukaryotic expression plasmids for 21p (21AM-pCIneo), 29p (29AMK-pCIneo), and IE62 (62AM-pCIneo). DNA sequencing confirmed the expected constructions.
To construct promoter and reporter plasmids, various VZV gene promoters were inserted into pGL3Basic-Luc (Promega) to drive expression of the luciferase reporter gene. The 284-bp ORF20/ORF21 and the 220-bp ORF28/ORF29 intergenic DNA sequences were amplified using oligonucleotide primers tagged with either KpnI or MluI restriction endonuclease sites. PCR products were digested with KpnI and MluI and purified on agarose gels. Insertion of the PCR products into pGL3Basic-Luc generated reporter plasmids containing ORF 20 (p20-luc) and ORF 29 (p29-Luc) promoters. To generate reporter constructs for opposing promoters (p21-Luc and p29-Luc), the pGL3Basic-luc multiple cloning site was reversed by ligation of synthetic adaptors. DNA sequencing of all inserted PCR products confirmed amplification fidelity.
Transient transformation and reporter assay.
Vero cells (4 × 105) were seeded into 9.6-cm2 culture dishes and incubated overnight. Endotoxin-free supercoiled plasmid DNA was extracted by affinity chromatography (Qiagen). Each transfection consisted of reporter plasmid (300 ng), β-Gal expression plasmid DNA (300 ng; pCMV-βgal, Promega), and various amounts of VZV 21p, 29p, or IE62 expression vector. The CMV IE-3 promoter was used to drive all inserts (β-Gal, 21p, 29p, and IE62) to ensure equivalent expression from all cotransfected plasmids. Lipid-based (Lipofectamine-plus; GIBCO-BRL) transfection protocols were used as directed by the supplier. Cells were harvested 24 h after transfection and lysed in 100 μl of reporter lysis buffer (Promega). Each transfection assay was performed at least in duplicate, and for each independent transfection, the β-Gal and luciferase activities were assayed in replicate 30-μl aliquots of the culture lysate. To account for possible variations in transfection efficiency, luciferase activities were normalized with respect to β-Gal.
RESULTS
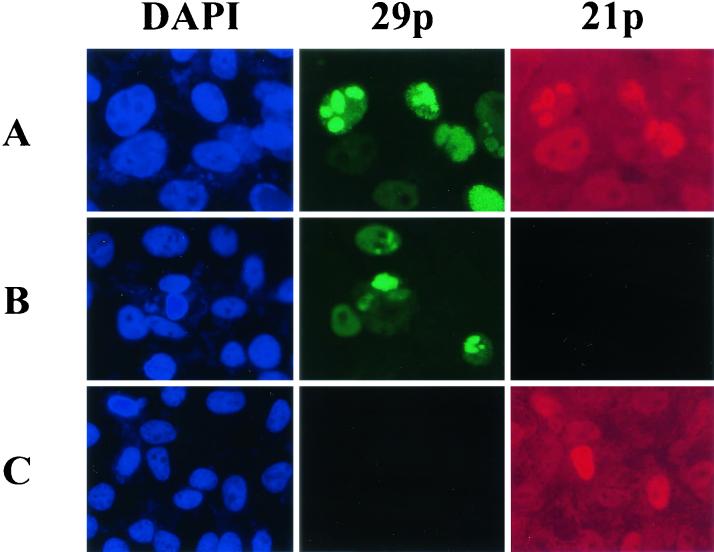
VZV genes 21p and 29p localize within VZV-infected cells.
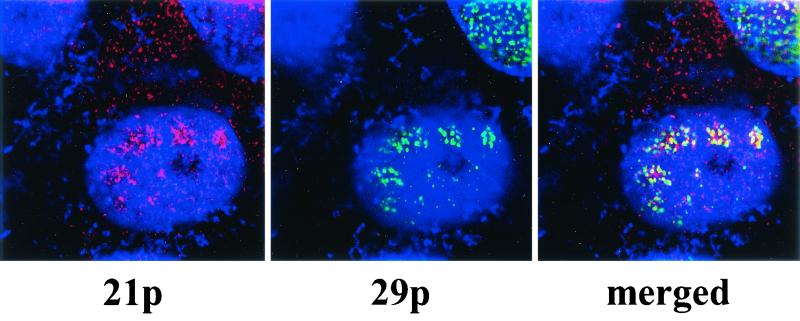
Two-color staining analysis with transmission UV microscopy to determine the cellular localization of 21p and 29p in VZV-infected Vero cells (Fig. 1) revealed 29p and 21p predominately within the nucleus; staining specificity was confirmed by the omission of anti-21p and anti-29p antibody. While transmission UV microscopy is sufficient for global analysis of the infected cells, confocal UV microscopy, which effectively slices the cell into 50-μm sections in the z axis, was used to more precisely determine the location of 21p and 29p (Fig. 2). Cy3-tagged 21p is seen in both the cytoplasm and nucleus (Fig. 2, left panel), while FITC-tagged 29p is located exclusively within the nucleus (center panel). The merged image (Fig. 2, right panel) shows 21p and 29p colocalization limited to internuclear punctate regions. This indicates that similar to their HSV-1 homologues, VZV 21p is both cytoplasmic and nuclear while 29p is exclusively nuclear. Further, within the nucleus, much of 21p is spatially associated with 29p.
FIG. 1.
Intracellular locations of VZV gene 21 protein (21p) and VZV gene 29 protein (29p) viewed by transmission UV microscopy. VZV-infected Vero cells were stained for cellular DNA in the nucleus (blue) with DAPI, FITC-tagged 29p (green), and Cy3-tagged 21p (red) as described in Materials and Methods. 29p and 21p are seen in punctuate intranuclear regions (row A), with 21p also present in the cytoplasm. Staining specificity was confirmed by the omission of anti-21p antibody (row B) and anti-29p antibody (row C).
FIG. 2.
Intracellular location of VZV 21p and 29p viewed by confocal UV microscopy. VZV-infected Vero cells were stained for DNA with DAPI (blue), Cy3-tagged 21p (red), and FITC-tagged 29p (green) as described in Materials and Methods. Note 21p in the nucleus and cytoplasm of infected cells (left panel) and 29p exclusively in the nucleus (middle panel). The right panel shows intranuclear colocalization of both 21p and 29p (yellow).
Putative phosphorylation of 21p and 29p.
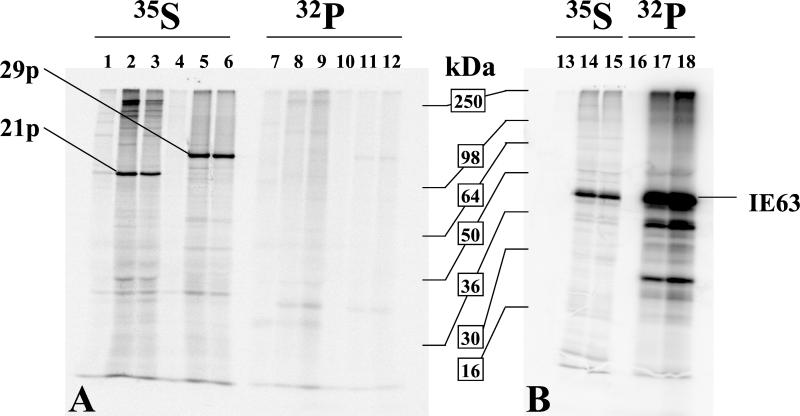
UL37, the HSV-1 homologue of VZV 21p, is stably phosphorylated in HSV-1-infected cells (2). Since the cellular localization of 21p and 29p mimics that of their HSV-1 homologues, we investigated whether the similarities might extend to the molecular level. Using the computer-assisted consensus motif search developed by Blom et al. (5), multiple potential phosphorylation sites in both 21p and 29p were identified, as were regions of homology between 21p and 29p for HSV-1 homologues UL37 and ICP8. To determine whether the candidate protein phosphorylation sites are utilized during productive infection, VZV-infected MeWo cells were labeled with [32P]- or [35S]methionine for 6 or 22 h, immunoprecipitated with anti-21p, anti-29p, or anti-IE63 serum, and resolved by SDS-PAGE (Fig. 3). For both labeling periods, the major immunoprecipitated proteins corresponded to the expected molecular masses of 21p (115 kDa) and 29p (132 kDa) deduced from the VZV DNA sequence (19). However, unlike its HSV-1 homologue, VZV 21p was not phosphorylated, and within the unsynchronized infected cells, 29p also was not significantly phosphorylated. Thus, despite putative phosphorylation sites in both proteins, neither is phosphorylated. Due to the difficulty in achieving synchronous infection, we determined that our 32P labeling was sufficient to label a VZV protein known to be phosphorylated. Immunoprecipitation of IE63 confirmed that the labeling conditions were compatible with protein phosphorylation. Thus, this VZV protein was synthesized and phosphorylated during both the 6- and 22-h labeling periods (Fig. 3B).
FIG. 3.
21p and 29p are not significantly phosphorylated in vivo. At 48 h postinfection, VZV-infected (lanes 2, 3, 5, 6, 8, 9, 11, 12, 14, 15, 17, and 18) and uninfected (lanes 1, 4, 7, 10, 13, and 16) MeWo cells were labeled with [35S]methionine (lanes 1 to 6 and 13 to 15) or 32P (lanes 7 to 12 and 16 to 18) for 6 h (lanes 2, 5, 8, 11, 14, and 17) or 22 h (lanes 1, 3, 4, 6, 7, 9, 10, 12, 13, 15, 16, and 18). At the end of the labeling period, protein extracts were prepared and immunoprecipitated as described in Materials and Methods with antisera directed against 21p (lanes 1 to 3 and 7 to 9), 29p (lanes 4 to 6 and 10 to 12), or IE63 (lanes 13 to 18). 21p and 29p were resolved by SDS-8% PAGE (panel A) and IE63 by SDS-12% PAGE (panel B). Phosphorimaging of immunoprecipitated 21p, 29p, and IE63 (40.5 kDa) demonstrated that all three proteins were synthesized during both the 6-h and 22-h labeling periods, but only IE63 was significantly phosphorylated. Two smaller phosphorylated proteins (lanes 17 and 18) are likely to be IE63 degradation products.
Pulse-chase labeling of 21p and 29p.
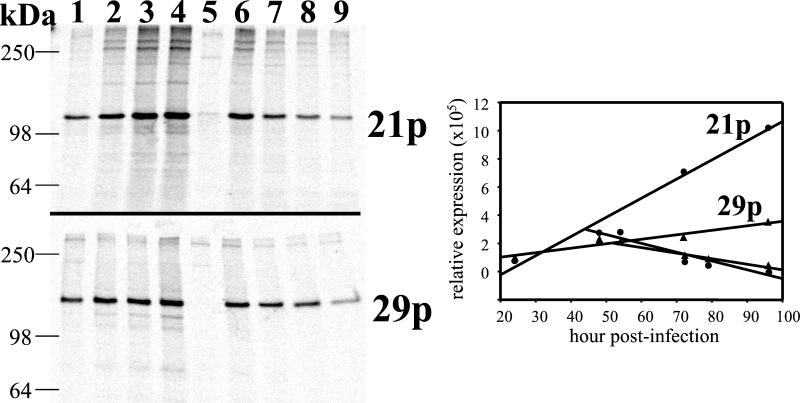
Within HSV-1-infected cells, UL37 is associated with ICP8 in the nucleus (2). Confocal analysis of VZV-infected cells indicates that a similar process may be occurring, in that 21p and 29p are colocalized within infected cell nuclei (Fig. 2). However, when VZV-infected cell protein extracts were analyzed by immunoprecipitation with sera directed against 21p or 29p, a 21p/29p complex was not seen (Fig. 3). Consequently, pulse-chase experiments were performed to determine if a transient 21p/29p complex could be detected. Figure 4 shows that 21p and 29p could be detected at all pulse and chase time points. However, no sample revealed coimmunoprecipitation of 21p with 29p. Use of milder cell lysis solutions consisting of phosphate-buffered saline containing 1% Brij 97 also showed no evidence of 21p/29p complex formation in VZV-infected cells (data not shown). Quantitation of 21p and 29p from the immunoprecipitation indicates that the rates of synthesis and degradation of both 21p and 29p are constant.
FIG. 4.
Pulse-chase labeling of 21p and 29p during productive VZV infection. At 24 h (lane 1), 48 h (lane 2), 72 h (lane 3), and 96 h (lane 4) postinfection, cells were labeled with [35S]methionine for 3 h and processed for immunoprecipitation of 21p or 29p. Lane 5 shows the results for uninfected cells labeled for 3 h beginning at 48 h after mock infection. The results depicted in lanes 6 to 9 indicate the stability of 21p and 29p. At 48 h postinfection, cultures were labeled with [35S]methionine for 3 h and chased with excess unlabeled methionine for an additional 6 h (lane 6), 24 h (lane 7), 31 h (lane 8), or 48 h (lane 9). Protein extracts were prepared and processed by immunoprecipitation to detect 21p or 29p. Both 21p and 29p were detected in all infected cells, but 21p and 29p did not coimmunoprecipitate in any sample. Quantitation of the immunoprecipitated proteins showed that the steady-state rates of synthesis and degradation for both 21p and 29p remained constant during the 96-h labeling period.
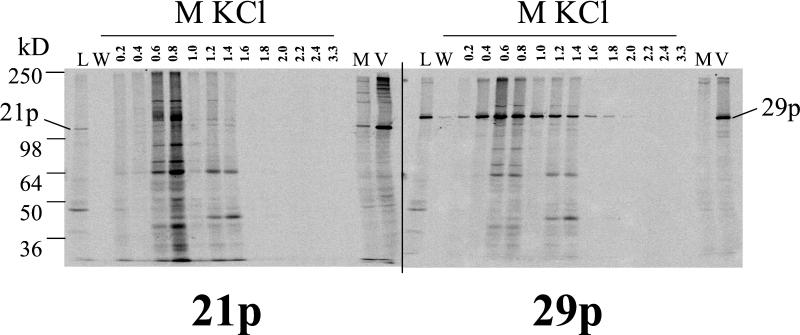
DNA binding activity of 21p and 29p.
The complex formed by HSV-1 UL37 and ICP8 elutes from DNA at salt concentrations characteristic of high-affinity DNA binding proteins (66). To determine whether VZV 21p and 29p form a DNA binding protein complex, nuclear proteins extracted from [35S]methionine-labeled VZV-infected cells were applied to single-stranded DNA agarose beads, extensively washed, eluted with increasing concentrations of KCl, immunoprecipitated with anti-21p or anti-29p serum, resolved by SDS-PAGE, and detected by phosphorimaging (Fig. 5). Consistent with the cellular locations of 21p and 29p, both proteins were present in nuclear extracts (Fig. 5, lanes L). The protein-loaded beads were extensively washed to remove unbound 21p and 29p (Fig. 5, lanes W). Along with specific immunoprecipitation of 21p from VZV-infected cells (Fig. 5, lanes V), the unadsorbed anti-21p serum used in this experiment also detected proteins of cellular origin (lanes M), which accounts for the nonspecific protein bands particularly evident in the 0.6 and 0.8 M KCl fractions. However, examination of the phosphorimage revealed no 21p present in any of the eluted fractions, while maximal elution of 29p occurred at 0.6 and 0.8 M KCl. The rabbit anti-29p serum detected 29p in VZV-infected cell lysates (Fig. 5, lanes V) with much less nonspecific cellular protein contamination (lanes M). These results indicate that unlike their HSV-1 counterparts, VZV 21p and 29p do not form a detectable DNA binding protein complex. VZV 29p bound to and eluted from the single-stranded DNA; therefore, if 21p/29p complex formation required single-stranded DNA, it was present in the assay.
FIG. 5.
DNA column elution profile of 21p and 29p. High-salt nuclear protein extract was prepared from [35S]methionine-labeled VZV-infected cells and loaded (lanes L) onto a DNA-agarose gel. After extensive washing (lanes W), proteins were eluted with increasing molar concentrations of KCl. The presence of 21p and 29p in each dialyzed and concentrated fraction was determined by immunoprecipitation, SDS-PAGE, and phosphorimaging. [35S]methionine-labeled protein extracts from mock (lanes M)- and VZV (lanes V)-infected cells controlled for the location of 21p and 29p. The maximum yield of 29p was obtained in the 0.6 M KCl fraction, whereas 21p was not detected in any fraction.
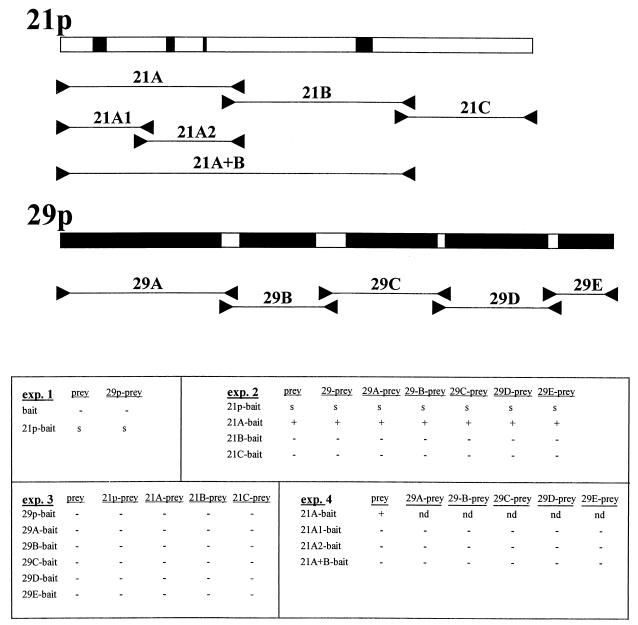
Two-hybrid analysis of 21p and 29p.
In a genetic approach developed to demonstrate specific protein-protein interactions (62), genes of interest are inserted in-frame into plasmids encoding either the DNA binding domain or the transcription-activation domain of the LexA yeast transcription factor. Interactions between the recombinant proteins are demonstrated through the induction of histidine prototrophism and confirmed by the induction of β-Gal activity. Figure 6 shows the results of four sets of plasmid construction-yeast transformations. The sequential transformation of L40 yeast cells with the bait and prey constructions yielded zeocin-resistant colonies that grew on minimal medium lacking tryptophan, indicating the presence of both plasmids. Isolated colonies were then assayed for (i) the presence of the expected insert by PCR analysis of extracted plasmid DNA; (ii) protein-protein interaction by growth on minimal medium lacking tryptophan, histidine, uracil, and lysine and containing zeocin (YC-WHUK+zeo); and (iii) protein-protein interaction by induction of β-Gal activity. In experiment 1, insertion of full-length 21p into the bait plasmid yielded slow-growing colonies with slight β-Gal activity independent of the prey plasmid DNA insert, indicating that 21p can induce low-level transcription when located correctly on the promoter. In this case, the correct location is supplied by the LexA binding domain of the bait plasmid. Because this slight prey-independent promoter activation precluded analysis of 21p/29p interaction, both 21p and 29p were subcloned for further analysis. The locations of subclones are schematically shown at the top of Fig. 6. All subclones were selected such that regions of homology between 21p and 29p and their HSV-1 homologues remained intact. Experiment 2 showed that the slight transactivation of the LexA promoter seen by 21p-bait was located in the first third of the protein and, like full-length 21p, was independent of the prey construct (Fig. 6). Rows 21B-bait and 21C-bait in Fig. 6 indicate that the remaining two-thirds of 21p did not interact with either full-length 29p or subclones of 29p.
FIG. 6.
Yeast two-hybrid analysis of 21p and 29p. Full-length ORFs 21p and 29p were inserted into pHybLex/Zeo (bait) or pYESTrp2 (prey) yeast expression plasmids. Oligonucleotide primers (between arrowheads) as listed in Table 1 were used to amplify regions within 21p and 29p, generating subclones of 21p and 29p for insertion into the bait and prey plasmids. The PCR primers were selected to be outside regions of homology between 21p and 29p and their analogous HSV-1 proteins (black boxes). The relative positions and nomenclatures for the subcloned fragments of 21p and 29p are listed below the respective proteins. The results of four independent yeast bait-prey transformation experiments (exp.) are shown. −, no growth on YC-WHUK+zeo plates and no β-Gal induction; +, growth on YC-WHUK+zeo plates and β-Gal induction; s, slow growth on YC-WHUK+zeo plates and slight β-Gal induction; nd, not done. The results indicate that full-length 21p contains slight intrinsic transcription activation capabilities, most likely located within the 21pA fragment of the protein. No 21p/29p interaction was demonstrated.
When bait and prey inserts were switched (experiment 3), 21A-prey showed no activation of LexA promoter (Fig. 6). Overall, experiment 3 shows that 29p and 21p do not form a detectable protein-protein interaction when expressed either as full-length proteins or as individual domains. Analysis of the transcriptional activation of 21A (experiment 4) showed that subcloning the 356 amino acids of 21A into 21A1 (175 amino acids) and 21A2 (189 amino acids) disrupted the 21A-dependent transcription activation of LexA when inserted into bait plasmids. Addition of 367 amino acid sequences downstream of 21A (plasmid 21A+B-bait) also abolished the ability of 21A-bait to activate transcription. Together, these results indicate that full-length 21p and 29p do not form a stable complex in yeast and neither do individual 21p and 29p domains; however, 21A does function in promoter activation when correctly placed within the responsive promoter.
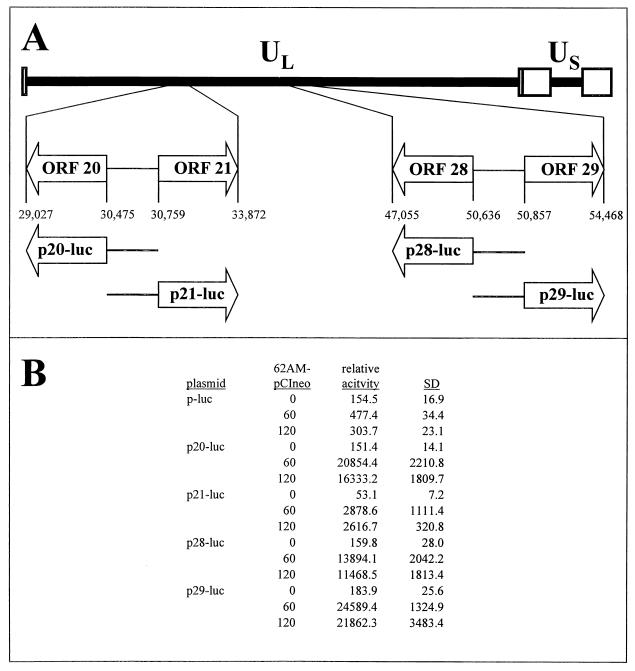
Effect of 21p and 29p on VZV gene promoter activity.
In latently infected human ganglia, VZV genes 21 and 29 are transcribed (14, 37, 53), whereas transcripts mapping to VZV genes 20 and 28 have not been detected during latency (14). Panel A of Fig. 7 shows the relative locations of ORFs 20, 21, 28, and 29 within the VZV genome. The promoters driving IE62-dependent transcription of genes 21, 28, and 29 have been located to the ORF 20/21 and ORF 28/29 intergenic regions (17, 54, 68). To place all promoters within the same reporter vector, the ORF 20/21 and ORF 28/29 intergenic regions were PCR amplified from VZV DNA and inserted into pGL3 Basic-luc such that each promoter controlled the synthesis of the luciferase reporter protein. Transient transfection of cells with the resulting reporter plasmids showed that each intergenic region contained no detectable intrinsic promoter activity. However, all promoter segments were induced by IE62 (Fig. 7, panel B). Relative activity values (average luciferase normalized to β-Gal activity) show the absolute magnitudes of the IE62 induction.
FIG. 7.
Cloning of gene 20, 21, 28, and 29 promoter reporter plasmids and their activation by IE62. (A) The 125-kbp VZV genome is composed of unique long (UL) and unique short (US) DNA segments, each bound by inverted repeat DNA sequences. The opposing ORFs 20 and 21, along with ORFs 28 and 29, are located within the UL segment (open boxes). The ORF 20/21 and ORF 28/29 intergenic regions were inserted into a promoterless luciferase reporter vector such that each region governed transcription of the luciferase reporter gene. (B) The resulting plasmids (p20-luc, p21-luc, p28-luc, and p29-luc) were used in transient transfection assays to determine intrinsic promoter activity and their response to induction by IE62. Data from at least duplicate experiments are presented as relative activity levels (luciferase activity normalized to β-Gal activity) to indicate the magnitude of promoter activity. Compared to the promoterless plasmid (p-luc), gene 20, 21, 28, and 29 promoters have no intrinsic activity but are induced by IE62. SD, standard deviation.
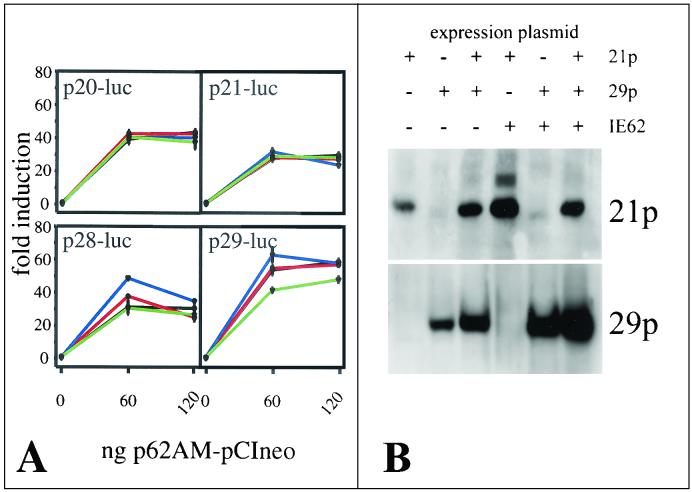
Figure 8A shows the effect of the presence of 21p and 29p on transcription activation of VZV gene 20, 21, 28, and 28 promoters by IE62. In these experiments, promoter activation, expressed as severalfold activation (relative promoter activity normalized to 0 ng of IE62), was used for ease in data interoperation. The presence of VZV protein 21p or 29p or both had no significant effect on the activation of gene 20 and 21 promoters by IE62 (Fig. 8A). The activation of gene 28 and 29 promoters was slightly enhanced by the presence of 29p at low (60 ng) 62AM-pCIneo concentrations. However, this augmentation was lost at higher (120 ng) 62AM-pCIneo concentrations. The presence of 21p had no significant effect on the ability of IE62 to activate transcription from the gene 28 and 29 promoters; however, the presence of both 21p and 29p reduced the activation of gene 29 promoter by IE62. Western blot analysis of transfected cells showed that 21p and 29p were synthesized in the presence or the absence of 62AM-pCIneo (panel B).
FIG. 8.
Effect of 21p and 29p on the transcriptional activation of VZV gene 20, 21, 28, and 29 promoters by IE62. In panel A, transient transfection experiments were performed to determine the effect of the presence of 21p (red lines), 29p (blue lines), or both proteins (green lines) on promoter activation (severalfold induction measured as relative activity levels normalized to 0 ng of 62AM-pCIneo) by IE62 (black lines). The presence of 21p has no effect on the induction of the promoter of gene 20, 21, 28, or 29 by IE62. The presence of 29p enhances activation of gene 28 and 29 promoters at low concentrations of IE62. The 29p-associated modulation of IE62 induction is diminished by 21p (green lines). In panel B, Western blot analysis of transfected cells showed that 21p and 29p were translated in the presence and in the absence of IE62.
DISCUSSION
Analysis of VZV gene regulation and latency is laden with obstacles. In humans, VZV latency is restricted to cranial, dorsal root, and autonomic ganglia, which are inaccessible during life. In rodents, some similarities to the latent infections seen in human ganglia have been shown but viruses do not reactivate (60, 61; reviewed in Rentier et al. [58]). Nevertheless, in both naturally infected human and experimentally infected rodent ganglia, VZV gene expression is restricted during latency (3, 38). Furthermore, VZV has a highly cell-associated nature in tissue culture and stable samples of high-titer, cell-free virus are unattainable. As a consequence, much of our understanding of VZV has been based on analogy to structurally homologous proteins in HSV-1. We have analyzed two VZV proteins (21p and 29p), whose transcripts are present in latently infected human and rodent ganglia (18, 38), to determine whether HSV-1 structural similarity relates to similarities of function.
Both VZV 21p and 29p partition within infected cells in a manner similar to that of their HSV-1 homologues, UL37 and ICP8. During productive HSV-1 infection, phosphorylated UL37 is chaperoned into the infected cell nucleus through an association with ICP8 and a UL37/ICP8 complex coelutes from single-stranded DNA columns (2, 66). Our search for an association between VZV 21p and 29p by coimmunoprecipitation, by KCl-dependent elution from single-stranded DNA-agarose beads, and by direct protein-protein interactions in yeast revealed no formation of 21p/29p complexes during productive infection. While it is interesting to speculate that an interaction between 21p and 29p occurs during latency, technical considerations preclude such experiments.
VZV 29p shares approximately 88% homology with ICP8 at the amino acid level, while the homology of VZV 21p to HSV-1 UL37 is less than 10%. The amino termini of HSV-1 UL37 and ICP8 have been implicated in the protein-protein interaction (F. Jenkins, personal communication). While 29p and ICP8 share homology at the amino terminus, 21p and UL37 are divergent at this location. Thus, one functional difference between 21p and UL37 might reside within the divergent amino termini. UL37 and 21p also differ with respect to phosphorylation; when extracted from HSV-1-infected cells, UL37 is stably phosphorylated, whereas in vivo-synthesized 21p is not significantly phosphorylated despite multiple predicted serine and threonine phosphorylation sites. It is intriguing to speculate that the divergence between 21p and UL37 at the amino terminus is also important with respect to the altered phosphorylation state of these proteins. However, the phosphorylation site of UL37 is unknown and thus, such comparison is premature. Also, protein phosphorylation may be cell-type dependent. In our assay, 21p and 29p phosphorylation was not detected; however, these proteins may be phosphorylated during latent infection in the human ganglion.
VZV 21p has no detectable intrinsic transcriptional activation capabilities with respect to its cognate promoter (68) (Fig. 7). In addition, the presence of 21p had no significant effect on the ability of IE62 to activate the promoters for VZV genes 20, 21, 28, and 29 (Fig. 8). However, two-hybrid yeast analysis of 21p showed that the protein had a modest ability to activate gene transcription when the protein was correctly tethered to the promoter. Subclonal analysis of 21p in yeast indicated that the transcriptional activation site associated with full-length 21p was situated within the 356 amino acids at the C terminus of the protein. When this 356-amino-acid segment of 21p was further subcloned, neither the resultant 176 amino-terminal amino acids nor 180-amino-acid fragment retained the ability to activate the LexA promoter. The lack of transcriptional activation of the LexA promoter by the full-length 21p or any of the subcloned fragments when inserted into the yeast prey plasmids suggests that the ability of 21p to activate transcription is dependent upon its tethering to the responsive promoter.
Previous transient transfection studies have shown that the presence of 29p affects the ability of IE62 to activate transcription of the gI promoter (8). Specifically, the presence of 29p was shown to stimulate induction of the gI promoter by IE62 in permissive cells while repressing IE62-induced activation of gI promoter in nonpermissive cells. In our study, we analyzed VZV gene 20, 21, 28, and 29 promoters, since the same intergenic regions appear to be differentially regulated in latently infected human ganglia. Our results indicate that both 21p and 29p have no intrinsic ability to activate transcription from the VZV gene promoters tested; however, the presence of 29p modestly enhances the ability of IE62 to activate transcription from gene 28 and 29 promoters, an effect observed at low concentrations of IE62. Thus, along with the ability of 29p to bind DNA, 29p may act in fine-tuning the regulation of VZV gene transcription by IE62. While it would be informative to determine the effect of the presence of 21p and 29p on other VZV promoters, in this study we limited our investigation to the bidirectional promoters for genes 20 and 21 and genes 28 and 29.
Acknowledgments
This work was supported in part by Public Health Service grants AG 06127 and NS 32623 from the National Institutes of Health.
We thank C. Grose for MeWo cells, F. Jenkins for helpful discussions, J. Bennett for aid in transmission UV microscopy, and J. MacManaman for aid in confocal microscopy. We also thank G. Owens, M. Burgoon, B. Hammack, and R. Mahalingam for reviewing the manuscript, Marina Hoffman for editorial review, and Cathy Allen for preparation of the manuscript.
REFERENCES
- 1.Abendroth, A., and A. Arvin. 1999. Varicella-zoster virus immune evasion. Immunol. Rev. 168:143-156. [DOI] [PubMed] [Google Scholar]
- 2.Albright, A. G., and F. J. Jenkins. 1993. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J. Virol. 67:4842-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annunziato, P. W., O. Lungu, and C. Panagiotidis. 2001. Varicella zoster virus in human and rat tissue specimens. Arch. Virol. Suppl. 17:135-142. [DOI] [PubMed] [Google Scholar]
- 4.Ben Ze'ev, A., R. Abulafia, and S. Bratosin. 1983. Herpes simplex virus and protein transport are associated with the cytoskeletal framework and the nuclear matrix in infected BSC-1 cells. Virology 129:501-507. [DOI] [PubMed] [Google Scholar]
- 5.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. [DOI] [PubMed] [Google Scholar]
- 6.Boehmer, P. E., M. S. Dodson, and I. R. Lehman. 1993. The herpes simplex virus type-1 origin binding protein. DNA helicase activity. J. Biol. Chem. 268:1220-1225. [PubMed] [Google Scholar]
- 7.Boehmer, P. E., and I. R. Lehman. 1993. Physical interaction between the herpes simplex virus 1 origin-binding protein and single-stranded DNA-binding protein ICP8. Proc. Natl. Acad. Sci. USA 90:8444-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucaud, D., H. Yoshitake, J. Hay, and W. Ruyechan. 1998. The varicella-zoster virus (VZV) open-reading frame 29 protein acts as a modulator of a late VZV gene promoter. J. Infect. Dis. 178:S34-S38. [DOI] [PubMed] [Google Scholar]
- 9.Bush, M., D. R. Yager, M. Gao, K. Weisshart, A. I. Marcy, D. M. Coen, and D. M. Knipe. 1991. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J. Virol. 65:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challberg, M. D. 1986. A method for identifying the viral genes required for herpesvirus DNA replication. Proc. Natl. Acad. Sci. USA 83:9094-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Y. M., and D. M. Knipe. 1996. A dominant mutant form of the herpes simplex virus ICP8 protein decreases viral late gene transcription. Virology 221:281-290. [DOI] [PubMed] [Google Scholar]
- 12.Clarke, P., T. Beer, R. Cohrs, and D. H. Gilden. 1995. Configuration of latent varicella-zoster virus DNA. J. Virol. 69:8151-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohrs, R., and H. Rouhandeh. 1982. Herpesvirus sylvilagus I. Polypeptides of virions and nucleocapsids. J. Virol. 41:1063-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohrs, R. J., K. Srock, M. B. Barbour, G. Owens, R. Mahalingam, M. E. Devlin, M. Wellish, and D. H. Gilden. 1994. Varicella-zoster virus (VZV) transcription during latency in human ganglia: construction of a cDNA library from latently infected human trigeminal ganglia and detection of a VZV transcript. J. Virol. 68:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohrs, R. J., M. B. Barbour, R. Mahalingam, M. Wellish, and D. H. Gilden. 1995. Varicella-zoster virus (VZV) transcription during latency in human ganglia: prevalence of VZV gene 21 transcripts in latently infected human ganglia. J. Virol. 69:2674-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohrs, R. J., M. Barbour, and D. H. Gilden. 1996. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 70:2789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohrs, R. J., M. Barbour, and D. H. Gilden. 1998. Varicella-zoster virus gene 21: transcriptional start site and promoter region. J. Virol. 72:42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohrs, R. J., J. Randall, J. Smith, D. H. Gilden, C. Dabrowski, K. H. van Der, and R. Tal-Singer. 2000. Analysis of individual human trigeminal ganglia for latent herpes simplex virus type 1 and varicella-zoster virus nucleic acids using real-time PCR. J. Virol. 74:11464-11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 20.Debrus, S., C. Sadzot-Delvaux, A. F. Nikkels, J. Piette, and B. Rentier. 1995. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J. Virol. 69:3240-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bruyn, K. A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857-868. [DOI] [PubMed] [Google Scholar]
- 22.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fierer, D. S., and M. D. Challberg. 1992. Purification and characterization of UL9, the herpes simplex virus type 1 origin-binding protein. J. Virol. 66:3986-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagan, W. M., A. G. Papavassiliou, M. Rice, L. B. Hecht, S. Silverstein, and E. K. Wagner. 1991. Analysis of the herpes simplex virus type 1 promoter controlling the expression of UL38, a true late gene involved in capsid assembly. J. Virol. 65:769-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao, M., and D. M. Knipe. 1991. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J. Virol. 65:2666-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilden, D. H., Y. Shtram, A. Friedmann, M. Wellish, M. Devlin, A. Cohen, N. Fraser, and Y. Becker. 1982. Extraction of cell-associated varicella-zoster virus DNA with Triton X-100-NaCl. J. Virol. Methods 4:263-275. [DOI] [PubMed] [Google Scholar]
- 27.Gilden, D. H., B. K. Kleinschmidt-DeMasters, J. J. LaGuardia, R. Mahalingam, and R. J. Cohrs. 2000. Neurologic complications of the reactivation of varicella-zoster virus. N. Engl. J. Med. 342:635-645. [DOI] [PubMed] [Google Scholar]
- 28.Grose, C., D. M. Perrotta, P. A. Brunell, and G. C. Smith. 1979. Cell-free varicella-zoster virus in cultured human melanoma cells. J. Gen. Virol. 43:15-27. [DOI] [PubMed] [Google Scholar]
- 29.Gupte, S. S., J. W. Olson, and W. T. Ruyechan. 1991. The major herpes simplex virus type-1 DNA-binding protein is a zinc metalloprotein. J. Biol. Chem. 266:11413-11416. [PubMed] [Google Scholar]
- 30.Guzowski, J. F., and E. K. Wagner. 1993. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J. Virol. 67:5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He, H., D. Boucaud, J. Hay, and W. T. Ruyechan. 2001. Cis and trans elements regulating expression of the varicella zoster virus gI gene. Arch. Virol. Suppl. 17:57-70. [DOI] [PubMed] [Google Scholar]
- 32.He, X., and I. R. Lehman. 2000. Unwinding of a herpes simplex virus type 1 origin of replication (OriS) by a complex of the viral origin binding protein and the single-stranded DNA binding protein. J. Virol. 74:5726-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He, X., and I. R. Lehman. 2001. An initial ATP-independent step in the unwinding of a herpes simplex virus type I origin of replication by a complex of the viral origin-binding protein and single-strand DNA-binding protein. Proc. Natl. Acad. Sci. USA 98:3024-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez, T. R., and I. R. Lehman. 1990. Functional interaction between the herpes simplex-1 DNA polymerase and UL42 protein. J. Biol. Chem. 265:11227-11232. [PubMed] [Google Scholar]
- 35.Kennedy, P. G., E. Grinfeld, and J. W. Gow. 1998. Latent varicella-zoster virus is located predominantly in neurons in human trigeminal ganglia. Proc. Natl. Acad. Sci. USA 95:4658-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy, P. G., E. Grinfeld, and J. W. Gow. 1999. Latent varicella-zoster virus in human dorsal root ganglia. Virology 258:451-454. [DOI] [PubMed] [Google Scholar]
- 37.Kennedy, P. G., E. Grinfeld, and J. E. Bell. 2000. Varicella-zoster virus gene expression in latently infected and explanted human ganglia. J. Virol. 74:11893-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy, P. G., E. Grinfeld, S. Bontems, and C. Sadzot-Delvaux. 2001. Varicella-zoster virus gene expression in latently infected rat dorsal root ganglia. Virology 289:218-223. [DOI] [PubMed] [Google Scholar]
- 39.Kinchington, P. R., G. Inchauspe, J. H. Subak-Sharpe, F. Robey, J. Hay, and W. T. Ruyechan. 1988. Identification and characterization of a varicella-zoster virus DNA-binding protein by using antisera directed against a predicted synthetic oligopeptide. J. Virol. 62:802-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinchington, P. R., J. K. Hougland, A. M. Arvin, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinschmidt-DeMasters, B. K., and D. H. Gilden. 2001. The expanding spectrum of herpesvirus infections of the nervous system. Brain Pathol. 11:440-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaGuardia, J. J., R. J. Cohrs, and D. H. Gilden. 1999. Prevalence of varicella-zoster virus DNA in dissociated human trigeminal ganglion neurons and nonneuronal cells. J. Virol. 73:8571-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leinbach, S. S., and J. F. Casto. 1983. Identification and characterization of deoxyribonucleoprotein complexes containing the major DNA-binding protein of herpes simplex virus type 1. Virology 131:274-286. [DOI] [PubMed] [Google Scholar]
- 44.Leinbach, S. S., and L. S. Heath. 1989. Characterization of the single-stranded DNA-binding domain of the herpes simplex virus protein ICP8. Biochim. Biophys. Acta 1008:281-286. [DOI] [PubMed] [Google Scholar]
- 45.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lungu, O., C. A. Panagiotidis, P. W. Annunziato, A. A. Gershon, and S. J. Silverstein. 1998. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc. Natl. Acad. Sci. USA 95:7080-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahalingam, R., M. Wellish, R. Cohrs, S. Debrus, J. Piette, B. Rentier, and D. H. Gilden. 1996. Expression of protein encoded by varicella-zoster virus open reading frame 63 in latently infected human ganglionic neurons. Proc. Natl. Acad. Sci. USA 93:2122-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahalingam, R., R. Lasher, M. Wellish, R. J. Cohrs, and D. H. Gilden. 1998. Localization of varicella-zoster virus gene 21 protein in virus-infected cells in culture. J. Virol. 72:6832-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makhov, A. M., P. E. Boehmer, I. R. Lehman, and J. D. Griffith. 1996. Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J. Mol. Biol. 258:789-799. [DOI] [PubMed] [Google Scholar]
- 50.McLauchlan, J. 1997. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J. Gen. Virol. 78:189-194. [DOI] [PubMed] [Google Scholar]
- 51.McLauchlan, J., K. Liefkens, and N. D. Stow. 1994. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J. Gen. Virol. 75:2047-2052. [DOI] [PubMed] [Google Scholar]
- 52.McNamee, E. E., T. J. Taylor, and D. M. Knipe. 2000. A dominant-negative herpesvirus protein inhibits intranuclear targeting of viral proteins: effects on DNA replication and late gene expression. J. Virol. 74:10122-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meier, J. L., R. P. Holman, K. D. Croen, J. E. Smialek, and S. E. Straus. 1993. Varicella-zoster virus transcription in human trigeminal ganglia. Virology 193:193-200. [DOI] [PubMed] [Google Scholar]
- 54.Meier, J. L., and S. E. Straus. 1995. Interactions between varicella-zoster virus IE62 and cellular transcription factor USF in the coordinate activation of genes 28 and 29. Neurology 45:S30-S32. [DOI] [PubMed] [Google Scholar]
- 55.Pevenstein, S. R., R. K. Williams, D. McChesney, E. K. Mont, J. E. Smialek, and S. E. Straus. 1999. Quantitation of latent varicella-zoster virus and herpes simplex virus genomes in human trigeminal ganglia. J. Virol. 73:10514-10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabkin, S. D., and B. Hanlon. 1990. Herpes simplex virus DNA synthesis at a preformed replication fork in vitro. J. Virol. 64:4957-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163-2177. [DOI] [PubMed] [Google Scholar]
- 58.Rentier, B., J. Piette, L. Baudoux, S. Debrus, P. Defechereux, M. P. Merville, C. Sadzot-Delvaux, and S. Schoonbroodt. 1996. Lessons to be learned from varicella-zoster virus. Vet. Microbiol. 53:55-66. [DOI] [PubMed] [Google Scholar]
- 59.Ruyechan, W. T., and A. C. Weir. 1984. Interaction with nucleic acids and stimulation of the viral DNA polymerase by the herpes simplex virus type 1 major DNA-binding protein. J. Virol. 52:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadzot-Delvaux, C., M. P. Merville-Louis, P. Delree, P. Marc, J. Piette, G. Moonen, and B. Rentier. 1990. An in vivo model of varicella-zoster virus latent infection of dorsal root ganglia. J. Neurosci. Res. 26:83-89. [DOI] [PubMed] [Google Scholar]
- 61.Sadzot-Delvaux, C., S. Debrus, A. Nikkels, J. Piette, and B. Rentier. 1995. Varicella-zoster virus latency in the adult rat is a useful model for human latent infection. Neurology 45:S18-S20. [DOI] [PubMed] [Google Scholar]
- 62.Sato, T., M. Hanada, S. Bodrug, S. Irie, N. Iwama, L. H. Boise, C. B. Thompson, E. Golemis, L. Fong, H. G. Wang, et al. 1994. Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc. Natl. Acad. Sci. USA 91:9238-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schenk, P., S. Pietschmann, H. Gelderblom, G. Pauli, H. Ludwig, and P. Schenck. 1988. Monoclonal antibodies against herpes simplex virus type 1-infected nuclei defining and localizing the ICP8 protein, 65K DNA-binding protein and polypeptides of the ICP35 family. J. Gen. Virol. 69:99-111. [DOI] [PubMed] [Google Scholar]
- 64.Schmitz, J. B., A. G. Albright, P. R. Kinchington, and F. J. Jenkins. 1995. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology 206:1055-1065. [DOI] [PubMed] [Google Scholar]
- 65.Shelton, L. S., M. N. Pensiero, and F. J. Jenkins. 1990. Identification and characterization of the herpes simplex virus type 1 protein encoded by the UL37 open reading frame. J. Virol. 64:6101-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shelton, L. S., A. G. Albright, W. T. Ruyechan, and F. J. Jenkins. 1994. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J. Virol. 68:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilcock, D., and D. P. Lane. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349:429-431. [DOI] [PubMed] [Google Scholar]
- 68.Xia, D., and S. E. Straus. 1999. Transcript mapping and transregulatory behavior of varicella-zoster virus gene 21, a latency-associated gene. Virology 258:304-313. [DOI] [PubMed] [Google Scholar]
- 69.Zhong, L., and G. S. Hayward. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol. 71:3146-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]