Abstract
The possible biochemical factors able to affect the in vitro expression of the high-risk human papillomavirus type 16 (HPV16) E7 oncoprotein have been analyzed. Evidence is provided that E7 mRNA stability is increased and, conversely, transcript translation is inhibited by binding to a 32-kDa protein from rabbit reticulocyte lysate; sequence analysis identified the 32-kDa binding protein as rabbit α1-globin protein; and interaction between rabbit α1-globin and E7 mRNA occurs through the 6-mer peptide SEQIKA present in human cytokeratin 7 protein. The in vitro data were confirmed by the occurrence of HPV16 E7 mRNA-cytokeratin 7 binding in squamous cervical cancer SiHa cells.
Cervical cancer is the second leading cause of death from cancer in women worldwide. Molecular and epidemiological studies have strongly implicated the sexually transmitted high-oncogenic-risk human papillomaviruses (HPVs) as a causative agent (39). HPVs have a unique position in viral carcinogenesis studies. Unlike other oncoviruses (e.g., simian virus 40 and polyomavirus) that infect a number of different organs and tissues, HPVs infect human epithelial tissues. Moreover, the viral life cycle and the differentiation program of the host epithelial keratinocyte are tightly connected; whereas HPV genomes can replicate in various undifferentiated cell lines, it seems that viral transcription is specifically restricted to keratinocytes (10). Also, the long latency (decades) required for cervical cancer development after primary viral infection and the absence of tumor-specific modifications in the viral oncogenes imply the action of additional factors in the outcome of genital cancer. Therefore, evaluation of the factors that affect viral gene expression is fundamental to clarifying the mechanism of cervical carcinogenesis.
This laboratory is investigating the factors that regulate the expression of the high-risk HPV type 16 (HPV16) E7 oncoprotein that is believed to play a major role in cervical neoplasia (7, 38). The ability of high-risk HPVs to contribute to malignant progression seems to depend on the expression of viral E6 and E7 oncoproteins, known to inactivate two cellular tumor suppressor gene products, p53 and retinoblastoma protein (35). In addition, deregulated expression of E7 is usually accompanied by disruption of the viral repressor E2 in cervical cancer (24, 31). Thus, a complex interplay of trans factors and cis sequences appears to be important to HPV16 E7 expression and its association with human uterine malignancy (22).
This study describes step by step the experimental progression that has led to individuation of the HPV16 E7 mRNA interaction with a 6-mer amino acid peptide, SEQIKA, present in rabbit α1-globin and human cytokeratin 7 protein sequences. It is shown that the SEQIKA fragment acts as a modulator of HPV16 E7 mRNA translation and stability.
MATERIALS AND METHODS
Templates and mRNA synthesis.
HPV16 E7 cDNA coding sequence devoid of the 5′ and 3′ untranslated regions (UTRs) (9) and control c-myc (2) and eIF-4E (18) templates were used. Uncapped mRNAs were generated from linearized cDNAs that had been subcloned into the pBluescript BKS transcription vector under T7 promoter control. In vitro transcription was carried out with the RiboMAX System (Promega, Milan, Italy).
RNA radiolabeling.
Substrate mRNAs (or enzymatic mRNA digests) were 5′-end labeled to a specific activity of at least 10,000 cpm/fmol with [γ-32P]ATP and the Ready-To-Go T4 polynucleotide kinase system (Amersham Pharmacia Biotech, Milan, Italy).
RNA decay and RNase digestion reactions.
The in vitro mRNA decay assay (29) was carried out at 30°C in 50-μl reaction mixtures with 5 to 10 ng of radiolabeled HPV16 E7 (or c-myc or eIF-4E) mRNA in a buffer containing 10 mM Tris-acetate (pH 7.5), 10 mM creatine phosphate, 2 mM dithiothreitol, 1 mM ATP, 0.4 mM GTP, 0.8 mM magnesium acetate, 100 mM potassium acetate, 1 μg of creatine phosphokinase, 0.1 mM spermine, 10 U of RNase inhibitor, and 8 μl of rabbit reticulocyte lysate (Amersham Pharmacia). After incubation for the required time, 5′-32P-labeled RNA was phenol extracted, subjected to 4% polyacrylamide gel electrophoresis (PAGE) at 35 mA for 10 to 20 min, and detected by autoradiography.
Susceptibility to RNase A was assayed (6). Radiolabeled RNA (10 μg) was incubated with 1 ng of pancreatic RNase A for 15 min at 37°C. To obtain an unfolded, linear, accessible mRNA structure, the viral transcript was heated at 90°C for 1 min. Following digestion with pancreatic RNase A, 5′-end-labeled RNA fragments were subjected to 20% PAGE and detected by autoradiography.
RNA and peptide electrophoretic mobility shift assays.
In the RNA electrophoretic mobility shift assay (REMSA) (34), 32P-labeled mRNA (1 μg) was first incubated in binding buffer (20 mM potassium HEPES [pH 8.0], 400 mM ammonium acetate, 10 mM magnesium acetate, 0.01% [vol/vol] Nonidet P-40, 5% [vol/vol] glycerol plus RNase inhibitor [100 U]) for 5 min at 37°C and then incubated with 5 μl of rabbit reticulocyte lysate (Amersham Pharmacia) for 10 min at 37°C. In addition, assays were run with proteinase K (0.4 mg/ml of incubation mixture). Then samples were electrophoresed (35 mA, 6 h, 4°C) through a 5% native polyacrylamide gel with 50 mM potassium HEPES (pH 8.0)-1 mM magnesium acetate-0.01% (vol/vol) Nonidet P-40 as the running buffer. Protein-labeled RNA complex was visualized by autoradiography.
When the peptide electrophoretic mobility shift assay (PEMSA) was carried out in order to define peptide-mRNA binding, unlabeled mRNA (1 μg) was incubated with biotinylated peptide (10 μg) in binding buffer and electrophoresed (35 mA, 1 h, 4°C), and the biotinylated peptide-mRNA complex was visualized by enhanced chemiluminescence (ECL) reaction (ECL kit; Amersham). Biotinylated SEQIKA, GEQIKA, SELIKA, SEQILA, and LPGALS peptides were obtained from Primm Srl, Milan, Italy.
Human, bovine, rat, rabbit, sheep, pig, and pigeon hemoglobins used in gel shift experiments (25 μg/assay) were from Sigma Aldrich Co., Milan, Italy.
HPV16 E7 mRNA-binding protein isolation.
HPV16 E7 mRNA-binding protein was purified by the magnetic DNA affinity procedure (15) with monodispersed supermagnetic particles. In this approach, the viral transcript was hybridized in 500 mM NaCl-33 mM NaH2PO4 · H2O-3.3 mM EDTA (pH 7.4)-0.1% sodium dodecyl sulfate (SDS) with the biotinylated 20-mer oligonucleotide 5′-TTATGGTTTCTGAGAACAGA, complementary to the 3′ end of E7 mRNA, and synthesized by Perkin Elmer, Monza, Italy. Hybrid coupling to streptavidin-coated Dynabeads M-280 and the subsequent biomagnetic separation were carried out by following faithfully the manufacturer's instructions (Unipath Dynal, Oslo, Norway). The E7 mRNA-coated beads were washed, mixed with rabbit reticulocyte lysate (about 1.5 × 107 mRNA-coated beads/100 μl of lysate), and incubated for 30 min at 0°C. The unbound proteins remained in solution and were saved to be used as a depleted reticulocyte lysate preparation. After magnetic separation, the bound protein component was removed from the Dynabeads by elution buffer (20 mM Tris-HCl [pH 8.0], 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol, 0.01% SDS, 1 M NaCl) and analyzed by SDS-12.5% PAGE. Prestained molecular weight protein markers were from Bio-Rad Laboratories Srl, Milan, Italy.
p32 partial sequencing.
Following elution from Dynabeads, the bound material was electrophoresed on SDS-12.5% PAGE and revealed by Coomassie R-250 staining. The gel portion corresponding to the p32 band was cut out and processed for internal sequencing. Additionally, N-terminal sequencing was carried out following gel electroblotting onto a polyvinylidene difluoride nylon membrane (Bio-Rad). Sequencing was done on the ABI-Perkin Elmer automatic sequencing instrument model 477A (Primm Srl).
Computer-assisted similarity analyses.
The N-terminal and internal fragment sequences were analyzed for sequence similarity and full-length alignments by FASTA search program (pir.georgetown.edu/pirwww/search/fasta) (25). Rabbit α1-globin sequence similarity to the human proteome was analyzed with the PIR, FASTA, BLAST, GenBank, SWISS-PROT, and PRINTS sequence analysis programs (1, 3-5).
Translational assay and HPV16 E7 oncoprotein immunodetection.
KpnI-linearized template was subjected to an in vitro translation assay with the ECL translation system from Amersham Pharmacia. The translation reaction was carried out according to the supplier's instructions with the following modifications: 5 μg of E7 mRNA was used, the RNase inhibitor amount was tripled, and 1 μg each of the protease inhibitors leupeptin, pepstatin A, aprotinin, and chymostatin (Boehringer Mannheim, Monza, Italy) was added. Following SDS-12% PAGE, the biotinylated translation products were electroblotted onto a polyvinylidene difluoride membrane and detected with streptavidin conjugated with horseradish peroxidase and ECL reagent (Amersham Pharmacia). Prestained low-molecular-weight protein markers were from Bio-Rad.
Membrane that had been analyzed for biotinylated translation products was blocked with 10% (vol/vol) ethanolamine in phosphate-buffered saline (PBS)-0.1% (vol/vol) Tween 20. Mouse anti-E7 monoclonal antibody (MAb) raised to HPV16 E7 (ED17, catalog no. sc-6981; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) was used to detect mRNA E7 translation product in immunoblots (dilution, 1:500) in PBS-0.1% (vol/vol) Tween 20-10% (vol/vol) ethanolamine. The unrelated mouse anti-GD3 ganglioside MAb MG22 (17) was used as a control. Immunoblots were developed with sheep anti-mouse immunoglobulin horseradish peroxidase-conjugated antibodies (dilution, 1:2,000; Santa Cruz Biotechnology, Inc.) as the secondary antibody with the chemiluminescence assay (ECL Western blotting analysis system; Amersham Pharmacia).
Mouse anti-p32 and antipeptide PAbs.
Anti-p32 polyclonal antibodies (PAbs) were raised in this laboratory by using C57BL/6J female mice, 2 months old. The E7 mRNA-binding protein was isolated from the rabbit reticulocyte lysate by a magnetic DNA affinity procedure and chromatographed by SDS-12% PAGE. Then 100-μl portions of gel material containing 32-kDa protein (about 0.2 to 0.3 μg) were injected into each mouse intraperitoneally. A booster immunization was administered 30 days later. Immune serum was collected 10 days later. Antipeptide polyclonal antisera against the SEQIKA and REWVPFACRE peptide sequences were raised in BALB/c mice by injecting the synthetic peptides coupled to keyhole limpet hemocyanin. The antipeptide antisera were obtained from Primm Srl. Following partial purification by three successive precipitations with 21% ammonium sulfate, mouse PAbs were used at a concentration of 50 μg/gel shift assay.
Analysis of in vivo E7 mRNA-CK7 interaction.
SiHa and 3T3 cell lines were obtained from the Interlab Cell Line Collection, Genoa, Italy. SiHa cells were grown in Earle's balanced saline solution plus 1% nonessential amino acids and 1 mM NaP, and 3T3 cells were grown in Dulbecco's modified Eagle's medium. Media were further supplemented with 10% calf serum and 2 mM l-glutamine. Cells were lysed directly in the flasks with precooled lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.02% sodium azide, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, 100 μg of pepstatin A, 1 μg of leupeptin, and 400 U of RNasin per ml). Cell lysate was centrifuged at 10,000 rpm for 5 min, dialyzed against sterile H2O, and made 500 mM NaCl, 33 mM NaH2PO4 · H2O, 3.3 mM EDTA (pH 7.4), and 0.1% SDS. Cell lysate (55 mg of total protein, 35-ml total volume) was incubated with streptavidin-coated Dynabeads M-280 (1 ml, i.e., 6.7 × 108 beads) that had been coupled with the biotinylated 20-mer oligonucleotide 5′-TTATGGTTTCTGAGAACAGA, complementary to the 3′ end of E7 mRNA. After 30 min of incubation at room temperature, the Dynabeads were repeatedly washed and the bead-bound material was eluted with elution buffer according to the manufacturer's instructions and electrophoresed in SDS-12% PAGE. Then the gel was silver stained or electroblotted onto nitrocellulose for immunoassays with MAbs raised against CK7 (ICN, Aurora, Ohio) and CK4, CK14, and CK17 (LabVision Corp., Fremont, Calif.).
RESULTS
In vitro HPV16 E7 mRNA half-life.
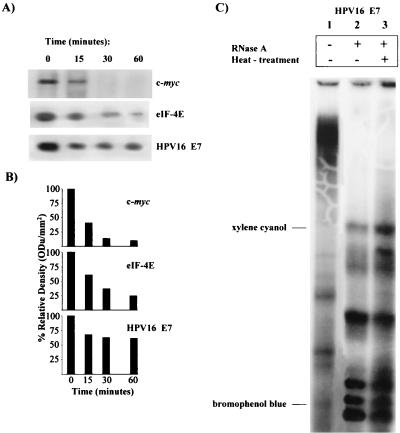
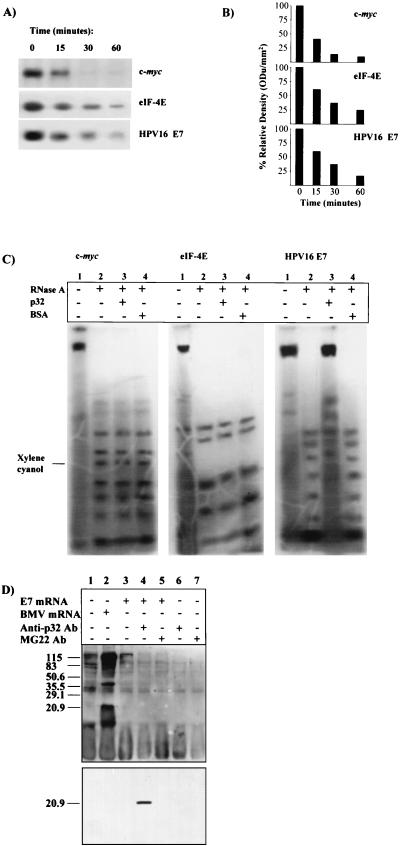
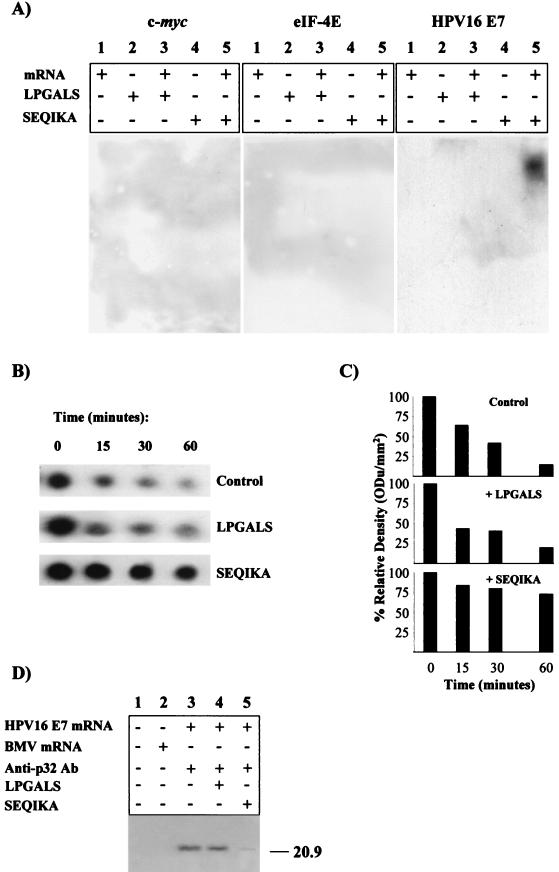
HPV16 E7 mRNA is scarcely translatable in vitro in a rabbit reticulocyte lysate system (9) under canonical experimental conditions (29). To understand the molecular basis of E7 untranslatability, the viral mRNA decay rate was studied. Transcript stability is a major determinant of mRNA abundance and consequently can affect the expression level of the relative encoded protein (30). The short-lived transiently expressed c-myc mRNA and the constitutively expressed mRNA for eIF-4E, a translational machinery protein, were used as controls. Figures 1A and B indicate that HPV16 E7 mRNA was very stable compared to both c-myc and eIF-4E transcripts. The c-myc mRNA degradation rate was in agreement with the half-life of 30 min or less reported in the literature (21). Thus, the lack of translatability of the E7 transcript could not be explained by a high mRNA degradation rate.
FIG. 1.
E7 mRNA in vitro half-life and nuclease accessibility. (A) In vitro half-life assay of E7 mRNA. RNA transcript was incubated for 0 to 60 min at 30°C in decay reaction mixtures containing rabbit reticulocyte lysate. RNA was phenol extracted and electrophoresed in 4% nondenaturing polyacrylamide gels. c-myc and eIF-4E mRNAs were used as controls. (B) Quantification of mRNA half-life by densitometric analysis. ODu, optical density units. (C) E7 mRNA secondary structure and nuclease accessibility: effect of heat treatment on digestion pattern of HPV16 E7 mRNA by RNase A. Lanes: 1, nontreated E7 mRNA; 2, RNase A-digested E7 mRNA; 3, RNase digest of preheated E7 mRNA.
HPV16 E7 mRNA secondary structure and accessibility to RNase A.
The stability of mRNA is controlled by the secondary structure; hairpins, loops, and other structured motifs can affect the accessibility of mRNA to nuclease attack (as well as to the translational machinery), thus determining the final protein expression (11, 23). It has to be noted that E7 cDNA coding sequence devoid of the 5′ and 3′ UTRs was purposely used in order to eliminate possible effects due to mRNA cis instability motifs (19, 21). Here, the possibility was considered that nuclease attack might be inhibited by structured motifs in the HPV16 E7 mRNA coding sequence. As a matter of fact, the complex secondary structure of the viral transcript was predicted by using an RNAfold algorithm (http://www.tbi.univie.ac.at/c/s.dll/RNAfold.cgi) (13). The free energy of the thermodynamic ensemble was −111.40 kcal/mol at 30°C, and it was of interest that the translational initiation codon was involved in a hairpin structure. Therefore, it was reasonable to hypothesize that mRNA folding hampered both nuclease attack and translational reaction. On the other hand, RNase experiments on unfolded, accessible E7 mRNA were not consistent with this hypothesis. In fact, Fig. 1 illustrates that similar fragment patterns were obtained by digesting nonheated (Fig. 1C, lane 2) and heat-treated (Fig. 1C, lane 3) unfolded E7 mRNA with pancreatic RNase A.
Binding of a 32-kDa protein to HPV16 E7 mRNA.
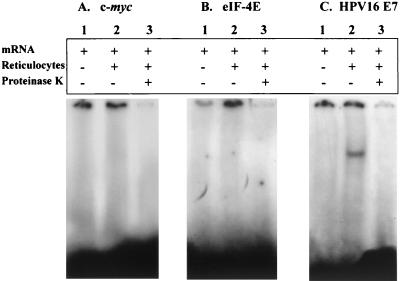
A number of multifunctional proteins appear to participate in gene expression regulation through translational silencing or activation. Moreover, careful analysis of the HPV16 E7 mRNA decay reaction (Fig. 1A and B) indicated a bimodal decay with an initially short half-life, indicated by the rapid decrease in abundance of the mRNA after the first time point, followed by a fraction of mRNA that is resistant to decay. Thus, a search was undertaken for possible trans factors (33) able to explain the bimodal decay behavior seen in vitro. In this approach, the REMSA was performed on the 32P-labeled viral transcript following incubation with rabbit reticulocyte lysate. A parallel sample containing proteinase K was run as a control. Figure 2C, lane 2, shows the presence of an mRNA-protein complex that could be detected following 5% PAGE. No gel shift signal was evident when the reaction mixture was supplemented with proteinase K (Fig. 2C, lane 3), documenting the protein nature of the complex. No protein complex was evident when c-myc or eIF-4E mRNA was used (Fig. 2A and B, respectively, lane 2).
FIG. 2.
Gel retardation experiments: analysis of interaction between HPV16 E7 mRNA and rabbit reticulocyte lysate. Radiolabeled c-myc (A), eIF-4E (B), and HPV16 E7 (C) mRNAs were incubated in binding buffer (lanes 1), binding buffer plus rabbit reticulocyte lysate (lanes 2), or binding buffer plus rabbit reticulocyte lysate and proteinase K (lanes 3) and electrophoresed in 5% PAGE.
Preliminary purification of the binding protein was carried out by the magnetic DNA affinity technique. HPV16 E7 mRNA was hybridized with a biotinylated 20-mer oligonucleotide complementary to its 3′ end, bound to streptavidin-coated Dynabeads, and incubated with rabbit reticulocyte lysate. The bound material was eluted from Dynabeads, electrophoresed by SDS-12% PAGE, and analyzed by silver staining. The remaining unbound solution was used as depleted reticulocyte lysate preparation. Figure 3, lane 2, shows that the component eluted from E7 mRNA-coated beads was positive to silver staining and migrated as a 32-kDa polypeptide (p32). The p32 component was absent in the depleted reticulocyte lysate preparation (Fig. 3, lane 3).
FIG. 3.
A p32 component from rabbit reticulocyte lysate binds to E7 mRNA-conjugated Dynabeads. For SDS-PAGE characterization of the E7 mRNA-binding protein purified by magnetic affinity, rabbit reticulocyte lysate was incubated in binding buffer with HPV16 E7 mRNA-activated magnetic beads. Following washings, the reticulocyte component fraction bound to Dynabeads was eluted and electrophoresed in SDS-12.5% PAGE. The gel was silver stained. Lanes: 1, whole rabbit reticulocyte lysate used for magnetic separation; 2, protein eluted with 1 M NaCl from the activated magnetic beads; 3, unbound proteins. Numbers on the left indicate positions of molecular mass standards (in kilodaltons).
Inhibition of HPV16 E7 mRNA degradation and translation by p32 from rabbit reticulocyte lysate.
The effect of p32 on HPV16 E7 mRNA degradation was analyzed with the p32-depleted reticulocyte lysate preparation. Figures 4A and B show that E7 mRNA stability was markedly reduced when assayed in the p32-depleted reticulocyte lysate (see, for comparison, the half-life behavior in the experiments reported in Fig. 1A and B). In parallel, Fig. 4C documents that p32 addition protected E7 mRNA but not c-myc or eIF-4E mRNAs from RNase A digestion. The protection was specific, since addition of bovine serum albumin (BSA) did not prevent the nuclease reaction. Finally, the possible role of p32 in regulating HPV16 E7 mRNA expression was confirmed by the striking translational signal obtained by carrying out translation in the presence of anti-p32 Abs (Fig. 4D, lower part, lane 4). The conclusion was reached that binding of E7 mRNA to a p32 component from rabbit reticulocyte lysate controls HPV16 E7 expression by preventing both degradation and translation of the viral transcript.
FIG. 4.
Effect of rabbit reticulocyte p32 on HPV16 E7 mRNA half-life, RNase A digestion, and translation. (A) Depletion of p32 promotes in vitro E7 mRNA decay. c-myc, eIF-4E, and HPV16 E7 mRNA decay was assayed with p32-depleted rabbit reticulocyte lysate. (B) Quantification of mRNA half-life by densitometric analysis. ODu, optical density units. (C) The p32 component protects E7 mRNA from RNase attack. c-myc, eIF-4E, and HPV16 E7 mRNAs were incubated with (lanes 2) RNase A, (lanes 3) RNase A plus p32 eluted from the magnetic beads, or (lanes 4) BSA (control); (lanes 1) nontreated mRNA. (D) Anti-p32 PAbs promote HPV16 E7 mRNA translation in complete rabbit reticulocyte lysate. Upper part, biotinylated translation pattern obtained following electroblotting onto a polyvinylidene difluoride membrane. Lower part, the same membrane probed with anti-E7 MAb. The positions of prestained molecular size markers are indicated (in kilodaltons).
Computer-assisted identification of p32 as rabbit α1-globin.
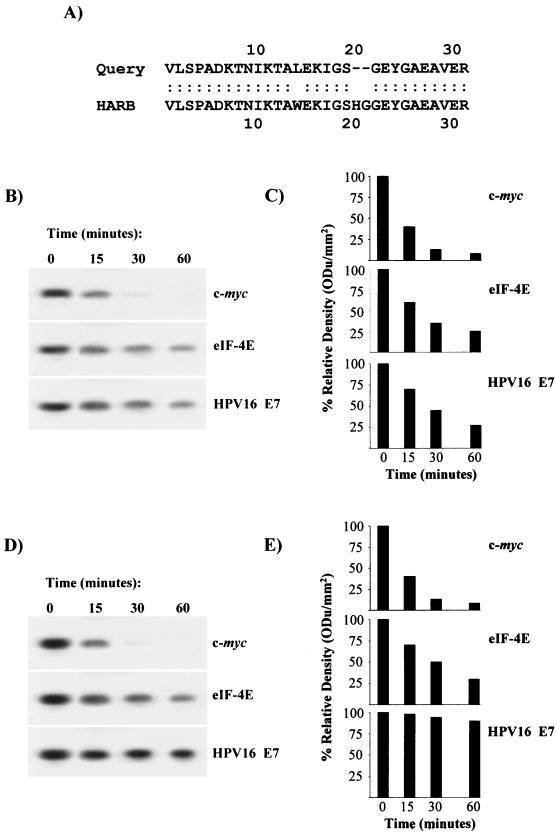
As a successive step, the 32-kDa mRNA-binding protein was identified. Following binding to and elution from Dynabeads, the bound protein was electrophoresed on SDS-12.5% PAGE, and the p32 fraction was subjected to both N-terminal and internal sequencing. Two fragment sequences (N-terminal, VLSPADKTNIKTALEKIGS; internal, GEYGAEAVER) were analyzed for sequence similarity by a full-length alignment search program (pir.georgetown.edu/pirwww/search/fasta) (5, 25). The similarity analysis indicated 90.323% identity of the fragments in a 31-amino-acid overlap to the rabbit hemoglobin α1 chain (Fig. 5A). The computational result was experimentally validated with rabbit hemoglobin in the mRNA decay reaction illustrated in Fig. 5D and E. It can be seen that addition of rabbit hemoglobin but not BSA to the p32-depleted reticulocyte lysate remarkably stabilized HPV16 E7 mRNA. Hemoglobin had no effect on c-myc or eIF-4E transcript half-life (Fig. 5B and C).
FIG. 5.
Identification of rabbit reticulocyte p32 as α1-globin. (A) Pir FASTA search for sequence similarity and full-length alignment of two peptide fragment motifs obtained by N-terminal and internal sequencing of p32. HARB, hemoglobin α1 chain, rabbit. Smith-Waterman score, 155. (B) Addition of BSA to p32-depleted rabbit reticulocyte lysate does not promote E7 mRNA stability. c-myc, eIF-4E, and HPV16 E7 mRNA decay was assayed by using the reticulocyte unbound proteins remaining in solution after incubation with E7 mRNA-activated magnetic beads plus control protein BSA. (C) Quantification of mRNA half-life as in panel B by densitometric analysis. (D) Addition of rabbit hemoglobin to p32-depleted rabbit reticulocyte lysate promotes E7 mRNA stability. c-myc, eIF-4E, and HPV16 E7 mRNA decay was assayed with the reticulocyte unbound proteins remaining in solution after incubation with activated magnetic beads plus rabbit hemoglobin (25 μg). (E) Quantification of mRNA half-life as in panel D by densitometric analysis. ODu, optical density units.
Computer-assisted similarity analysis.
Then it was asked whether the rabbit globin-E7 mRNA interaction found in vitro might be of importance to the biology of HPV infections. To this end, a search was undertaken for a related human protein by carrying out sequence similarity analysis between the rabbit α1-globin sequence and the human proteome. By utilizing the PIR Peptide Match program sequence analysis program (pir.georgetown.edu/pirwww/search/patmatch) (5), the rabbit α1-globin sequence (SWISS-PROT accession number P01948) was dissected into 6-mer motifs offset by one residue (i.e., overlapping by five residues: VLSPAD, LSPADK, SPADKT, etc.) that were used as sequence probes in computer-assisted similarity analyses. The results of the computational analysis are reported in Table 1.
TABLE 1.
Linear 6-mer sequence probes shared by rabbit α1-globin protein and the human proteome
| α1-Globin 6-mer probe | No. of matches to human proteome | Human protein or sequence |
|---|---|---|
| VLSPAD | 1 | VAV2 |
| GSHGGE | 1 | Telencephalin precursor |
| SHGGEY | 1 | Telencephalin precursor |
| GGEYGA | 1 | HNF-3/fork-head homolog-3 |
| SEQIKA | 1 | Keratin, type II cytoskeletal 7 (CK7) |
| KKVSEA | 1 | Tubby related protein 2 (TULP2) |
| KVSEAL | 1 | Pregnancy-specific beta-1 glycoprotein |
| EALTKA | 1 | SmcY |
| KAVGHL | 1 | Methionine synthase |
| GHLDDL | 1 | ABC transporter MOAT-B isoform |
| DLPGAL | 1 | ADP-ribosylation factor-like protein 2 |
| LPGALS | 3 | ADP-ribosylation factor-like protein 2, paxillin beta, paxillin gamma |
| PGALST | 1 | ALRa |
| LSTLSD | 1 | Low-affinity immunoglobulin G receptor CD16 |
| LSDLHA | 1 | Lymphocyte phosphatase-associated P protein |
| HKLRVD | 1 | 60S ribosomal protein L1 (L4) |
| KFLANV | 1 | Amino acid transporter-1 (CAT-1) |
| VSTVLT | 1 | Cationic amino acid transporter 3 |
| STVLTS | 1 | Cyclic GMP-stimulated phosphodiesterase |
| TVLTSK | 1 | Neuroendocrine convertase 2 precursor |
| VLTSKY | 1 | Ras GTPase-activating protein-related protein |
| LTSKYR | 1 | p60 katanin mRNA |
Acute lymphoblastic leukemia-1-related protein.
It can be seen that the rabbit α1-globin protein presents similarity to 6-mer portions of a number of human proteins. In particular, the following two fragments appeared potentially interesting: SEQIKA, because of its presence in CK7, a marker of glandular epithelium (27); and LPGALS, which is present in the focal adhesion cytoskeletal protein paxillin. The corresponding biotinylated peptides were synthesized and used in PEMSA experiments and mRNA decay-translation reactions in order to individuate possible specific peptide-viral mRNA interactions. PEMSA results are shown in Fig. 6A. It is clearly evident that the rabbit α1-globin52-57/human CK791-96 SEQIKA fragment did interact with HPV16 E7 mRNA. The interaction was peptide and mRNA specific, since no interaction was found with the cytoskeletal paxillin LPGALS peptide or the control c-myc or eIF-4E mRNA. In addition, Fig. 6B and C show that the 6-mer peptide SEQIKA stabilized HPV16 E7 mRNA when incubated in the p32-depleted reticulocyte lysate. Likewise, the SEQIKA fragment restored the translational inhibition removed by anti-p32 PAbs in the in vitro translational assay carried out with complete rabbit reticulocyte lysate (Fig. 6D).
FIG. 6.
Rabbit α1-globin52-57/human CK791-96 SEQIKA binds to, prevents decay of, and blocks translation of HPV16 E7 mRNA. (A) SEQIKA fragment binds to E7 mRNA. PEMSA experiments were run by incubating unlabeled c-myc, eIF-4E, or HPV16 E7 mRNA with biotinylated control peptide LPGALS or SEQIKA. Following electrophoresis, the biotinylated peptide-mRNA complex was visualized by ECL. (B) SEQIKA fragment promotes E7 mRNA stability. c-myc, eIF-4E, and HPV16 E7 mRNA decay was assayed in vitro with the p32-depleted rabbit reticulocyte lysate obtained following incubation with E7 mRNA-conjugated magnetic beads plus the 6-mer peptide SEQIKA or LPGALS. (C) Quantification of mRNA half-life by densitometric analysis. ODu, optical density units. (D) SEQIKA fragment prevents HPV16 E7 mRNA translation promoted by anti-p32 PAbs in complete rabbit reticulocyte lysate. Translation reaction mixtures were preincubated with anti-p32 PAbs (50 μg) and/or peptide (10 μg) for 10 min prior to mRNA addition. Translation reactions were electrophoresed in SDS-12% PAGE. Following electroblotting onto a polyvinylidene difluoride membrane, E7 oncoprotein expression was analyzed by Western blotting with anti-E7 MAb.
HPV16 E7 mRNA-SEQIKA binding: definition of consensus amino acid sequence.
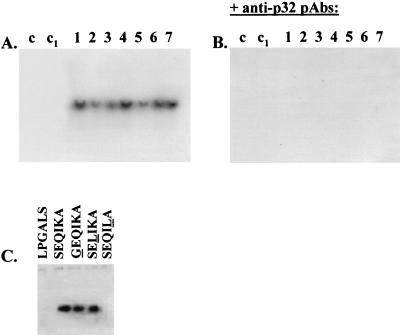
To gain some preliminary insights into the mode of rabbit α1-globin52-57/human CK791-96 SEQIKA action, I tested the mRNA-binding ability of hemoglobins having double and triple amino acid substitutions inside the α1-globin52-57 motif. Table 2 describes the hemoglobins that were assayed. It can be seen that the common α1-globin52-57 motif is the sequence SXQXKX, where S52, Q54, and K56 are invariant amino acid residues in the hemoglobins analyzed. Then, the hemoglobins described in Table 1 were tested by REMSA performed on the 32P-labeled viral transcript. Figure 7A illustrates that all of the hemoglobins listed in Table 1 were able to bind HPV16 E7 mRNA, indicating that the conserved globin motif S52-Q54-K56 was sufficient for the interaction with the transcript. Anti-p32 PAbs suppressed hemoglobin binding to HPV16 E7 mRNA (Fig. 7B).
TABLE 2.
Hemoglobins used in HPV16 E7 mRNA gel shift assay
FIG. 7.
HPV16 E7 mRNA-SEQIKA binding: definition of the consensus amino acid sequence. (A) Hemoglobins with double and triple amino acid substitutions inside the α1-globin52-57 SEQIKA motif and numbered from 1 to 7 as described in Table 2 were assayed for binding to radiolabeled HPV16 E7 mRNA by gel retardation experiments. In the controls, mRNA was incubated in binding buffer (lane c) and binding buffer plus BSA (25 μg) (lane c1). (B) Binding assays were supplemented with 50 μg of mouse anti-p32 PAbs and run as in panel A. (C) PEMSA experiments were run by incubating unlabeled HPV16 E7 mRNA with biotinylated control peptide LPGALS, binding peptide SEQIKA, and SEQIKA-derived peptides GEQIKA, SELIKA and SEQILA, each hosting a single amino acid change. The changed residues are underlined. Following electrophoresis, the biotinylated peptide-mRNA complex was visualized by ECL.
The peptide binding sequence was further investigated by synthesizing three biotinylated 6-mer peptides changed at the three invariant positions S52, Q54, and K56, i.e., GEQIKA, SELIKA, and SEQILA, with the changed residues in italics. The three substituted 6-mer peptides were then used in the PEMSAs reported in Fig. 7C. It can be seen that the K amino acid residue was essential for peptide binding to HPV16 E7 mRNA.
In vivo HPV16 E7 mRNA-CK7 interaction.
Analysis of in vivo interaction of E7 mRNA with the rabbit α1-globin52-57/human CK791-96 SEQIKA sequence was performed in the HPV16-positive cancer SiHa cell line from a human cervix, with the normal 3T3 cell line as a control. The level of CK7 in the two cell lysates is shown in Fig. 8. The interaction assay was carried out with magnetic beads conjugated with the biotinylated 20-mer oligonucleotide 5′-TTATGGTTTCTGAGAACAGA, complementary to the 3′ end of E7 mRNA, as bait. Oligonucleotide-coated beads were incubated with SiHa cell lysate and then eluted with the high-salt elution buffer.
FIG. 8.
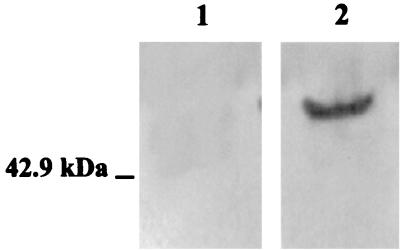
Expression of CK7 in squamous cervical cancer SiHa cell lysate. Equal amounts of 3T3 (lane 1) and SiHa (lane 2) cell lysate (10 μg) were separated by SDS-12% PAGE, and CK7 expression was determined by Western blot analysis with anti-CK7 MAb.
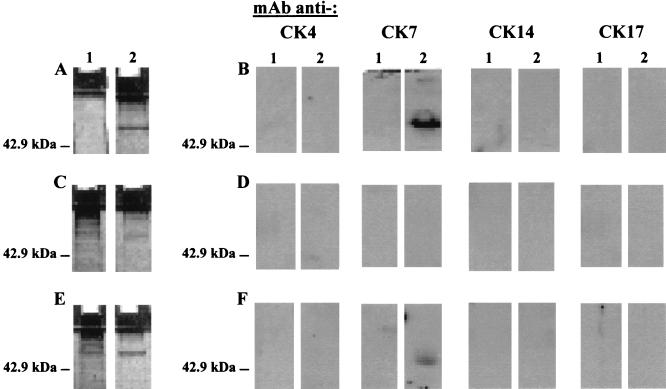
Figure 9A documents that the eluted material had a molecular mass of 54 kDa in SDS-12.5% PAGE and was positive for silver staining. Figure 9B shows that the eluate material did react with a mouse MAb raised against CK7. No signal was detected with control MAbs against CK4, a 59-kDa protein that may be detected in squamous cell carcinomas (36); CK14, a polypeptide of 50 kDa expressed in squamous cell carcinomas (26); or CK17, a polypeptide of 46 kDa predominant in basal cell carcinomas (16). Moreover, Fig. 9D illustrates that eluate reactivity to the anti-CK7 MAb involved the SEQIKA sequence, since it was abolished by pretreatment of SiHa cell lysate with mouse anti-SEQIKA PAbs. Unrelated control anti-REWVPFACRE PAbs that were at hand in the laboratory did not abolish the reactivity to the anti-CK7 MAb (Fig. 9F). No reactivity was observed in an eluate obtained following incubation of oligonucleotide-coated beads with the 3T3 cell lysate (Fig. 9B, D, and F, lanes 1).
FIG. 9.
HPV16 E7 mRNA-CK7 interaction in squamous cervical cancer SiHa cell lysate. (A) SDS-PAGE characterization of the silver-stained material bound to and eluted from oligonucleotide-activated beads following incubation with 3T3 (lane 1) or SiHa (lane 2) cell lysate. (B) Western blot analysis of reactivity to MAbs raised against human CK4, CK7, CK14, or CK17 of material bound to and eluted from oligonucleotide-activated beads following incubation with 3T3 (lanes 1) or SiHa (lanes 2) cell lysate. (C and D) Analyses as in panels A and B, respectively, for cell lysates that had been treated with 50 μg of anti-SEQIKA PAbs. (E and F) Analyses as in panels A and B, respectively, for cell lysates that had been treated with 50 μg of anti-REWVKFAKPCRE PAbs.
DISCUSSION
The HPV16 E6 and E7 early genes are responsible for initiation of viral DNA replication and are believed to play a major role in virus-induced cervical neoplasia (38). Proliferation and malignant phenotype of human cervical carcinoma cell cultures have been shown to depend on continuous expression of high-risk HPV oncogenes E6 and E7 (37). Consequently, regulation of early gene expression has been studied intensively at the transcriptional level, and a number of both cis and trans regulatory factors have been found and characterized (28). On the other hand, little is known about the translational regulation of E6 and E7 transcripts, possibly also because of the inefficient in vitro translation of HPV E7 mRNA in a rabbit reticulocyte lysate translation system (9).
The present study reports three main findings. It explains E7 mRNA untranslatability in vitro as being mediated by a specific interaction between the rabbit α1-globin peptide fragment SEQIKA and the viral transcript; it shows that the E7 mRNA-binding SEQIKA sequence is also present in the human CK7 protein; and it demonstrates that the HPV16 E7 mRNA-CK7 interaction does occur in HPV16-positive squamous cancer SiHa cells.
These results have several important implications. mRNA-binding proteins generally inhibit mRNA translation and, in parallel, stabilize the transcripts. This twofold regulatory mechanism has the scope of protecting and storing particular mRNAs to be expressed at defined developmental stages (8, 30). In this context, the E7 mRNA-CK791-96 SEQIKA fragment interaction might contribute to explaining the protection of E7 mRNA during the differentiation program of the host keratinocyte. As a matter of fact, CK7 expression occurs in epithelial cells in parallel to differentiation (12).
Moreover, CK7 is specifically localized in glandular (also called columnar) epithelium (27). In the well-documented natural history of invasive squamous cell carcinoma of the cervix, it is a literature datum that the majority of invasive cervical neoplasms arise at the transition between the stratified squamous epithelium and the simple epithelium at the squamo-columnar junction. The cervical squamo-columnar junction is a region of cellular instability, and at certain critical physiological periods (i.e., pregnancy and menopause), the cells of the junctional epithelium undergo a process described as squamous metaplasia, in which the original simple cervical epithelium is replaced by squamous epithelium, with a concomitant change in the cytokeratin profile. The area in which metaplastic epithelium is found is known as the transformation zone (32). Most cervical carcinomas arise in this transformation zone (20); it is this transformation zone that must be biopsied to exclude carcinoma or precancerous lesions. This has led to the belief that neoplastic changes occur as a result of the action of oncogenic agents on metaplastic epithelium, resulting in a transformation event (14). In this scenario, the spatial-temporal E7 protection ensured by CK7 might be a key event to understanding the transformation zone as the election area for the cervical carcinogenesis process.
In conclusion, the experiments reported here suggest new mechanisms for the regulation of HPV16 E7 expression which may be important during viral latency and activation and show that a previously hypothesized factor(s) might be the concomitant necessary cause for cervical cancer development following HPV infection (39). The study of the precise determinants and mechanism of interaction between the CK791-96 SEQIKA fragment and HPV16 E7 mRNA might help in clarifying some aspects of cervical carcinogenesis and delineating possible peptide-based therapeutic approaches.
Acknowledgments
The precious help of C. Cacciapaglia, T. Giannini, L. Renna, and V. Cataldo is gratefully acknowledged.
This study was funded by the European Community.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amati, B., S. Dalton, M. W. Brooks, T. D. Littlewood, G. I. Evan, and H. Land. 1992. Transcriptional activation by the human c-myc oncoprotein in yeast requires interaction with Max. Nature 359:423-426. [DOI] [PubMed] [Google Scholar]
- 3.Apweiler, R. 2000. Protein sequence databases, p. 31-71. In F. M. Richards, D. S. Eisenberg, and P. S. Kim (ed.), Advances in protein chemistry. Academic Press, New York, N.Y. [DOI] [PubMed]
- 4.Bairoch, A., and R. Apweiler. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28:45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, W. C., J. S. Garavelli, Z. Hou, H. Huang, R. S. Ledley, P. B. McGarvey, H. W. Mewes, B. C. Orcutt, F. Pfeiffer, A. Tsugita, C. R. Vinayaka, C. Xiao, L.-S. L. Yeh, and C. Wu. 2001. Protein Information Resource: a community resource for expert annotation of protein data. Nucleic Acids Res. 29:29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branch, A. D., B. J. Benenfeld, and H. D. Robertson. 1989. RNA fingerprinting. Methods Enzymol. 180:130-154. [DOI] [PubMed] [Google Scholar]
- 7.Crish, J. F., F. Bone, S. Balasubramanian, T. M. Zaim, T. Wagner, J. Yun, E. A. Rorke, and R. L. Eckert. 2000. Suprabasal expression of the human papillomavirus type 16 oncoproteins in mouse epidermis alters expression of cell cycle regulatory proteins. Carcinogenesis 21:1031-1037. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, D., R. Lehmann, and P. D. Zamore. 1995. Translational regulation in development. Cell 81:171-178. [DOI] [PubMed] [Google Scholar]
- 9.De Pasquale, C., and D. Kanduc. 1998. Modulation of HPV16 E7 translation by tRNAs in eukaryotic cell-free translation systems. Biochem. Mol. Biol. Int. 45:1005-1009. [DOI] [PubMed] [Google Scholar]
- 10.Desaintes, C., and C. Demeret. 1996. Control of papillomavirus DNA replication and transcription. Semin. Cancer Biol. 7:339-347. [DOI] [PubMed] [Google Scholar]
- 11.de Smit, M. H., and J. van Duin. 1994. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J. Mol. Biol. 244:144-150. [DOI] [PubMed] [Google Scholar]
- 12.Eckert, R. L., J. F. Crish, and N. A. Robinson. 1997. The epidermal keratinocyte as a model for the study of gene regulation and cell differentiation. Physiol. Rev. 77:397-424. [DOI] [PubMed] [Google Scholar]
- 13.Felciano, R. M., R. O. Chen, and R. B. Altman. 1997. RNA secondary structure as a reusable interface to biological information resources. Gene 190:GC59-GC70. [DOI] [PubMed] [Google Scholar]
- 14.Ferenczy, A., and B. Winkler. 1987. Cervical intra-epithelial neoplasia and condyloma, p. 177-218. In R. Kurman (ed.), Blaustein's pathology of the female genital tract, 2nd ed. Springer Verlag, New York, N.Y.
- 15.Gabrielsen, O. S., and J. Huet. 1993. Magnetic DNA affinity purification of yeast transcription factor. Methods Enzymol. 218:508-525. [DOI] [PubMed] [Google Scholar]
- 16.Guelstein, V. I., T. A. Tchypysheva, V. D. Ermilova, L. V. Litvinova, S. M. Troyanovsky, and G. A. Bannikov. 1988. Monoclonal antibody mapping of keratins 8 and 17 and of vimentin in normal human mammary gland, benign tumors, dysplasias and breast cancer. Int. J. Cancer 42:147-153. [DOI] [PubMed] [Google Scholar]
- 17.Hellström, I., V. Brankovan, and K. E. Hellström. 1985. Strong antitumor activities of IgG3 antibodies to a human melanoma-associated ganglioside. Proc. Natl. Acad. Sci. USA 82:1499-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaramillo, M., J. Pelletier, I. Edery, P. J. Nielsen, and N. Sonenberg. 1991. Multiple mRNAs encode the murine translation initiation factor eIF-4E. J. Biol. Chem. 266:10446-10451. [PubMed] [Google Scholar]
- 19.Jeon, S., and P. F. Lambert. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 92:1654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, L. D., C. L. Easterday, H. Gore, and A. T. Hertig. 1964. The histogenesis of carcinoma in situ of the uterine cervix. Cancer 17:213-229. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. R., and M. D. Cole. 1987. Rapid cytoplasmatic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol. Cell. Biol. 7:4513-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitasato, H., H. Delius, H. zur Hausen, K. Sorger, F. Rösl, and E. M. de Villiers. 1994. Sequence rearrangements in the upstream regulatory region of human papillomavirus type 6: are these involved in malignant transition? J. Gen. Virol. 75:1157-1162. [DOI] [PubMed] [Google Scholar]
- 23.Kozak, M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol. Cell. Biol. 9:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lees, E., K. Osborn, L. Banks, and L. Crawford. 1990. Transformation of primary BRK cells by human papillomavirus type 16 and EJ-ras is increased by overexpression of the viral E2 protein. J. Gen. Virol. 71:183-193. [DOI] [PubMed] [Google Scholar]
- 25.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perkins, W., I. Campbell, I. M. Leigh, and R. M. MacKie. 1992. Keratin expression in normal skin and epidermal neoplasms demonstrated by a panel of monoclonal antibodies. J. Cutan. Pathol. 19:476-482. [DOI] [PubMed] [Google Scholar]
- 27.Ramaekers, F., C. van Niekerk, L. Poels, E. Schaafsma, A. Huijsmans, H. Robben, G. Schaart, and P. Vooijs. 1990. Use of monoclonal antibodies to keratin 7 in the differential diagnosis of adenocarcinomas. Am. J. Pathol. 136:641-655. [PMC free article] [PubMed] [Google Scholar]
- 28.Rösl, F., and E. Schwarz. 1997. Regulation of E6 and E7 oncogene transcription, p. 25-70. In M. Tommasino (ed.), Papillomaviruses in human cancer: the role of E6 and E7 oncoproteins. Landes Bioscience, Austin, Tex.
- 29.Ross, J. 1994. Analysis of messenger RNA turnover in cell-free extracts from mammalian cells, p. 107-133. In S. J. Higgins and B. D. Hames (ed.), RNA processing: a practical approach, vol. 2. IRL Press, Oxford, England. [Google Scholar]
- 30.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Perez, A. M., S. Soriano, A. R. Clarke, and K. Gaston. 1997. Disruption of the human papillomavirus type 16 E2 gene protects cervical carcinoma cells from E2F-induced apoptosis. J. Gen. Virol. 78:3009-3018. [DOI] [PubMed] [Google Scholar]
- 32.Singer, A., and J. A. Jordan. 1976. The anatomy of the cervix, p. 132-143. In J. A. Jordan and A. Singer (ed.), The cervix. W. B. Saunders & Co., London, England.
- 33.Stutz, A., B. Conne, J. Huarte, P. Gubler, V. Volkel, P. Flandin, and J. D. Vassalli. 1998. Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes Dev. 12:2535-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talbot, S. J., and S. Altman. 1994. Gel retardation analysis of the interaction between C5 protein and M1 RNA in the formation of the ribonuclease P holoenzyme from Escherichia coli. Biochemistry 33:1399-1405. [DOI] [PubMed] [Google Scholar]
- 35.Tommasino, M., and L. Crawford. 1995. Human papillomavirus E6 and E7: proteins which deregulate the cell cycle. Bioessays 17:509-518. [DOI] [PubMed] [Google Scholar]
- 36.van Muijen, G. N., D. J. Ruiter, W. W. Franke, T. Achtstatter, W. H. Haasnoot, M. Ponec, and S. O. Warnaar. 1986. Cell type heterogeneity of cytokeratin expression in complex epithelia and carcinomas as demonstrated by monoclonal antibodies specific for cytokeratins nos. 4 and 13. Exp. Cell Res. 162:97-113. [DOI] [PubMed] [Google Scholar]
- 37.von Knebel Doeberitz, M., C. Rittmüller, D. Glitz, H. zur Hausen, and M. Dürst. 1992. Inhibition of tumorigenicity of cervical cancer cells in nude mice by HPV18 E6-E7 antisense RNA. Int. J. Cancer 51:831-834. [DOI] [PubMed] [Google Scholar]
- 38.Zehbe, I., F. Ciccolini, M. Dell'Orco, C. De Pasquale, F. Zaccaro, V. Albarani, A. Marchini, D. Kanduc, and M. Tommasino. 1998. E7 protein of human papillomaviruses and its interaction with cellular pathways, p. 97-107. In P. Bannasch, D. Kanduc, S. Papa, and J. M. Tager (ed.), Cell growth and oncogenesis. Birkhäuser Verlag, Basel, Switzerland.
- 39.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]