Abstract
We generated recombinant vesicular stomatitis viruses (VSV) expressing genes encoding hybrid proteins consisting of the extracellular domains of hepatitis C virus (HCV) glycoproteins fused at different positions to the transmembrane and cytoplasmic domains of the VSV G glycoprotein (E1G and E2G). We show that these chimeric proteins are transported to the cell surface and incorporated into VSV virions efficiently. We also generated VSV recombinants in which the gene encoding the VSV G protein was deleted and replaced by one or both of the E1G and E2G genes, together with a green fluorescent protein gene. These ΔG viruses incorporated E1G and E2G proteins at levels approximately equivalent to the normal level of VSV G itself, or about 1,200 molecules of each protein per virion. Given the potency of VSV recombinants as vaccines in other studies, this high-level expression and incorporation of HCV proteins into virions could be very important for development of an HCV vaccine. Despite the presence of E1G and E2G proteins at high levels in the virions, these virions did not infect cell lines that have been reported to support at least a low level of HCV infection and replication.
Hepatitis C virus (HCV), the major cause of non-A, non-B viral hepatitis, was first identified in 1989 (3) and has infected approximately 170 million people worldwide (34), including about 4 million in the United States (1). HCV infection leads to chronic hepatitis, cirrhosis, and hepatocellular carcinoma and is now the leading cause of liver transplantation in the United States (7). Current treatment for HCV infection leads to sustained viral clearance in only about 50% of patients (24), and no vaccine is available to prevent new infections.
HCV is a positive-stranded RNA virus with a genome of 9.6 kb encoding a polyprotein precursor of 3,000 amino acids (19). The polyprotein is proteolytically cleaved into 10 distinct products: the structural proteins (C, E1, E2, and P7) and several nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B). E1 and E2 are type I transmembrane proteins which are highly glycosylated and associate to form a noncovalently linked heterodimer (6). The C-terminal hydrophobic regions of both proteins contain signals that are responsible for retaining these proteins in the endoplasmic reticulum (4, 5, 8). Deletions in these C-terminal regions and replacement with foreign transmembrane and cytoplasmic domains result in the expression of both E1 and E2 at the cell surface. E2 binds with high affinity to CD81, a tetraspanin protein expressed on various cell types including hepatocytes and B lymphocytes (23). However, definitive evidence that CD81 is an HCV receptor has not yet been obtained.
Vesicular stomatitis virus (VSV) is a nonsegmented negative-strand RNA virus and the prototype of the rhabdovirus family. VSV infection in animals induces strong cellular and humoral immunity to its own proteins and to additional proteins encoded by recombinant viruses (12, 20, 26, 35). VSV has an 11-kb RNA genome of negative (noncoding) sense and replicates in the cytoplasm, where it has only RNA intermediates in replication. It synthesizes five subgenomic mRNAs encoding five structural proteins (28). VSV recombinants can be recovered from DNA copies, and foreign genes can be expressed at high levels from multiple sites in the genome (11, 14, 17, 31).
It has been reported that cells transiently expressing HCV E1 or E2 chimeric proteins containing the VSV G transmembrane and cytoplasmic domains generate VSV pseudotypes when infected with a VSV mutant (ts045) that is temperature sensitive for G protein transport to the cell surface (16). Pseudotypes formed with either the E1G or E2G constructs were reported to infect mammalian cell lines, including cell lines not derived from the liver. Sera from HCV-infected chimpanzees were reported to neutralize the infectivity of the pseudotypes, while an anti-VSV neutralizing antibody did not (16). This result is in apparent conflict with another report showing that expression of both E1G and E2G hybrid proteins was required to obtain a low-level cell-cell fusion detectable in a highly sensitive assay (33). A recent study from the same group reported that infectious VSV(HCV) pseudotypes could be generated in a cell line expressing E1G and E2G proteins together but not in a cell line expressing E1G or E2G protein alone (21). These pseudotypes showed the highest infectivity on a human hepatoma cell line (HepG2), but infectivity was not neutralized by specific antibodies to E1 and E2 or by sera from HCV-infected animals.
In our study, we show that several HCV E1 and E2 chimeric glycoproteins composed of ectodomains of E1 and E2 and the transmembrane and cytoplasmic domains of VSV G can be expressed from live virus recombinants on the cell surface. The proteins are incorporated at high levels into VSV virions in the presence or absence of the VSV glycoprotein.
MATERIALS AND METHODS
Cells and cell lines used.
VSV recombinants were recovered on BHK-21 cells. Cells for testing infectivity of VSV recombinants were BHK-21, COS-7, HEK 293, CHO-K1, and primary human hepatocytes. The primary hepatocytes were isolated by enzymatic collagenase perfusion through slices of liver tissue and were cultured in hormonally defined media on collagen-coated dishes (18).
Plasmid construction.
Sequences of E1 and E2 genes were obtained from a subclone of an infectious clone of HCV (13) called pBRTM/HCV1-809con. This plasmid contains the capsid, E1, E2, and p7 genes of HCV. The DNA sequences encoding the putative ectodomains of E1 and E2 and the preceding signal sequences were joined to the VSV G transmembrane and cytoplasmic tail sequences by using an overlapping PCR technique. An ATG translation initiation codon preceded by an A in the −3 position was included before each signal sequence from the preceding gene. For the E2661G construct, the VSV G transmembrane and tail sequences were amplified from pBSSK-G with forward primer 5′-GGAAG ACAGG GACAG GTCCG AGGGT TGGTT CAGTA GTTGG AAAAGC and the VSV G reverse primer 5′-CGCGG GATCCG CTAGCA GGATT TGAGT TACTTTCC, containing an NheI restriction site (underlined) and a BamHI restriction site (boldfaced). For the E2717G construct, the forward fusion primer was 5′-CGTCC TGGGC CATTAA GTGGG AGGGT TGGTT CAGTAG TTGGA AAAGC and the reverse primer was the VSV G reverse primer given above. Italicized sequences correspond to 5′ extensions of HCV-E2 sequence necessary for fusion of the E2 and VSV sequences. The products from these amplifications were purified and used in a second round of PCR using pBRTM/HCV as a template. The product was amplified by using VENT polymerase (New England Biolabs, Beverly, Mass.) with forward primer 5′-CCACG CGTCTC GAGACCATGGTGGGGAACTGGGCGAAGG (underlining indicates an XhoI restriction site) and the VSV G reverse primer described above. The ATG codon corresponds to amino acid 364 in the HCV polyprotein and to amino acid 173 in the preceding E1 protein. For cloning into pBSSK, the purified products were digested with XhoI-BamHI. The resulting clones were designated pBS-E2661G and pBS-E2717G and were used to verify expression of the proteins using the vaccinia virus-T7 system (9). For cloning into pVSV-XN2 for recovery of infectious VSV, the inserts were excised with XhoI-NheI and ligated into pVSV-XN2 (31), which had been digested with the same enzymes. The resulting clones were designated pVSVXN-E2661G and pVSVXN-E2717G. To generate VSV recombinants lacking VSV G, pVSVXN-E2661G and pVSVXN-E2717G were digested with MluI and XhoI and religated after a fill-in reaction using T4 polymerase. (New England Biolabs) The resulting clones were designated pVSVΔG-E2661G and pVSVΔG-E2717G.
The E1352G construct was also prepared by the overlapping PCR technique described above. The VSV G transmembrane and tail sequences were amplified by using the forward primer 5′-GGACA TGATC GCTGG TGCT CAC GGTTG GTTCA GTAGT TGGAA AAGC (italicized sequence corresponds to overlap of the HCV E1 sequence). The forward primer used in the second round of amplification was 5′-GGGTA CCGTCGAC ACGCGT ACCATG GGTTG CTCTT TCTCT ATCTTCC (with the SalI site boldfaced and the MluI site underlined). The underlined ATG is followed by the E1 signal coding sequence from the preceding capsid gene starting at Gly 171. The purified fragment was digested with SalI and NheI and ligated into pVSV-XN2, which had been previously digested with XhoI-NheI. The resulting plasmid was designated pVSVXN-E1352G. To generate a plasmid for derivation of a clone lacking VSV G, this plasmid was digested with MluI and religated as described above to generate pVSVΔG-E1352G.
To construct pVSVΔG-E1352G-E2661G and pVSVΔG-E1352G-E2717G, the E1G gene was amplified from pVSVXN-E1352G by using the forward primer 5′-TATCA CGCTC GACAC GCGTA CCATG GGTTGC, containing an MluI site (underlined), and the reverse primer 5′-CCCGG GGTCG ACGA TTGC TGTTA GTTTT TTTCA TAAAA ATTAA AAACTC, with a SalI site (underlined) and a VSV transcription stop-start sequence (italicized). The product was purified, digested with MluI and SalI, and ligated into pVSVXN-E2661G and pVSVXN-E2717G which had been previously digested with MluI and XhoI to remove VSV G.
pVSVΔG-E1340G-E2711G-GFP was constructed by using an overlapping PCR technique. VSV G transmembrane and tail sequences were amplified by PCR from pVSVMXA2 by using the forward fusion primer 5′-GGTAG GGTCA AGCAT CGCGT CCAAA AGCTC TATTG CCTCT TTTTTC and the VSV G reverse primer given above. The italicized sequence corresponds to the 5′ extension of HCV-E2. The product was gel purified and used in a second round of PCR with pBRTN/HCV as a template. The product was amplified with the forward primer 5′-CCACG CGTCT CGAGA CCATG GTGGG GAACT GGGCG AAGG (boldface indicates an XhoI restriction site) and the VSV G reverse primer described above. For cloning into pBSSK, the product was purified and digested with XhoI and BamHI. The resulting clone was designated pBSE2711G. The product was excised with XhoI and NheI and cloned into pVSVXN-GFP to produce pVSVXN-E2711G-GFP.
E1340G was constructed as described above with the following changes. The VSV G transmembrane and tail with the 5′ extension of HCV E1 was amplified by PCR with the forward fusion primer 5′-GTGGT AGCTC AGCTG CTCCG GATCA AAAG CTCTA TTGCC TCTTTTTTC (with the 5′ E1 extension italicized). The E1G reverse primer was 5′-CCCGG GGTCG ACGAT TGC TGTTAG TTTTT TTCATA AAAAT TAAAA ACTC (the transcription stop-start is italicized, and the SalI site is boldfaced). The product was purified and amplified by PCR as described above by using the forward primer 5′-CACGC TCGAC ACGCG TACCA TGGGT TGCTC TTTCT CTATC TTCC (boldface indicates a MluI restriction site) and the E1G reverse primer described above. The product was gel purified, digested with MluI and SalI, and cloned into pVSVΔG-E2711G-GFP which had been previously digested with MluI and XhoI.
The pCAGGS vector (22) was used for transient expression of complementing E1G and E2G proteins. To generate the pCAGGS-E1340G vector, the E1340G PCR product described above was amplified with the forward primer CACGC TCGAC GAATT CACCAT GGGTT GCTCT TTCTC TATCTTCC, containing an EcoRI restriction site (boldfaced). The reverse primer, containing a BglII site (boldfaced), was CCCTC GAGAGATCTT TACTT TCCAA GTCGG TTCAT CTC. This PCR product was digested with EcoRI and BglII, purified, and cloned into pCAGGS cut with EcoRI and BglII. To construct pCAGGS-E2711G, the PCR product described above was digested with XhoI and BamHI and cloned into pCAGGS digested with XhoI and BglII.
Virus recoveries.
Recombinant VSVs were recovered using established methods (17, 32). Briefly, baby hamster kidney (BHK) cells were plated to 40% confluency on 10-cm tissue culture plates. The cells were infected with vTF7-3 (9), a recombinant vaccinia virus that expresses T7 RNA polymerase at a multiplicity of infection (MOI) of 10. After incubation at 37°C for 1 h, each dish was transfected with 3 μg of pBS-N, 5 μg of pBS-P, 1 μg of pBS-L, and 10 μg of a plasmid encoding one of the full-length recombinants described above. Transfections were performed using a cationic liposome reagent (27). Cells were then incubated at 37°C for 48 h. Cell supernatants were then passed through a 0.2-μm-pore-size filter to remove vaccinia virus and were then applied to BHK cells for an additional 48 h at 37°C. Recovery of infectious virus was confirmed by scanning BHK cell monolayers for VSV cytopathic effect and, in some recoveries, for green fluorescent protein (GFP) fluorescence. Stocks of each virus were grown from single plaques on BHK cells. These stocks were then stored at −80°C.
Infectious virus was recovered from pVSVΔG plasmids by the same procedure, with the following modifications. BHK cells were transfected as described above with the addition of 4 μg of pBS-G. After 48 h at 37°C, supernatants were filtered and passaged onto BHK-G cells (32) that had been induced to express G 12 h previously. Stocks of these viruses were also grown from single plaques on induced BHK-G cells.
Immunofluorescence microscopy.
BHK cells were plated to near-confluency on coverslips and infected with recombinant wild-type VSV (rwtVSV), VSVE1G, VSVE2661G, or VSVE1G-E2661G. After 6 h at 37°C, cells were fixed in 3% paraformaldehyde. Coverslips were then washed in phosphate-buffered saline (PBS) containing 10 mM glycine (PBS-glycine). Coverslips were incubated with a 1:1,000 dilution of a monoclonal antibody to E1 or a 1:75 dilution of a monoclonal antibody to E2 (Austral Biologicals, San Ramon, Calif.) followed by a 1:50 dilution of a fluorescein isothiocyanate-conjugated goat anti-mouse antibody. (Jackson Research, West Grove, Pa.). Coverslips were then mounted on slides, and cells were observed and photographed by using a Nikon Microphot-FX microscope with a 40× objective and a SPOT digital camera.
Metabolic labeling of infected cells and virus.
BHK cells at about 50% confluency were infected with wild-type VSV and recombinants at an MOI of 10 in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum. Four to six hours postinfection, the medium was removed. The cells were then labeled in methionine-free DMEM containing 100 μCi of [35S]methionine. To prepare labeled cell extracts, the radioisotope was added in 1.0 ml of medium for 1 to 2 h at 37°C. The medium was removed, and the cells were first washed with PBS and then lysed in 0.5 ml of detergent solution.(1% Nonidet P-40 [NP-40],0.4% deoxycholate, 50 mM Tris-HCl [pH 8.0], 62.5 mM EDTA). To prepare labeled virus, 100μCi of [35S]methionine was added to cells 4 h postinfection in 1 ml of methionine-free DMEM supplemented with 1% fetal bovine serum. Labeling was continued for 16 to 18 h at 37°C. The culture medium was transferred to an Eppendorf tube and clarified by for 2 min. Supernatants were then layered onto a solution of 20% sucrose in TE buffer (10 mM Tris [pH 7.4]-1 mM EDTA) and centrifuged at 180,000 × g for 1 h at 4°C in a Beckman SW50.1 rotor. Virus pellets were resuspended in 40 μl of TE buffer. Virus and cell lysate samples were analyzed by electrophoresis on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel.
Immunoprecipitations and endoglycosidase F digestion.
Labeled purified virus was dissolved in 200 μl of radioimmunoprecipitation assay (RIPA) buffer (0.1% SDS, 1% deoxycholate,1% NP-40, and 0.15 M NaCl in 10 mM Tris [ph7.4]) and incubated at 37°C for 15 min. Pansorbin (30 μl) (Calbiochem, La Jolla, Calif.) was added, and samples were incubated for 20 min at 37°C. Samples were clarified by centrifugation at 10,000 rpm in an Eppendorf microcentrifuge for 2 min and then transferred to new tubes. A polyclonal rabbit anti-VSV G tail antibody (2 μl) was added, and samples were incubated for 1 h at 37°C. Pansorbin (30 μl) was added, and samples were incubated for 30 min at 37°C. Samples were pelleted, washed three times with ice-cold RIPA buffer, and spun dry in an Eppendorf microcentrifuge. The pellet was resuspended in Laemmli sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by exposure to a PhosphorImager screen (Molecular Dynamics).
For endoglycosidase F digestion, the dry sample pellets were resuspended in 40 μl of denaturing buffer (5% SDS-10% β-mercaptoethanol) and incubated at 100°C for 10 min. Samples were clarified by centrifugation in an Eppendorf microcentrifuge for 2 min and then transferred to new tubes. To each sample a 1/10 volume of 10% NP-40-10× G7 buffer (0.5 M sodium phosphate [pH 7.5]) was added. Peptide N-glycosidase F (1 μl) was added, and samples were incubated at 37°C for 1 h. Samples were boiled in Laemmli sample buffer and analyzed by SDS-PAGE.
Complementation and neutralization assays.
BHK cells (5 × 106 per 10-cm plate) were transfected with either pCAGGS-E1340G or PCAGGS-E2711G, or with both plasmids, by using Lipofectamine (GIBCO/BRL). After a 3-h incubation at 37°C, the medium was replaced with 10 ml of 10% DMEM. At 18 h posttransfection, cells were infected with 1 ml of VSVΔG (108 PFU/ml) in 3 ml of serum-free DMEM. Cells were incubated for 2 h at 37°C, and then the medium was removed and replaced with 10 ml of 10% DMEM. After a 24-h incubation at 37°C, the medium samples containing the VSV E1-, E2-, or E1E2-complemented virus stocks were collected.
For neutralization assays, BHK and HepG2 cells were plated at 5 × 105 cells per 35-mm-diameter dish. Viruses (106 to 108 PFU) were neutralized for 1 h at 37°C with mouse monoclonal antibodies to VSV G (I1 and I14) at a final dilution of 1:100. Neutralized virus samples were added to cells and incubated overnight at 37°C. Cells were examined for expression of GFP by fluorescence microscopy.
RESULTS
Chimeric HCV E1G and E2G glycoproteins.
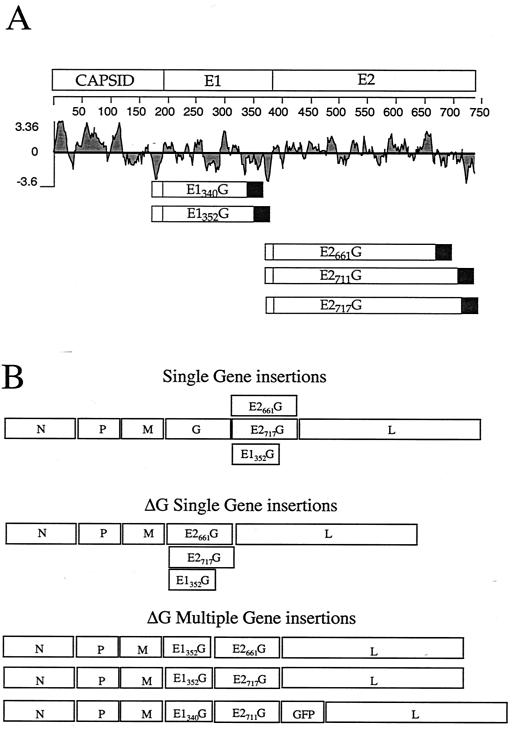
Carboxy-terminal truncations of the E1 and E2 proteins of HCV have been shown to allow the glycoproteins to be expressed on the cell surface (4, 5, 8, 16) and induce cell-cell fusion (33). Based on this information, the amino acid positions of the E1 and E2 glycoproteins were chosen for the chimeric gene constructs. As shown in Fig. 1A, protein sequences in the HCV polyprotein are numbered starting with the first amino acid of the capsid protein. The capsid comprises amino acids 1 to 191, E1 comprises amino acids 192 to 383, and E2 comprises amino acids 384 to 746. The E1 (E1340G and E1352G) and E2 (E2661G, E2711G, and E2717G) constructs encode the E1 and E2 protein sequences out to the indicated amino acid followed by the VSV G transmembrane and cytoplasmic domains. All constructs include the natural signal sequence from the upstream HCV gene preceded by an ATG initiation codon with an A in the −3 position. These chimeric gene products were made by using an overlapping PCR technique described in Materials and Methods and were cloned into the pBSSK vector (Stratagene). The constructs were then expressed under the control of the bacteriophage T7 promoter in BHK cells infected with a recombinant vaccinia virus (vTF7-3) that expresses T7 RNA polymerase (9). Protein expression on the cell surface was confirmed by indirect immunofluorescence (data not shown).
FIG. 1.
Diagrams of chimeric E1340G, E1352G, E2661G, E2711G, and E2717G glycoprotein constructs and VSV recombinants. (A) Diagram of each glycoprotein construct relative to the position in the HCV polyprotein. The signal sequence preceding E1 or E2 (open boxes) was appended to the N terminus of each construct, and the VSV transmembrane and tail sequences (filled boxes) were appended to the C terminus. The hydrophobicity of the protein sequence is indicated by the Kyte-Doolittle plot (15). (B) Gene diagrams of the nine recombinant VSVs generated in this study. The positions of the VSV genes encoding the N, P, M, G, and L proteins are indicated, along with the positions of the HCV or GFP genes, in each of the recombinants.
Recovery of recombinant VSVs expressing chimeric HCV E1 and E2 glycoproteins.
To generate VSV recombinants expressing the HCV/VSV chimeric glycoproteins, we started with a plasmid DNA vector that allows derivation of VSV recombinants expressing foreign genes (31). The VSV recombinants were then generated by using standard procedures involving expression of the full-length antigenomic recombinant VSV RNA in cells also expressing the VSV N, P, and L proteins. Recombinant VSVs expressing E1G, E2G, or both glycoproteins were recovered as described in Materials and Methods. In addition, we prepared recombinants that expressed E1G, E2G, or both glycoproteins in the absence of the VSV glycoprotein. Because these VSVΔG constructs do not encode G, we propagated them using a complementing BHK cell line that expresses VSV G protein (32). A diagram of the recombinant VSVs generated in this study is given in Fig. 1B.
Recombinant VSVs express the HCV E1 and E2 chimeric glycoprotein genes.
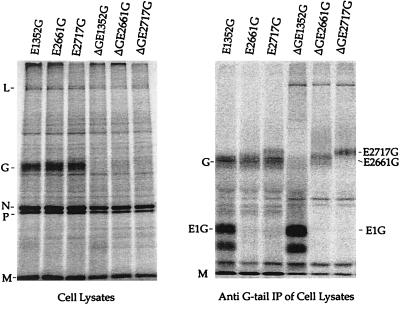
To determine if recombinant VSVs expressed the chimeric glycoproteins, we first analyzed six recombinants that encoded E1352G, E2661G, or E2717G separately. These recombinants were generated with or without (ΔG) the VSV G gene present. BHK cells were infected with either VSV-E1352G, VSV-E2661G, VSV-E2717G, VSVΔG-E1352G, VSVΔG-E2661G, or VSVΔG-E2717G and were labeled for 1 h with [35S]methionine. Total-cell lysates were analyzed first by SDS-PAGE without immunoprecipitation (Fig. 2, left panel). Cells infected with the recombinants containing G showed the five VSV proteins L, G, N, P, and M, while VSVΔG-infected cells lacked the G band. Because the E1G and E2G proteins were not readily apparent on the gel of the total lysate, the labeled cell lysates were immunoprecipitated with a polyclonal anti-VSV G cytoplasmic tail antibody (10). This precipitation (Fig. 2, right panel) showed additional bands of the sizes expected for the E1 (36 kDa, including 5 N-linked glycans) and E2 (64 or 70 kDa, with 10 N-linked glycans) chimeric glycoproteins (Fig. 2, right panel). Note that the E2 G proteins run with or just slower than VSV G (65 kDa), which is also precipitated by the G tail antibody, and therefore these proteins are best seen in the ΔG constructs.
FIG. 2.
Analysis of proteins encoded by VSV recombinants. BHK cells were infected with either VSV-E1352G, VSV-E2661G, VSV-E2717G, VSVΔG-E1352G, VSVΔG-E2661G, or VSVΔG-E2717G at an MOI of 10 and labeled with [35S]methionine. (Left) Cell lysates were analyzed by SDS-10% PAGE. The positions of the five VSV proteins L (241 kDa), G (63 kDa), N (47 kDa), P (30 kDa), and M(27 kDa) are indicated. (Right) BHK cell extracts were immunoprecipitated with a rabbit anti-VSV G tail antibody and analyzed by SDS-10% PAGE. VSV M protein aggregates and appears at substantial levels in all precipitates.
Protein expression on the cell surface.
To determine if the chimeric E1G and E2G glycoproteins expressed by recombinant VSVs were transported to the cell surface, BHK cells were infected with either VSV-wt, VSV-E1352G, or VSV-E2661G, fixed, and incubated with antibodies to either E1, E2, or VSV G followed by fluorescent secondary antibodies. Fluorescence microscopy showed that cells infected with wt VSV stained only with an antibody to VSV G (Fig. 3A) and not with a mixture of anti-E1 and anti-E2 monoclonal antibodies (Fig. 3B). Cells infected with VSV-E1352G were stained with anti-E1 (Fig. 3C) but not with anti-E2 (Fig. 3D). Cells infected with VSV-E2661G stained with an antibody to E2 (Fig. 3F) but not with an antibody to E1 (E). The pattern of the protein labeling clearly outlined the cell surface, indicating that the chimeric glycoproteins expressed from recombinant VSVs were transported to the plasma membrane.
FIG. 3.
Cell surface expression of chimeric E1G and E2G glycoproteins encoded by VSV recombinants. Cells were infected with either VSV-wt (A and B), VSV-E1352G (C and D), VSV-E2661G (E and F), or VSVΔGE1340G-E2711G (G and H) for 6 h, fixed, and labeled with either monoclonal antibodies to VSV G (A), a mixture of antibodies to E1 and E2 (B), an antibody to E1 alone (C, E, and G), or an antibody to E2 alone (D, F, and H), followed by fluorescein isothiocyanate-labeled goat anti-mouse antibodies. Cells were photographed with a Nikon Microphot FX fluorescence microscope equipped with a 40× Planapochromat objective and a SPOT digital camera. Exposure times for positive and negative samples were the same.
Incorporation of HCV E1 and E2 chimeric proteins into virus particles.
Initial experiments showed that the E1G and E2G proteins were readily incorporated into VSV recombinants expressing VSV G (data not shown). We next wanted to determine if E1 and E2 chimeric proteins could be incorporated into ΔG VSV particles budding from the cell surface. BHK cells were infected either with wt VSV or with G-pseudotyped ΔG VSV recombinants expressing the E1352G or E2661G protein or both. Infected cells were labeled with [35S]methionine for 18 h, and labeled virus was purified from the tissue culture supernatant and analyzed by SDS-PAGE (Fig. 4, left panel). Purified virus from cells infected with VSV showed five VSV proteins, as indicated (Fig. 4, lane 1). Virus from cells infected with the VSVΔG recombinants (Fig. 4, lanes 2 to 4) did not contain VSV G protein but instead had additional broad bands with the mobilities expected for heterogeneously glycosylated E1352G and E2661G. We performed immunoprecipitation of detergent-disrupted purified ΔG viruses using an antibody to the VSV G cytoplasmic tail to verify the identities of the proteins (Fig. 4, right panel, lanes 5 to 7). These results showed that broad bands precipitated from the VSVΔG-E1352G and VSVΔG-E2661G viruses did contain the G tail epitope.
FIG. 4.
Incorporation of E1G and E2G chimeric glycoproteins into VSV virions. BHK cells were infected for 4 h with VSV-wt, VSVΔG-E1352G, VSVΔG-E2661G, or VSVΔG-E1352G-E2661G. The cells were then labeled overnight with [35S]methionine, and virus was purified from the supernatant. Purified virus was analyzed by SDS-PAGE, and the positions of VSV proteins as well as E1352G and E2661G are indicated (left panels). Immunoprecipitations of proteins from the lysates by using the anti-G tail antibody are shown in the right panel.
To determine the molar ratios of E1G and E2G relative to VSV N or M protein in purified virus, we quantitated the gels in Fig. 4 as well as other gels and corrected for the methionine content of the proteins. The results showed that the numbers of E1G or E2G molecules incorporated into virions were 80 to 110% of the number of G protein molecules in wt virions. Because VSV G and E2G proteins comigrate substantially, we were not able to quantify E2G incorporation in the presence of G, although it appeared to be present at levels at least 50% of those in ΔG particles (data not shown). Although the example shown is only for the E1352 and E2661 proteins (Fig. 4), very similar levels of incorporation into virions were observed for all ΔG constructs described in Fig. 1.
The broad bands of E1352G and E2661G result from glycosylation.
The E1352G and E2661G in virions electrophoresed as broad, diffuse bands that were substantially more heterogeneous than those seen after a 1-h pulse-label of infected cells (Fig. 2). To confirm that this size heterogeneity was due to glycosylation and to verify the molecular weights of the core proteins, we digested the proteins with endoglycosidase F prior to electrophoresis. The glycoproteins from [35S]methionine-labeled VSV or the indicated recombinants were immunoprecipitated with an anti-VSV G tail antibody, treated or mock treated with endoglycosidase F, and analyzed by SDS-PAGE (Fig. 5). The core proteins migrated as much sharper bands and had mobilities consistent with the expected molecular weights of 39,000 (E2G) and 25,000 (E1G).
FIG. 5.
Endoglycosidase F digestion of E1G and E2G chimeric glycoproteins from purified virions. [35S]methionine-labeled virions of VSV-wt, VSVΔG-E1352G, VSVΔG-E2661G, and VSVΔG-E1352G-E2661G were immunoprecipitated with an anti-VSV G tail polyclonal antibody and then treated (+) or mock treated (−) with endoglycosidase F to remove N-linked glycans. Digests were then analyzed by SDS-12% PAGE. The mobilities of the VSV G protein and the E1352G and E2661G proteins with carbohydrate and without carbohydrate (−CHO) are indicated. The VSV M protein is present in all immunoprecipitates.
Lack of infectivity of the VSVΔG/HCV viruses in the absence of VSV G.
To determine if the VSVΔG/E1E2 viruses were able to infect cells through interactions involving the E1 and E2 proteins, we prepared virus stocks initially by growth in complementing BHK G cells. These stocks were then used to infect BHK cells not expressing VSV G so that virions with only E1G and E2G could be analyzed. We next tested the infectivities of the VSVΔG-E1352G-E2661G and VSVΔG-E1352G-E2717G viruses on several cell lines including BHK (hamster), COS-7 (monkey), HEK 293 (human embryonic kidney), HepG2 (human hepatoma), and human primary liver cells. Infection was assessed either with preincubation of virions with a neutralizing monoclonal antibody to VSV G (I1) or without the antibody. We could detect a low level of infection in all cells in the absence of I1 preincubation, but no consistent infection after preincubation with I1 (data not shown). Infection was assessed either by indirect immunofluorescence for N protein or by GFP expression in those constructs also encoding GFP.
The neutralization of a low level of infectivity by I1 suggests that traces of VSV G from the input virus are recycled through the BHK cells onto the virions. This result was observed previously with a VSV/human immunodeficiency surrogate virus (2). Our initial results indicated that even high levels of the particular E1G and E2G proteins constructs we had made were not able to facilitate entry of the VSV recombinants into mammalian cells.
After we had obtained these results, it was reported by Takikawa et al. that E1G and E2G proteins with slightly different fusion points in E1 and E2 had a low level of fusion activity with HepG2 cells (33). We therefore generated a recombinant VSVΔG-E1340G-E2711G-GFP virus with fusion points in E1G and E2G identical to those described in that report. Cells infected with this virus expressed both the E1G and E2G proteins on the cell surface as determined by indirect immunofluorescence (Fig. 3G and H). To examine the proteins produced by this recombinant virus, we analyzed [35S]methionine-labeled virus by SDS-PAGE (Fig. 6). As expected, VSVΔG-E1340G-E2711G-GFP expressed the four VSV proteins excluding G. When immunoprecipitated with the anti-VSV G tail antibody, two proteins of the sizes expected for E1340G and E2711G were clearly visible. Although this virus clearly incorporated E1G and E2G proteins into the virions, we were unable to detect specific infection due to the HCV hybrid proteins as in the experiments described above.
FIG. 6.
Proteins encoded by VSVΔG-E1340G-E2711G-GFP. BHK cells were infected with either wt VSV or VSVΔG-E1340G-E2711G-GFP for 4 h and were then labeled for 18 h with 100 μCi of [35S]methionine. Purified virus was prepared and disrupted with detergent, and proteins were immunoprecipitated with an anti-VSV G tail antibody. Samples were analyzed by SDS-PAGE. The relative positions of VSV proteins, as well as E1340G and E2711G, are indicated.
We also tried to evaluate infectivity using pseudotypes of both E1340G and E2711G. BHK cells were transfected with the high-level expression vector pCAGGS (22). Cells were transfected with pCAGGS-G, pCAGGS-E1340G, and pCAGGS-E2711G constructs either individually or in combination. The cells were then infected with a high-titer stock of VSVΔG-GFP to obtain VSV(HCV) pseudotypes. HepG2 and BHK cells were infected either with nonneutralized pseudotypes as controls or with pseudotypes that had been neutralized with anti-VSV serum. Pseudotype particles containing E1340G, E2G711G, or a combination of E1340G and E2711G failed to show any infectivity on any of the cell lines tested.
DISCUSSION
We have found that HCV E1G and E2G hybrid glycoproteins whose intracellular retention signals have been deleted can be expressed by recombinant VSVs and incorporated at high levels into VSV virions. These viruses have been engineered either with or without the VSV G protein gene. In the absence of G protein, the levels of E1G and E2G proteins incorporated into the budding particles are similar to the levels of VSV G protein incorporated into wt VSV virions, as many as 1,200 molecules of each protein per virion.
VSV recombinants expressing foreign viral glycoproteins induce potent and protective immune responses to influenza and measles viruses after single intranasal vaccinations in animal models (25, 26, 30). This protection is based largely on production of neutralizing antibodies. Control of simian immunodeficiency virus replication and prevention of AIDS have also been achieved in a monkey model by vaccination with VSV vectors carrying env and gag genes (29). At least the initial protection in this model is dependent on potent induction of cytotoxic T lymphocytes (29). Given the lack of a vaccine for HCV and relatively little understanding of the types of immune responses that might confer protection, VSV vectors expressing the E1 and E2 glycoproteins as well as other HCV proteins are ideal candidates for vaccine studies.
One surprising aspect of our findings is that the VSVΔG recombinants expressing E1G and E2G constructs separately or together did not show any specific infectivity on a variety of cell lines, including a human HepG2 liver cell line, or on primary human liver cells. A low level of infectivity was detected in each case, but this was neutralized by a monoclonal antibody to VSV G, indicating that it was due to carryover of traces of VSV G after passage of G-pseudotyped recombinants through BHK cells. We do not know why our results with recombinant viruses are different from those of others using pseudotyped VSV (16, 21). The glycoprotein genes used as starting material are identical to those present in an infectious HCV clone (13), and the sequences of each construct were shown to match those of the infectious clone. We even generated a ΔG recombinant with the same E2G and E1G fusion points reported to yield infectious pseudotypes on HepG2 cells (21), but we still did not obtain infectivity on HepG2 cells. The infectivity reported by that group apparently required production of VSV pseudotypes on a CHO cell line expressing E1G and E2G, and it is possible that there is something important about long-term expression of the E1G and E2G proteins in the cell line. Long-term expression might promote better folding of the ectodomains of proteins that normally reside in the endoplasmic reticulum before incorporation into HCV virions.
Other possibilities worthy of future investigation are varying the carbohydrate modification on the E1G and E2G proteins by use of inhibitors of specific steps of glycosylation or cell lines defective in specific steps, or expression of additional membrane proteins of HCV. It is also possible that the viruses we have constructed would be able to infect a cell type that is permissive for HCV but that we have not yet found the appropriate cell type.
Acknowledgments
This work was supported by NIH grant AI24345 to J.K.R., NIH grants AI40034 and CA57973 to C.M.R., and the Greenberg Medical Research Foundation.
REFERENCES
- 1.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 2.Boritz, E., J. Gerlach, J. E. Johnson, and J. K. Rose. 1999. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J. Virol. 73:6937-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 4.Cocquerel, L., S. Duvet, J. C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubuisson, J. 2000. Folding, assembly and subcellular localization of hepatitis C virus glycoproteins. Curr. Top. Microbiol. Immunol. 242:135-148. [DOI] [PubMed] [Google Scholar]
- 7.Fishman, J. A., R. H. Rubin, M. J. Koziel, and B. J. Periera. 1996. Hepatitis C virus and organ transplantation. Transplantation 62:147-154. [DOI] [PubMed] [Google Scholar]
- 8.Flint, M., and J. A. McKeating. 1999. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J. Gen. Virol. 80:1943-1947. [DOI] [PubMed] [Google Scholar]
- 9.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan, J. L., C. E. Machamer, and J. K. Rose. 1985. Glycosylation allows cell-surface transport of an anchored secretory protein. Cell 42:489-496. [DOI] [PubMed] [Google Scholar]
- 11.Haglund, K., J. Forman, H. G. Krausslich, and J. K. Rose. 2000. Expression of human immunodeficiency virus type 1 Gag protein precursor and envelope proteins from a vesicular stomatitis virus recombinant: high-level production of virus-like particles containing HIV envelope. Virology 268:112-121. [DOI] [PubMed] [Google Scholar]
- 12.Kim, S. K., D. S. Reed, S. Olson, M. J. Schnell, J. K. Rose, P. A. Morton, and L. Lefrancois. 1998. Generation of mucosal cytotoxic T cells against soluble protein by tissue-specific environmental and costimulatory signals. Proc. Natl. Acad. Sci. USA 95:10814-10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 14.Kretzschmar, E., L. Buonocore, M. J. Schnell, and J. K. Rose. 1997. High-efficiency incorporation of functional influenza virus glycoproteins into recombinant vesicular stomatitis viruses. J. Virol. 71:5982-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 16.Lagging, L. M., K. Meyer, R. J. Owens, and R. Ray. 1998. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J. Virol. 72:3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, A. P., M. A. Roque, D. J. Beck, and D. L. Kaminski. 1992. Isolation and culturing of hepatocytes from human livers. J. Tissue Cult. Methods 14:139-146. [Google Scholar]
- 19.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-10041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 20.Masopust, D., V. Vezys, A. L. Marzo, and L. Lefrancois. 2001. Preferential localization of effector memory cells in nonlymphoid tissue. Science 291:2413-2417. [DOI] [PubMed] [Google Scholar]
- 21.Matsuura, Y., H. Tani, K. Suzuki, T. Kimura-Someya, R. Suzuki, H. Aizaki, K. Ishii, K. Moriishi, C. S. Robison, M. A. Whitt, and T. Miyamura. 2001. Characterization of pseudotype VSV possessing HCV envelope proteins. Virology 286:263-275. [DOI] [PubMed] [Google Scholar]
- 22.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 23.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 24.Reichard, O., R. Schvarcz, and O. Weiland. 1997. Therapy of hepatitis C: alpha interferon and ribavirin. Hepatology 26:108S-111S. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, A., L. Buonocore, R. Price, J. Forman, and J. K. Rose. 1999. Attenuated vesicular stomatitis viruses as vaccine vectors. J. Virol. 73:3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts, A., E. Kretzschmar, A. S. Perkins, J. Forman, R. Price, L. Buonocore, Y. Kawaoka, and J. K. Rose. 1998. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J. Virol. 72:4704-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 28.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication, p. 1221-1244. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 29.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 30.Schlereth, B., J. K. Rose, L. Buonocore, V. ter Meulen, and S. Niewiesk. 2000. Successful vaccine-induced seroconversion by single-dose immunization in the presence of measles virus-specific maternal antibodies. J. Virol. 74:4652-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnell, M. J., L. Buonocore, M. A. Whitt, and J. K. Rose. 1996. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 70:2318-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnell, M. J., J. E. Johnson, L. Buonocore, and J. K. Rose. 1997. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell 90:849-857. [DOI] [PubMed] [Google Scholar]
- 33.Takikawa, S., K. Ishii, H. Aizaki, T. Suzuki, H. Asakura, Y. Matsuura, and T. Miyamura. 2000. Cell fusion activity of hepatitis C virus envelope proteins. J. Virol. 74:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. [PubMed] [Google Scholar]
- 35.Zinkernagel, R. M., B. Adler, and J. J. Holland. 1978. Cell-mediated immunity to vesicular stomatitis virus infections in mice. Exp. Cell Biol. 46:53-70. [DOI] [PubMed] [Google Scholar]