Abstract
The expression of reporter genes driven by the same human elongation factor 1α (EF1α) promoter in murine leukemia virus (MLV)- and human immunodeficiency virus type 1 (HIV-1)-based vectors was studied in either transfected or virally transduced cells. The HIV-1 vectors consistently expressed 3 to 10 times higher activity than the MLV vectors at both the RNA and protein levels. The difference was not attributable to transcriptional interference, alternative enhancer/silencer, or differential EF1α intron splicing. Based on nuclear run-on assays, both vectors exhibited similar EF1α transcriptional activity. The reduced RNA levels of MLV vectors could not be explained by the decrease in RNA half-lives. Southern analysis of proviral DNA indicated that both HIV-1 and MLV vectors efficiently propagated the EF1α intron in the transduced cells. To decipher the discrepancy in transgene expression between MLV and HIV-1 vectors, the role of RNA 3′-end processing was examined using a sensitive Cre/lox reporter assay. The results showed that MLV vectors, but not HIV-1 vectors, displayed high frequencies of readthrough of the 3′ polyadenylation signal. Interestingly, the polyadenylation signal of a self-inactivating (SIN) HIV-1 vector was as leaky as that of the MLV vectors, suggesting a potential risk of oncogene activation by the lentiviral SIN vectors. Together, our results suggest that an efficient polyadenylation signal would improve both the efficacy and the safety of these vectors.
The development of viral vectors involves extensive deletion of viral sequences, and these modifications may affect viral RNA processing and stability as well as vector efficiency. The efficiency of gene transduction can be improved by incorporating additional viral elements and by employing strong internal promoters and/or regulatory elements in the viral vectors. In eukaryotic cells, the majority of the transcribed RNA is not efficiently processed for nuclear export, and that accumulation and translation of many species of cellular RNA in the cytoplasm are rate limiting and dependent on the presence of appropriate introns, nuclear export signals, and/or polyadenylation tails (9, 28). Because of this, there is a growing interest in understanding posttranscriptional processing and transport of RNAs derived from gene transfer vectors in order to improve expression of transgenes in these vector systems.
Oncoretroviral and lentiviral vectors combine some essential features for long-term gene transfer. The ability to stably integrate into the host genome and the lack of immune reactivity make these vectors popular in gene therapy studies (3, 7, 26, 32). Lentiviral vectors have overcome some major limitations of the murine oncoretroviral vector system by transducing nondividing cells in vitro and in vivo. The in vivo expression of the lentiviral transgene has been reported to last for periods longer than 6 months (22, 29). Several studies have shown better expression in different primary tissue cultures, including human hematopoietic stem cells, with lentiviral vectors than with oncoretroviral vectors (4, 7, 8, 34). Thus, in addition to nuclear accessibility, lentiviral vectors may have other advantages over oncoretroviral vectors.
In the retroviral genomes, several genetic elements have been shown to be of exceptional importance for posttranscriptional processing and transport of viral RNA; these include the splice sites, the cis-acting repressive sequences or inhibitory/instability RNA sequences, and the constitutive RNA transport elements or Rev-responsive elements (7, 14, 25, 33, 37). Cells infected with either oncoretroviral or lentiviral vectors are presumably equally capable of transcribing transgenes from an identical internal promoter.
Here we compared oncoretroviral and lentiviral vectors carrying an identical intron-containing promoter-reporter gene cassette and found that the efficiency of the lentiviral vectors was consistently 3- to 10-fold higher than that of the oncoretroviral vectors. The evidence supported that the weak 3′ poly(A) signal of the murine leukemia virus (MLV) vector significantly reduced cytoplasmic RNA accumulation of the transgene. Interestingly, the self-inactivating (SIN) vectors of both MLV and human immunodeficiency virus type 1 (HIV-1) displayed high frequencies of 3′ RNA readthrough. This finding highlights the importance of the poly(A) signals for efficacy and safety improvement of all retroviral vectors.
MATERIALS AND METHODS
Plasmid construction.
The oncoretroviral (MLV) and lentiviral vectors (HIV-1 and HIV-1 SIN) used for this study (see Fig. 1) were constructed as described previously (8, 19). All HIV-1 SIN vectors (pTY) have a 3′ bovine growth hormone polyadenylation signal (bGHpA) inserted behind the 3′ truncated long terminal repeat (LTR) (19). An enhanced green fluorescent protein (eGFP) expression plasmid, pHEFeGFP, was constructed by ligating the NotI-digested pHEF with a NotI-digested eGFP fragment derived from the humanized eGFP construct obtained from the Vector Core of UF Powell Gene Therapy Center (39). The pTYEFeGFP was made by inserting an eGFP fragment (XhoI-EcoRI) from pTVdl.EFeGFP into pTYEFnlacZ, replacing the nuclear lacZ (nlacZ) gene. pTVdl.EFeGFP was generated by replacing the nlacZ fragment (XhoI-EcoRI) of pTVdl.EFnlacZ with the eGFP fragment (XhoI-EcoRI) isolated from pHEFeGFP. The MLV gag-pol construct was based on pcDNA3.1/Zeo(+) (Invitrogen) with the cytomegalovirus immediate-early promoter replaced by the human elongation factor 1α (EF1α) promoter. The MLV SIN vector was generated by deleting NheI to XbaI in the 3′ LTR. The MLV vector pLFG was made by replacing SpeI in the 5′ untranslated leader to NotI in the polylinker of pLLL with the corresponding sequences of pMFG, which contains an extended packaging signal and a 3′ splice site (23, 30). pLLL-EFeGFP was constructed by digesting pTYEFeGFP with HindIII to obtain the EFeGFP fragment and cloned into the HindIII site of pLLL. pLFGEFnlacZ was constructed by inserting EFnlacZ from pTVΔEFnlacZ into pLFG, and the MLV SIN vector, pmLFGEFnlacZ, was made by replacing the wild-type (wt) 3′ U3 with the deleted U3 of the MLV SIN vector. pLPA, which contains a bovine growth hormone polyadenylation signal (bGHpA) in the 3′ LTR, was made by a four-fragment ligation approach using a 2,437-bp NdeI-EcoRI fragment from pUC19 as backbone, a 2,328-bp EcoRI-NotI fragment of pmLFG-EFnlacZ containing the 5′ LTR and leader, a 4,700-bp NotI-SalI fragment of the same plasmid containing the EFnlacZ gene cassette, and a 500-bp SalI-NdeI containing a modified LTR-bGHpA fragment. The 500-bp LTR-bGHpA fragment was generated by a double PCR and confirmed by DNA sequencing.
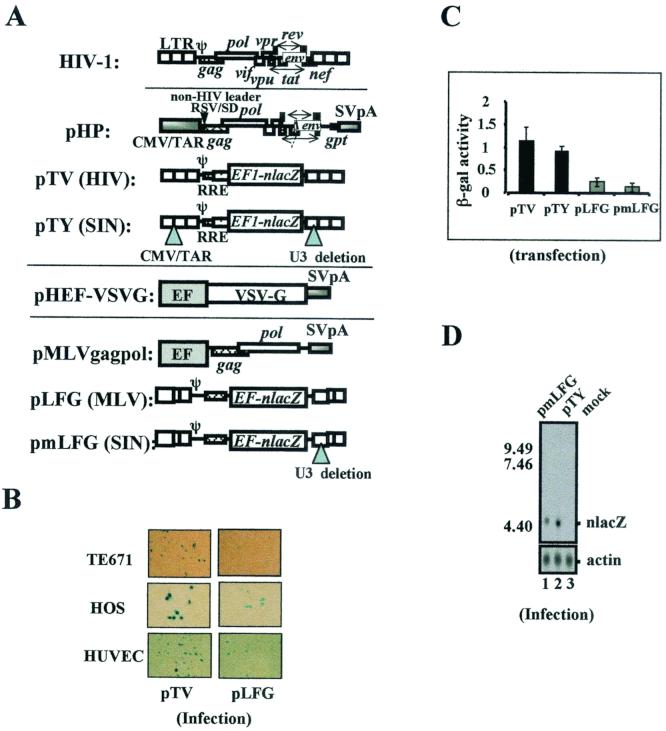
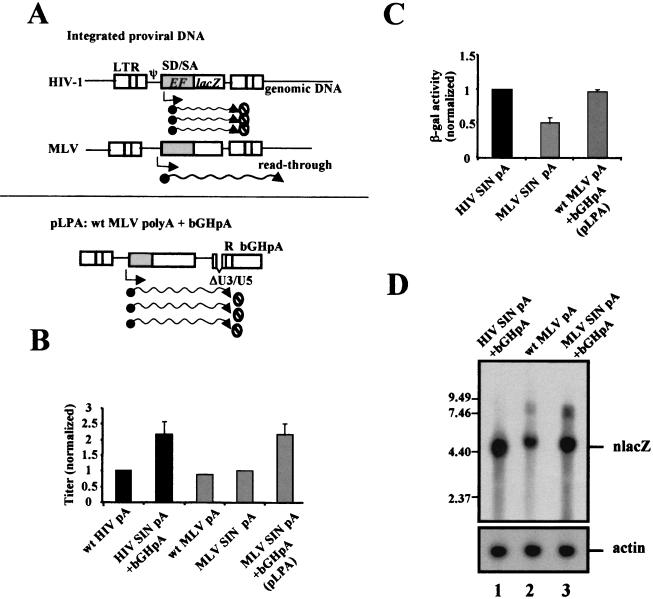
FIG. 1.
Schematic presentation of oncoretroviral and lentiviral vector constructs and differential transgene expression efficiencies. (A) HIV-1 genome organization and the HP/TV vector and MLV vector constructs. Both the wt LTR and the SIN LTR vectors are shown. (B) Staining of nlacZ activity in different human cell types transduced with wt MLV or HIV-1 vectors. The TE671 and HOS cells were transduced at 0.1 MOI and the HUVEC cells were transduced at 1 MOI, based on vector titers determined on TE671 cells. (C) Quantitative analysis of β-galactosidase (β-gal) activity of TE671 cells transfected with either HIV (pTV), HIV SIN (pTY), MLV (pLFG), or MLV SIN (pmLFG) vectors. A control plasmid, pHEFeGFP, was included to normalize transfection efficiency. (D) Northern analysis of cells transduced with MLV SIN or HIV-1 nlacZ SIN vectors (MOI = 6). TE671 cells were subjected to RNA harvesting 9 days after infection. The full-length viral RNA was not detected for these SIN vectors.
pflox and the Cre expression plasmids were gifts of X.-D. Fu (University of California, San Diego). For the establishment of a Cre/lox reporter cell line, pfloxnlacZ was constructed via a five-fragment ligation approach using the following restriction enzyme-digested and gel-purified DNA fragments: SalI-XbaI (∼1,485-bp EF1α promoter) of pHEF-VSVG, XbaI-BamHI (∼3,660-bp; tk-PGK-neo genes) of pflox, BamHI-AatII (654-bp; gene for N-terminal nlacZ) of pSP72 nlacZpA, AatII-HindIII (2,728 bp, nlacZ and SV40pA) of pSP72 nlacZpA, and the vector backbone HindIII-SalI (∼3,000 bp) of pflox. pHEFnlCre, a nuclear-localizing Cre expression plasmid, was generated by PCR amplification of the Cre gene using a 5′ primer containing the simian virus 40 (SV40) large-T-antigen nuclear localization signal and a modified eukaryotic translation initiation codon.
The poly(A) readthrough reporter construct was made by cloning a P2-Cre-pA fragment into the 3′ end of pLPA. P2-Cre-pA contains an internal ribosomal entry site (P2) from poliovirus (a gift of N. Sonenberg), nlCre, and an SV40 poly(A) signal. Additional P2-Cre-pA readthrough constructs were generated by inserting wt MLV LTR, MLV SIN LTR, wt HIV-1 LTR, or HIV-1 SIN LTR in place of U3-bGHpA of the pLPA-P2-Cre-pA construct via a two-step subcloning process.
Cell culture, virus production, and titration.
TE671 (human rhabdomyosarcoma) cells were obtained from the European Collection of Cell Culture (Wiltshire, United Kingdom), and HOS cells were obtained from the NIH AIDS Research and Reference Reagent Program (catalog no. 3519), contributed by N. Landau. The HEK293 cells were obtained from American Type Culture Collection, and the 293T cells were obtained from H. Goldstein. The primary human umbilical vessel endothelial cell (HUVEC) culture was a gift of M. Segal, prepared from donor human umbilical cord with University of Florida Institutional Review Board approval. The HUVEC culture was maintained in endothelial cell growth medium-2 with necessary growth factors purchased from Clonetics (BioWhittaker). The other cells were maintained in Dulbecco's modified Eagle's medium (Mediatech or Life Technologies, Gibco BRL) supplemented with 10% fetal bovine serum (Gibco BRL) and 100 units of penicillin-streptomycin (Gibco BRL)/ml at 37°C, 5% CO2. Viral vectors were generated by cotransfecting TE671 or 293T cells with the helper and transducing plasmids as previously described (8). In addition, TE671 and HEK293 cells were transfected by GeneJammer (Stratagene) and Superfect (Qiagen), respectively, following the manufacture's protocols using 1/10 of the plasmid DNA normally required for the calcium phosphate transfection protocol. These transfection protocols generate vector titers in the range of 105 to ∼106/ml in TE671 or 293T cells. After DNA cotransfection, virus supernatants were harvested every 12 h for 2 days. Virus supernatant was filtered using a 0.22- or 0.45-μm-pore-size low-protein binding filter (Millex-HV; Millipore) and stored in aliquots at −80°C until use. Vesicular stomatitis virus envelope protein (VSV-G)-pseudotyped vectors were routinely concentrated 20 to 50 times by centrifugation in a table-top microfuge at full speed for 2.5 h (20,000 × g) and resuspended by vortexing at 4°C for 4 h to overnight. The virus titer was determined using TE671 cells as previously described (8).
Generation of Cre/lox reporter cell lines and single-cell cloning.
The Cre/lox reporter cell line TE26 was established by transfecting TE671 with pfloxnlacZ and selected with G418 for 7 to 10 days. Single-cell clones were tested for Cre responsiveness. Stable transfectants of MLV and HIV-1 vectors were generated by transfecting 293 cells with 8 μg of either pTYEFeGFP or pLLLEFeGFP/well using the calcium phosphate-DNA coprecipitation procedure and propagated for 7 to 10 days. The eGFP-expressing single-cell clones were identified under a fluorescence microscope, individually picked, and expanded. This process was repeated once to obtain pure clonal lineages of cells.
Quantitative LacZ and eGFP reporter gene assays.
The eGFP expression was quantified by a fluorescence assay developed as follows. The cells were collected (48 h after transfection) in phosphate-buffered saline (PBS) containing 2.5 mM EDTA and washed twice with PBS, and different numbers of cells were dotted on 3 MM paper. The paper was left in the dark at room temperature for 5 min to briefly dry, and fluorescence intensity was measured using the Molecular Dynamics PhosphorImager (Storm 486). The LacZ enzyme assay was performed using the Promega β-galactosidase kit. For quantitative analysis of β-galactosidase activity in transfected cells, cells were cotransfected with pHEFeGFP and lysed 48 h after transfection, and 2 to 5 μl of the lysate was dotted on 3 MM paper for fluorescence assay prior to determination of β-galactosidase activity. The amount of lysates used for the β-galactosidase enzyme assay was normalized according to the transfection efficiencies determined by the eGFP control.
Genomic DNA slot blot and Southern analysis.
Genomic DNA was harvested using Promega's genomic DNA harvesting kit, and 2 and 4 μg of genomic DNA of each cell line was fixed on the GeneScreen membrane (NEN Life Science, Boston, Mass.) using a slot blotting device (Schleicher & Schuell Inc., Keene, N.H.). Different amounts (0.0125 to 0.6 ng) of pHEFeGFP DNA were used as standards. The DNA slot blot was then hybridized with a 32P-labeled 810-bp eGFP probe derived from XbaI-EcoRI digestion of pTY-EFeGFP and random primed using Stratagene's Prime-it II kit. Southern analysis was performed as previously described (8).
Northern analysis of cytoplasmic RNA and slot blot analysis of virion RNA.
Cytoplasmic RNA was harvested using a Qiagen RNAeasy kit, and Northern analyses were carried out as previously described (11). Virion RNA was harvested by precipitating the viral particles with 5% polyethylene glycol (PEG 8000) overnight, and RNA was extracted using TRI reagent (BRL) and precipitation. Quantification of the Northern or the slot blots was done using a Fuji phosphorimager, and the blots were also exposed to X-ray films for documentation. The radioactive probes for the RNA analyses were prepared as described above. For rehybridization, the blots were stripped by boiling for 10 min twice in double-distilled water with 0.5% sodium dodecyl sulfate.
Nuclear run-on assays.
The cell nuclei were harvested 40 h after transfection or from 2 × 106 to 3 × 106 stably transfected cell clones as described previously (24) with the following modifications. Cells were washed twice in cold PBS and collected using a cell scraper. Nuclei were obtained by resuspending cells in an NP-40 lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) in a 1-ml volume followed by 50 and 100 strokes for TE671 and 293 cells, respectively, using a Dounce homogenizer. The nuclei were pelleted, gently resuspended in 100 μl of glycerol storage buffer (50 mM HEPES [pH 7.5], 4 mM MnCl2, 1 mM MgCl2, 0.1 mM EDTA, 5 mM dithiothreitol, 50% glycerol), immediately frozen in liquid nitrogen, and stored at −80°C for later use. Nuclear DNA was transcribed in vitro, providing radioactive labeled [32P]UTP by adding an equal volume of 2× reaction buffer (10 mM Tris HCl [pH 8.0], 5 mM MgCl2, 0.3 M KCl, 5 mM dithiothreitol, 1 mM concentrations each of ATP, GTP, and CTP, and 200 μCi of an 800 Ci/mmol stock) to the thawed nuclei followed by 30 min of incubation at room temperature. Radioactively labeled RNA was purified with TRI reagent (BRL) according to the manufacturer's instruction, precipitated with ethanol, and resuspended in diethylpyrocarbonate-treated water containing 10% formamide. The labeled RNA was hybridized to a slot blot containing 0.15 μg of different DNA fragments UV cross-linked to the membrane.
RESULTS
Differential transgene expression of MLV versus HIV-1 vectors.
MLV- and HIV-1-based vectors, including both wt LTR and U3 deletion (SIN) vectors, were constructed to carry the same internal human EF1α promoter and the nuclear lacZ reporter gene (Fig. 1A). Transduction of different cells, including TE671, HOS, and HUVEC, with these vectors at the same multiplicity of infection (MOI) of both vectors showed consistent higher nlacZ expression with the HIV-1 vectors (Fig. 1B). To analyze the differential transgene expression quantitatively and to see if the difference between MLV and HIV-1 vectors was caused by transcriptional interference by the upstream LTR, we examined the internal EF1α promoter activity of both MLV SIN and HIV-1 SIN vectors by viral transduction using established cell lines and primary human cell cultures, including TE671 cells, K562 cells (human lymphoid cells), and human peripheral blood lymphocytes. HIV SIN and MLV SIN vectors have reduced LTR transcription (the LTR promoter-driven full-length RNA but not the internal promoter) in the transduced cells because of the U3 deletion (19) (Fig. 1D). The lentiviral SIN vectors (pTY) contain a bovine growth hormone poly(A) signal (bGHpA) cloned behind the 3′-truncated LTR (Fig. 2A). However, this bGHpA signal is not propagated in the progeny virus and therefore has no effect in the transduced cells. These results showed that the MLV SIN vectors, compared to the HIV-1 SIN vectors, exhibited reduced β-galactosidase activity similar to that of the wt MLV vectors (Fig. 1C). This was verified when a different reporter gene, eGFP, was used in either transiently or stably transfected cells (data not shown).
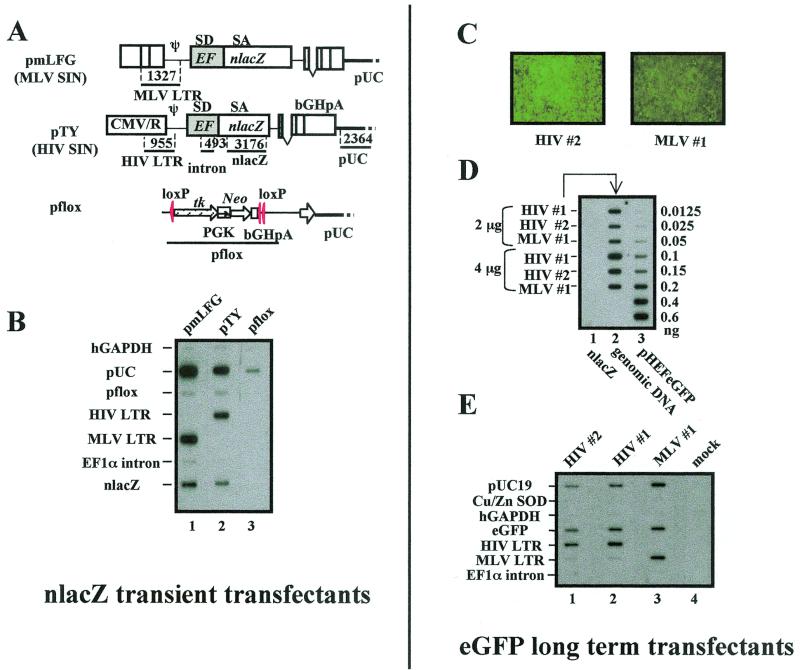
FIG. 2.
Nuclear run-on analyses of MLV and HIV-1 vectors in transiently and stably transfected cells. Nuclear run-on reactions were performed using the nuclei harvested from transiently transfected TE671 cells (40 h) or from stable HEK293 single-cell clone transfectants. The 32P-labeled RNA probes were used to hybridize to the DNA fragments fixed on the nitrocellulose membrane. (A) Detailed schematic diagram of MLV SIN and HIV-1 SIN vectors and the DNA fragments used for the nuclear run-on assay. (B) Nuclear run-on analysis of cells transfected with pmLFG (MLV SIN vector), pTY (HIV-1 SIN vector), or a control pflox plasmid in TE671 cells; lane 1, pmLFG plus pflox; lane 2, pTY plus pflox; lane 3, pflox. The radioactive signals were quantified, and the pixel values (PSL-background) were determined: for hGAPDH/MLV, 83; for hGAPDH/HIV, 78; for pflox/MLV, 78; for pflox/HIV, 76; for HIV LTR/MLV, 0; for HIV LTR/HIV, 119; for MLV LTR/MLV, 157; for MLV LTR/HIV, 0; for nlacZ/MLV, 116; and for nlacZ/HIV, 92. (C) HEK293 single-cell clones stably transfected with pTYEFeGFP (HIV #2) and pLLLEFeGFP (MLV #1) under a fluorescence microscope. (D) Determination of copy number of integrated DNA in cloned 293 cells. The DNA slot blot was hybridized to an eGFP probe, and the copy number of integrated DNA was determined based on the control amounts of pHEFeGFP shown in lane 3. Lane 1, 2 and 4 μg of integrated nlacZ genomic DNA as a negative control; lane 2, 2 and 4 μg of genomic DNA of the three different stable eGFP transfectants, HIV #1 and #2 and MLV #1, as depicted. The radioactive signals representing the LTR promoters and the internal EF1α promoter were quantified, and the pixel values (PSL-background) were determined: for eGFP/HIV #2, 44; for eGFP/HIV #1, 64; for eGFP/MLV #1, 63; for HIV LTR/HIV #2, 67; for HIV LTR/HIV #1, 98; for MLV LTR/MLV #1, 73. (E) Nuclear run-on analysis of integrated MLV and HIV-1 vectors. The nuclei were harvested from 106 stable HEK293 transfectants of HIV #1 and #2 and MLV #1 and used for the assay. Different DNA fragments were blotted on the membrane to study the activities of different promoters, as depicted. SD, splice donor site; SA, splice acceptor site.
To determine whether viral enhancers or silencers played a role, different deletion constructs of HIV-1 and MLV transducing vectors were generated, and the EF1α-nlacZ activity was analyzed. We observed a moderate increase in the reporter gene activity when the 5′ LTR and leader sequences of the MLV vector were deleted. Overall, there was little to no significant difference in lacZ activity between the wt and the LTR/leader deletion mutants at both the RNA and protein levels of both MLV and HIV-1 vectors (data not shown). Northern analyses of virally transduced cells showed that cells infected with HIV-1 vectors (MOI = 6) consistently expressed higher RNA levels than those infected with the MLV vectors (Fig. 1D, relative nlacZ RNA amounts, 0.34, 1, and 0.0 for lanes 1, 2, and 3, respectively).
Analyses of transcriptional and posttranscriptional RNA kinetics of MLV and HIV-1 vectors.
The different levels of EF1α transcripts between MLV and HIV-1 vectors could be due to transcriptional or posttranscriptional regulation. To directly measure the transcriptional activity, a quantitative nuclear run-on assay for nascent RNA synthesis was performed. We studied both transiently transfected and long-term-transfected cells because this would allow for evaluation of the EF1α promoter in both extrachromosomal and chromosomal states. We first performed a transient transfection study. As both wt and SIN retroviral vectors use the same wt 5′ LTR in transfected cells, TE671 cells were cotransfected with either the HIV-1 SIN (pTY) or the MLV SIN (pmLFG) vector, together with a control plasmid, pflox (Fig. 2A), and 40 h later, the nuclei were isolated and radioactively labeled UTP was incorporated in the nuclear run-on reaction. The transcriptional activity of any given promoter was determined by the corresponding amount of 32P-labeled RNA that hybridized to the specific DNA fragments (Fig. 2A) cross-linked onto a nylon membrane (Fig. 2B). The results showed that the transcriptional activity of the EF1α promoter of the MLV vector was slightly higher than that of the HIV-1 vector (Fig. 2B, lane 1 versus lane 2). This could be due to the activity of upstream promoters (the 5′ LTR). However, judged by the transcriptional activities of the upstream LTR of these two vectors (Fig. 2B, HIV LTR versus MLV LTR) and across the EF1α intron region, no significant difference of the internal EF1α promoter activities of these two vectors was noted. No marked difference was seen for the control human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and pflox promoters. The LTR activities were detected for both vectors, and as expected, the HIV-1 LTR activity was low because the transactivator tat was not included in the transfection. DNA fragments of the pUC19 plasmid backbone detected slightly higher levels of transcription in the MLV (pmLFG) vector-transfected cells than in the HIV-1 (pTY) vector-transfected cells, suggesting possible transcription or 3′ RNA readthrough into the plasmid backbone.
To examine transcriptional activity in stable transfectants, a second nuclear run-on assay was carried out using stably transfected cell clones. Single-cell clones of either MLV or HIV-1 vector containing equal copy number of integrated EF1α-eGFP DNA were examined (Fig. 2C; copy number was determined by genomic DNA slot blot [Fig. 2D]). pUC19 DNA and housekeeping Cu/Zn SOD and human GAPDH cDNAs were included as controls. The results demonstrated similar RNA transcriptional levels for the internal EF1α promoter (eGFP) with all three transfectants, and again, the MLV vector (with 5′ wt MLV LTR) displayed a slightly higher transcriptional rate than the HIV-1 vectors (pTY and HIV #1 and #2, with 5′ wt LTR) (Fig. 2E), in accordance with the transient transfection results (Fig. 2B).
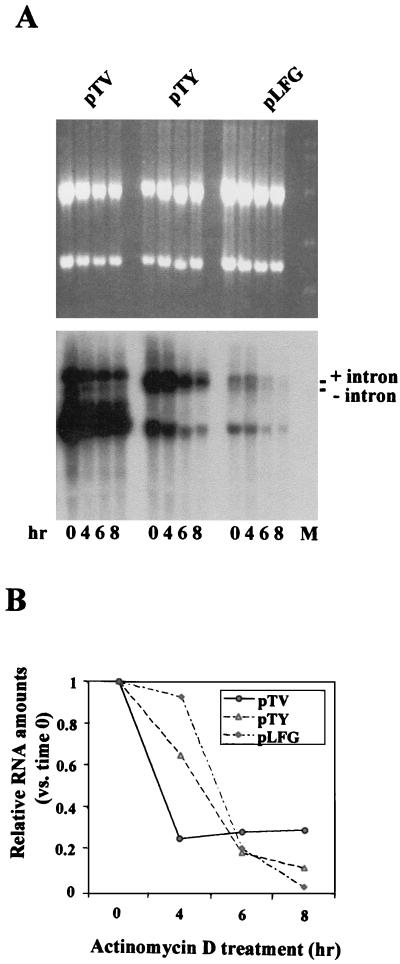
Since cytoplasmic RNA expression is affected by efficiencies of promoter function, RNA processing, and RNA stability, we investigated the half-lives of MLV and HIV-1 vector transcripts in the cytoplasm. TE671 cells were transfected with HIV-1 and MLV vectors, together with a helper construct, pHP and pMLVgagpol, respectively, and the VSV-G plasmid DNA, similar to the condition used for viral vector production. The transfected cells were treated with actinomycin D (2 μg/ml) to block RNA synthesis 48 h after DNA addition. The cytoplasmic RNA was harvested at different time points after actinomycin D addition (0, 4, 6, and 8 h) and analyzed by Northern blotting using a lacZ probe. The results showed that although HIV-1 (wt and SIN) vectors displayed higher RNA levels of both the genome size and the internal EF1α transcripts (3 to 10 times higher than that of the MLV vectors) (Fig. 3, comparing with Fig. 1D), HIV-1 and MLV vectors exhibited similar RNA half-lives (3 to 5 h, for both the viral and the EF1α transcripts) with no evidence of increased stability towards the HIV-1 transcripts. The full-length, EF1α intron-containing viral transcripts (Fig. 3A blot) were also detected in the cytoplasm of both MLV and HIV vector-transfected cells, suggesting that both vectors efficiently transported full-length, intron-containing viral RNA from the nucleus to the cytoplasm.
FIG. 3.
RNA stability kinetics of MLV and HIV-1 vectors. TE671 cells were transfected with oncoretroviral and lentiviral vectors, including helper and VSV-G constructs, using the GeneJammer method as described in Materials and Methods. The cells were treated with actinomycin D 48 h after DNA addition, and cytoplasmic RNA was harvested 0, 4, 6, and 8 h later. (A) RNA gel stained with ethidium bromide and subjected to Northern blot hybridization using a lacZ probe. Two species of full-length HIV-1 and MLV RNA were detected and determined to be EF1α intron-containing and intronless transcripts (+ and −intron, respectively). (B) RNA stability kinetics (full length) after quantitation with a phosphorImager (Molecular Dynamics).
Propagation of the EF1α intron in the viral genomes by both lentiviral and oncoretroviral vectors.
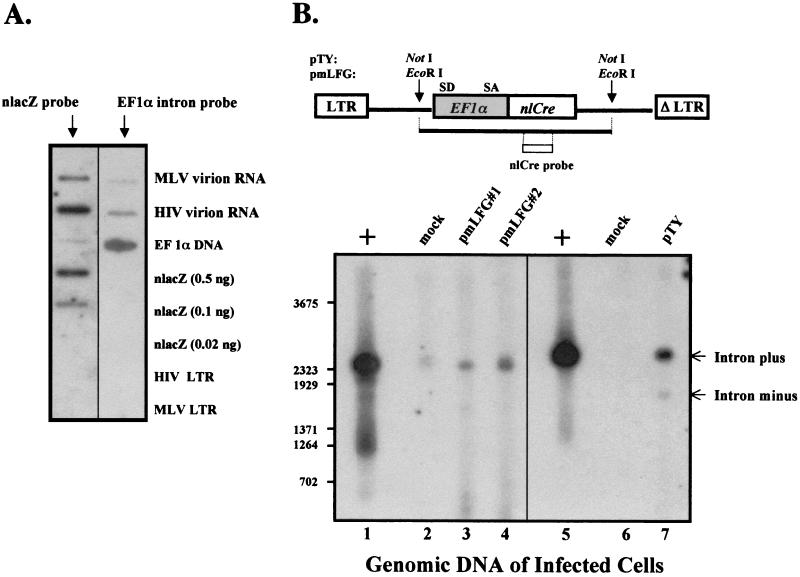
The full-length, EF1α intron-containing viral transcripts of both MLV and HIV vectors (Fig. 3A blot; additional data not shown) were detected in the cytoplasms of transfected cells, suggesting that both vectors efficiently transported full-length, intron-containing viral RNA from the nucleus to the cytoplasm. Our previous studies have shown that the EF1α intron is critical to its promoter activity (L.-J. Chang, unpublished data). To see if the viral particles of both vectors contained the EF1α intron sequence, virion RNA was examined by RNA slot blotting. TE671 cells were cotransfected with the viral vector DNA and all of the helper constructs for virus production, viral particles were collected from the culture supernatants, and the titer of virus was determined as described in Materials and Methods. Equivalent infectious units of MLV and HIV-1 vector particles were subjected to RNA extraction and extensive DNase digestion to remove residual amounts of contaminating DNA. The virion RNA was quantitatively analyzed by slot blotting using either the nlacZ or the EF1α intron probe alongside controls, as illustrated in Fig. 4A. The result showed that both HIV-1 and MLV packaged the EF1α intron-containing viral RNA, as detected by the EF1α intron probe, but at a reduced level for the MLV vectors (Fig. 4A). This could be due to different stabilities of the different vector RNAs in the viral particles. Nevertheless, one possible explanation of the reduced MLV vector activity is that the EF1α intron-containing RNA is inefficiently propagated during infection. To test this, we examined the integrated proviral DNA in the transduced cells by Southern blotting. TE671 cells were infected with either MLV or HIV-1 vectors (carrying an EF1α-nlCre reporter gene), and after several passages, the genomic DNAs were harvested and subjected to Southern blot analysis. The results showed that both HIV-1 and MLV preferentially preserved the EF1α intron in the transduced cells (Fig. 4B, lanes 3 and 7), indicating that differential expression of transgene in transduced cells was not due to differential EF1α intron splicing.
FIG. 4.
Analyses of EF1α intron-containing virion RNA and proviral DNA. (A) Slot blot analysis of virion RNA. The RNA was extracted from equivalent infectious units of virus particles and applied to the slot blots. Also included was control DNA representing different regions of the vector sequence. The probes were derived from the reporter gene nlacZ and the EF1α intron sequences and used for hybridization as indicated. The radioactive signals representing RNA in HIV-1 and MLV virions were quantified, and the pixel values (PSL-background) were determined: for MLV/lacZ, 30; for HIV-1/lacZ, 100; for MLV/EF1α intron, 6; for HIV-1/EF1α intron, 16. (B) Southern analysis of integrated proviral DNA. TE671 cells were infected with MLV SIN or HIV-1 SIN vectors for 12 days at the same MOI (=5), and the genomic DNA was harvested and digested with restriction enzymes that cleaved the nlCre gene as indicated. Two different infected cultures were included for the MLV vectors. The vector DNA was digested with the same enzymes and used as a positive control (+). The Southern blot was hybridized with an nlCre probe as depicted in the diagram. SD, splice donor site; SA, splice acceptor site.
A strong poly(A) signal up-regulates the MLV transgene expression.
Results of the above-described experiments indicated that both vectors efficiently propagated the EF1α intron. Thus, differential intron splicing and packaging could not resolve the discrepancy of the different vector efficiencies. Retroviral transcription initiates at the 5′ R, bypasses the 5′ poly(A) signal, and proceeds through to the poly(A) signal in the 3′ R, and in the vectors, the internal promoter uses the same 3′ poly(A) signal for transcriptional termination. Therefore, a possible explanation for the different transgene activities of MLV and HIV-1 vectors is that the HIV-1 poly(A) signal is more efficient than the MLV poly(A) signal, which results in the frequent MLV RNA readthrough and degradation during processing as illustrated in Fig. 5A. Since the conserved AAUAAA poly(A) motif is located in the R region and the R region is preserved in both the wt and SIN vectors of both MLV and HIV-1 vectors, it would be interesting to see if the viral poly(A) motif played a role in the transgene function. To test this, we inserted behind the 3′ LTR of the MLV SIN vector a strong poly(A) signal, the bovine growth hormone poly(A) (bGHpA), as illustrated in Fig. 5A (pLPA). Analysis of this new poly(A) construct showed a consistent increase in titer of vector by DNA cotransfection compared with wt and SIN MLV vectors (Fig. 5B). This result supported that the addition of a strong poly(A) signal increases MLV vector efficiency. More importantly, in transient transfections, the pLPA vector displayed reporter gene activity similar to that of the HIV-1 vectors (Fig. 5C). The increased activity correlated with increased EF1α nlacZ RNA levels as shown by Northern analysis (Fig. 5D).
FIG. 5.
Improved titer and transgene expression of MLV vector after polyadenylation modification. (A) A model of poly(A) usage of integrated MLV and HIV-1 vectors. The model shows that the MLV 3′ poly(A) is leaky and produces unstable readthrough RNA, and the insertion of a strong poly(A) signal (bGHpA) reduces the readthrough frequency (pLPA). (B) The titer increase of MLV SIN vector containing bGHpA (pLPA). Viral vectors were generated and the titer was determined as described in Materials and Methods. The titer of the HIV-1 vector with wt LTR (HIV) was arbitrarily set at 1.00. HIV, wt HIV-1 poly(A) (pA); HIV SIN, HIV-1 SIN LTR with wt HIV-1 poly(A) and the additional bGHpA; MLV SIN, MLV SIN LTR with wt MLV poly(A); pLPA, MLV SIN with wt MLV poly(A) and the additional bGHpA. (C) Increased transgene expression of pLPA in transfected cells. The β-galactosidase enzyme assay was performed using cells transfected with different vectors using pHEFeGFP as internal control. The expression of wt HIVpA (pTY) was arbitrarily set at 1.00, and the error bars were generated from three independent experiments. (D) Northern analysis of cytoplasmic RNA of MLV wt poly(A) versus MLV bGHpA vectors in transfected cells. wt HIVpA + bGHpA (pTY), wt MLVpA (pLFG), and wt MLV pA + bGHpA (pLPA) were transfected into TE671 cells without the helper plasmids, and cytoplasmic RNA analyzed by Northern blotting using nlacZ probe. The full-length RNA of the HIV-1 vector (lane 1) was not visible (but could be detected in longer exposure) because the transactivator tat gene was not included. The sizes of pLPA RNA with U3/U5 deletion were slightly smaller than that of the corresponding pLFG RNA (lanes 2 versus lane 3). The hybridization probe was removed from the blot, and the blot was rehybridized with an actin probe for RNA loading control. SD, splice donor site; SA, splice acceptor site.
Quantitative analyses of transcriptional readthrough of different viral LTRs.
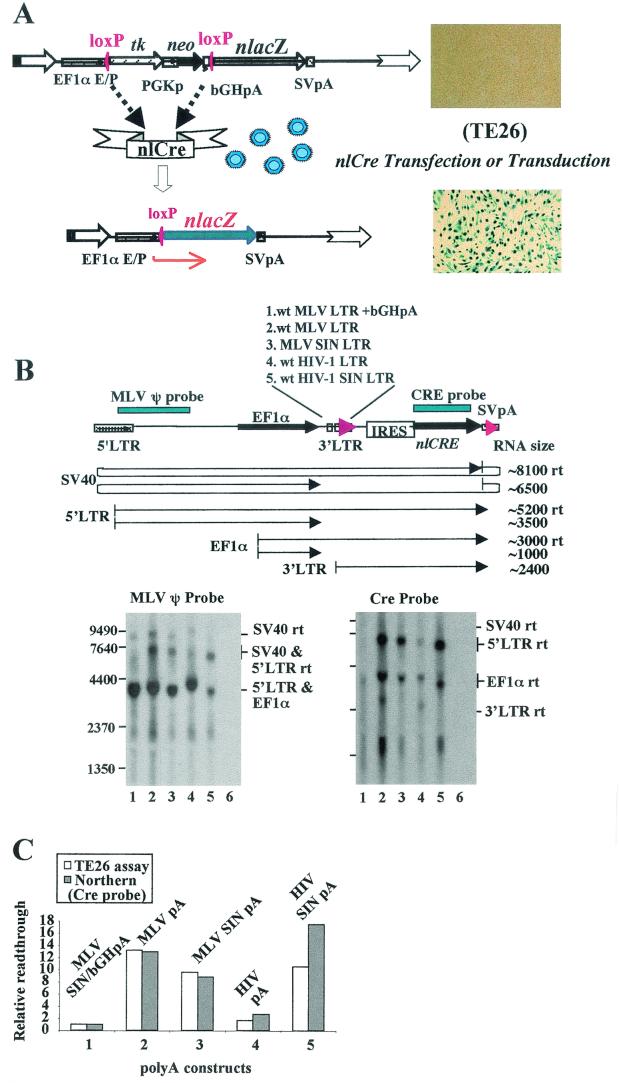
The increased reporter gene expression of MLV vectors with a strong 3′ poly(A) insertion did not directly prove that the MLV 3′ poly(A) was leaky. Since the readthrough RNA could be hard to quantify, we designed a sensitive reporter gene assay using the Cre/loxP recombination system. A loxP/nlacZ reporter cell line, TE26, was established by transfecting TE671 cells with pfloxnlacZ, which carries an nlacZ reporter gene 3′ to a loxP-flanked selective marker gene cassette (Fig. 6A). TE26 cells do not express nlacZ unless the nlCre recombinase is present in the nucleus. In the presence of nlCre, nlacZ will be activated, providing a quantitative assay for nlCre expression. To examine the 3′ readthrough, we inserted an IRES-nlCre-SV40 poly(A) reporter gene cassette downstream of the 3′ LTR in an MLV construct to stabilize the readthrough RNA and to allow internal translation of nlCre (Fig. 6B). To test the readthrough function of the native MLV and HIV-1 poly(A) signals, we recloned the wt LTR and the SIN LTR of both MLV and HIV-1 vectors into this reporter construct. The bGHpA signal of the HIV-1 SIN vector (pTY) was not included in order to test the native HIV-1 SIN poly(A) mimicking the transduced proviral genome. A series of different poly(A) signals including the wt MLV LTR (MLVpA), MLV SIN LTR (MLV SIN pA), wt HIV-1 LTR (HIV pA), and HIV-1 SIN LTR (HIV SIN pA, without bGHpA) were cloned into the readthrough IRES-nlCre reporter construct (shown in Fig. 6B). MLV bGHpA (MLV SIN bGHpA) was also included to artificially introduce a strong poly(A) signal to terminate transcription. The plasmid DNA was transfected into TE671 cells, and 40 h later, the cytoplasmic RNA was harvested and analyzed by Northern blotting using either an MLV packaging signal (ψ) probe or an nlCre probe as depicted at the top of Fig. 6B. The results illustrated that efficient readthrough was detected for both wt and SIN poly(A) constructs of MLV (Fig. 6B, wt MLVpA, lane 2, and MLV SIN pA, lane 3), and to our surprise, for the HIV-1 SIN poly(A) construct as well (HIV SIN pA, lane 5). The MLV SIN LTR displayed somewhat reduced readthrough activity compared with the wt MLV LTR (Fig. 6C), suggesting that the 3′ U3 sequence of the MLV LTR may play a role in the leakiness of the 3′ poly(A). The MLV LTR containing bGHpA and the wt HIV-1 LTR both exhibited very low readthrough activity (lanes 1 and 4, respectively, Fig. 6B). These RNA readthrough data corroborated well with results of the nlCre functional assay whereby TE26 cells were transfected with different amounts of poly(A) readthrough reporter DNA (Fig. 6C and data not shown).
FIG. 6.
Quantitative analysis of 3′ readthrough of MLV and HIV-1 wt and SIN vectors. (A) The Cre/loxP reporter cell line (TE26). TE26 is a cell clone that displays high sensitivity to Cre-mediated loxP recombination and no background lacZ activity. (B) The schematic structure of the IRES-nlCre-pA reporter constructs and Northern analyses of normal and readthrough (rt) RNA transcripts. The sizes of predicted RNA species are illustrated. Northern analyses of TE671 cytoplasmic RNA of different poly(A) constructs were performed using two hybridization probes, an MLV ψ probe and a Cre probe. Lane 1, MLV LTR with bGHpA; lane 2, wt MLV LTR; lane 3, MLV SIN LTR; lane 4. wt HIV-1 LTR; lane 5, HIV-1 SIN LTR (without bGHpA); lane 6, mock transfection. (C) Quantitative measurement of readthrough RNA and nlCre activity. TE26 cells were transfected with different amounts of plasmid DNA in three independent transfections, and 36 and 60 h later, cells were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and the number of blue nucleated cells was counted under an inverted microscope. The relative amounts of readthrough RNA of panel B were determined by a phosphorimager and plotted together with the corresponding nlCre activity of different constructs against MLV SIN + bGHpA (pLPA), which was arbitrarily set at 1.0.
DISCUSSION
The level of transgene expression is often a limiting factor to the success of gene therapy in clinical trials (5, 21). Human EF1α promoter is a strong housekeeping promoter with an intact intron (20) and has been shown to function efficiently in lentiviral vectors in many cell types including human hematopoietic CD34+ cells (8, 31). When comparing HIV-1 vectors with MLV vectors carrying the same EF1α internal promoter, we consistently observed higher reporter gene activities with the HIV-1 vectors. A simple explanation would be that the HIV-1 LTR is not active without transactivator Tat in the transduced cells, and therefore, there is no transcriptional interference from upstream LTR, as there would be with the MLV vectors. This possibility was quickly excluded because increased expression was also observed with SIN vectors of MLV and HIV-1 which lack functional LTRs; therefore, the differential expression cannot be explained simply by transcriptional interference of upstream LTR upon retroviral integration. In this study, we have carefully investigated the contributions of a variety of factors in order to understand the regulation of retroviral transgene expression.
The 3′ untranslated region (3′ UTR) of both vectors may carry signals critical for nuclear export, polyadenylation, subcellular targeting, translation, and degradation of viral mRNA (10). In addition, sequences upstream of the internal promoter in the oncoretroviral vectors possess multiple cis-acting elements that may be responsible for regulation of transcription (35). Such elements do not appear to be present in the lentiviral genome, as no significant difference was found among different HIV-1 deletion constructs. Negative cis-regulatory elements in the LTR and in the primer binding site of MLV have been reported (6, 15). Our finding of a modest increase in MLV transgene expression when the 5′ leader sequence was deleted is consistent with those studies (data not shown); however, this negative regulation accounts for only part of the observed difference in transgene expression compared with the HIV-1 vectors.
The nuclear run-on assay and the analysis of cytoplasmic RNA stability kinetics both confirmed that the internal EF1α promoter was equally, if not more, active in cells transfected with the MLV and with the HIV-1 vectors. These results suggested that the differential transgene expression of MLV and HIV-1 vectors was not due to differences in promoter function and cytoplasmic RNA stability. It is interesting that although both wt and SIN vectors of HIV-1 exhibited similar transgene activity (Fig. 1), the HIV-1 wt vector (pTV) consistently produced more EF1α transcript than the SIN vector (Fig. 3). However, the HIV-1 SIN vector (pTY) consistently produced more genomic RNA than did the wt vector (pTV), as evident in Fig. 3 (also data not shown). The latter effect is probably due to the modification of the 5′ LTR in the SIN vector (pTY), which may contribute to an increased transcription of the pTY viral RNA and interference of the internal EF1α promoter. Northern analysis of cytoplasmic and virion RNA and Southern analysis of proviral DNA showed that both HIV-1 and MLV vectors expressed the EF1α intron-containing RNA in the cytoplasm and in the transduced cells (Fig. 4). Interestingly, there appeared to be a selective packaging and propagation advantage for the intron-containing viral genomes by both MLV and HIV-1 vectors, because intron-minus proviral DNA was detected at much lower ratios than those reflected in the cytoplasm.
Differential usage of the two poly(A) signals in the 5′ and 3′ R of the HIV-1 genome has been shown to be regulated by the 5′ splice site, functioning as a poly(A) suppressor, the U3 sequences, and the 3′ upstream sequence element (USE) upstream of the 3′ R functioning as a poly(A) enhancer (1, 13, 36, 38). Efficient usage of the 3′ poly(A) of HIV-1 appears to require U5 sequence as well as an adjacent poly(A) hairpin structure (2, 12). This is consistent with our previous report that HIV-1 SIN vectors containing USE (most of it except for several nucleotides in the 5′ USE) display a 20% increase in vector titer, whereas the U5 deletion results in a titer reduction by more than 50% (19).
In the avian leucosis virus (ALV) genome, sequences regulating RNA splicing in gag and env have been shown to enhance the 3′ poly(A) usage (27). It has been shown that the 3′ readthrough occurs at approximate 15% efficiency with the ALV. However, little has been reported on the regulation of the MLV poly(A) usage (17). Analysis of the poly(A) signal in the 5′ LTR of the MLV genome suggests that the 5′ MLV poly(A) signal is intrinsically weak compared with the 5′ poly(A) signal of the HIV-1 LTR (16). Here we directly demonstrated that the 3′ MLV poly(A) signal is leaky, and the IRES-Cre reporter gene study showed that readthrough of the MLV poly(A) is 3 to 8 times more frequent than the bovine growth hormone poly(A) signal (Fig. 6). While the wt HIV-1 LTR exhibited high poly(A) termination efficiency, similar to that of the bovine growth hormone poly(A), the HIV-1 SIN poly(A) was as leaky as the MLV poly(A) (Fig. 6). Although the poly(A) of the HIV-1 SIN vector was leaky, it did not appear to diminish the viral transgene activity, as with the MLV vectors. The reason could be that the HIV-1 SIN poly(A) signal is qualitatively different from that of the MLV counterpart, and it is also possible that the HIV-1 SIN poly(A) is somewhat more compatible with the transcriptional machinery of the human cells. This reduced 3′ termination efficiency would cause RNA instability during transgene expression if no alternative poly(A) signal is encountered further downstream in the transduced cells. Frequent transcriptional readthrough could pose increased risk of activating or capturing cellular oncogenes 3′ to the viral integration site. A large readthrough RNA (11.2 kb) has been found to be packaged into ALV carrying a 3′ poly(A) mutation which potentially increases the risk of retroviral transduction of oncogenes (18).
Our study suggests that MLV, like ALV, carries a leaky 3′ poly(A) signal and has the potential to mobilize cellular sequences due to the frequent 3′ poly(A) readthrough. Ironically, lentiviral SIN vectors are considered safer than lentiviral vectors carrying wt LTRs. The finding of high readthrough frequency of HIV-1 SIN vectors argues that lentiviral SIN vectors may impose oncogenic risk similar to that with MLV vectors. For gene therapy applications with humans using either oncoretroviral or lentiviral vectors, it will be important to improve the 3′ poly(A) signals to increase transgene expression efficiency and to decrease the risk of activation of harmful cellular genes.
Acknowledgments
A.-K.Z. and S.S. contributed equally to this study.
We thank X.-D. Fu for pflox and Cre plasmids, H. Nick for hGAPDH and Cu/Zn SOD cDNA constructs, and M. Stoltzfus and P. Lapis for critical comments.
This work is supported by a grant from National Institutes of Health (HL-59412).
REFERENCES
- 1.Ashe, M. P., P. Griffin, W. James, and N. J. Proudfoot. 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 9:3008-3025. [DOI] [PubMed] [Google Scholar]
- 2.Bohnlein, S., J. Hauber, and B. R. Cullen. 1989. Identification of a U5-specific sequence required for efficient polyadenylation within the human immunodeficiency virus long terminal repeat. J. Virol. 63:421-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchschacher, G. L., Jr., and F. Wong-Staal. 2000. Development of lentiviral vectors for gene therapy for human diseases. Blood 95:2499-2504. [PubMed] [Google Scholar]
- 4.Case, S. S., M. A. Price, C. T. Jordan, X. J. Yu, L. Wang, G. Bauer, D. L. Haas, D. Xu, R. Stripecke, L. Naldini, D. B. Kohn, and G. M. Crooks. 1999. Stable transduction of quiescent CD34(+)CD38(-) human hematopoietic cells by HIV-1-based lentiviral vectors. Proc. Natl. Acad. Sci. USA 96:2988-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavazzana-Calvo, M., S. Hacein-Bey, G. de Saint Basile, F. Gross, E. Yvon, P. Nusbaum, F. Selz, C. Hue, S. Certain, J. L. Casanova, P. Bousso, F. L. Deist, and A. Fischer. 2000. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288:669-672. [DOI] [PubMed] [Google Scholar]
- 6.Challita, P. M., D. Skelton, A. El-Khoueiry, X. J. Yu, K. Weinberg, and D. B. Kohn. 1995. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J. Virol. 69:748-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, L.-J., and E. E. Gay. 2001. The molecular genetics of lentiviral vectors—current and future perspectives. Curr. Gene Ther. 1:237-251. [DOI] [PubMed] [Google Scholar]
- 8.Chang, L.-J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6:715-728. [DOI] [PubMed] [Google Scholar]
- 9.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 10.Conne, B., A. Stutz, and J. D. Vassalli. 2000. The 3′ untranslated region of messenger RNA: a molecular "hotspot' for pathology? Nat. Med. 6:637-641. [DOI] [PubMed] [Google Scholar]
- 11.Cui, Y., T. Iwakuma, and L. J. Chang. 1999. Contributions of viral splice sites and cis-regulatory elements to lentivirus vector function. J. Virol. 73:6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das, A. T., B. Klaver, and B. Berkhout. 1999. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 73:81-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeZazzo, J. D., J. E. Kilpatrick, and M. J. Imperiale. 1991. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3′ end formation. Mol. Cell. Biol. 11:1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerman, M., and M. H. Malim. 1998. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science 280:1880-1884. [DOI] [PubMed] [Google Scholar]
- 15.Feuer, G., M. Taketo, R. C. Hanecak, and H. Fan. 1989. Two blocks in Moloney murine leukemia virus expression in undifferentiated F9 embryonal carcinoma cells as determined by transient expression assays. J. Virol. 63:2317-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furger, A., J. Monks, and N. J. Proudfoot. 2001. The retroviruses human immunodeficiency virus type 1 and Moloney murine leukemia virus adopt radically different strategies to regulate promoter-proximal polyadenylation. J. Virol. 75:11735-11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guntaka, R. V. 1993. Transcription termination and polyadenylation in retroviruses. Microb. Rev. 57:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman, S. A., and J. M. Coffin. 1987. Efficient packaging of readthrough RNA in ALV: implications for oncogene transduction. Science 236:845-848. [DOI] [PubMed] [Google Scholar]
- 19.Iwakuma, T., Y. Cui, and L. J. Chang. 1999. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology 261:120-132. [DOI] [PubMed] [Google Scholar]
- 20.Kim, D. W., T. Uetsuki, Y. Kaziro, N. Yamaguchi, and S. Sugano. 1996. Use of the human elongation factor 1α promoter as a versatile and efficient expression system. Gene 91:217-223. [DOI] [PubMed] [Google Scholar]
- 21.Kohn, D. B., G. Bauer, C. R. Rice, J. C. Rothschild, D. A. Carbonaro, P. Valdez, Q. Hao, C. Zhou, I. Bahner, K. Kearns, K. Brody, S. Fox, E. Haden, K. Wilson, C. Salata, C. Dolan, C. Wetter, E. Aguilar-Cordova, and J. Church. 1999. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood 94:368-371. [PubMed] [Google Scholar]
- 22.Kordower, J. H., M. E. Emborg, J. Bloch, S. Y. Ma, Y. Chu, L. Leventhal, J. McBride, E. Y. Chen, S. Palfi, B. Z. Roitberg, W. D. Brown, J. E. Holden, R. Pyzalski, M. D. Taylor, P. Carvey, Z. Ling, D. Trono, P. Hantraye, N. Deglon, and P. Aebischer. 2000. Neurodegeneration prevented by lentiviral vector delivery of GDNF in primate models of Parkinson's disease. Science 290:767-773. [DOI] [PubMed] [Google Scholar]
- 23.Krall, W. J., D. C. Skelton, X.-J. Yu, I. Riviere, P. Lehn, R. C. Mulligan, and D. B. Kohn. 1996. Increased levels of spliced RNA account for augmented expression from the MFG retroviral vector in hematopoietic cells. Gene Ther. 3:37-48. [PubMed] [Google Scholar]
- 24.Laine, R. O., N. F. Shay, and M. S. Kilberg. 1994. Nuclear retention of the induced mRNA following amino acid-dependent transcriptional regulation of mammalian ribosomal proteins L17 and S25. J. Biol. Chem. 269:9693-9697. [PubMed] [Google Scholar]
- 25.Malim, M. H., S. Bohnlein, R. Fenrick, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. Functional comparison of the Rev trans-activators encoded by different primate immunodeficiency virus species. Proc. Natl. Acad. Sci. USA 86:8222-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, A. D. 1997. Development and applications of retroviral vectors, p. 437-473. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 27.Miller, J. T., and C. M. Stoltzfus. 1992. Two distant upstream regions containing cis-acting signals regulating splicing facilitate 3′-end processing of avian sarcoma virus RNA. J. Virol. 66:4242-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minvielle-Sebastia, L., and W. Keller. 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 11:352-357. [DOI] [PubMed] [Google Scholar]
- 29.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 93:11382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson, D., J. F. Elliott, and L.-J. Chang. 1995. Retroviral vector with a CMV-IE/HIV-TAR hybrid LTR gives high basal expression levels and is upregulated by HIV-1 Tat. Gene Ther. 2:269-278. [PubMed] [Google Scholar]
- 31.Salmon, P., V. Kindler, O. Ducrey, B. Chapuis, R. H. Zubler, and D. Trono. 2000. High-level transgene expression in human hematopoietic progenitors and differentiated blood lineages after transduction with improved lentiviral vectors. Blood 96:3392-3398. [PubMed] [Google Scholar]
- 32.Somia, N., and I. M. Verma. 2000. Gene therapy: trials and tribulations. Nat. Rev. Genet. 1:91-99. [DOI] [PubMed] [Google Scholar]
- 33.Stoltzfus, C. M. 1988. Synthesis and processing of avian sarcoma retrovirus RNA. Adv. Virus Res. 35:1-38. [DOI] [PubMed] [Google Scholar]
- 34.Sutton, R. E., H. T. Wu, R. Rigg, E. Bohnlein, and P. O. Brown. 1998. Human immunodeficiency virus type 1 vectors efficiently transduce human hematopoietic stem cells. J. Virol. 72:5781-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang, H., K. L. Kuhen, and F. Wong-Staal. 1999. Lentivirus replication and regulation. Annu. Rev. Genet. 33:133-170. [DOI] [PubMed] [Google Scholar]
- 36.Valsamakis, A., S. Zeichner, S. Carswell, and J. C. Alwine. 1991. The human immunodeficiency virus type 1 polyadenylylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc. Natl. Acad. Sci. USA 88:2108-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varmus, H. 1988. Regulation of HIV and HTLV gene expression. Genes Dev. 2:1055-1062. [DOI] [PubMed] [Google Scholar]
- 38.Weichs an der Glon, C., J. Monks, and N. J. Proudfoot. 1991. Occlusion of the HIV poly(A) site. Genes Dev. 5:244-253. [DOI] [PubMed] [Google Scholar]
- 39.Zolotukhin, S., M. Potter, W. W. Hauswirth, J. Guy, and N. Muzyczka. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]