Abstract
Human rhinoviruses (HRV) of the minor receptor group use several members of the low-density lipoprotein receptor superfamily for cell entry. These proteins are evolutionarily highly conserved throughout species and are almost ubiquitously expressed. Their common building blocks, cysteine-rich ligand binding repeats about 40 amino acids in length, exhibit considerable sequence similarity. Various numbers of these repeats are present in the different receptors. We here demonstrate that HRV type 1A (HRV1A) replicates in mouse cells without adaptation. Furthermore, although closely related to HRV2, it fails to bind to the human low-density lipoprotein receptor but recognizes the murine protein, whereas HRV2 binds equally well to both homologues. This difference went unnoticed due to the presence of other receptors, such as the low-density lipoprotein receptor-related protein, which allow species-independent attachment. The species specificity of HRV1A reported here will aid in defining amino acid residues establishing the contact between the viral surface and the receptor.
Human rhinoviruses (HRVs) constitute a large genus within the family Picornaviridae and are the major cause of common cold infections (for a review on picornaviruses, see reference 43). Their icosahedral capsid is composed of 60 copies each of the viral proteins VP1, VP2, VP3, and VP4. The protein shell encases an RNA genome of positive polarity which is translated into a polyprotein upon arrival in the cytoplasm. The precursor protein is cleaved autocatalytically and cotranslationally by virally encoded proteases, giving rise to the capsid proteins and to the nonstructural proteins involved in replicative functions.
Apart from one exception (HRV type 87 [HRV87]), the 102 serotypes are divided into a major and a minor group based on specific interaction with their cellular receptors, intercellular adhesion molecule 1 (ICAM-1; the major group) and members of the low-density lipoprotein receptor (LDLR) family (the minor group). Amino acid sequence information for the entire capsid protein region is available for 11 different serotypes (http://www.iah.bbsrc.ac.uk/virus/picornaviridae/SequenceDatabase/Index.html), and the three-dimensional structures of three major-group and two minor-group serotypes have been solved at atomic resolution (21, 38, 42, 49, 54). Medium-resolution structures from complexes of the major-group viruses HRV14 and HRV16 with soluble ICAM-1 (22, 39) as well as of the minor-group virus HRV2 and a soluble fragment of the very-low-density lipoprotein receptor (VLDLR) (14) have been obtained by image reconstruction from cryoelectron microscopy data. Whereas ICAM-1 binds inside the canyon, a cleft encircling the fivefold axes of icosahedral symmetry, the HRV2 receptor complex revealed that the BC loop and the HI loop of VP1 were largely covered by two of the three repeats of the recombinant VLDLR fragment VLDLR1-3, encompassing ligand binding repeats 1 to 3 only (41).
The LDLR family comprises the LDLR proper, VLDLR, LDLR-related protein (LRP), and megalin, along with several other membrane proteins (for a review see references 11 and 47). Their ligand binding domains at the N terminus of the molecule are composed of 7, 8, 31, and 35 imperfect direct cysteine rich repeats, respectively, which are followed and/or interrupted by YWTD-EGF domain pairs (20). The structure of the repeats is maintained by three disulfide bonds and a Ca2+ ion, as deduced from X-ray crystallography of LDLR repeat 5 (9) and from nuclear magnetic resonance structure determination of repeat 1 (7), repeat 2 (6), and repeats 5 and 6 (36, 37). The structures of concatemers of repeats 1 and 2 (24) and repeats 5 and 6 (1) suggest that individual repeats fold and move independently with respect to each other. Based on multiple alignments of the repeats derived from LDLR, VLDLR, and LRP, four groups with common ancestor sequences were defined; their sequential arrangement differs in, e.g., LDLR and VLDLR (45).
Whereas LDLR is specific for lipoproteins containing apo-B and apo-E, the other receptors are promiscuous and bind a large number of structurally and functionally unrelated ligands (34). The high sequence similarity of the repeats within a given molecule, within different receptors, and within species raises the question of how most of the ligands maintain their specificity for one particular receptor. On the other hand, attachment of HRV2 to LDLR, VLDLR, and LRP expressed in a number of cell lines of various origins indicated very low specificity of the interaction. Uncapher and colleagues (48) demonstrated binding of minor-group viruses to a large number of species, and we have shown that HRV2 binds human LDLR, LRP (17), and VLDLR from chickens (12) and humans (29).
As far as the virus is concerned, only the tripeptide sequence Thr-Glu-Lys (TEK) within the HI loop and the dipeptide sequence Tyr-Asn (YN) within the BC loop of VP1 are conserved at least in those minor-group HRVs whose sequences are known (8). Apart from these, the amino acid residues within the receptor's footprint are largely divergent, and the basis of receptor recognition is not understood.
Despite attaching to mouse cells, rhinoviruses have been reported to grow in human or primate cells but not in mouse cells. Only a time-consuming adaptation procedure resulted in HRV2 variants (HRV2L) which were able to grow in mouse cells (53). From these and other experiments, involving transfection of HRV39 RNA into mouse cells and isolation of variants able of replicating in the mouse (26), it was concluded that proteins encoded in the genomic P2 region (namely 2B, 2C, and the precursor 2BC) are responsible for a change in host range.
We report here on the investigation of viral infection and replication of mouse-adapted HRV2L and wild-type HRV1A in a mouse fibroblast cell line (M1) and in cell lines with various gene disruptions rendering them LRP−/− (M2), LDLR−/− (M3), or LRP−/− LDLR−/− (M4) (13, 19, 50). We observed that wild-type HRV1A was able to replicate in mouse cells without adaptation; this is in marked contrast to most, if not all, other minor-group HRVs. Furthermore, HRV1A distinguishes between mouse and human LDLR. This property will be instrumental for the identification of amino acid residues involved in viral receptor recognition.
MATERIALS AND METHODS
Materials.
All chemicals were purchased from Sigma (St. Louis, Mo.) unless specified otherwise; enzymes were from New England Biolabs (Beverly, Mass.), tissue culture media and supplements as well as Lipofectamine were from GibcoBRL (Gaithersburg, Md.). Tissue culture plates and flasks were from Costar (Cambridge, Mass.), and loose dishes were from Iwaki Glass (Chibaken, Japan).
Cells and viruses.
Simian virus 40 large-T-antigen-immortalized murine wild-type fibroblasts or fibroblasts with LDLR, LRP, or LDLR and LRP gene disruptions (M cell lines) (Table 1) were kindly provided by Joachim Herz (Department of Molecular Genetics, Southwestern Medical Center at Dallas, Dallas, Tex.) (13, 19). Cells were grown in monolayers in Dulbecco's modified essential medium supplemented with 5% fetal calf serum (FCS), 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. HeLa-H1 cells (Flow Laboratories) were maintained in minimal essential medium containing 10% FCS, l-glutamine, and antibiotics as described above. All viruses were grown in HeLa-H1 cells, which are termed HeLa cells here for simplicity. To up-regulate LDLR expression, cells were incubated in medium containing 10% delipidized FCS (Dipro, Wiener Neudorf, Austria) for 2 days; they were then seeded into appropriate culture plates containing the same medium supplemented with 1 μg of lovastatin (Merck, Darmstadt, Germany) per ml and incubated for one more day.
TABLE 1.
Genotypes of M cell lines
| Cell line | Genotype
|
|
|---|---|---|
| LDLR | LRP | |
| M1 | +/+ | +/+ |
| M2 | +/+ | −/− |
| M3 | −/− | +/+ |
| M4 | −/− | −/− |
HRV serotypes 1A and 2 were originally obtained from the American Type Culture Collection (Rockville, Md.). The mouse-adapted variant of HRV2 (HRV2L) was kindly provided by Fay H. Yin and Nancy B. Lomax (Central Research and Development Department, E.I. du Pont de Nemours and Company, Wilmington, Del.) (26, 53).
Membrane preparation and analysis of receptor proteins.
Cell monolayers were washed twice with phosphate buffered saline (PBS), harvested with a cell scraper, resuspended in PBS, and pelleted at 1,200 rpm in a Heraeus Megafuge 1.0. Cell pellets were resuspended in lysis buffer (200 mM Tris-maleate [pH 6.5], 2 mM CaCl2, 0.5 mM phenylmethylsulfonyl fluoride, 2.5 μM leupeptin) and disrupted with a Dounce homogenizer at 4°C or by three freeze-thaw cycles. Cell membranes and cytoplasm were separated by ultracentrifugation at 70,000 rpm in a Beckman Optima TLX ultracentrifuge for 40 min at 4°C. Membranes collected in the pellet were solubilized in lysis buffer containing 1% Triton X-100 (Merck) for 10 min at 4°C. Insoluble material was removed by ultracentrifugation (70,000 rpm, 40 min, 4°C). Protein concentration in the extracts was estimated from Coomassie-stained sodium dodecyl sulfate (SDS)-8% polyacrylamide gels after electrophoresis under nonreducing conditions. Similar amounts of total protein were then used in Western blot and virus overlay blot analyses.
In order to verify that the respective cell lines expressed the receptors as expected, membrane proteins were separated on SDS-polyacrylamide gels under nonreducing conditions and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, Mass.). The membranes were blocked with PBS containing 2% Tween 20 (blocking buffer) for 1 h, incubated with the appropriate antibodies diluted in PBS containing 0.1% Tween 20 (incubation buffer) for 1 h, and washed three times with incubation buffer for 10 min. Rabbit anti-human LDLR antiserum (a kind gift from J. Nimpf, Vienna, Austria) or chicken immunoglobulin Y (IgY) (25 mg/ml), prepared from eggs of a hen immunized with the ligand binding domain of recombinant human LDLR (30) were used at a 1:5,000 dilution, and LDLR was revealed by further incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit or alkaline phosphatase (AP)-conjugated goat anti-chicken antibody, respectively (both from Southern Biotechnologies, Birmingham, Ala.). The Myc-tagged human single-chain antibody scFv7 (0.51 mg/ml), which is directed against the chicken homologue of mammalian VLDLR and cross-reacts with mouse and human LRP (16), was used at a 1:2,000 dilution; as a secondary antibody, the mouse anti-Myc antibody 9E10 was employed at 1 μg/ml followed by HRP-conjugated goat anti-mouse antibody (Bio-Rad Laboratories, Hercules, Calif.). Enzyme-coupled antibodies were always used at a final dilution of 1:5,000. HRP was detected with the ECL kit from Amersham Pharmacia, and AP was detected with 66 μl of nitroblue tetrazolium chloride (50 mg/ml in 70% dimethylformamide) and 33 μl of 5-bromo-4-chloro-3-indolylphosphate (50 mg/ml in H2O) in 10 ml of AP buffer (100 mM Tris-HCl [pH 9.6], 100 mM NaCl, 5 mM MgCl2).
For virus overlay blots, membranes were blocked with Tris-buffered saline containing 2 mM CaCl2 (TBS-Ca) and 2% Tween 20 (blocking buffer), probed with 1.5 × 104 cpm of 35S-labeled HRV1A or HRV2 per ml in 10 ml of TBS-Ca containing 0.1% Tween 20 (incubation buffer) for 1.5 h and washed three times with incubation buffer for 10 min each. The membranes were dried and autoradiographed for 24 h on Kodak MR films. Virus was radiolabeled with [35S]methionine in vivo and purified as described previously (35).
Infection assays.
To determine the susceptibilities of the respective cell lines to HRV infection, cells were seeded into six-well plates the day before infection. Growth medium was replaced by infection medium (IM; minimal essential medium or Dulbecco's modified essential medium for HeLa and the M cell collection, respectively, containing 2% FCS, 30 mM MgCl2, antibiotics, and glutamine as described above), and the cells were challenged at a multiplicity of infection (MOI) of 1 for 1.5 h at 34°C. Cells were washed with PBS, with HEPES buffer (140 mM KCl, 10 mM HEPES [pH 5.3]), and again with PBS to remove surface-bound virus and were further incubated at 34°C. Samples taken at different times postinfection (p.i.) were used to determine the virus titers as described below.
Determination of virus titers.
Infected cells in six-well plates were broken by three cycles of freezing and thawing, debris was removed, and serial 10-fold dilutions of the supernatants were prepared in IM. Samples were then transferred onto subconfluent monolayers of HeLa cells grown in 96-well culture plates containing 100 μl of IM. Following incubation at 34°C for 5 days, cells were stained with 0.1% crystal violet (in water) for 20 min. The tissue culture infectious dose which infects 50% of the cells was calculated as described by Blake and O'Connell (3).
Binding assays.
Cells were seeded into 24-well culture plates and grown to near confluence. Cell monolayers were washed with PBS and incubated with 200 μl of IM containing 104 cpm of 35S-labeled HRV1A or HRV2 for 20 min at 34°C with gentle agitation. The supernatants were collected, and cell layers were washed with 300 μl of PBS to remove unbound virus; the wash and the supernatants were combined. Cell layers were trypsinized and suspended in IM to a final volume of 500 μl. Supernatants and cell suspensions, respectively, were mixed with scintillation cocktail (Readysafe; Beckman Coulter, Fullerton, Calif.), radioactivity was determined in a liquid scintillation counter (Tricarb; Packard, Meriden, Conn.), and the percentage of cell-associated virus was calculated.
Preadsorption of antisera.
To avoid unspecific reactions with cellular proteins, the rabbit anti-HRV1A hyperimmune serum, a kind gift from Dan Pevear, ViroPharma, Exton, Pa., was preadsorbed on HeLa cells and M1 cells, respectively. Cells were scraped from the tissue culture flasks, resuspended, washed with PBS, and fixed with 2% paraformaldehyde (PFA) for 1 h at 25°C. Fixed cells were suspended in rabbit anti-HRV1A hyperimmune serum diluted 1:100 in PBS (3 × 107 cells/ml) and incubated for 2 h at room temperature and then overnight at 4°C with gentle agitation. The supernatant was cleared from cell debris by centrifugation and stored in aliquots at −20°C until further use.
Expression of human LDLR in M4 cells.
The cDNA coding for full-length human LDLR was excised from pTZ1 (30), a derivative of pLDLR-2 (51), and inserted into the eukaryotic expression vector pEF-Puro.PL3, which is derived from pEF-BOS (32) and was a kind gift of J. C. Renauld, Ludwig Institute for Cancer Research, Brussels, Belgium, to obtain pEF-LDLR. A truncated form of the LDLR, where a stop codon was introduced at amino acid 807 to disrupt the clathrin-coated-pit localization signal, was generated by PCR and introduced into the same vector to give pEF-LDLR806. Both plasmids were transfected stably in M4 cells by using Lipofectamine. After selection in puromycin (2 μg/ml), individual clones were isolated and screened for LDLR and LDLR806 expression by immunoblotting with chicken anti-LDLR antibody.
Analysis of infected cells by immunofluorescence.
Cells grown on coverslips overnight were infected with virus at an MOI of 40 for 45 min at 34°C, washed with PBS, and fixed with 3% PFA in PBS for 15 min at room temperature. After complete inactivation of PFA by incubation with 50 mM NH4Cl for 10 min and three washes with PBS, cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min followed by three more washes. Blocking was performed with 5% FCS in PBS for 10 min. Viral particles were detected by incubation for 45 min with the appropriate antibodies diluted in 1% FCS in PBS. HRV1A was revealed with preadsorbed rabbit anti-HRV1A hyperimmune serum (1:2) followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibody (1:200), and HRV2 was detected with mouse monoclonal antibody 8F5 raised against HRV2 (46) diluted 1:200 and FITC-conjugated goat anti-mouse antibody (1:400). After the cells had been embedded in Vectashield mounting medium (Vector Laboratories, Burlingame, Calif.), preparations were viewed with a confocal microscope (TCS NT; Leica, Heidelberg, Germany). To emphasize the accumulation of the virus in the perinuclear area, single sections through the middle of the cells were recorded.
Immunoaffinity chromatography and deglycosylation of LDLR.
Chicken anti-LDLR IgY (5 mg/ml of settled gel) was covalently linked to CNBr-activated Sepharose (Amersham Pharmacia) according to the manufacturer's protocol by incubation of antibody with the beads in 2 volumes of coupling buffer (100 mM NaHCO3 [pH 8.3], 500 mM NaCl) overnight at 4°C with agitation; remaining active groups were blocked with 100 mM Tris-HCl (pH 8.0) for 2 h at room temperature, and the beads were transferred into a column. The column was washed three times alternating between 100 mM sodium acetate (pH 4.0) and 100 mM Tris-HCl (pH 8.0), both containing 500 mM NaCl, and equilibrated with incubation buffer (0.5× lysis buffer containing 1% Triton X-100). Membrane extracts of HeLa cells were prepared as described above and diluted 1:10 in incubation buffer, and material corresponding to about 3 × 107 cells was then loaded onto 0.5 ml of IgY-Sepharose. Bound LDLR was eluted with incubation buffer containing 1 M NH3. The eluate was concentrated to half the volume, and NH3 was evaporated with a Speedvac. After the addition of 0.2% SDS and boiling for 5 min, N-linked oligosaccharides were removed by incubation with 1 U of N-glycosidase F (Boehringer Mannheim) per ml for 17 h at 37°C.
RESULTS
Properties of M cells.
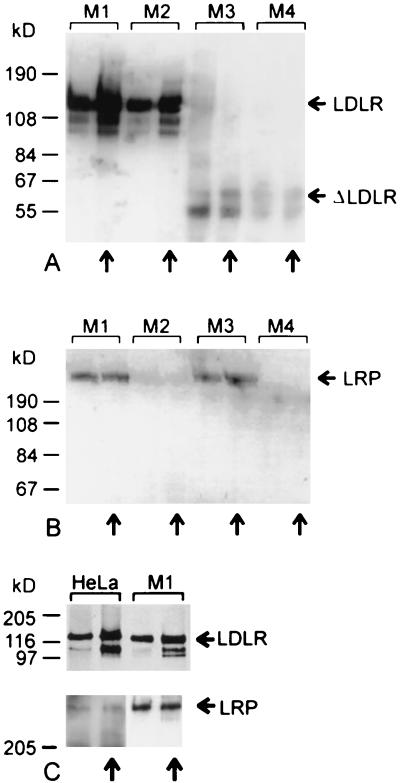
The simian virus 40 large-T-antigen-immortalized mouse fibroblasts used in this work originated from gene disruption experiments and resulted in LRP−/− (M2), LDLR−/− (M3), and LRP−/− LDLR−/− (M4) cell lines (Table 1); a control wild-type line (M1) was also available. First, we verified whether the receptor expression patterns corresponded to those described elsewhere (13, 19, 50). Membrane proteins were extracted from each cell line, and expression of the respective receptors was assayed by Western blot analysis using antibodies specific for LDLR and LRP (Fig. 1). M1 expresses LDLR and LRP, M2 expresses only LDLR, M3 expresses only LRP, and M4 expresses neither. To confirm the identity of the 120-kDa band reacting with rabbit antiserum directed against human LDLR, cells were grown under conditions which lead to up-regulation of LDLR expression (4, 23). This resulted in a significant increase in the intensity of the 120-kDa band in M1 and M2 cells, confirming its identity with LDLR (Fig. 1A). In membrane extracts of M3 and M4 cells, sometimes a weak band with an approximate molecular mass of 60 kDa and reacting with LDLR-specific antibodies was noticeable (Fig. 1A [ΔLDLR]). This presumably corresponds to truncated LDLR resulting from gene disruption (19). The presence of LRP was verified with the single-chain antibody scFv7, which was originally selected for binding to VLDLR from chicken ovaries but cross-reacts with human LRP (16). The expression pattern in the M cell lines corresponded to that expected from the genotypes. In accordance with earlier reports (31), no up-regulation of LRP expression was apparent (Fig. 1B). In a separate experiment, LDLR and LRP expression levels in HeLa cells grown under the same conditions as those used for Fig. 1A and B were compared. Figure 1C indicates that receptor expression was regulated similarly in mouse and human cells.
FIG. 1.
Western blot analysis of membrane proteins from HeLa and M cell lines grown under normal conditions or under conditions of up-regulation of LDLR expression (↑). Membrane proteins extracted from approximately 106 cells were separated on SDS-polyacrylamide gels with 8% (A; C, top), 4.5 to 18% (B), and 4.5 to 10% (C, bottom) acrylamide under nonreducing conditions and electrophoretically transferred to PVDF membranes. After blocking with 2% Tween 20 in PBS, LDLR was revealed with antibodies against human LDLR followed by enzyme-conjugated secondary antibodies. LRP was detected with Myc-tagged scFv7 followed by mouse anti-Myc antibody 9E10 and HRP-conjugated goat anti-mouse antibody. Note that in the bottom gel in panel C, twice the amount of membrane proteins was used for HeLa cells compared to M1 cells, and the exposure times for detection using chemiluminescence were 1.5 min for HeLa cells and 10 s for M1 cells.
Mouse-adapted HRV2L and HRV1A grow in murine fibroblasts.
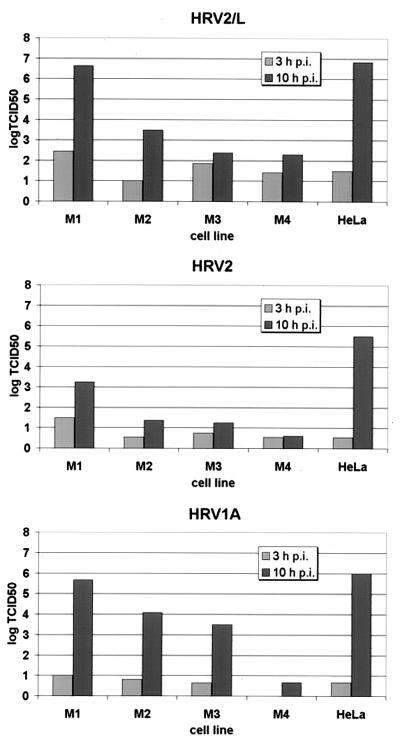
We then tested whether HRV2L, a variant of HRV2 which was adapted to replicate in mouse L cells (52, 53), was able to grow in the mouse fibroblasts of the M cell collection. For control purposes, wild-type HRV2 and another minor-group virus, HRV1A, were tested in parallel. Cells were infected with an MOI of 1 for 90 min at 34°C, nonattached virus was washed away, and incubation was continued. As seen from the increase in viral titer from 3 to 10 h p.i., HRV2L replicated in all cells, although to different extents, whereas the wild-type virus attained much lower titers in these cells (Fig. 2). The low but clearly discernible replication of HRV2L in M4 cells might be a consequence of fluid phase uptake of virus, which is consistent with the presence of a low number of infected cells, as seen by fluorescence microscopy (data not shown). To our surprise it turned out that HRV1A behaved similarly to HRV2L in attaining comparable titers. At 10 h p.i. HRV1A and HRV2L infection led to lysis of HeLa cells, whereas only a slight cytopathic effect was seen in M1 cells, at 10 h, followed by lysis at 24 h p.i. Wild-type HRV2 lysed HeLa cells at 24 h p.i. HRV1A thus appears to be an exception, as HRV29, HRV30, HRV47, HRV49, and HRV62, other minor-group viruses tested in parallel, were not able to replicate in M1 cells to any significant extent (data not shown).
FIG. 2.
Replication of mouse-adapted HRV2L, HRV2 wild type, and HRV1A in HeLa and M cell lines. Cells grown to about 50% confluence in six-well plates were infected with the respective viruses at an MOI of 1 for 1.5 h. Nonadsorbed virus was removed by repeated washing, and the cells were incubated in IM for the times indicated. Cells were then broken by three freeze-thaw cycles, and the virus titer was determined by endpoint dilution. TCID50, 50% tissue culture infective dose.
HRV1A and HRV2 exhibit differences in binding to murine and human receptors.
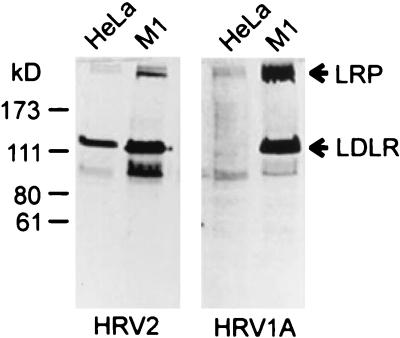
From the amino acid sequence similarity between the murine and human homologues of the LDLR family (e.g., 78% for LDLR) and extensive cross-reaction of antibodies, virus binding to mouse receptors was expected to be similar to that of human receptors. Whereas major-group HRVs bind only to primate cells, minor-group viruses attach to cells of a number of other species (48). Nevertheless, we felt it necessary to determine whether HRV2 and HRV1A bound equally well to the mouse and human receptors. Therefore, virus overlay blots were performed with proteins extracted from M1 cells and compared with proteins from HeLa cells. As seen in Fig. 3, HRV2 indeed attached to human as well as to murine LDLR. The band corresponding to LRP was, however, much weaker in HeLa cells than mouse cells. This is consistent with the smaller amount of LRP present in HeLa cells than in mouse fibroblasts (Fig. 1C) (33). Surprisingly, the binding pattern of HRV1A and HRV2 differed markedly. In contrast to HRV2, HRV1A did not bind to human LDLR, whereas binding to the mouse LDLR was similar for both serotypes.
FIG. 3.
Virus binding to membrane proteins of HeLa and M1 cells. Proteins from membrane extracts were separated on SDS-8% polyacrylamide gels under nonreducing conditions and electrophoretically transferred to PVDF membranes. The membranes were blocked with TBS-Ca containing 2% Tween 20 and probed with 1.5 × 104 cpm of 35S-labeled HRV2 and HRV1A per ml in 10 ml of TBS-Ca-0.1% Tween 20 for 1.5 h. Membranes were autoradiographed for 24 h on Kodak MR film.
It should be mentioned that virus overlay blots showed some weak binding of HRV1A to human LDLR when large amounts of receptor were present (data not shown). This indicates that the human receptor is recognized but with very low affinity. It is in agreement with our previous finding that HRV1A could be neutralized by recombinant human LDLR fragments (30). The present results suggest that HRV1A (and possibly other minor-group viruses) discriminates between LDLR homologues of different species, as the expression level of LDLR was comparable in the two cell lines (Fig. 1C). In addition to the bands corresponding to LDLR and to LRP, both serotypes also bound to various extents to a 95-kDa protein (Fig. 3). This presumably corresponds to VLDLR or is unspecific.
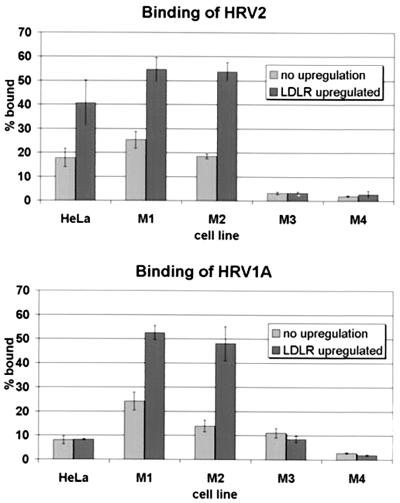
In order to substantiate these findings, virus binding to HeLa cells and to the collection of M cell lines was assayed with cells grown under conditions which lead to up-regulation of LDLR expression. Cells were seeded into 24-well culture plates, and the following day binding and internalization of radiolabeled virus were determined at 34°C by incubation for 20 min. Figure 4 shows that HRV1A bound less to HeLa cells than HRV2, whereas more HRV1A was bound in the case of M3 cells. This is reminiscent of the stronger binding of HRV1A to LRP seen in the virus overlay blot in Fig. 3. However, whereas HRV2 binding was clearly increased upon up-regulation of LDLR expression in HeLa, M1, and M2 cells, for HRV1A, this effect was seen in the mouse cells but was absent from HeLa cells. This strongly suggests that HRV1A attachment closely follows the expression level of mouse LDLR but fails to do so in the case of human LDLR. As expected from the absence of both receptors from M4 cells, only background binding was observed regardless of the viral serotype tested. It is noteworthy that HRV1A binds less well than HRV2 to HeLa cells (Fig. 4) but replicates to similarly high titers (Fig. 2). This indicates either that a receptor different from LDLR (presumably LRP) is very efficient in HRV1A internalization and/or uncoating or that another receptor exists which is unable to bind virus in a virus overlay assay.
FIG. 4.
Binding and internalization of HRV2 and HRV1A into HeLa cells and M cell lines. Cells grown to near confluence in 24-well plates were washed with PBS and incubated with 200 μl of IM containing 104 cpm of 35S-labeled virus for 20 min at 34°C with gentle agitation. Nonassociated virus was removed by washing with PBS, and the amount of cell-associated virus as well as virus in the combined wash and supernatant was determined by scintillation counting. Radioactivity in the cell pellet is given as percentage of total counts. The means of three parallel experiments ± standard deviations are shown.
It is worth mentioning that the small increase in LDLR expression upon up-regulation (Fig. 1 and 4) failed to give rise to a detectable increase in viral titer in HeLa or in M1 cells (data not shown). This is most probably due to the different accuracies of the assays; binding tests pick up differences in the percentage range, whereas 50% tissue culture infective dose tests instead cover orders of magnitude.
HRV1A does not bind to human LDLR expressed in mouse M4 cells.
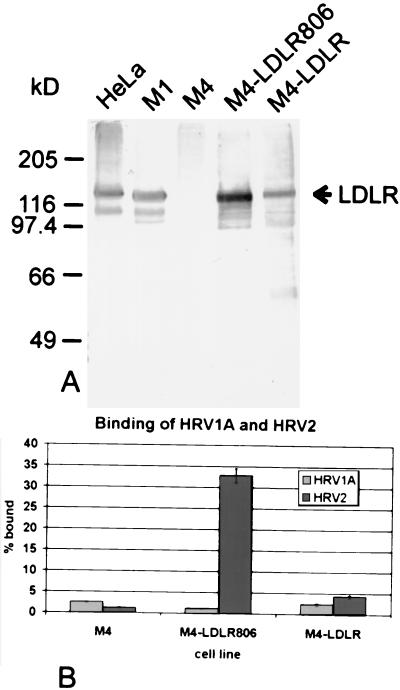
In order to confirm the absence of binding of HRV1A to human LDLR, this receptor was expressed in M4 cells. The entire human LDLR cDNA and a construct in which the tyrosine codon of the NPVY internalization signal was replaced by a stop codon (termed LDLR806) were introduced into pEF-Puro.PL3, and the plasmids were transfected into M4 cells. Cells stably expressing the foreign proteins were selected with puromycin, and clones M4-LDLR and M4-LDLR806 were isolated. These cells were originally created to study virus internalization (Snyers et al. submitted). Membrane proteins were prepared, and the clones were compared with HeLa, M1, and M4 cells with respect to expression of the receptor protein by Western blotting (Fig. 5A). LDLR806 was present in larger amounts than LDLR, presumably because of lower internalization and degradation. Next, virus binding was investigated. HRV2 attached with high efficiency to M4-LDLR806, whereas HRV1A binding was comparable to nontransfected M4 cells. Similarly, binding of HRV1A to M4-LDLR and to M4 cells was identical, whereas significantly more HRV2 (P > 99.7% as determined by the t test) was bound to the cells expressing the recombinant receptor (Fig. 5B).
FIG. 5.
Expression of human LDLR in M4 cells and virus binding. M4 cells were transfected with pEF-LDLR and pEF-LDLR806, respectively, and M4-LDLR and M4-LDLR806, which stably express the respective proteins, were selected. (A) Membrane extracts were prepared, and expression was verified by Western blotting with chicken IgY raised against human LDLR and cross-reacting with mouse LDLR followed by AP-conjugated goat anti-chicken antibody and substrate. For control purposes, Western blotting was also carried out with membrane extracts from HeLa, M1, and M4 cells in parallel. (B) Cells grown in 24-well plates to near confluence were incubated with 104 cpm of 35S-labeled viruses in 200 μl of IM at 34°C for 20 min, and virus binding plus internalization was determined as for Fig. 4. The means of three independent experiments ± standard deviations are shown.
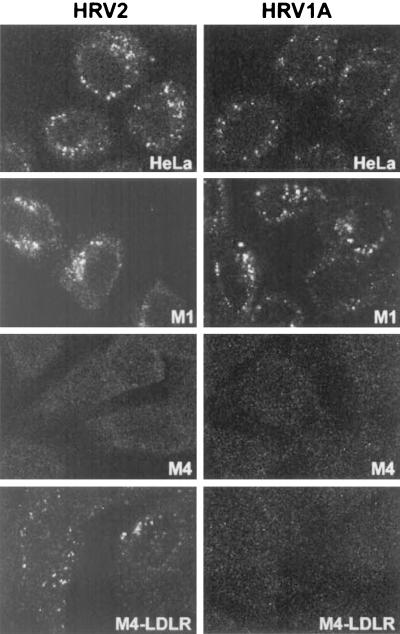
Since binding of HRV2 to M4-LDLR was low, we wanted to confirm by a different method that these cells were indeed able to bind and internalize HRV2 but not HRV1A. M4 cells and M4-LDLR cells were incubated with HRV2 and HRV1A at an MOI of 40 for 45 min at 34°C, fixed, and prepared for indirect immunofluorescence microscopy. Again, for comparison, the same experiments were also carried out in parallel with HeLa, M1, and M4 cells. As seen in Fig. 6, HRV2 was internalized into HeLa, M1, and into M4-LDLR cells. In contrast, HRV1A was internalized into HeLa and M1 cells but not into M4-LDLR cells. As expected, M4 cells failed to take up either of the virus serotypes. In all cases where internalization had taken place, the virus was seen to accumulate in the perinuclear region, indicative of its localization in late endosomes and lysosomes (18).
FIG. 6.
HRV2 but not HRV1A is internalized into mouse M4 cells expressing human LDLR. HeLa, M1, M4, and M4-LDLR cells were grown on coverslips and incubated with the respective viruses at an MOI of 40 for 45 min at 34°C. Cells were washed, fixed with paraformaldehyde, and permeabilized with Triton X-100, and virus was revealed with rabbit antiserum against HRV1A and monoclonal antibody 8F5 against HRV2, followed by FITC-conjugated goat IgG directed against the virus-specific antibodies. The images show a single section through the middle of the cells to emphasize the accumulation of the virus in perinuclear structures. Cells were viewed with a Leica TCS NT confocal microscope.
Species-specific glycosylation is not responsible for receptor discrimination.
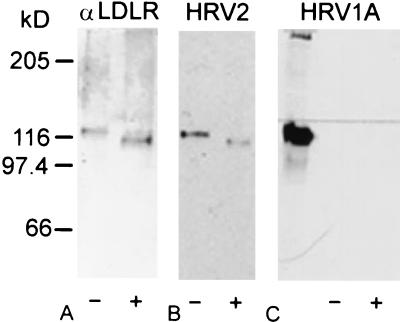
There are three potential N-glycosylation sites in the ligand binding domain of the human LDLR and two in the mouse LDLR. Based on direct analysis of glycopeptides prepared from human LDLR extracted from A-431 epidermoid carcinoma cells, Cummings and colleagues (5) presented evidence that at least one but not more than two of the potential glycosylation sites were used. This is in accordance with expression of recombinant human LDLR fragments encompassing repeats 2 and 3, 3 and 4, and 4 and 5 in Sf9 insect cells (30). Using Western blotting with concanavalin A, we observed that the site within repeat 4 was glycosylated whereas that within repeat 2 was not (data not shown), although the nature of the oligosaccharide certainly differs from that present in the human cells (5). The glycosylation site in repeat 4 is absent from the mouse homologue, as the human NSS sequence at position 157 is replaced by KSS in the mouse. We therefore wondered whether the species-specific recognition of the receptor by HRV1A was based on differences in the glycosylation; glycosylation might reduce the accessibility of repeat 4 in the human homologue. Human LDLR was purified from HeLa membrane extracts by immunoaffinity column chromatography on chicken anti-LDLR IgY covalently attached to Sepharose. Bound material was eluted and subjected to digestion with N-glycosidase (Fig. 7), which completely removes oligosaccharides attached to asparagine but does not attack O-glycosylation sites (28), and analyzed by Western blotting with LDLR-specific IgY and for virus binding by virus overlay blots with radioactive HRV2 and HRV1A (Fig. 7). Incubation with the IgY fraction shows a decrease in Mr of the LDLR which is consistent with the removal of the N-linked oligosaccharide chains from the protein (25). Whereas HRV2 bound equally well to the glycosylated and the unglycosylated forms of LDLR, no binding to either preparation was seen for HRV1A. For control purposes, a membrane extract of M1 cells was run in parallel, indicating strong binding of HRV1A (Fig. 7C, leftmost lane).
FIG. 7.
Glycosylation is not involved in receptor discrimination of HRV1A. Human LDLR from HeLa cell plasma membrane extracts was purified by affinity chromatography on anti-LDLR IgY covalently attached to Sepharose and subjected to enzymatic deglycosylation. Mock-incubated LDLR (−) and LDLR incubated with N-glycosidase F (+) were run on an SDS-8% polyacrylamide gel under nonreducing conditions, and the proteins were electrotransferred to PVDF membranes and probed with anti-LDLR IgY (A), 105 cpm of radiolabeled HRV2 (B), and HRV1A (C). Bound antibodies were revealed with AP-conjugated goat anti-IgY followed by substrate, radioactive virus was revealed by exposure to X-ray film overnight. For control purposes, a membrane extract from M1 cells was also run on the gel in panel C (leftmost lane).
Attempts to block glycosylation in vivo by addition of tunicamycin at 5 μg/ml for 24 h revealed that the drug was toxic to M1 cells; while the HeLa cells appeared to grow normally, no immunoreactive LDLR was detectable in the cell membranes. Reduction of the tunicamycin concentration to 1 μg/ml resulted in the appearance of an additional band migrating ahead of the native receptor for both cell types as seen in SDS-polyacrylamide gel electrophoresis. Virus overlay blots again showed no attachment of HRV1A to any form of the human receptor. HRV2 bound to the glycosylated form of the receptor only. This is taken to indicate that correct folding of the protein was impaired in the presence of the drug (data not shown).
DISCUSSION
In this report we demonstrate that HRV1A binds much more strongly to the mouse than the human homologue of LDLR. Using enzymatic digestion and inhibition with tunicamycin, we showed that glycosylation was not involved. This species specificity is remarkable, as the high evolutionary conservation of the LDLR and of other members of the family results in recognition of ligands from various species. Minor-receptor-group HRVs have been found to attach to a number of different cell types, (48), presumably via several representatives of the LDLR family, including LDLR, VLDLR, and LRP, as usually more than one type of receptor is present on the cell membrane. This might be why the lack of recognition of human LDLR by HRV1A has remained unnoticed.
We also discovered that HRV1A multiplied in a mouse fibroblast cell line with an efficiency similar to that of HRV2L, a mouse-adapted variant. It was believed that despite being taken up into the cell, minor-group HRVs could not replicate in species other than humans. This property of HRV1A thus appears to be exceptional, as all of the other minor-group HRVs tested failed to grow in murine cells.
Upon reconstruction of electron cryomicroscopy images of a complex between HRV2 and VLDLR1-3, two repeats were seen to cover the surface-exposed BC and HI loops of VP1 (14), and image reconstructions between a fusion protein containing only repeats 2 and 3 suggested that repeat 1 of the former construct was not involved in binding. In addition, we have evidence that single repeats also exhibit some affinity for HRV2 (unpublished data). Comparison of the footprint of the VLDLR on HRV2 with that predicted for HRV1A, based on the amino acid sequence and the X-ray structure of this serotype, revealed only marginal similarity. As noted earlier (8, 21, 40), no more than the tripeptide TEK in the HI loop and the dipeptide YN in the BC loop are conserved within all minor-group serotypes sequenced so far. However, in contrast to foot-and-mouth disease virus, where the interaction between the RGD tripeptide in the GH loop of VP1 with αvβ3 integrin (2) can be inhibited with synthetic peptides (10), the interaction of HRVs with their receptors cannot be blocked by a peptide including the TEK sequence (our unpublished observations). Therefore, the epitope recognized by the receptor must be conformational rather than sequential, and it is highly probable that amino acid residues in the vicinity of the conserved peptide sequences are involved. It is noteworthy that HRV1A appears to have a more extended positive surface charge cluster in the star-shaped dome at the fivefold symmetry axes than HRV2.
The determination of the three-dimensional structure of repeat 5 of human LDLR by X-ray crystallography (9) revealed that the carboxylates of four acidic amino acids are directed inwards, chelating a Ca2+ ion, rather than being exposed at the surface. Nevertheless, it is still believed that charge complementarity with the cationic surface is the main ligand binding determinant (27). Could it be that the distribution of negative surface charges is different between human and mouse LDLR? A comparison of the entire ligand binding domain reveals too many amino acid differences to allow the identification of residues possibly involved in discrimination. However, considering that repeat 5 of LDLR is most important for recognition of LDL and β-VLDL, with its deletion reducing binding of these two ligands to less than 8% and 50%, respectively (44), and that mutations resulting in familial hypercholesterolemia are concentrated in this repeat (15), we speculate that this repeat also contributes most to the interaction with the viruses. Site-directed mutagenesis of human LDLR fragments in combination with binding experiments will help to elucidate the nature of the residues involved in the interaction between the different serotypes and LDLR. Furthermore, studies adapting HRV1A to grow in M4 mouse cells expressing the human LDLR (M4-LDLR) are under way. Analysis of mutations resulting in adaptation might also allow us to identify the amino acid residues implicated in receptor discrimination.
Acknowledgments
We thank Joachim Herz for the collection of M cells, Dan Pevear for the antiserum against HRV1A, and Johannes Nimpf for rabbit anti-LDLR antiserum.
This work was supported by a grant from the Austrian Virology Foundation and the Austrian Science Foundation (14503-MOB).
REFERENCES
- 1.Beglova, N., C. L. North, and S. C. Blacklow. 2001. Backbone dynamics of a module pair from the ligand-binding domain of the LDL receptor. Biochemistry 40:2808-2815. [DOI] [PubMed] [Google Scholar]
- 2.Berinstein, A., M. Roivainen, T. Hovi, P. W. Mason, and B. Baxt. 1995. Antibodies to the vitronectin receptor (integrin αvβ3) inhibit binding and infection of foot-and-mouth disease virus to cultured cells. J. Virol. 69:2664-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake, K., and S. O'Connell. 1993. Virus culture, p. 81-122. In D. R. Harper (ed.), Virology labfax. Blackwell Scientific Publications, London, United Kingdom.
- 4.Chan, P. C., R. Lafreniere, and H. G. Parsons. 1997. Lovastatin increases surface low density lipoprotein receptor expression by retarding the receptor internalization rate in proliferating lymphocytes. Biochem. Biophys. Res. Commun. 235:117-122. [DOI] [PubMed] [Google Scholar]
- 5.Cummings, R. D., S. Kornfeld, W. J. Schneider, K. K. Hobgood, H. Tolleshaug, M. S. Brown, and J. L. Goldstein. 1983. Biosynthesis of N- and O-linked oligosaccharides of the low density lipoprotein receptor. J. Biol. Chem. 258:15261-15273. [PubMed] [Google Scholar]
- 6.Daly, N. L., J. T. Djordjevic, P. A. Kroon, and R. Smith. 1995. Three-dimensional structure of the second cysteine-rich repeat from the human low-density lipoprotein receptor. Biochemistry 34:14474-14481. [DOI] [PubMed] [Google Scholar]
- 7.Daly, N. L., M. J. Scanlon, J. T. Djordjevic, P. A. Kroon, and R. Smith. 1995. Three-dimensional structure of a cysteine-rich repeat from the low-density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 92:6334-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duechler, M., S. Ketter, T. Skern, E. Kuechler, and D. Blaas. 1993. Rhinoviral receptor discrimination: mutational changes in the canyon regions of human rhinovirus types 2 and 14 indicate a different site of interaction. J. Gen. Virol. 74:2287-2291. [DOI] [PubMed] [Google Scholar]
- 9.Fass, D., S. Blacklow, P. S. Kim, and J. M. Berger. 1997. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature 388:691-693. [DOI] [PubMed] [Google Scholar]
- 10.Fox, G., N. R. Parry, P. V. Barnett, B. McGinn, D. J. Rowlands, and F. Brown. 1989. The cell attachment site on foot-and-mouth disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid). J. Gen. Virol. 70:625-637. [DOI] [PubMed] [Google Scholar]
- 11.Gliemann, J. 1998. Receptors of the low density lipoprotein (LDL) receptor family in man. Multiple functions of the large family members via interaction with complex ligands. Biol. Chem. 379:951-964. [PubMed] [Google Scholar]
- 12.Gruenberger, M., R. Wandl, J. Nimpf, T. Hiesberger, W. J. Schneider, E. Kuechler, and D. Blaas. 1995. Avian homologs of the mammalian low-density lipoprotein receptor family bind minor receptor group human rhinovirus. J. Virol. 69:7244-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herz, J., D. E. Clouthier, and R. E. Hammer. 1992. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell 71:411-421. [DOI] [PubMed] [Google Scholar]
- 14.Hewat, E. A., E. Neumann, J. F. Conway, R. Moser, B. Ronacher, T. C. Marlovits, and D. Blaas. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobbs, H. H., M. S. Brown, and J. L. Goldstein. 1992. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1:445-466. [DOI] [PubMed] [Google Scholar]
- 16.Hodits, R. A., J. Nimpf, D. M. Pfistermueller, T. Hiesberger, W. J. Schneider, T. J. Vaughan, K. S. Johnson, M. Haumer, E. Kuechler, G. Winter, and D. Blaas. 1995. An antibody fragment from a phage display library competes for ligand binding to the low density lipoprotein receptor family and inhibits rhinovirus infection. J. Biol. Chem. 270:24078-24085. [DOI] [PubMed] [Google Scholar]
- 17.Hofer, F., M. Gruenberger, H. Kowalski, H. Machat, M. Huettinger, E. Kuechler, and D. Blaas. 1994. Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. USA 91:1839-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber, M., M. Brabec, N. Bayer, D. Blaas, and R. Fuchs. 2001. Elevated endosomal pH in HeLa cells overexpressing mutant dynamin can affect infection by pH-sensitive viruses. Traffic 2:727-736. [DOI] [PubMed] [Google Scholar]
- 19.Ishibashi, S., M. S. Brown, J. L. Goldstein, R. D. Gerard, R. E. Hammer, and J. Herz. 1993. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J. Clin. Investig. 92:883-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon, H., W. Meng, J. Takagi, M. J. Eck, T. A. Springer, and S. C. Blacklow. 2001. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat. Struct. Biol. 8:499-504. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S., T. J. Smith, M. S. Chapman, M. G. Rossmann, D. C. Pevear, F. J. Dutko, P. J. Felock, G. D. Diana, and M. A. McKinlay. 1989. Crystal structure of human rhinovirus serotype-1A (Hrv1A). J. Mol. Biol. 210:91-111. [DOI] [PubMed] [Google Scholar]
- 22.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowal, R. C., J. Herz, J. L. Goldstein, V. Esser, and M. S. Brown. 1989. Low density lipoprotein receptor-related protein mediates uptake of cholesteryl esters derived from apoprotein E-enriched lipoproteins. Proc. Natl. Acad. Sci. USA 86:5810-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurniawan, N. D., A. R. Atkins, S. Bieri, C. J. Brown, I. M. Brereton, P. A. Kroon, and R. Smith. 2000. NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor. Protein Sci. 9:1282-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemp, D., A. Haselbeck, and F. Klebl. 1990. Molecular cloning and heterologous expression of N-glycosidase F from Flavobacterium meningosepticum. J. Biol. Chem. 265:15606-15610. [PubMed] [Google Scholar]
- 26.Lomax, N. B., and F. H. Yin. 1989. Evidence for the role of the P2 protein of human rhinovirus in its host range change. J. Virol. 63:2396-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LundKatz, S., P. M. Laplaud, M. C. Phillips, and M. J. Chapman. 1998. Apolipoprotein B-100 conformation and particle surface charge in human LDL subspecies: implication for LDL receptor interaction. Biochemistry 37:12867-12874. [DOI] [PubMed] [Google Scholar]
- 28.Maley, F., R. B. Trimble, A. L. Tarentino, and T. H. Plummer, Jr. 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180:195-204. [DOI] [PubMed] [Google Scholar]
- 29.Marlovits, T. C., C. Abrahamsberg, and D. Blaas. 1998. Very-low-density lipoprotein receptor fragment shed from HeLa cells inhibits human rhinovirus infection. J. Virol. 72:10246-10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marlovits, T. C., T. Zechmeister, M. Gruenberger, B. Ronacher, H. Schwihla, and D. Blaas. 1998. Recombinant soluble low density lipoprotein receptor fragment inhibits minor group rhinovirus infection in vitro. FASEB J. 12:695-703. [DOI] [PubMed] [Google Scholar]
- 31.Medh, J. D., S. L. Bowen, G. L. Fry, S. Ruben, M. Andracki, I. Inoue, J. M. Lalouel, D. K. Strickland, and D. A. Chappell. 1996. Lipoprotein lipase binds to low density lipoprotein receptors and induces receptor-mediated catabolism of very low density lipoproteins in vitro. J. Biol. Chem. 271:17073-17080. [DOI] [PubMed] [Google Scholar]
- 32.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mucci, D., J. Forristal, D. Strickland, R. Morris, D. Fitzgerald, and C. B. Saelinger. 1995. Level of receptor-associated protein moderates cellular susceptibility to pseudomonas exotoxin A. Infect. Immun. 63:2912-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neels, J. G., I. R. Horn, B. M. M. vandenBerg, H. Pannekoek, and A. J. vanZonneveld. 1998. Ligand-receptor interactions of the low density lipoprotein receptor-related protein, a multiligand endocytic receptor. Fibrinolysis Proteolysis 12:219-240. [Google Scholar]
- 35.Neubauer, C., L. Frasel, E. Kuechler, and D. Blaas. 1987. Mechanism of entry of human rhinovirus 2 into HeLa cells. Virology 158:255-258. [DOI] [PubMed] [Google Scholar]
- 36.North, C. L., and S. C. Blacklow. 2000. Solution structure of the sixth LDL-A module of the LDL receptors. Biochemistry 39:2564-2571. [DOI] [PubMed] [Google Scholar]
- 37.North, C. L., and S. C. Blacklow. 1999. Structural independence of ligand-binding modules five and six of the LDL receptor. Biochemistry 38:3926-3935. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira, M. A., R. Zhao, W. M. Lee, M. J. Kremer, I. Minor, R. R. Rueckert, G. D. Diana, D. C. Pevear, F. J. Dutko, M. A. McKinlay, and M. G. Rossmann. 1993. The structure of human rhinovirus 16. Structure 1:51-68. [DOI] [PubMed] [Google Scholar]
- 39.Olson, N. H., P. R. Kolatkar, M. A. Oliveira, R. H. Cheng, J. M. Greve, A. McClelland, T. S. Baker, and M. G. Rossmann. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA 90:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reischl, A., M. Reithmayer, G. Winsauer, R. Moser, I. Goesler, and D. Blaas. 2001. Viral evolution toward change in receptor usage: Adaptation of a major group human rhinovirus to grow in ICAM-1-negative cells. J. Virol. 75:9312-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ronacher, B., T. C. Marlovits, R. Moser, and D. Blaas. 2000. Expression and folding of human very-low-density lipoprotein receptor fragments: neutralization capacity toward human rhinovirus HRV2. Virology 278:541-550. [DOI] [PubMed] [Google Scholar]
- 42.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 43.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott Raven Publishers, Philadelphia, Pa.
- 44.Russell, D. W., M. S. Brown, and J. L. Goldstein. 1989. Different combinations of cysteine-rich repeats mediate binding of low density lipoprotein receptor to two different proteins. J. Biol. Chem. 264:21682-21688. [PubMed] [Google Scholar]
- 45.Sappington, T. W., and A. S. Raikhel. 1998. Ligand-binding domains in vitellogenin receptors and other LDL-receptor family members share a common ancestral ordering of cysteine-rich repeats. J. Mol. Evol. 46:476-487. [DOI] [PubMed] [Google Scholar]
- 46.Skern, T., C. Neubauer, L. Frasel, P. Gruendler, W. Sommergruber, W. Zorn, E. Kuechler, and D. Blaas. 1987. A neutralizing epitope on human rhinovirus type 2 includes amino acid residues between 153 and 164 of virus capsid protein VP2. J. Gen. Virol. 68:315-323. [DOI] [PubMed] [Google Scholar]
- 47.Strickland, D. K., M. Z. Kounnas, and W. S. Argraves. 1995. LDL receptor-related protein: a multiligand receptor for lipoprotein and proteinase catabolism. FASEB J. 9:890-898. [DOI] [PubMed] [Google Scholar]
- 48.Uncapher, C. R., C. M. Dewitt, and R. J. Colonno. 1991. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology 180:814-817. [DOI] [PubMed] [Google Scholar]
- 49.Verdaguer, N., D. Blaas, and I. Fita. 2000. Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300:1179-1194. [DOI] [PubMed] [Google Scholar]
- 50.Willnow, T. E., and J. Herz. 1994. Genetic deficiency in low density lipoprotein receptor-related protein confers cellular resistance to Pseudomonas exotoxin A. Evidence that this protein is required for uptake and degradation of multiple ligands. J. Cell Sci. 107:719-726. [PubMed] [Google Scholar]
- 51.Yamamoto, T., C. G. Davis, M. S. Brown, W. J. Schneider, M. L. Casey, J. L. Goldstein, and D. W. Russell. 1984. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell 39:27-38. [DOI] [PubMed] [Google Scholar]
- 52.Yin, F. H., and N. B. Lomax. 1986. Establishment of a mouse model for human rhinovirus infection. J. Gen. Virol. 67:2335-2340. [DOI] [PubMed] [Google Scholar]
- 53.Yin, F. H., and N. B. Lomax. 1983. Host range mutants of human rhinovirus in which nonstructural proteins are altered. J. Virol. 48:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, R., D. C. Pevear, M. J. Kremer, V. L. Giranda, J. A. Kofron, R. J. Kuhn, and M. G. Rossmann. 1996. Human rhinovirus 3 at 3.0 angstrom resolution. Structure 4:1205-1220. [DOI] [PubMed] [Google Scholar]