Abstract
The hepatitis C virus (HCV)-encoded NS5B protein is an RNA-dependent RNA polymerase which plays a substantial role in viral replication. We expressed and purified the recombinant NS5B of an HCV genotype 3a from Esherichia coli, and we investigated its ability to bind to the viral RNA and its enzymatic activity. The results presented here demonstrate that NS5B interacts strongly with the coding region of positive-strand RNA, although not in a sequence-specific manner. It was also determined that more than two molecules of polymerase bound sequentially to this region with the direction 3′ to 5′. Also, we attempted to determine the initiation site(s) of de novo synthesis by NS5B on X RNA, which contains the last 98 nucleotides of HCV positive-strand RNA. The initiation site(s) on X RNA was localized in the pyrimidine-rich region of stem I. However, when more than five of the nucleotides of stem I in X RNA were deleted from the 3′ end, RNA synthesis initiated at another site of the specific ribonucleotide. Our study also showed that the efficiency of RNA synthesis, which was directed by X RNA, was maximized by the GC base pair at the penultimate position from the 3′ end of the stem. These results will provide some clues to understanding the mechanism of HCV genomic RNA replication in terms of viral RNA-NS5B interaction and the initiation of de novo RNA synthesis.
Hepatitis C virus (HCV) is the major causative agent of human viral hepatitis in most cases of non-A non-B hepatitis (5, 10). Nearly 80% of infections develop into chronic hepatitis, of which 10 to 20% progress to cirrhosis and 1 to 5% progress to hepatocellular carcinoma (22). However, neither a vaccine to prevent HCV infection nor a specific treatment for liver disease caused by chronic HCV infection is yet available. Thus, there is an urgent need to develop HCV-specific antiviral agents to counteract this human pathogen.
HCV is classified as a separate genus in the family Flaviviridae (21). The virion is an enveloped virus containing a positive-strand RNA genome of 9.5 kb. The RNA genome consists of a 5′ untranslated region (UTR), an open reading frame (ORF), and a 3′ UTR (5, 6, 8). The order of the gene products of the single ORF is NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH, and this polyprotein is subsequently processed by host and viral proteases into 10 separate proteins. Among the six nonstructural proteins, the NS5B protein was recognized to be an RNA-dependent RNA polymerase (RdRp), which is the key enzyme for viral replication. Since the full-length HCV NS5B protein has poor solubility for purification, the recombinant NS5B is generally expressed on a large scale by removing the C-terminal 21 amino acid residues (28). This C-terminal region is dispensable for RdRp activity, and even more significant, this truncation positively affects its enzymatic activity (25). The HCV 3′ UTR, which plays a major role in the initiation of RNA replication after viral infection, consists of three elements: a variable region, a poly(U-U/C) tract, and a 98-nucleotide (nt) X region. The 98-nt X RNA is highly conserved among HCV isolates and contains three stem-loop structures with high stability in its secondary structure (11, 15, 23).
For the RdRp reaction, the NS5B protein can catalyze the various viral or nonviral RNA templates in vitro only if the RNA templates meet some requirements (3, 14, 17-19, 30). However, it is necessary that NS5B discriminate the HCV genomic RNA from other RNAs preferentially and catalyze its substrate for viral amplification in vivo. There have been diverse efforts to show evidence of the molecular interaction between HCV RNA and NS5B. It was suggested that the coding region of HCV genomic RNA, but not the 3′ UTR (or X RNA), is important for binding to NS5B polymerase (4). Consistent with this result, there have been additional studies which suggested that HCV NS5B interacts with X RNA with little specificity and that it has no clear preference to utilize the X RNA as a template in the RdRp reaction (16, 28). On the other hand, some attempts have been made to elucidate the biological function of X RNA for binding to NS5B and to discover the role of X RNA as a cis-acting element in viral replication. These proposed that the 3′ UTR, which contains X RNA, is absolutely required for in vivo infectivity and viral replication (15, 29). In addition, it has been emphasized that X RNA interacts with HCV NS5B specifically and that the RNA synthesis is initiated selectively from a specific nucleotide on this RNA (19).
To reconcile these controversial issues, we investigated the interaction between the NS5B polymerase and the viral RNA, and we studied the template requirements for efficient RNA synthesis on X RNA. In this report, we will present the following conclusions: NS5B binds to its genomic RNA in the coding region sequentially from the 3′ end in increasing amounts, but it does not bind in a sequence-specific manner; de novo synthesis on X RNA initiates in the pyrimidine-rich region of stem I; and the transcription efficiency is controlled by the nucleotide composition or the secondary structure of stem I. We suggest that stem I has the dual functions of regulating the reaction efficiency and determining the initiation sites of de novo RNA synthesis. Finally, the results show the definite requirements of the X RNA template for the RdRp reaction at specific sites. Future advances in our understanding of the interaction of HCV NS5B and the viral RNA, as well as the initiation mechanism of RNA synthesis, should aid in the development of highly specific and potent inhibitors of viral RdRp as anti-HCV agents and in the construction of the new viral replicon generating high copies of HCV genome.
MATERIALS AND METHODS
Expression and purification of recombinant NS5B.
The plasmid pLysN was kindly produced and provided by B.-L. Seong (Yeonsei University, Seoul, Korea) by the insertion of the gene for the N-terminal 154 amino acids of Escherichia coli lysyl-tRNA synthetase (lysS) and the histidine polypeptide gene into pGEMEX-1 (Promega). The NS5B gene of the type 3a HCV isolate, which has a 20-amino-acid coding region truncation from the 3′ end, was subcloned into pLysN, and this plasmid was designated pLysN-NS5B. pLysN-NS5B expresses LysN-GSGSG-(His)10-ΔNS5B as a fusion protein (unpublished data). For expressing the recombinant proteins, each plasmid of pLysN and pLysN-NS5B was transformed into the E. coli strains BL21pLysS(DE3) and HMS174(DE3), respectively. The transformants were cultured in 2 ml of Luria-Bertani medium with 50 μg of ampicillin/ml at 37°C overnight. The cultures were then diluted in 200 ml of Luria-Bertani medium with 50 μg of ampicillin/ml and incubated at 37°C until the optical density at 600 nm reached 0.6 to 0.8. These cultures were then induced overnight with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C. The cells expressing the recombinant proteins were harvested by centrifugation. The pellet was suspended in 6 ml of a CE buffer (50 mM Na3PO4 [pH 6.8], 100 mM NaCl, 5 mM dithiothreitol [DTT], 1 mM EDTA, 10% glycerol, 250 mM sucrose, and 0.2% sodium azide) with 1 mM phenylmethylsulfonyl fluoride, and after sonication, the supernatant was collected for protein purification. The LysN protein expressed from pLysN was purified using the metal affinity resin (TALON; Clontech) as specified by the manufacturer. For purifying the LysN-NS5B protein expressed from pLysN-NS5B, the supernatant fraction from the crude cell extract was mixed with a half volume of saturated ammonium sulfate. After centrifugation, the supernatant was equilibrated in CE buffer by dialysis overnight. The dialyzed fraction was mixed with 2 ml of SP-Sepharose (Pharmacia Biotech), and the protein was allowed to adsorb to the beads for 1 h at 4°C. The beads were then washed thoroughly with CE buffer by increasing the concentration of NaCl up to 450 mM. The recombinant LysN-NS5B protein was eluted with 4 ml of CE buffer containing 1 M NaCl. The eluted fractions were collected and dialyzed against CE buffer at 4°C. For further purification, the product from SP-Sepharose was applied to a Superdex 200 column (Amersham Pharmacia Biotech) equilibrated with CE buffer. The identity of the protein was determined by Coomassie blue staining or Western blot analysis using rabbit anti-LysN antibody, anti-penta-His antibody (Qiagen), or patient serum infected with HCV. Finally, the concentration of the protein was measured by the Bradford method.
Preparation of RNAs for in vitro assay.
The plasmid pSP3a was constructed by cloning the gene corresponding to NS3 to the 3′ UTR of a type 3a HCV isolate into pBR322 (New England Biolabs) (unpublished data). However, the gene for the 3′ UTR based on the sequence of a type 1b isolate was synthesized artificially and ligated to the end of the coding region because the sequence corresponding to the 3′ UTR of a type 3a isolate was not yet elucidated clearly. For preparation of RNAs for in vitro assay, various DNA fragments were amplified from pSP3a using Deep Vent DNA polymerase (New England Biolabs) for the PCR. The construction of the RNAs for the binding assay and the RdRp assay is shown in Fig. 1, and the primer sets for PCR amplification are summarized in Table 1. The PCR products were identified on the agarose gel and eluted with a Qia-quick PCR purification kit (Qiagen). Each forward primer was designed to contain the T7 promoter sequence for in vitro transcription by T7 RNA polymerase. The in vitro transcription reaction was carried out using a MEGAscript T7 kit (Ambion). The RNA transcripts were identified and purified by electrophoresis on a 5% polyacrylamide-7 M urea gel. The concentrations of the RNAs were determined by measuring the optical density at 260 nm.
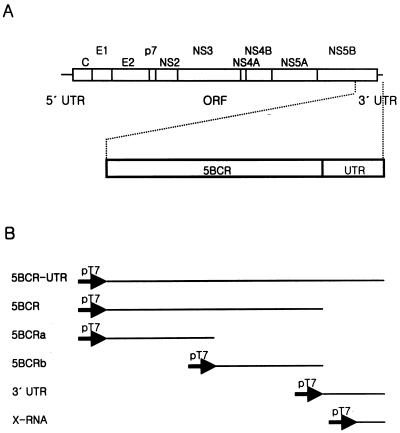
FIG. 1.
HCV genome structure and RNA templates for in vitro assay. (A) The HCV genome organization is presented with 5′ and 3′ UTRs (solid lines) and the ORF (open box). The polyprotein cleavage products are indicated. A detailed view of the RNA template domains containing a part of NS5B and the 3′ UTR is drawn below the full viral genome. 5BCR, one-third of NS5B from the 3′ terminus (nt 8760 to 9405 of the genome); UTR, 3′ UTR (nt 9406 to 9588 of the genome). (B) Summary of template constructs used in this study. All of the RNAs were transcribed directly from the PCR products which were amplified from plasmid pSP3a using the specific primers. The solid arrows indicate the promoter for the T7 RNA polymerase.
TABLE 1.
Oligonucleotides used for PCR amplification
| Primer | Sequence (5′ to 3′)a | Positionb | Sensec |
|---|---|---|---|
| T7-8760 | ATTAATACGACTCACTATAGGGGAGGAGGTACTATTAC | 8760-8778 | + |
| T7-9080 | ATTAATACGACTCACTATAGGGTCGCGGGGACACTCAGG | 9080-9099 | + |
| T7-9406 | ATTAATACGACTCACTATAGGGCTGGTAAGATAACACTCCA | 9406-9425 | + |
| T7-9491 | ATTAATACGACTCACTATAGGGTGGCTCCATCTTAGCCC | 9491-9509 | + |
| 9079 | TATTGAGCTCTACTGGAGAGTAAC | 9056-9079 | − |
| 9405 | TCACCGAGCTGGCAGGAGAAAGATG | 9381-9405 | − |
| 9588 | ACTTGATCTGCAGAGAGGCCAGTATC | 9563-9588 | − |
In the chimeric oligonucleotides, the T7 promoter sequence and extra-G sequence for high efficiency of in vitro transcription are shown in italics.
In the HCV type-3a nucleotide sequence. The numbering includes the poly(U-U/C) tract of the 3′ UTR, which is peculiar to the individual isolate used in our study.
The polarities of HCV-specific primers are indicated as either the HCV genome RNA sense (+) or its complement (−).
To block the 3′-OH group of RNAs, the templates were treated with NaIO4 (Sigma), extracted with phenol-chloroform-isoamyl alcohol, and precipitated with ethanol (3). After being washed several times with 70% ethanol, the RNAs were eluted by gel filtration using Quick Spin RNA columns (Roche Molecular Biochemicals). The degree of oxidation was measured by labeling the RNA templates, before and after treatment with NaIO4, with [α-32P]pCp (NEN Life Science Products) and T4 RNA ligase (Promega) according to the manufacturers' instructions.
Labeling of RNA.
For labeling the 5′ ends of RNAs with 32P, the transcripts were dephosphorylated with shrimp alkaline phosphatase (Roche Molecular Biochemicals) and phosphorylated with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (3,000 Ci/mmol; NEN Life Science Products). The internally labeled RNAs were obtained by transcription in the presence of [α-32P]CTP (3,000 Ci/mmol; NEN Life Science Products), as recommended in the protocols and applications guide (Promega). All of the labeled RNAs were purified from the denaturing gel electrophoresis.
Preparation of RNA ladder.
To generate the alkaline-hydrolyzed ladder as a marker, the end-labeled RNA and 1 μg of E. coli tRNA were incubated in 10 μl of 50 mM Na2CO3-NaHCO3 buffer (pH 9.5) at 95°C for 5 min. The hydrolyzed RNA was mixed with an equal volume of gel loading buffer II (Ambion).
To make a G ladder with RNase T1, the end-labeled RNA and 1 μg of E. coli tRNA were denatured in 10 μl of 25 mM sodium citrate containing urea dye (7 M urea, 1 mM EDTA, 0.03% xylene cyanol, and 0.03% bromophenol blue). It was digested by 0.1 to 0.5 U of RNase T1 (Ambion) at 55°C for 15 min, and then it was loaded directly onto a denaturing polyacrylamide gel.
Gel mobility shift assay.
The internally 32P-labeled 5BCR RNA (2 fmol) was mixed with the unlabeled competitor RNA, and then it was denatured in the binding buffer (10 mM HEPES [pH 7.6], 0.3 mM MgCl2, 40 mM KCl, 5% glycerol, and 1 mM DTT) by heating it at 75°C for 3 min and quickly chilling it on ice for 5 min. Then the renatured RNA mixture was incubated with 0.8 pmol of LysN or LysN-NS5B in a 20-μl reaction volume at 30°C for 15 min. After incubation, half of the mixture was loaded directly onto a nondenaturing 4% polyacrylamide (79:1 acrylamide-bisacrylamide) gel. The gels were run at 10 to 13 mA for 2 h by cooling them with flowing water in a 0.25× Tris-borate-EDTA electrophoresis buffer. After electrophoresis, the gel was fixed, vacuum dried, and subjected to autoradiography.
Footprinting assay.
The RNA (20 fmol) 32P-labeled at its 3′ end was folded in a 10-μl binding buffer by heating it at 75°C for 3 min and quickly cooling it on ice for 5 min. Then it was incubated with increasing amounts of NS5B protein at 30°C for 15 min. After incubation, the RNA was digested with 0.1 U of RNase T1 (Ambion) at 30°C for 15 min. Samples were mixed with equal volumes of gel loading buffer II, denatured at 75°C for 2 min, and then analyzed by electrophoresis on a 5% denaturing sequencing gel.
RdRp activity assay.
The RdRp reactions were performed in a 15-μl volume with 20 mM HEPES (pH 8.0), 1.5 mM MnCl2, 100 mM ammonium acetate, 1 mM DTT, 500 μM GTP, 250 μM (each) ATP and UTP, 5 μM CTP, 10 μCi of [α-32P]CTP (NEN Life Science Products), 12 pmol of RNA template, 10 U of recombinant RNasin (Promega), and 1 to 3 μmol of NS5B. The reaction mixtures were incubated at 30°C for 2 h, extracted with phenol-chloroform-isoamyl alcohol, and then ethanol precipitated. The synthesized RdRp products were separated by electrophoresis on 4 to 8% denaturing polyacrylamide (19:1 acrylamide-bisacrylamide)-7 M urea gels.
RESULTS
Expression and purification of recombinant HCV NS5B.
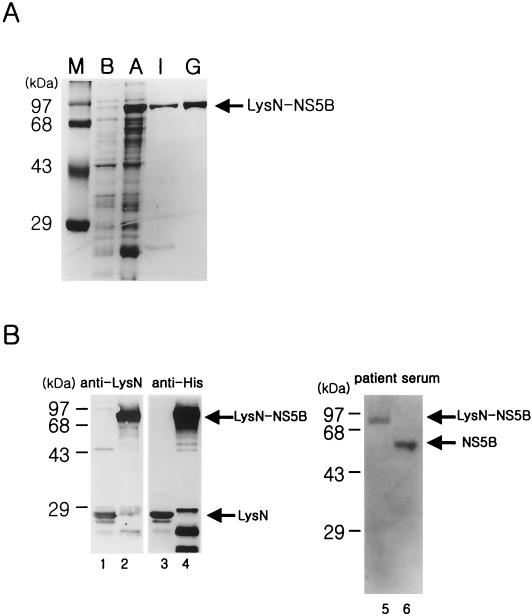
The HCV NS5B protein was expressed from the plasmid pLysN-NS5B in E. coli by induction with IPTG (see Materials and Methods). The poor solubility of this protein was resolved by truncating 20 amino acids from the C terminus and by introducing the N-terminal domain of LysS protein (LysN) as a fusion partner. The soluble fraction of recombinant NS5B was eluted by SP-Sepharose ion-exchange chromatography. For further purification, the partially purified protein was applied to a Superdex 200 column as a gel filtration step. The LysN-NS5B protein of 83 kDa was homogeneous, as judged by Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 2A). The identity of the recombinant protein was confirmed by Western blot analysis (Fig. 2B). The LysN protein of 21 kDa and the LysN-NS5B protein of 83 kDa, eluted from a metal affinity column and an ion-exchange column, respectively, were identified with rabbit anti-LysN antibody and the commercial anti-His antibody (Fig. 2B, lanes 1 to 4). The identity of the final purified protein was also confirmed by immunoassay with the HCV-infected patient serum. The highly purified NS5B protein expressed in the insect cell was used as a positive control (Fig. 2B, lanes 5 and 6).
FIG. 2.
Expression of recombinant HCV NS5B protein in E. coli. (A) The HCV LysN-NS5B protein eluted from an ion-exchange and a gel filtration column was subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and visualized by Coomassie blue staining. Lanes: M, size marker; B, cell extract before induction; A, cell extract after induction with IPTG; I, eluted fractions from an ion-exchange column; G, eluted fraction from a gel filtration column. (B) Western blot analysis of the purified proteins was performed with various antibodies. Lanes 1 and 3, LysN protein from a metal affinity column detected with rabbit anti-LysN antibody and anti-His antibody, respectively; lanes 2 and 4, partially purified LysN-NS5B protein from an ion-exchange column with rabbit anti-LysN antibody and anti-His antibody, respectively; lane 5, finally purified LysN-NS5B protein expressed in E. coli with patient serum; lane 6, purified NS5B protein generated from a baculovirus expression system in the insect cells as a positive control. Both of the proteins in lanes 5 and 6 were purified from a gel filtration column.
The interaction between HCV NS5B polymerase and viral RNA.
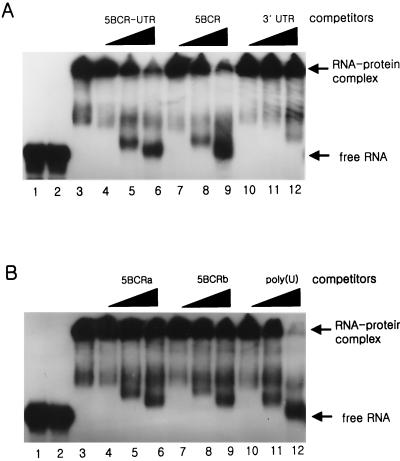
To determine whether the HCV genomic RNA interacts with NS5B, we first performed the gel mobility shift assay (GMSA) (Fig. 3). The probe and the competitor RNAs for this assay were synthesized by in vitro transcription directly from the PCR products (Table 1) (see Materials and Methods), and their localization on the viral genome is summarized in Fig. 1. Because it was reported that the 3′ end of the NS5B coding region RNA bound to NS5B with strong affinity, we decided to confirm the interaction between them (4). In our experiment, the target RNA for the binding assay was extended farther upstream from the previously reported RNA and was designed to contain the final one-third of the NS5B coding region. From the GMSA, it was observed that the viral RNA bound to the NS5B protein but not to LysN, which was used as a fusion partner at the N terminus of the NS5B protein (Fig. 3A, lanes 2 and 3). Most of the RNA-protein complex was retarded at the loading well on the native polyacrylamide gel. It might be that the complex was formed by the assembly of multiple molecules of protein to the probe RNA. To compare the binding affinity of this coding region RNA with that of the 3′ UTR, we performed a competition assay with the unlabeled 5BCR-UTR, 5BCR, and 3′ UTR RNAs (Fig. 3A). The products of the complexes were drastically diminished by increasing amounts of the unlabeled competitor of the 5BCR-UTR or 5BCR RNA with similar efficiencies (Fig. 3A, lanes 4 to 9). In contrast, the 3′ UTR showed a poor competition effect for the interaction of 5BCR RNA and NS5B (Fig. 3A, lanes 10 to 12). This result indicates that the 5BCR RNA rather than the 3′ UTR binds to NS5B protein preferentially and that the 3′ UTR has little affect on the interaction between them either in cis or in trans.
FIG. 3.
Gel mobility shift assay of HCV NS5B protein and the viral RNA. The binding assay was conducted with HCV NS5B protein and the 5BCR RNA as a probe by increasing the amounts of competitor RNAs. (A) The competitor RNAs were the unlabeled 5BCR-UTR, 5BCR, and 3′ UTR RNAs. (B) The competitor RNAs were the 5BCRa, 5BCRb, and homopolymer of poly(U) RNAs. Lanes 1, labeled 5BCR RNA as a marker for free RNA; lanes 2, probe RNA with LysN; lanes 3, probe RNA with NS5B. In the mixture of NS5B and the probe RNA, the unlabeled competitor RNAs of 1- (lanes 4, 7, and 10), 5- (lanes 5, 8, and 11), and 10-fold (lanes 6, 9, and 12) molar excess to the probe RNA were added. The products of the RNA-protein complex and free RNA are indicated at the right of each gel.
To determine the binding region in the 5BCR RNA in more detail, it was divided into two molecules, 5BCRa and 5BCRb RNAs, with similar nucleotide lengths. The competition assay with these competitor RNAs showed that both of them still maintained a strong affinity for NS5B (Fig. 3B, lanes 4 to 9). However, it was difficult to decide which segment of the two molecules interacted with NS5B with a higher affinity. Previously, Cheng et al. suggested that the 3′-end RNA of the NS5B coding region, apparently corresponding to the 5BCRb RNA in our experiments, is especially important for interaction with NS5B (4). Therefore, it was surprising to find that another coding region RNA, the 5BCRa RNA, also bound strongly to NS5B. Our results show that the NS5B polymerase interacts with the coding region of HCV genomic RNA at multiple binding sites. Moreover, since it has been reported that poly(U) RNA binds to NS5B with the highest efficiency among the homopolymeric RNAs, we carried out the competition assay with poly(U) RNA of an average length of 600 nt to investigate its affinity to NS5B (16) (Fig. 3B). It was observed that both the 5BCR and 5BCR-UTR RNAs interacted with the RdRp as strongly as the poly(U) RNA did.
Mapping of binding sites on the 5BCRa and 5BCRb RNAs by NS5B.
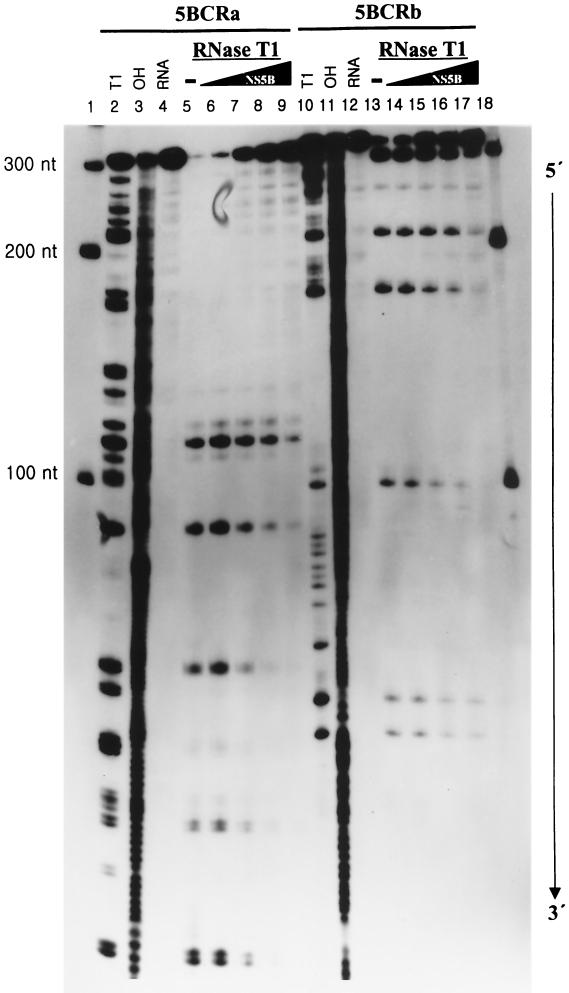
To specify the NS5B-binding sites on the 5BCRa and 5BCRb RNAs, we performed the footprinting assay by treating the RNA-protein complex with RNase T1 (Fig. 4). Each of the 3′-end-labeled RNAs was folded to make it adopt stable secondary structures by heating and cooling processes in the binding buffer. It was incubated with NS5B protein in increasing amounts, and then the RNA-protein complex was treated with RNase T1, which digests the G residues on the single-stranded region. As expected from the previous results of GMSA, the RdRp protein bound to both the 5BCRa and 5BCRb RNAs. However, the recognition site for HCV NS5B was not localized in any specific region on these RNAs. The protected sites ranged from about 100 up to 200 nt from the 3′ end of each RNA template. Even though the recognition sites on the RNAs were widely distributed, they were extended in a 3′-to-5′ direction with increasing amounts of protein. From this result, it can be expected that more than two molecules of the NS5B protein bind to its genomic RNA in a sequential mode. This property of HCV NS5B might make it difficult to select the cis-acting element on the genomic RNA with a high affinity in vitro.
FIG. 4.
Footprinting analysis of 5BCRa and 5BCRb RNAs with NS5B. The 3′-end-labeled RNAs (20 fmol) were incubated at 30°C for 15 min in the absence (lanes 4 and 12) or in the presence (lanes 5 and 13) of RNase T1. The probe RNA was incubated with the NS5B protein by increasing its amount (lanes 6 and 14, 50 fmol; lanes 7 and 15, 100 fmol; lanes 8 and 16, 200 fmol; lanes 9 and 17, 400 fmol of protein) and then digested by RNase T1. The RNA fragments were then resolved on an 8 M urea-5% polyacrylamide gel. Lanes 1 and 18, RNA size markers; lanes 2 and 10, RNase T1-digested products under denaturing conditions; lanes 3 and 11, fragments generated by partial alkaline hydrolysis. The numbers on the left side of the gel indicate the length of the RNA marker.
De novo synthesis of RNA by NS5B polymerase on viral RNA templates.
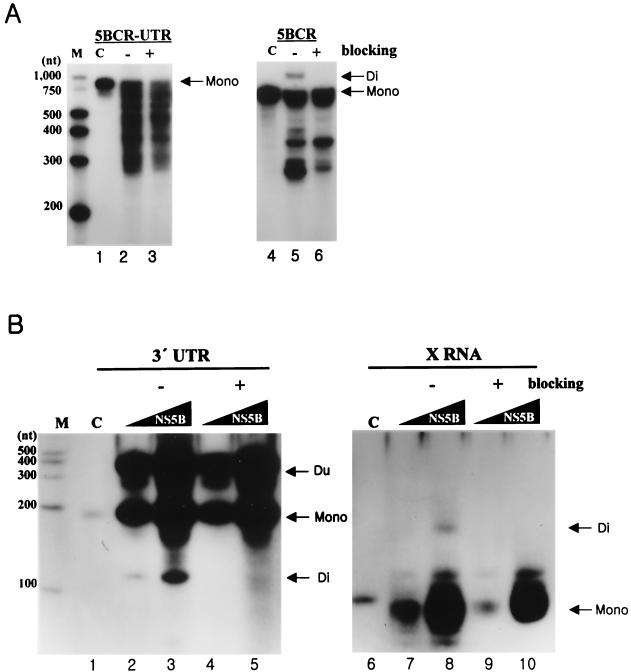
In addition to the binding assay, the question of whether the purified recombinant NS5B could direct RNA synthesis on the viral genomic RNA was addressed. It had been previously reported that HCV NS5B produces two different major RNA products, the monomer and the dimer. The monomer-size RNA, which is identical in length to the input RNA template, can be generated by de novo initiation or by the addition of some extra nucleotides at the 3′ terminus of the template. The dimer-size RNA is generated by a copy-back mechanism (3, 17, 19, 30). We performed the RdRp reaction on the 5BCR UTR and 5BCR RNAs (Fig. 5A). As expected, the NS5B protein synthesized the transcripts regarded as the monomer or the dimer on the denaturing gel (Fig. 5A, lanes 2 and 5). To determine that the monomer is a product of a de novo initiation mechanism, the 3′ end of each RNA template was oxidized with sodium periodate. Our result indicates that the monomer-size RNA was synthesized not by TNTase activity or by a copy-back mechanism but by a de novo initiation mechanism directly (Fig. 5A, lanes 3 and 6). Moreover, the 5BCR RNA directed synthesis of the dimer RNA, which migrated more slowly than the input RNA. However, the amount of the dimer product of the 5BCR-UTR RNA was too small to detect on the denaturing gel (Fig. 5A, lane 2). In an additional experiment, the dimer product of the 5BCR-UTR RNA was generated as the concentration of protein increased. The reaction efficiency for the monomer and the dimer depended on the specific ratio of the template RNA to the protein (data not shown). From the results, it can be proposed that the efficiency of the RdRp reaction by a copy-back or a de novo synthesis mechanism is dependent on three factors: the nucleotide sequence composition, the secondary structure of the 3′ end of the template RNA, and the concentration of protein relative to the substrate. In this case, the reaction efficiency with the 5BCR-UTR RNA was lower than that with the 5BCR RNA. This means that the 3′-UTR RNA does not serve as a good template for the RdRp reaction.
FIG. 5.
Analysis of RdRp reaction products from viral RNAs as templates. (A) The RdRp reaction on 5BCR-UTR RNA and 5BCR RNA. The template of each product is indicated at the top of the panel. The reaction was performed with 0.1 pmol of RNA template and 0.1 to 0.3 pmol of NS5B by adding a recombinant NTP mixture of 500 μM GTP, 250 μM ATP, 250 μM UTP, 100 μM CTP, and 0.5 μCi of CTP in the 15-μl reaction volume. Lane M, size marker; lanes 1 and 4, internally labeled input templates; lanes 2 and 5, RdRp products without (−) blocking the 3′-OH group of the template RNA; lanes 3 and 6, RdRp products with (+) blocking of the 3′-OH group of the template RNAs. Mono, the monomer RNA; Di, the dimer RNA. (B) RdRp reaction on 3′ UTR RNA and X RNA. The template of each product is indicated at the top of the gel. The RdRp reaction was performed with 12 pmol of RNA template by increasing the amounts of NS5B from 1 (lanes 2, 4, 7, and 9) to 3 pmol (lanes 3, 5, 8, and 10). Other contents for RNA synthesis are given in Materials and Methods. Lane M, size marker; lanes 1 and 6, internally labeled input template; lanes 2, 3, 7, and 8, labeled products without (−) blocking the 3′-OH group of the template RNAs; lanes 4, 5, 9, and 10, labeled products with (+) blocking of the 3′-OH group of the template RNAs. Mono, the monomer RNA; Di, the dimer RNA; Du, the duplex product by hybridization of the monomer and the template RNAs.
Some reports have suggested that NS5B utilizes not only its genomic RNA but also non-HCV genomic RNA as a template (3, 18, 19, 30). However, we know that the 3′ end of the HCV genomic RNA is indispensable for the NS5B polymerase to recognize it for amplification of the viral RNA in vivo, even though it is a poor element for interacting with the NS5B protein and for directing RNA synthesis. To determine whether this recombinant LysN-NS5B can also initiate RNA synthesis on the separate 3′-end fragments of viral RNA, we examined the RdRp reaction on the 3′-UTR RNA and X RNA, respectively (Fig. 5B). The NS5B protein could synthesize the major RNA products by de novo initiation. When the 3′-UTR RNA was used as a template, an additional band migrating more slowly than that of the input RNA was detected on the denaturing gel (Fig. 5B, lanes 2 to 5). This product is suspected to be a duplex because of a tight hybridization of the transcripts and the excess templates in the reaction mixture. A faster-migrating product than that of the input RNA is regarded as intramolecularly initiated by a copy-back mechanism. This dimer product disappeared after the 3′ OH of the RNA template was modified. As in our results, this faster mobility of the dimer product than that of the monomer was also detected in previous reports (3, 17). Also, on the sequencing gel, the monomer and the dimer were identified with the precise sizes expected (data not shown).
In the case of X RNA, the length of the major de novo synthesis product was shorter than the 98 nt of the input RNA (Fig. 5B, lanes 7 to 10). This result is consistent with the previous experimental results of Oh et al. (19). They proposed that this product is a de novo synthesis product initiated from the internal region. In our experiments, NS5B also synthesized a dimer product about 160 nt in length on X RNA by a copy-back mechanism (Fig. 5B, lane 8). These results mean that the 3′ UTR or X RNA can direct RNA synthesis at a specific initiation site(s), even though they meet few template requisites and have low affinity to the NS5B protein.
RNA synthesis is initiated by HCV NS5B from nucleotides C86 and U87 on stem I of X RNA.
We tried to determine the initiation site(s) of de novo synthesis on the 3′ UTR or X RNA, because the synthesis of negative-strand viral RNA begins at this region. However, the products from the 3′ UTR RNA diffused on the sequencing gel, so we could not determine its dominant initiation point(s) (data not shown). This result seems to imply that RNA synthesis occurs by slippage or by the template switching mechanism on the poly(U-U/C) tract (1, 13). However, it is believed that the initiation site(s) of de novo synthesis on the 3′ UTR is identical to that of X RNA because the stable secondary structure of X RNA is known to be conserved in the entire 3′ UTR (11).
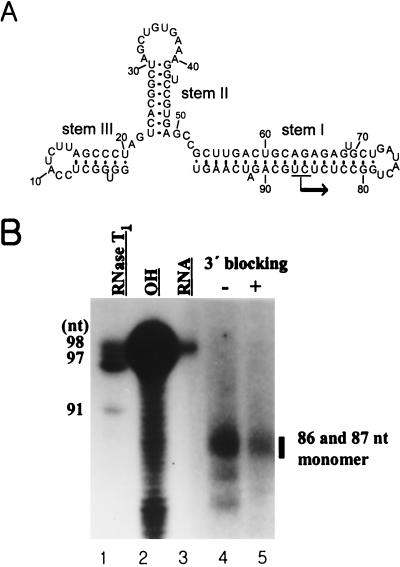
Previously, it was reported that HCV NS5B initiated de novo RNA synthesis at the 3′ end of X RNA and generated a product of the same length as the input RNA (30). In contrast, other evidence was also reported showing that the length of the major product of de novo synthesis on X RNA is 78 nt, which is smaller than the 98 nt of the template RNA (19). In this case, it was suggested that RNA synthesis was initiated not at the 3′ end but at nt 78 of loop I. From these controversial results, we intended to elucidate the identity of the RdRp reaction product and its initiation point(s) on X RNA. First, we analyzed the secondary structure of X RNA by RNase mapping (data not shown). As expected, X RNA showed a conformation identical to that of those previously reported (11, 15, 23). The template X RNA has a stable secondary structure with three stems and loops (Fig. 6A). As mentioned above, the RdRp product of de novo synthesis on X RNA was smaller than the input RNA (Fig. 5B, lanes 6 to 10). We assessed the exact size of this product by analyzing it on the sequencing gel (Fig. 6B). Surprisingly, the transcripts from X RNA showed the major products to be 86 and 87 nt long. Also, we could infer that they were generated by de novo synthesis because they were still produced after 3′-OH modification of the template RNA. However, this modification reduced the reaction efficiency somewhat. Interestingly, the initiation nucleotide complementary to these sites corresponds to GTP or ATP. Additionally, the 78- and 82-nt-long products were also detected from the RNA polymerization reaction, but the quantities were not dominant.
FIG. 6.
Determination of the initiation sites of RNA synthesis on X RNA by HCV NS5B. (A) Secondary structure of X RNA. The major initiation sites of RNA synthesis are indicated by the arrow on stem I. (B) An 8 M urea-5% polyacrylamide gel electrophoresis autoradiogram showing the RNA product of HCV NS5B using the X RNA template. The bar indicates the major RdRp products on the gel. The numbers on the left give the length of the 5′-labeled X RNA partially digested by RNase T1 (lane 1). The numbers on the right give the lengths of the RNA synthesis products of NS5B. Lane 2, partial alkaline hydrolysis of the 5′-labeled X RNA; lane 3, template X RNA labeled internally with [α-32P]CTP as a marker; lanes 4 and 5, RdRp product on the X RNA template before (−) and after (+) the 3′ end was blocked.
Stem I of X RNA regulates the initiation specificity of RNA synthesis by NS5B.
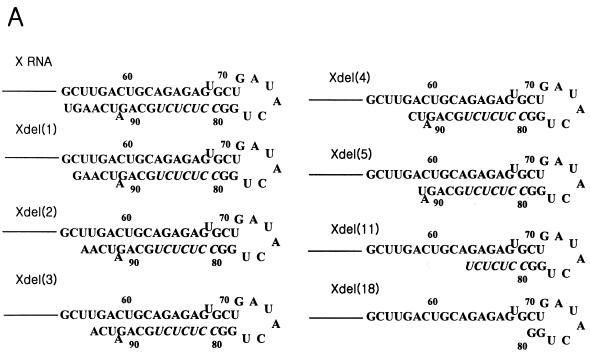
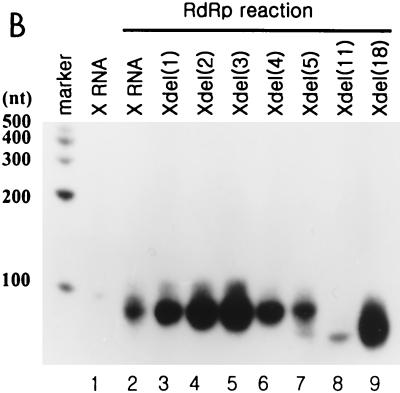
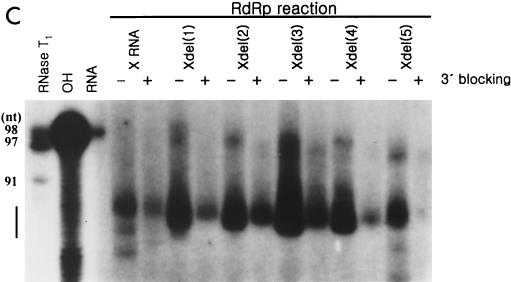
To confirm that de novo RNA synthesis was selectively initiated, we prepared the deletion mutants by removing nucleotides from the 3′ end of X RNA (Fig. 7A). The RdRp reaction on each RNA was carried out, and their products were analyzed on the slab gel or sequencing gel (Fig. 7B and C). The mutants, which had up to 5 nt serially deleted from the 3′ end generated products nearly equivalent to that of wild-type X RNA (Fig. 7B, lanes 2 to 7). However, de novo synthesis on the deletion mutant Xdel (11) and Xdel (18) RNAs showed that the initiation site shifted farther from the 3′ end, and they generated reaction products shorter than that of X RNA or the Xdel (1) to Xdel (5) RNAs (Fig. 7B, lanes 8 and 9). In the case of the Xdel (5) RNA template, the RdRp product, like that of Xdel (11) or Xdel (18) RNA, was also detected, but with weak intensity, on the gel (Fig. 7B, lanes 7 to 9). We expect that the length of stem I in particular plays an important role in regulating the specificity of the RdRp reaction. Furthermore, the deletion mutants had different transcription efficiencies. Among the Xdel (1) to Xdel (5) deletion mutants, Xdel (3) RNA directed the most efficient RdRp reaction. The transcription efficiency of this mutant was even higher than that of wild-type X RNA. This means that the length and the specific nucleotide sequence of stem I determine not only the specificity but also the efficiency of viral RNA replication.
FIG. 7.
RdRp reaction on the deletion mutants of X RNA by HCV NS5B. (A) Sequences of stem I of the deletion mutant RNAs. The nucleotides in italics indicate the pyrimidine-rich region in stem I. (B) RdRp reaction products of wild-type X RNA and its deletion mutants on a 7 M urea-8% polyacrylamide gel. The length of the marker RNA is indicated at the left of the gel. Lane 1, template X RNA with internally incorporated 32P; lanes 2 to 9, RdRp products of X, Xdel (1), Xdel (2), Xdel (3), Xdel (4), Xdel (5), Xdel (11), and Xdel (18) RNAs, respectively. (C) An 8 M urea-5% polyacrylamide gel electrophoresis autoradiogram showing the RNA products of HCV NS5B using X RNA and the deletion mutant RNA templates before (−) or after (+) treatment with NaIO4. RNase T1, 5′-labeled X RNA partially digested by RNase T1; OH, partial alkaline hydrolysis of 5′-labeled X RNA; RNA, template X RNA labeled internally with [α-32P]CTP as a marker. The numbers on the left give the length of the RNA, and the bar at the left of the gel indicates the pyrimidine-rich region of X RNA.
To define the exact sizes of the replication products of some deletion mutants and to elucidate that they were generated only by a de novo synthesis mechanism, the RdRp reaction was carried out before or after modifying the 3′ ends of the template RNAs and their products were analyzed on the sequencing gel (Fig. 7C). The reaction products of these deletion mutants showed similar mobilities on a denaturing polyacrylamide slab gel, but their lengths were slightly different on a sequencing gel. The major products from X RNA were 86 and 87 nt long, those from Xdel (1) RNA were 84 and 85 nt, those from Xdel (2) RNA were 81 to 84 nt, those from Xdel (3) RNA were 84 to 86 nt, those from Xdel (4) RNA were 82 and 83 nt, and those from Xdel (5) RNA were 83 and 84 nt. Altogether, the initiation sites of RNA polymerization for these products were concentrated in the pyrimidine-rich region (nt 81 to 87) of stem I. This result strongly suggests that this region offers the initiation sites for NS5B. It is also correlated with the fact that NS5B can direct de novo synthesis on poly(C) and poly(U) RNA templates at high concentrations (≥50 μM) of GTP and ATP without the oligonucleotide primer (17). We propose that the initiation site(s) on the viral RNA is not fixed at a single site but wobbles in the pyrimidine-rich bloc. Moreover, all of the RNA templates which were modified at their 3′ OH with NaIO4 directed RNA synthesis of products 1 to 2 nt longer than the products from the unmodified RNA templates (Fig. 7C). The reason for this result has not yet been elucidated; however, we expect that the ribose 3′ OH of the 3′-terminal nucleotide may also regulate RdRp reaction specificity.
The position of the GC base pair at the terminus of stem I determines the efficiency of the RdRp reaction.
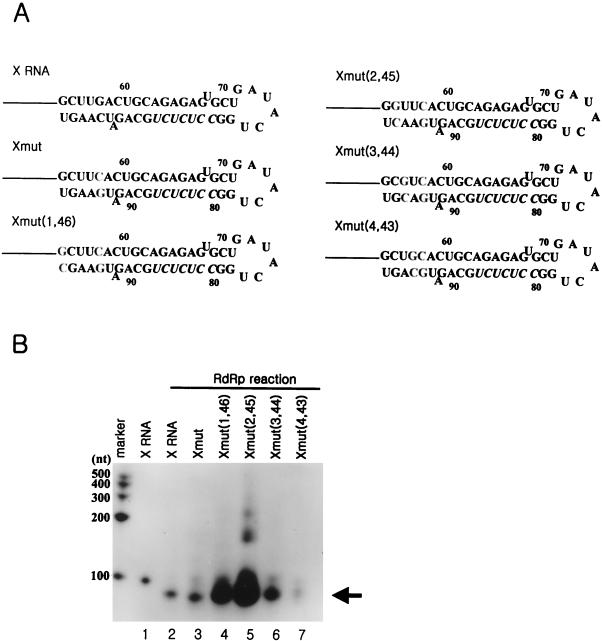
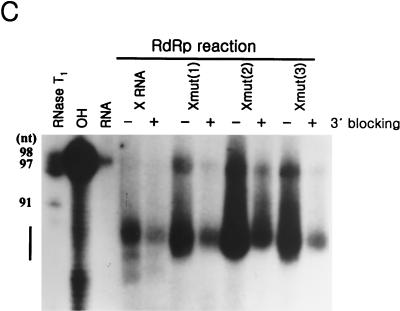
In the RdRp reactions with some of the deletion mutants of X RNA, which direct the transcription initiation in the pyrimidine-rich region, it was observed that the reaction efficiency was maximized at the Xdel (3) RNA. However, this RdRp reaction product suddenly disappeared if >5 nt was deleted (Fig. 7B). From the deletion analysis of X RNA, it could be suggested that the positions of some sequences might play an important role in determining the efficiency of initiation for RNA synthesis. We speculated that one of the most important elements might be the GC base pair at the fifth position from the end of stem I, because deletion of this nucleotide reduced the amount of reaction product and induced initiation at another site. To detect the effect caused by this GC base pair, it was changed complementarily and then arranged at the first to the fifth positions from the end of stem I by serial PCR mutations (Fig. 8A). We identified by computer prediction that all of these mutants contained secondary structures identical to that of X RNA, and they were used for directing the RNA polymerization reaction by the HCV NS5B polymerase (Fig. 8B). The reaction efficiency of the Xmut RNA was similar to that of the wild-type X RNA; however, Xmut (1,46), Xmut (2,45), and Xmut (3,44) directed the RNA synthesis with higher efficiency than X RNA did. Among these mutants, Xmut (2,45) was the most adjustable RNA template for directing de novo RNA synthesis. As the GC base pair shifted to the inner side from the second position at the end of stem I, the reaction efficiencies of Xmut (3,44) and Xmut (4,43) drastically decreased. We expect that although the RdRp reaction does not initiate at the GC base pair in the penultimate position from the end of the stem, this element is essential to satisfy the template requirements for the NS5B protein through a direct or an indirect interaction.
FIG. 8.
effect of the position of the GC base pair at stem I on the RdRp reaction. (A) Sequence identities of the mutant RNAs which were generated by shifting the GC base pair from the end to the inside of stem I. The nucleotides in boldface indicate the mutated sequences. Xmut RNA, as a control RNA template, was produced by reversing the GC base pair at the fifth position of the terminus of stem I. Xmut (1,46) RNA was generated from Xmut RNA by regenerating the GC base pair at the end of stem I; Xmut (2,45) RNA was generated from Xmut RNA by regenerating the GC base pair at the second position from the end of stem I; Xmut (3,44) RNA was generated from Xmut RNA by regenerating the GC base pair at the third position from the end of stem I; Xmut (4,43) RNA was generated from Xmut RNA by regenerating the GC base pair at the fourth position from the end of stem I. (B) RdRp reaction products using the mutant RNA templates which contain the GC base pair at different sites of stem I. The length of the marker RNA is indicated at the left of the gel. The monomer RNA produced by de novo synthesis is indicated by the arrow. Lane 1, internally labeled X RNA template; lane 2, RdRp product directed by X RNA; lanes 3 to 7, RdRp products of Xmut, Xmut (1,46), Xmut (2,45), Xmut (3,44), and Xmut (4,43) RNAs, respectively.
The efficiency of the RdRp reaction was affected by the secondary structure of stem I of X RNA.
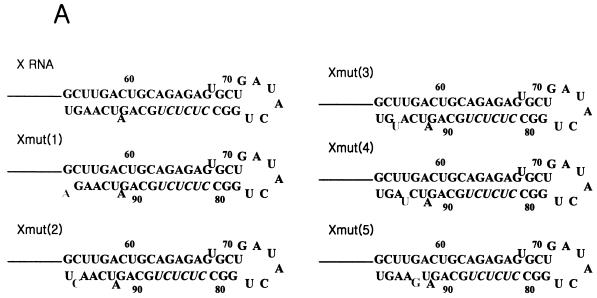
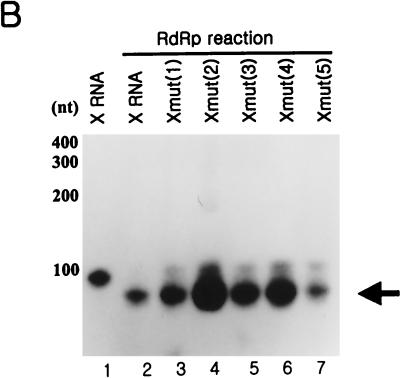
To research the structural effect of the template RNA for directing de novo synthesis, we prepared some mutant RNAs on X RNA by creating the bulge at the terminal region of stem I serially. In these mutant RNAs, the nucleotides of a nonviral sequence were introduced by replacing the wild-type sequence with the complementary one for breaking the base pairs (Fig. 9A). All of them were tested for generating the radiolabeled products by HCV NS5B (Fig. 9B). As expected, they were able to direct de novo initiation, and the RdRp products directed from them had sizes similar to that of X RNA. Despite these findings, these RdRp products had different reaction efficiencies. The Xmut (2) template in particular directed RNA synthesis with the highest efficiency. This template was generated by replacing G with C at nt 97 and finally making the bulge at the penultimate position from the end of stem I. Therefore, we speculated that both the bulge structure and the nucleotide C at this point might play critical roles in increasing the synthesis efficiency of NS5B. In addition, the bulge at the fourth position of the Xmut (4) RNA also efficiently directed the proper initiation of RNA synthesis. In order to determine the precise initiation sites of the de novo RNA synthesis of some mutants, we analyzed the RdRp products on the sequencing gel, and we detected that the RNA synthesis on these mutant RNAs initiated in the pyrimidine-rich region (Fig. 9C). Finally, we propose that the bulge positioned at the second and fourth nucleotide pairs from the end of the stem efficiently enhances the RdRp reaction.
FIG. 9.
RdRp reaction of the point mutants of X RNA with HCV NS5B. (A) Nucleotide sequences in stem I of the mutant RNAs. The characters in boldface represent the elements for rendering the bulge structure. The nucleotides in italics indicate the pyrimidine-rich region in stem I. (B) Products of RdRp reaction on wild-type X RNA and its point mutants on a 7 M urea-8% polyacrylamide gel. The length of the marker RNA is indicated at the left of the gel. Lane 1, template X RNA internally labeled with 32P; lanes 2 to 7, RdRp products of X, Xmut (1), Xmut (2), Xmut (3), Xmut (4), and Xmut (5) RNAs, respectively. (C) An 8 M urea-5% polyacrylamide sequencing gel electrophoresis autoradiogram showing the de novo synthesis products of X RNA and the point mutant RNAs before (−) or after (+) treatment with NaIO4. The template RNAs for directing RNA synthesis by NS5B are indicated at the top of the gel. RNase T1, 5′-labeled X RNA partially digested by RNase T1; OH, partial alkaline hydrolysis of the 5′-labeled X RNA; RNA, template X RNA labeled internally with [α-32P]CTP as a marker. The numbers on the left give the length of the RNA, and the bar at the left of the gel indicates the pyrimidine-rich region of X RNA.
DISCUSSION
Several reports have explained the biological activity of HCV NS5B on the various RNA templates. These recent studies increased the understanding of the mechanism of HCV replication, in spite of the lack of a consistent virus culture system. In this study, we presented the interaction between HCV RNA and its polymerase and the template requirements of HCV NS5B for more efficient RNA synthesis on the minimized viral RNA.
In the binding assay, HCV NS5B bound to its coding region RNA (named 5BCR RNA) competitively. It was previously reported that the fragment of the latter half of this RNA had a stronger affinity than the 3′ UTR with the RdRp (4). Interestingly, we detected that not only the latter half but also the first half of the 5BCR RNA bound to the HCV polymerase with similar affinities. Moreover, the binding domain on the RNA was not localized at a specific site. Thus, we determined the following: the polymerase interacted with the viral RNA through a wide range of 100 to 200 nt with a preference for 3′ to 5′. This nonspecific but strong interaction is consistent with the general characteristic of HCV polymerase of not discriminating its genomic RNA from other RNAs as its template.
Nevertheless, we expected the binding affinity between HCV NS5B and its coding region RNA to be changed by introducing the 3′ UTR at its 3′ end. This is because it is well known that a homopolymer of poly(U) RNA has a property of strong binding to the NS5B polymerase, and the HCV genomic RNA contains U-rich sequences at its poly(U-U/C) tract of the 3′ UTR. However, the 3′ UTR did not affect the interaction between the coding region RNA and NS5B. The reason for this unexpected result was speculated to be that the arrangement of the U content of the 3′ UTR is not successive because of tandem C or G distribution. On the other hand, poly(U) has the pure-U content of 600 nt. In other words, the lengths of the successive U contents may determine the affinity of the 3′ UTR to viral RdRp. Thus, the possibility that some 3′-UTR RNAs with enough continuous U repeats bind to viral polymerase with high affinity cannot be excluded. Furthermore, we speculate that the biological role of the 3′ UTR is to regulate the viral-RNA affinity to NS5B for initiating de novo synthesis of the negative-strand RNA. Thus, since its affinity is not strong enough to initiate RNA synthesis actively, this may reduce the efficiency of HCV replication by a weak interaction, and finally, it may contribute to the progression and help conserve the chronic state of the liver disease caused by HCV.
From the enzymatic-activity study, we confirmed that the recombinant LysN-NS5B protein, whose NS5B part was constructed by deletion of 20 amino acids at the C terminus, was active and could direct de novo synthesis on the viral RNA. In the RdRp reaction with the 5BCR or 5BCR-UTR RNA, the smaller RNAs, in addition to the full-length product, were also detected as a ladder pattern on the denaturing gel. These additional products might be generated by the scattered internal initiation on the viral RNA template. Interestingly, the presence of the 3′ UTR RNA did not cause any changes in the efficiency of directing the RdRp reaction in vitro. This finding means that the 3′ UTR (or X RNA) has not evolved as the optimal template for the viral polymerase. However, these RNAs could direct de novo RNA synthesis and transcription initiation at 86C and 87U of the pyrimidine-rich region on stem I of X RNA, predominantly. This result was surprising, because it was previously determined that the RNA synthesis on X RNA initiated at the first nucleotide (78U) of the single strand from the 3′ end (19). In our experiment, the additional 78-nt product was also detected, but the initiation at this site was not major.
Recently, a report proposed that the template for HCV NS5B required the stem structure, the single-stranded region at the 3′ end, and the cytidylate (as an initiation nucleotide) in this 3′ single-stranded region (14). However, in considering these requisites of the RNA template for the RdRp reaction, we found that X RNA does not satisfy all of these requirements in its secondary structure or nucleotide composition. This means that any other requirements may be applied to X RNA for the RdRp reaction. According to our experimental results, the native X RNA was appropriate for the NS5B substrate, even though it contains the blunt-ended stem and not the single-stranded region at its 3′ terminus. In this case, C or U in the internal region of the stem was recognized as the initiation nucleotide. Thus, it can be suggested that a nucleotide in the double-stranded region is also available as the initiation site on the template, but the extended deletion of >5 nt from the 3′ end moved the initiation site to another position. Additionally, it was found that if the GC base pair resided at the penultimate position from the end of the stem, the efficiency of the RdRp reaction was maximized. It is speculated that C of the GC base pair offers the interaction site to an RNA binding pocket of NS5B, although it does not serve as an initiation nucleotide. Another finding was that the bulge structure at the second or fourth position from the end of the stem also enhanced the efficiency of RNA synthesis. These data suggest that there are two functional roles of stem I of X RNA: one is to determine the initiation site, and the other is to regulate the efficiency of de novo synthesis.
Despite these findings, it remains an open question whether the immature products are also generated during viral replication. If this internal initiation product is generated, there should be a tool by which the immature product can be repaired to the full-length viral RNA to maintain the genetic identity. The following has been proved regarding the turnip crinkle virus (TCV) transcripts of satC: the TCV transcripts of satC with a deletion of the motif are repaired to wild type in vivo by RdRp-mediated extension of abortively synthesized oligoribonucleotide primers complementary to the 3′ end of the TCV genomic RNA (9). Further study should be undertaken to elucidate if this repair system works with the HCV NS5B polymerase in vivo.
Otherwise, the possibility must be considered that another driving force exists which induces de novo RNA synthesis at the 3′ end of the viral RNA in HCV-infected cells and the maintenance of the full-length genomic RNA. In other words, cellular and/or viral factors may participate in the initiation of an RdRp reaction. Some recent studies have revealed that the cellular proteins, for example, polypyrimidine tract-binding and ribosomal proteins, specifically interact with X RNA and then regulate the translation of the viral polyprotein or viral replication (7, 12, 26, 27). It was also proposed that the HCV NS5B polymerase interacts with16S and 23S cellular RNAs to regulate viral genomic translation (24). Alternatively, the HCV NS3 helicase may unwind X RNA as the adjustable structure for initiating RNA synthesis at the right end of the positive-strand viral RNA (2, 20). Undoubtedly, our report will provide further insight for understanding the biological mechanism of recognition of the viral RNA by the HCV NS5B polymerase and the initiation preference for RNA synthesis on the HCV genomic RNA.
REFERENCES
- 1.Arnold, J. J., and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 274:2706-2716. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, R., and A. Dasgupta. 2001. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3′-terminal sequences of viral positive- and negative-strand RNA. J. Virol. 75:1708-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, J. C., M. F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 6.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, J. W., and R. W. Moyer. 2000. The effects of the conserved extreme 3′-end sequence of hepatitis C virus (HCV) RNA on the in vitro stabilization and translation of the HCV RNA genome. J. Hepatol. 33:632-639. [DOI] [PubMed] [Google Scholar]
- 8.Francki, R. I. B., C. M. Fauquet, D. L. Knudson, and F. Brown. 1991. Classification and nomenclature of viruses: the fifth report of the international committee on taxonomy of viruses. Arch. Virol. Suppl. 2:223-233. [Google Scholar]
- 9.Guan, H., and A. E. Simon. 2000. Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: an activity involved in 3′-end repair of viral RNAs. Proc. Natl. Acad. Sci. USA 97:12451-12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houghton, M. 1996. Hepatitis C virus, p. 1035-1058. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 11.Ito, T., and M. M. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, T., S. M. Tahara, and M. M. Lai. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 20.Paolini, C., R. De Francesco, and P. Gallinari. 2000. Enzymatic properties of hepatitis C virus NS3-associated helicase. J. Gen. Virol. 81:1335-1345. [DOI] [PubMed] [Google Scholar]
- 21.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-959. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 22.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, et al. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka, T., K. Sugiyama, M. Ikeda, A. Naganuma, A. Nozaki, M. Saito, K. Shimotohno, and N. Kato. 2000. Hepatitis C virus NS5B RNA replicase specifically binds ribosomes. Microbiol. Immunol. 44:543-550. [DOI] [PubMed] [Google Scholar]
- 25.Tomei, L., R. L. Vitale, I. Incitti, S. Serafini, S. Altamura, A. Vitelli, and R. De Francesco. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol. 81:759-767. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchihara, K., T. Tanaka, M. Hijikata, S. Kuge, H. Toyoda, A. Nomoto, N. Yamamoto, and K. Shimotohno. 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 71:6720-6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood, J., R. M. Frederickson, S. Fields, and A. H. Patel. 2001. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 75:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 29.Yanagi, M., M. St Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]