Abstract
The utility of the present generation of adenovirus (Ad) vectors for gene therapy applications could be improved by restricting native viral tropism to selected cell types. In order to achieve modification of Ad tropism, we proposed to exploit a minor component of viral capsid, protein IX (pIX), for genetic incorporation of targeting ligands. Based on the proposed structure of pIX, we hypothesized that its C terminus could be used as a site for incorporation of heterologous peptide sequences. We engineered recombinant Ad vectors containing modified pIX carrying a carboxy-terminal Flag epitope along with a heparan sulfate binding motif consisting of either eight consecutive lysines or a polylysine sequence. Using an anti-Flag antibody, we have shown that modified pIXs are incorporated into virions and display Flag-containing C-terminal sequences on the capsid surface. In addition, both lysine octapeptide and polylysine ligands were accessible for binding to heparin-coated beads. In contrast to virus bearing lysine octapeptide, Ad vector displaying a polylysine was capable of recognizing cellular heparan sulfate receptors. We have demonstrated that incorporation of a polylysine motif into the pIX ectodomain results in a significant augmentation of Ad fiber knob-independent infection of CAR-deficient cell types. Our data suggest that the pIX ectodomain can serve as an alternative to the fiber knob, penton base, and hexon proteins for incorporation of targeting ligands for the purpose of Ad tropism modification.
Human adenovirus (Ad) includes at least 47 viral serotypes grouped into six distinct subgroups (A to F) and represents a large family of nonenveloped viruses containing a linear double-stranded DNA genome of approximately 36 kb (26). Ad is composed of multiple copies of 11 structural proteins, 7 of which (II, hexon; III, penton base; IIIa; IV, fiber; VI; VIII; and IX) form the icosahedral capsid, while the other 4 (V, VII, μ, and tp) are packaged with the DNA genome in the viral particle (VP) core (28). Studies of the mechanism of Ad infection have revealed that penton base and fiber proteins are responsible for recognition of cellular receptors and therefore determine viral tropism. Ad infection is initiated by the binding of the globular knob domain of the fiber with the primary cellular receptor (19, 27). A fiber receptor for Ad of subgroups A, C, D, E, and F has been identified as the coxsackievirus group B and Ad receptor, called CAR (3, 24, 29). The major histocompatibility complex-1 α2 subunit was reported as the cellular receptor for subgroup C Ad (15), in addition to CAR and a sialic acid-containing glycoprotein as a receptor moiety for Ad37 of subgroup D (2). Following binding to the fiber receptor, RGD motifs within penton base interact with αvβ integrins and facilitate virus internalization via receptor-mediated endocytosis (13, 21, 32).
Ad is widely used as a vector for both in vitro and in vivo gene delivery due to its ability to infect a variety of cell types (37). However, patterns of viral receptor expression vary between different tissues (10), predicating their susceptibilities to Ad infection. The increased knowledge of the Ad capsid structure combined with an understanding of the biology of virus interaction with cellular receptors has facilitated the development of targeted Ad vectors (6, 7). Genetic engineering of Ad capsid proteins to incorporate targeting ligands has been employed to generate Ad vectors with novel viral tropism that can overcome the limited infectivity associated with deficiency of viral receptors (18, 31). Several heterologous peptide ligands have been successfully engineered into the HI loop (8, 23, 36) and C terminus of fiber (33, 35), the L1 loop of hexon (30), and the RGD loop of penton base (34), resulting in markedly increased efficiency of Ad infection in a variety CAR-deficient cell types. However, the structural properties of the surface-exposed loops of capsid proteins make them suitable only for incorporation of constrained heterologous sequences, and addition of ligands to the C terminus of fiber apparently has size limitations (35).
The present study evaluates the utility of Ad capsid protein IX (pIX) (4, 5) for the purpose of viral tropism modification via genetic incorporation of heterologous peptides. pIX is a minor component of Ad capsid that stabilizes hexon-hexon interactions (11) and is also essential for viral DNA packaging (12). Recent studies have demonstrated that the C terminus of pIX is exposed on the outer surface of the viral capsid (1, 25), suggesting that it could be used as a novel locale for incorporation of targeting ligands. To assess the feasibility of incorporating heterologous sequences into pIX, we engineered Ad vectors encoding recombinant pIX containing either eight consecutive lysines or a polylysine sequence following a C-terminal Flag octapeptide. Here, we demonstrate that modified pIX is incorporated into mature Ad virions and displays Flag-containing carboxy-terminal extensions which are accessible for binding. Incorporation of polylysine, a heparan sulfate binding motif, resulted in significant augmentation of knob-independent Ad infection of CAR-deficient cell types, illustrating compatibility of pIX ectodoman ligand additions with Ad tropism modification strategies. Our results suggest that the pIX ectodoman may be used for targeting ligand incorporation as an alternative to Ad fiber knob, hexon, and penton base.
MATERIALS AND METHODS
Cells.
The 293 human kidney cell line transformed with Ad5 DNA was purchased from Microbix (Toronto, Ontario, Canada). The human breast cancer cell lines AU-565 and GI-101A established from mammary gland adenocarcinomas and human endothelial HUVEC cells were from the American Type Culture Collection (Manassas, Va.). All cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2 in the recommended growth media supplied by Mediatech (Herndon, Va.) containing 10% fetal bovine serum (HighClone, Logan, Utah) and 2 mM glutamine. Infection of the cells with Ad was carried out in infection medium containing 2% fetal bovine serum.
Construction of recombinant plasmids.
To incorporate Flag peptide-coding sequence into the 3′ end of the pIX gene, the oligonucleotides 5′-CTG CCG ATT ATA AGG ATG ACG ATG ACA AGT and 5′-ACT TGT CAT CGT CAT CCT TAT AAT CGG CAG were used. The oligonucleotides were annealed to form a DNA duplex, ligated with BsrGI-DraI (716 bp) and DraI-BstXI (581 bp) DNA fragments of the Ad genome, and then cloned between BsrGI and BstXI sites in the pShuttle plasmid (14), generating pShlpIXflag. To introduce a unique restriction site into the 3′ end of the pIX gene, PCR primers NheU (5′-CGA TGA CAA GCT AGC CAT AAA TAA AAA ACC AGA CTC TG) and NheL (5′-TTA TGG CTA GCT TGT CAT CGT CAT CCT TAT AAT CGG) were designed containing NheI recognition sites (underlined) and substituting the stop codon for a Leu codon. Two DNA fragments, 165 and 236 bp long, were amplified using the following pairs of primers: Ad3904pIX.F (5′-AGT TGA CGG CTC TTT TGG CAC A) and NheL, and NheU and Ad4230pIX.R (5′-ATG AAG CTC TGC AGT GGT GCT ACC T), respectively, using pShlpIXflag as a template. The DNA fragments were purified and then joined by a second PCR using the Ad3904pIX.F and Ad4230pIX.R primers. The resultant PCR fragment was digested with MfeI and PflMI, and the 277-bp DNA fragment was ligated with the MfeI-PflMI fragment of the pShuttle plasmid, generating pShlpIXNhe. To incorporate the sequence encoding Leu Gly Ser Ala Ser Ala followed by eight consecutive lysines and a stop codon into the 3′ end of the pIXflag gene, the oligonucleotides 5′-CTA GGA TCC GCA TCC GCA AAG AAA AAG AAG AAA AAG AAA AAG TAA and 5′-CTA GTT ACT TTT TCT TTT TCT TCT TTT TCT TTG CGG ATG CGG ATC were synthesized. The oligonucleotide duplex containing NheI-compatible 5′-cohesive ends was cloned into NheI-digested pShlpIXNhe, generating the pShlpIX8K plasmid. To construct a reporter gene expression cassette, MluI-HindIII fragment DNA (661 bp) containing the human cytomegalovirus (CMV) immediate-early promoter was isolated from the pcDNA3 plasmid (Invitrogen, Carlsbad, Calif.) and cloned between the MluI and HindIII sites in the pGL3-Basic vector (Promega, Madison, Wis.), generating pGL3hCMV. The expression cassette containing the CMV-driven firefly luciferase gene was excised from pGL3hCMV with Acc65I and SalI and cloned into the pShuttle, pShlpIXflag, pShlpIXNhe, and pShlpIX8K plasmids between the Acc65I and XhoI sites, generating the pSlLuc, pSlLucIXflag, pSlLucIXNhe, and pSlLucIX8K shuttle vectors, respectively. All plasmids were sequenced to confirm the correct orientation and structure of cloned oligonucleotides and DNA fragments. The constructed plasmids were used for homologous recombination with the pAdEasy-1 vector to create Ad genomes as recommended for the AdEasy system (14). The resultant plasmids, designated pAdLuc, pAdLucIXflag, pAdLucIXpK, and pAdLucIX8K, were used to generate Ad5 vectors containing modified pIX genes and the CMV-driven luciferase gene in place of the E1 region of the Ad genome.
Viruses.
Replication-incompetent Ad5 vectors, AdLuc, AdLucIXflag, AdLucIXpK, and AdLucIX8K, were generated by transfection of 293 cells with PacI-digested pAdLuc, pAdLucIXflag, pAdLucIXpK, and pAdLucIX8K plasmids, respectively, as described elsewhere (14). Recombinant Ad vectors were propagated on 293 cells and purified by centrifugation on CsCl gradients by standard protocols. The titers of physical and infectious VPs were determined by the methods of Maizel et al. (20) and Mittereder et al. (22), respectively. The VP/PFU ratios determined for AdLuc, AdLucIXflag, AdLucIXpK, and AdLucIX8K were 55, 100, 220, and 55, respectively. Radiolabeled Ads were made by adding 50 μCi of [methyl-3H]thymidine (Amersham Pharmacia Biotech, Piscataway, N.J.) per ml of the cell medium at 20 h postinfection. The infected cells were harvested at 50 h postinfection, and the viruses were purified as described above. The specific activities of the labeled viruses ranged from 1.6 × 10−5 to 8.0 × 10−5 cpm/VP.
Protein electrophoresis and Western blotting.
Samples of CsCl-purified virions were boiled in Laemmli buffer and subjected to 4 to 20% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to separate the viral capsid proteins. For Western blot analysis, electrophoretically resolved viral proteins were transferred to polyvinylidene difluoride membranes and probed with anti-Flag M2 monoclonal antibody (MAb) (Sigma, St. Louis, Mo.). Bound murine immunoglobulin G was detected with a secondary alkaline phosphatase-conjugated goat anti-mouse antibody (Sigma).
ELISA.
Solid-phase binding enzyme-linked immunosorbent assays (ELISAs) were performed as follows. CsCl-purified virions diluted in 50 mM NaHCO3 (pH 9.6) to concentrations ranging from 1.2 × 108 to 1.0 × 1010 VP/ml were immobilized in triplicate in Nunc-Maxisorp ELISA plate wells (100-μl/well). The wells were blocked with phosphate-buffered saline (PBS) (10 mM NaH2PO4, 10 mM KH2PO4 [pH 7.4], and 136 mM NaCl) containing 0.05% Tween 20 and 2% bovine serum albumin (BSA) and then washed with PBS containing 0.05% Tween 20. Anti-Flag M2 MAb (Sigma) diluted in blocking buffer to 2 μg/ml was added to the wells in 100-μl aliquots. After a 1-h incubation at room temperature, the wells were washed and incubated for 45 min with a 1:5,000 dilution of secondary goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (Sigma). The plates were developed using the signal-producing reagent p-nitrophenyl phosphate (Sigma), and absorbance was measured in a microtiter plate reader set at 405 nm. The results are presented as mean absorbance ± standard deviation (SD).
Radiolabeled Ad binding assay.
Binding of radiolabeled Ad vectors to heparin beads or cells was assayed as follows. Suspensions of ceramic HyperD M hydrogel composite beads (Sigma) coated with heparin were washed twice with PBS containing 1% BSA, and 30-μl aliquots of 50% bead slurry were transferred to 1.5-ml tubes. Fifty-microliter aliquots of 3H-labeled Ad vectors containing 1010 VP (∼2 × 105 to 8 × 105 cpm) were then added, and the tubes were incubated with shaking to allow binding. After a 1-h incubation at room temperature, the beads were washed twice with PBS by centrifugation. The supernatant containing unbound virus was aspirated, and the pellets were resuspended in EcoLume scintillation cocktail (ICN Biomedicals, Costa Mesa, Calif.).
Confluent cells were released with EDTA, washed once with PBS, pelleted, and resuspended to a final concentration of 107/ml in binding medium (Dulbecco's modified Eagle's medium-F12, 20 mM HEPES, 0.5% BSA). One hundred-microliter cell aliquots were mixed with 50 μl of Ad5 knob protein (60 μg/ml), heparin (9 mg/ml), or PBS in 5-ml test tubes and incubated for 30 min at 4°C. Aliquots of 3H-labeled Ad vectors (∼2 × 105 to 8 × 105 cpm) were then added, and incubation was continued for 1 h to allow binding. The cells were diluted with 4 ml of binding buffer and centrifuged. The supernatant containing unbound virus was aspirated, and the cell pellets were solubilized in scintillation cocktail. Radioactivity was measured in a liquid scintillation analyzer (Packard, Downers Grove, Ill.).
Gene transfer assay.
Cell monolayers grown in a 24-well plate (3 × 105 to 5 × 105 cells/well) were incubated with 0.2 ml of Ad5 knob protein (50 μg/ml), heparin (3 mg/ml), or PBS at room temperature. After a 15-min incubation, 200-μl aliquots of Ad vector were added to the cells at a multiplicity of infection of 100 VP/cell and allowed to be internalized for 30 min. Then the infection medium was aspirated, and the cells were washed with PBS and incubated in growth medium at 37°C to allow reporter gene expression. The cells were lysed 20 h postinfection, and luciferase activity was analyzed by using the Promega luciferase assay. For the virion thermostability assay, samples of Ad vectors were incubated at 45°C for different time intervals and then used to infect 293 cells at a multiplicity of infection of 20 VP/cell. Luciferase activity in infected cells was analyzed 18 h postinfection as described above.
RESULTS
Generation of pIX-modified Ad vectors.
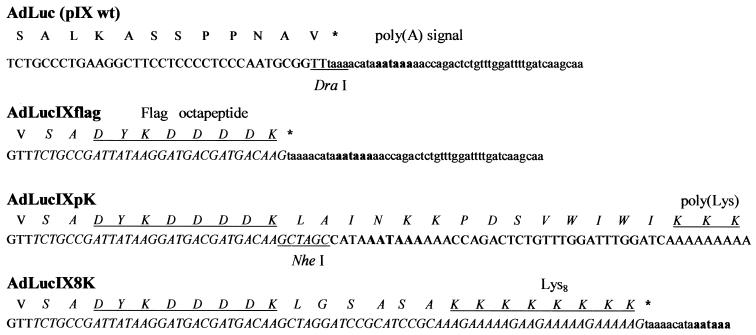
The recombinant Ad vector AdLucIXflag, encoding pIX containing a C-terminal Flag octapeptide (DYKDDDDK) and a luciferase reporter gene, was constructed using the AdEasy vector system (14). To incorporate different heterologous peptides into the C terminus of pIX, we introduced a unique NheI restriction site in the shuttle plasmid following the Flag coding sequence. The recombinant Ad generated using this shuttle vector was designated AdLucIXpK. Incorporation of an NheI site caused a deletion of the pIX stop codon and should have resulted in translation of mRNA containing a poly(A) tail. To validate this, we carried out reverse transcription-PCR using total mRNA isolated from AdLucIXpK-infected cells as a template. Sequencing of PCR products confirmed the presence of 39 nucleotides, including a poly(A) signal downstream of the Flag coding sequence followed by multiple adenosines (Fig. 1). To derive the AdLucIX8K vector encoding pIX incorporating a carboxy-terminal Flag epitope along with eight consecutive lysines (Lys8), the corresponding DNA duplex was cloned into the shuttle plasmid NheI site, restoring the deleted stop codon. Thus, we have constructed three Ad vectors containing a pIX gene modified to introduce heterologous peptide sequences, including the Flag epitope for detection purposes. Both Lys8 and poly(Lys) incorporated sequences serve as heparin-binding domains known to target Ad vectors to broadly expressed heparan sulfate-containing cellular receptors (33, 35).
FIG. 1.
Schema of Ad pIX modifications. To generate the AdLucIXflag vector, the Flag octapeptide (DYKDDDDK) coding sequence was introduced into the DraI site of the wild-type pIX gene. Introduction of the NheI site following the Flag coding sequence in AdLucIXpK caused a pIX stop codon deletion and translation through the poly(A) signal, resulting in incorporation of a poly(Lys) tail into the carboxy terminus of pIX. Cloning of the DNA sequence encoding eight consecutive lysines resulted in restoration of the stop codon and incorporation of a Lys8 peptide into the C terminus of pIX. Modified DNA and protein sequences of pIX are designated by italics. Translated amino acid sequences are presented in capital letters. Restriction sites are underlined, and the poly(A) signal is indicated in boldface. Incorporated Flag, poly(Lys), and Lys8 peptide sequences are underlined.
The presence of modified pIX in Ad capsid.
The incorporation of modified pIX containing heterologous peptides into viral capsids was demonstrated by Western blotting. The virions of AdLucIXflag, AdLucIX8K, and AdLucIXpK purified by CsCl gradient centrifugation were denatured by being boiled, and the capsid proteins were separated by SDS-PAGE. The AdLuc vector containing the wild-type pIX gene was used as a negative control. The probing of electrophoretically resolved viral capsomers of AdLucIXflag and AdLucIX8K using an anti-Flag M2 MAb detected the presence of protein bands with molecular masses of 15.7 and 17.2 kDa, as expected for pIXflag and pIX8K, respectively (Fig. 2). The presence of poly(Lys) sequences in AdLucIXpK capsids was confirmed by the shift of its electrophoretic mobility compared to pIXflag and pIX8K. An increase in the molecular mass of pIXpK by approximately 3 kDa compared to pIXflag corresponds to an increase in protein length of at least 25 amino acids.
FIG. 2.
Western blot analysis of pIX-modified Ad vectors. Samples of CsCl-purified AdLuc (pIXwt), AdLucIXflag (pIXflag), AdLucIX8K (pIX8K), and AdLucIXpK (pIXpK) were boiled in Laemmli loading sample buffer and separated by 4 to 20% gradient SDS-PAGE. Electrophoretically resolved viral proteins were transferred to polyvinylidene difluoride membranes, probed with anti-Flag M2 MAb, and detected with secondary alkaline phosphatase-conjugated goat anti-mouse antibodies. The numbers on the left indicate the molecular masses of marker proteins (lane MW) in kilodaltons.
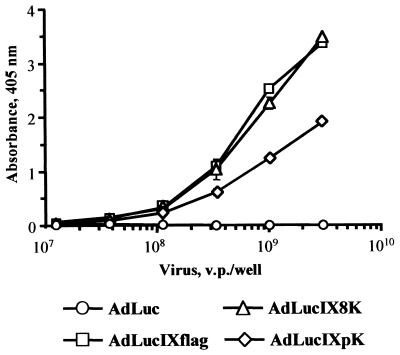
To test if peptides incorporated into modified pIX are displayed on the surfaces of VPs, we performed an ELISA. Purified AdLucIXflag, AdLucIX8K, AdLucIXpK, and control AdLuc virions were immobilized on ELISA plates and probed with an anti-Flag MAb. AdLucIXflag and AdLucIX8K revealed the same level of interaction with the anti-Flag MAb, while AdLucIXpK showed somewhat less efficient binding (Fig. 3). These results demonstrate that Flag peptides introduced into the pIX carboxy terminus are accessible for recognition by antibodies, strongly suggesting that both the Flag epitope and downstream peptide sequences are displayed on the surfaces of these recombinant Ad capsids.
FIG. 3.
Presentation of modified pIX C terminus on Ad capsid. Dilutions of CsCl-purified AdLuc, AdLucIXflag, AdLucIX8K, and AdLucIXpK were immobilized on an ELISA plate and probed with anti-Flag M2 MAb. Anti-Flag MAb bound to the VPs was detected with alkaline phosphatase-conjugated secondary antibody. Each point represents the cumulative mean ± SD of triplicate determinations. Some error bars depicting SDs are smaller than the symbols.
Accessibility of pIX-incorporated peptides for binding.
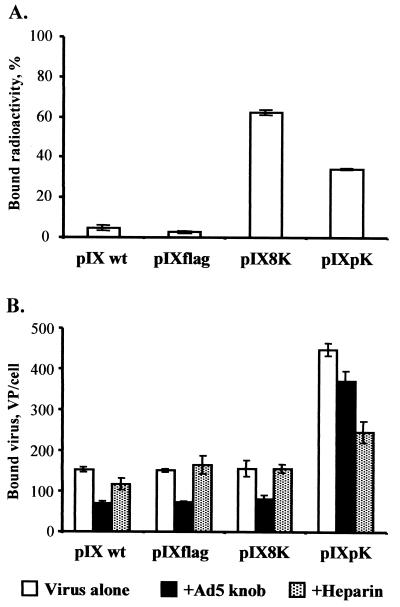
We used binding assays to confirm that lysine sequences incorporated into AdLucIX8K and AdLucIXpK are capable of binding heparan sulfates. First, we tested the ability of Lys8 and poly(Lys) peptides to effect binding of radioactively labeled AdLucIX8K or AdLucIXpK to heparin-coated beads. As shown in Fig. 4A, both viruses demonstrated markedly increased binding to heparin molecules compared to AdLuc and AdLucIXflag control viruses.
FIG. 4.
Accessibility of pIX-incorporated ligands for binding. (A) Binding of pIX-modified Ad to heparin-coated beads. Suspensions of heparin-coated ceramic beads were incubated with 1010 VP (2 × 105 to 8 × 105 cpm) of 3H-labeled AdLuc (pIX wt), AdLucIXflag (pIXflag), AdLucIX8K (pIX8K), and AdLucIXpK (pIXpK). The beads were washed by centrifugation to remove unbound virions, and then bound radioactivity was calculated as a percentage of input radioactivity for each Ad sample. Each bar represents the cumulative mean ± SD of triplicate determinations. (B) Binding of pIX-modified Ad to AU-565 cells. Aliquots of AU-565 cells (106) were preincubated separately with Ad5 knob protein, heparin, or PBS (Virus alone). Samples of radiolabeled AdLuc (pIX wt), AdLucIXflag (pIXflag), AdLucIX8K (pIX8K), and AdLucIXpK (pIXpK) containing 1010 VP (2 × 105 to 8 × 105 cpm) were added to the cells and incubated for 1 h at 4°C. Bound radioactivity was determined after washing the cell samples by centrifugation, and the VP/cell ratio was calculated for each Ad vector. Each bar represents the cumulative mean ± SD of triplicate determinations.
To characterize the ability of AdLucIX8K and AdLucIXpK to recognize heparan sulfate-containing cell surface receptors, we carried out binding assays with AU-565 cells in the presence or absence of heparin or Ad5 knob protein. The results presented in Fig. 4B show that AdLucIXpK exhibits a 2.5-fold higher cell-binding efficiency than AdLuc and AdLucIXflag control viruses (P < 0.001). In contrast to binding to immobilized heparin, AdLucIX8K did not show any improvement of binding to heparan sulfate cell surface receptors. The presence of free heparin blocked 45% of AdLucIXpK cell binding (P < 0.001) but did not affect the binding of the control viruses to the cells. Addition of Ad5 knob blocked 50% of control virus binding (P < 0.004) while having relatively little effect (17%; P = 0.01) on AdLucIXpK. These data suggest that the poly(Lys) sequence displayed on AdLucIXpK capsid mediates Ad interaction with cellular heparan sulfate receptors, resulting in CAR-independent virus-cell binding.
Infection efficiency of pIX-modified Ad vectors.
We evaluated whether the ability of AdLucIXpK to bind heparan sulfate-containing receptors would result in improvement of Ad infection of CAR-deficient cell types. Our previous study showed that AU-565, GI-101A, and HUVEC cells are relatively refractory to Ad infection due to the low level of CAR on their surfaces (unpublished data). Using a gene transfer assay, we tested the ability of poly(Lys) displayed by pIX on the viral capsid to mediate Ad infection in the presence or absence of Ad5 knob protein. As illustrated in Fig. 5A, AdLucIXpK demonstrated 4.3-, 2.8-, and 2.1-fold enhancement of gene transfer to GI-101A, AU-565, and HUVEC cells, respectively, compared to AdLucIXflag control virus in the presence of Ad5 knob protein (P = 0.02). There were no significant differences in the levels of gene transfer detected following infection of CAR-positive 293 cells. To confirm that the observed augmentation of CAR-independent infection efficiency was due to the binding of AdLucIXpK to cellular heparan sulfates, we performed gene transfer assays in the presence of free heparin. As can be seen in Fig. 5B, gene transfer mediated by AdLucIXpK was blocked by heparin to a significantly greater extent (30 to 73%, depending on the cell line; P ≤ 0.05) than that mediated by control Ad vectors. Consistent with our cell-binding assays, AdLucIX8K did not show any augmentation in the level of infectivity.
FIG. 5.
Infection properties of pIX-modified Ad vectors. (A) Monolayers of GI-101A, AU-565, and HUVEC CAR-deficient cell lines and CAR-positive 293 cells were infected with pIX-modified AdLucIXflag, AdLucIX8K, AdLucIXpK, or control AdLuc vector in the presence or absence of Ad5 knob protein. The levels of luciferase activity were determined in lysates of infected cells 20 h postinfection. The results are presented as the percentages of luciferase activity detected in the cells infected in the presence of Ad5 knob protein calculated with respect to luciferase activity determined in the cells infected in the absence of knob (100%). Each bar represents the cumulative mean ± SD of triplicate determinations. (B) Cell monolayers were infected with AdLucIXflag, AdLucIX8K, AdLucIXpK, or control AdLuc vectors in the presence or absence of free heparin. The luciferase from cell lysate activities was analyzed 20 h postinfection. The results are presented as the percentages of luciferase activity detected in the cells infected in the presence of free heparin calculated with respect to luciferase activity determined in the absence of heparin (100%). Each bar represents the cumulative mean ± SD of triplicate determinations.
Thermostabilities of pIX-modified viruses.
Since pIX is involved in Ad capsid stabilization and is essential for proper viral DNA packaging, we tested the thermostabilities of pIX-modified Ad vectors to see whether the incorporated peptides affect virion structural integrity. The relative thermostabilities of pIX-modified viruses were compared indirectly by measuring Ad-mediated gene transfer efficiencies following incubation of virions at 45°C. This experiment shows that the level of luciferase expression achieved by AdLucIXpK is reduced twofold compared to those of AdLucIXflag, AdLucIX8K, and unmodified AdLuc (Fig. 6), indicating a decrease of AdLucIXpK thermostability. This finding is consistent with an increased VP/PFU ratio (220) observed for AdLucIXpK compared to both AdLucIX8K and AdLuc (VP/PFU ratio, 55).
FIG. 6.
Thermostabilities of pIX-modified Ad vectors. Aliquots of AdLucIXflag, AdLucIX8K, AdLucIXpK, or AdLuc vector were incubated at 45°C for different time intervals and then used to infect 293 cells. The results are presented as the percentages of luciferase activity detected in the cells infected with a heat-treated viral sample with respect to luciferase activity determined in the cells infected with untreated virus (100%). Each bar represents the cumulative mean ± SD of triplicate determinations. Some error bars depicting SDs are smaller than the symbols.
DISCUSSION
To develop a targeted Ad vector, it is necessary both to ablate native viral tropism and to introduce a novel specificity which will allow targeting of certain cell types otherwise refractory to Ad infection. Ablation of Ad binding to its natural receptors is a prerequisite for the generation of truly targeted Ad vectors and has been addressed recently (16, 17). Mutagenesis of both Ad5 fiber knob and penton has been employed to abrogate CAR- and integrin-dependent viral tropism and to improve the targeting potential of Ad vectors (9). The next step toward the generation of target-specific Ad is to redirect the virus to alternative receptors present on the cells of interest. In this regard, genetic modification of viral capsid proteins to incorporate targeting ligands has proven to be a rational approach for introducing novel cell-specific tropism and overcoming limitations associated with low levels of native receptors. Targeting peptide ligands have been introduced into the HI loop (8, 23, 36) or C terminus of Ad5 fiber protein (33, 35) and the L1 loop of hexon protein (30). Engineered Ad vectors bearing either polylysine (33), RGD, or NGR motifs (8, 23, 30, 35) or a transferrin receptor binding peptide (36) exhibited a marked increase of infection efficiency in a variety of CAR-deficient cell types via recognition of heparan sulfates, integrins, or the transferrin receptor, respectively. The limited number of genetic modifications of Ad capsid achieved so far reflects, in part, the limited availability of exploitable peptide ligands. In this regard, structural properties of the surface-exposed loops in capsid proteins apparently make them suitable only for constrained ligand sequences. Furthermore, carboxy-terminal extensions of fiber protein can affect correct folding of fiber trimers, thus interfering with virus assembly. Therefore, a major improvement in genetic Ad targeting strategies would be the identification of a capsid locale that allows incorporation of heterologous, high-affinity polypeptide ligands.
In this study, we evaluated the utility of pIX, a minor component of the Ad capsid, as an alternative site for ligand incorporation for the purpose of viral tropism modification. pIX functions as a cement protein stabilizing hexon-hexon interactions and is essential for viral DNA packaging. In addition to its structural contribution, pIX exhibits transcription-regulatory properties (25). It was recently shown for Ad serotypes 2, 3, and 5 that the C-terminal region of pIX is located on the virion surface (1, 25). We proposed to exploit this finding for pIX carboxy-terminal incorporation of heterologous polypeptides that could serve as ligands for receptor-specific Ad5 infection. Such incorporation of targeting ligands would ideally be achieved in such a manner as to allow both surface presentation and proper assembly of VPs. To evaluate this concept, we first engineered the AdLucIXflag vector encoding a carboxy-terminal extension of pIX consisting of the Flag octapeptide. We then established the surface localization of the Flag epitope in the context of assembled virions and its accessibility for binding with anti-Flag MAb using affinity column purification (data not shown). Ad vectors, AdLucIX8K and AdLucIXpK, were engineered to encode pIX C-terminal heparin-binding motifs consisting of either eight or multiple consecutive lysines. For detection of modified pIX ectodomain on the outer capsid surface, we placed a Flag peptide upstream of the lysine-containing sequences. The probing of generated viruses with anti-Flag MAb demonstrated that modified pIX is incorporated into viral capsids and displays Flag-containing additions on the surfaces of VPs. Importantly, both AdLucIX8K and AdLucIXpK were shown to bind heparin immobilized on ceramic beads, demonstrating that the pIX ectodomain displays peptide ligands accessible for binding. However, AdLucIX8K did not reproduce its ability to bind heparan-containing receptors on cellular membranes. These data suggest that the length of the C-terminal extension of pIX is important for positioning the ligand sequence distal to the Ad capsid surface, making it more accessible for interaction with cellular receptors. The use of the Ad fiber knob to inhibit virus-cell binding and subsequent gene transfer to CAR-deficient cell lines revealed significant augmentation of knob-independent infection mediated by AdLucIXpK compared to AdLucIXflag and AdLuc control viruses. On the other hand, both cell binding and gene transfer mediated by AdLucIXpK were blocked by free heparin to a higher degree than for control viruses. The observed augmentation of knob-independent infection achieved by AdLucIXpK is very likely due to the binding of pIX-incorporated polylysine to heparan sulfates on cellular membranes. These results suggest that pIX-mediated presentation of targeting ligands on the surfaces of Ad vector capsids may be compatible with Ad tropism modification strategies. To evaluate whether carboxy-terminal pIX modifications have an impact on the proper assembly of VPs, we tested the relative thermostabilities of generated Ad vectors. Comparison of gene transfer levels achieved by pIX-modified Ad vectors following incubation of virions at 45°C revealed that AdLucIXpK was less stable than AdLucIXflag, AdLucIX8K, and AdLuc. Consistent with a fourfold-increased VP/PFU ratio observed for AdLucIXpK compared to control Ad vectors, this result indicates that carboxy-terminal peptide extensions of pIX can have some impact on virion structural integrity. Considering both positive and negative aspects of pIX modification revealed in this report, we believe that pIX may have utility for genetic modifications of Ad capsid. Our study suggests that the pIX ectodoman may represent an attractive capsid locale, alternative to fiber knob, hexon, and penton base, for ligand incorporation for the purpose of Ad targeting.
Acknowledgments
We are grateful to Joel N. Glasgow for fruitful discussions and proofreading of the manuscript.
This work was supported by the following grants: N01 C0-97110, P50 CA89019, R01 CA86881, R01 CA74242, R01 CA68245, R01 CA90547, and R01 HL67962 from the National Institutes of Health and the Juvenile Diabetes Foundation to David T. Curiel.
REFERENCES
- 1.Akalu, A., H. Liebermann, U. Bauer, H. Granzow, and W. Seidel. 1999. The subgenus-specific C-terminal region of protein IX is located on the surface of the adenovirus capsid. J. Virol. 73:6182-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnberg, N., K. Edlund, A. H. Kidd, and G. Wadell. 2000. Adenovirus type 37 uses sialic acid as a cellular receptor. J. Virol. 74:42-48. [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 4.Boulanger, P., P. Lemay, G. E. Blair, and W. C. Russell. 1979. Characterization of adenovirus protein IX. J. Gen. Virol. 44:783-800. [DOI] [PubMed] [Google Scholar]
- 5.Burnett, R. M. 1985. The structure of the adenovirus capsid. II. The packing symmetry of hexon and its implications for viral architecture. J. Mol. Biol. 185:125-143. [DOI] [PubMed] [Google Scholar]
- 6.Curiel, D. T. 2000. Rational design of viral vectors based on rigorous analysis of capsid structures. Mol. Ther. 1:3-4. [DOI] [PubMed] [Google Scholar]
- 7.Curiel, D. T. 1999. Strategies to adapt adenoviral vectors for targeted delivery. Ann. N. Y. Acad. Sci. 886:158-171. [DOI] [PubMed] [Google Scholar]
- 8.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 72:9706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Einfeld, D. A., R. Schroeder, P. W. Roelvink, A. Lizonova, C. R. King, I. Kovesdi, and T. J. Wickham. 2001. Reducing the native tropism of adenovirus vectors requires removal of both CAR and integrin interactions. J. Virol. 75:11284-11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fechner, H., A. Haack, H. Wang, X. Wang, K. Eizema, M. Pauschinger, R. Schoemaker, R. Veghel, A. Houtsmuller, H. P. Schultheiss, J. Lamers, and W. Poller. 1999. Expression of coxsackie adenovirus receptor and alphav-integrin does not correlate with adenovector targeting in vivo indicating anatomical vector barriers. Gene Ther. 6:1520-1535. [DOI] [PubMed] [Google Scholar]
- 11.Furcinitti, P. S., J. van Oostrum, and R. M. Burnett. 1989. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 8:3563-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh-Choudhury, G., Y. Haj-Ahmad, and F. L. Graham. 1987. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 6:1733-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 14.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong, S. S., L. Karayan, J. Tournier, D. T. Curiel, and P. A. Boulanger. 1997. Adenovirus type 5 fiber knob binds to MHC class I alpha2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 16:2294-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 2000. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J. Virol. 74:2804-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 1999. Mutations in the DG loop of adenovirus type 5 fiber knob protein abolish high-affinity binding to its cellular receptor CAR. J. Virol. 73:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasnykh, V. N., J. T. Douglas, and V. W. van Beusechem. 2000. Genetic targeting of adenoviral vectors. Mol. Ther. 1:391-405. [DOI] [PubMed] [Google Scholar]
- 19.Louis, N., P. Fender, A. Barge, P. Kitts, and J. Chroboczek. 1994. Cell-binding domain of adenovirus serotype 2 fiber. J. Virol. 68:4104-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology 36:115-125. [DOI] [PubMed] [Google Scholar]
- 21.Mathias, P., T. Wickham, M. Moore, and G. Nemerow. 1994. Multiple adenovirus serotypes use alpha v integrins for infection. J. Virol. 68:6811-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittereder, N., K. L. March, and B. C. Trapnell. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 70:7498-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuguchi, H., N. Koizumi, T. Hosono, N. Utoguchi, Y. Watanabe, M. A. Kay, and T. Hayakawa. 2001. A simplified system for constructing recombinant adenoviral vectors containing heterologous peptides in the HI loop of their fiber knob. Gene Ther. 8:730-735. [DOI] [PubMed] [Google Scholar]
- 24.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosa-Calatrava, M., L. Grave, F. Puvion-Dutilleul, B. Chatton, and C. Kedinger. 2001. Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 75:7131-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shenk, T. 1996. Adenoviridae: the viruses and their replication, p. 2111-2148. In B. N. Fields, D. M. Knipe, et al. (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 27.Stevenson, S. C., M. Rollence, B. White, L. Weaver, and A. McClelland. 1995. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J. Virol. 69:2850-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart, P. L. 2002. Adenovirus structure, p. 1-18. In D. T. Curiel and J. T. Douglas (ed.), Adenoviral vectors for gene therapy. Academic Press, San Diego, Calif.
- 29.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vigne, E., I. Mahfouz, J. F. Dedieu, A. Brie, M. Perricaudet, and P. Yeh. 1999. RGD inclusion in the hexon monomer provides adenovirus type 5-based vectors with a fiber knob-independent pathway for infection. J. Virol. 73:5156-5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickham, T. J. 2000. Targeting adenovirus. Gene Ther. 7:110-114. [DOI] [PubMed] [Google Scholar]
- 32.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 33.Wickham, T. J., P. W. Roelvink, D. E. Brough, and I. Kovesdi. 1996. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat. Biotechnol. 14:1570-1573. [DOI] [PubMed] [Google Scholar]
- 34.Wickham, T. J., D. M. Segal, P. W. Roelvink, M. E. Carrion, A. Lizonova, G. M. Lee, and I. Kovesdi. 1996. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J. Virol. 70:6831-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickham, T. J., E. Tzeng, L. L. Shears II, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia, H., B. Anderson, Q. Mao, and B. L. Davidson. 2000. Recombinant human adenovirus: targeting to the human transferrin receptor improves gene transfer to brain microcapillary endothelium. J. Virol. 74:11359-11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, W. W. 1999. Development and application of adenoviral vectors for gene therapy of cancer. Cancer Gene Ther. 6:113-138. [DOI] [PubMed] [Google Scholar]