Abstract
Aura and Sindbis viruses are closely related alphaviruses. Unlike other alphaviruses, Aura virus efficiently encapsidates both genomic RNA (11.8 kb) and subgenomic RNA (4.2 kb) to form virus particles. Previous studies on negatively stained Aura virus particles predicted that there were two major size classes with potential T=3 and T=4 capsid structures. We have used cryoelectron microscopy and three-dimensional image reconstruction techniques to examine the native morphology of different classes of Aura virus particles produced in BHK cells. Purified particles separated into two components in a sucrose gradient. Reconstructions of particles in the top and bottom components were computed to resolutions of 17 and 21 Å, respectively, and compared with reconstructions of Sindbis virus and Ross River virus particles. Aura virus particles of both top and bottom components have similar, T=4 structures that resemble those of other alphaviruses. The morphology of Aura virus glycoprotein spikes closely resembles that of Sindbis virus spikes and is detectably different from that of Ross River virus spikes. Thus, some aspects of the surface structure of members of the Sindbis virus lineage have been conserved, but other aspects have diverged from the Semliki Forest/Ross River virus lineage.
Alphavirus is a genus within the family Togaviridae that consists of about 26 members. Alphaviruses are enveloped, positive-sense, single-strand RNA viruses, are transmitted by arthropods (primarily mosquitoes), and cause fever, rash, or encephalitis in humans or domestic animals (24). The alphaviruses can be grouped into three lineages based on comparisons of their genome sequences: the VEE/EEE (Venezuelan equine encephalitis/Eastern equine encephalitis virus) lineage, the SFV/RRV (Semliki Forest/Ross River virus) lineage, and the SINV (Sindbis virus) lineage (18). Aura virus, first isolated from Aedes serratus in Brazil and northern Argentina, is a member of the SINV lineage (21). There is no evidence for human disease caused by the virus.
The 11.8-kb alphavirus genome contains two domains. The 5′-terminal two-thirds encodes four nonstructural proteins required for RNA replication. The 3′ one-third encodes the structural proteins, which are expressed by means of a 4.2-kb subgenomic mRNA. These structural proteins are translated as a polyprotein that is subsequently cleaved into five proteins: the nucleocapsid protein (CP) (∼30 kDa); two envelope glycoproteins, E1 (∼52 kDa) and E2 (∼49 kDa); and two small peptides, E3 (∼10 kDa) and 6K (∼6 kDa) (19). The Aura virus structural proteins and those of SINV have amino acid sequence identity of 77% for CP, 56% for E2, and 61% for E1, for an average identity of 62% (21). These structural proteins have only 46% sequence identity with those of RRV or SFV, although the structural proteins of RRV and SFV have 75% sequence identity.
No atomic structure of an alphavirus has been determined because the best crystals thus far diffract X-rays to a resolution of only ∼30 Å (10). Low-resolution (20- to 30-Å) and moderate-resolution (9-Å) structures of several alphaviruses have been determined by means of cryoelectron microscopy (cryo-EM) and three-dimensional image reconstruction techniques (5, 13, 15, 16, 27). Cryo-EM studies on RRV (5), SFV (27), and SINV (16) demonstrated that alphaviruses are spherical, multilayered structures with an outer diameter of ∼710 Å. The viral genome is encapsidated in a closed protein shell, called the nucleocapsid core, and is composed of 240 copies of the CP protein arranged in a T=4 icosahedral lattice (26). The CP subunits are organized as pentameric and hexameric capsomers, which extend to a radius of 215 Å and project ∼45 Å above the base of the protein shell. This base contains the N terminus of the CP protein, which interacts with the viral RNA. It is not known if the base contains surface-accessible RNA or whether the RNA is simply encapsidated by this protein shell. However, the RNA in isolated nucleocapsid cores is sensitive to RNase digestion (11). Hence, the protein shell may not completely shield the genome. In virions, the viral nucleocapsid core is tightly enveloped by a lipid bilayer derived from the host. Glycoproteins penetrate the bilayer and form a one-to-one association with CP (5). Glycoproteins E1 and E2 form heterodimers that are present in a T=4 icosahedral lattice, organized as 80 trimeric spikes on the viral surface. Glycoprotein E2 forms the external protruding portion of each spike, whereas E1 lies roughly parallel to the membrane, forming an icosahedral scaffold (12, 17).
The assembly of alphavirus capsids requires RNA, as no example of nucleic acid-free virus assembly has been reported (25). In most alphaviruses, packaging of RNA is specific, and the genomic RNA is preferentially packaged because an RNA packaging signal is present only in the genomic RNA (23, 29). However, both the genomic RNA and subgenomic RNA of Aura virus are efficiently encapsidated to form virion particles, suggesting that a packaging signal for Aura virus RNA may lie in the region of the genome transcribed into subgenomic RNA (22). Two major size classes of Aura particles were found by electron microscopy in negatively stained preparations (20). One class contained particles with a diameter of ∼720 Å which are similar in size to the virions of other alphaviruses, whereas the second class contained particles with a diameter of ∼620 Å. The 720-Å-diameter particles were postulated to be T=4 particles that contain one copy of genomic RNA or up to three copies of subgenomic RNA. The 620-Å-diameter particles were postulated to contain only a single copy of subgenomic RNA and possibly to have T=3 symmetry. In addition, a small fraction of very small particles (∼360-Å diameter) was also observed (20).
In this paper we report the use of cryo-EM and three-dimensional image reconstruction (3) to examine the native morphology of Aura virus particles produced in BHK cells. We found that Aura virus particles separated into two components in a sucrose gradient. The upper component consisted of wild-type virions, the majority of which contain genomic RNA. In contrast, the lower component contained a mixture of genomic and subgenomic RNA-containing particles. Although 10 to 30% of the particles in the lower fraction appeared smaller in stained preparations, cryo-EM reconstructions revealed that Aura virus particles from both the top and bottom components of the sucrose gradient (AuraT and AuraB, respectively) are similar T=4 structures that resemble those of other alphaviruses.
MATERIALS AND METHODS
Virus propagation and purification.
Baby hamster kidney cells (BHK-15) used for propagation of Aura virus were grown in Eagle minimal essential medium supplemented with 10% fetal bovine serum. Large-scale production of virus was made by infecting BHK cells with virus at a multiplicity of infection of 1. Virus was harvested at 25 h postinfection and was first precipitated from the supernatant with polyethylene glycol after a brief centrifugation (5,000 × rpm for 10 min at 4°C in a Sorvall GSA rotor). The virus was resuspended in 1.5 ml of TNE buffer (50 mM Tris, 200 mM NaCl, 1 mM EDTA) with a pH of 7.6 and was sedimented on a freeze-thaw sucrose gradient for 1.25 h at 38,000 rpm at 4°C in a Beckman SW41 rotor. The purified virus was prepared by centrifugation on a 15 to 30% sucrose gradient at 36,000 rpm for 1.5 h at 4°C in a Beckman SW41 rotor. Two viral bands were discernible. The top and bottom bands, corresponding to AuraT and AuraB, respectively, were collected by side puncture. Each band was then separately pelleted by centrifugation in a Beckman 50.2 rotor for 2 h at 36,000 rpm at 4°C, and each pellet was resuspended in 20 to 40 μl of TNE buffer.
Cryo-EM and three-dimensional image reconstruction.
Small aliquots (3.5 μl) of Aura virus (AuraT and AuraB), SINV, and RRV samples were prepared for cryo-EM as described previously (1, 2, 5, 14). Images were recorded in a Philips CM200 field emission gun transmission electron microscope under low-dose conditions (10 to 13.5 e−/Å2) at a nominal magnification of ×38,000. Micrographs of AuraT, AuraB, SINV, and RRV, all of which displayed a uniform distribution of particles and ice thickness, were digitized at 7-μm intervals on a Zeiss-SCAI scanner. Each image was bin averaged to 14-μm pixels, corresponding to intervals of 3.68 Å at the specimen.
Images of individual virus particles were boxed and floated as is customary (3). The common-lines method (9) was used to compute initial reconstructions of each of the virus samples. These reconstructions were then used as models to determine and refine orientation and origin parameters for each virus image by means of the model-based polar-Fourier transform method (1). Orientation and origin refinement were monitored with real- and reciprocal-space correlation coefficients. The Fourier transform of each image was modified to correct in part for the effects of the microscope contrast transfer function. Eigen value spectra were used to assess the conditioning of the linear, least-squares equations. The resolution of each map was estimated by splitting the image data into two roughly equal sets and comparing the structure factors, calculated independently for each data set.
Difference maps of AuraT minus AuraB and of AuraT minus SINV were computed as described previously (17). The three-dimensional reconstructions of AuraB, SINV, and RRV were compared to the AuraT reconstruction by means of correlation coefficient analysis computed as a function of radius.
The absolute hand of the Aura virus structure was determined by means of tilt experiments (4). Simian virus 40 (T=7d lattice) was used as a control to establish a protocol for recording tilt-pair images on a Philips CM200 microscope at a magnification of ×38,000. Each particle image from the tilt data set was cross-correlated with the corresponding projections of a reconstruction and its enantiomorph. At 32-Å resolution, the reconstruction of AuraT of the correct hand exhibited an average correlation coefficient of 0.474 ± 0.161, compared to 0.128 ± 0.062 for that of the reconstruction of the incorrect hand. The corresponding correlation coefficients for the AuraB reconstructions with the correct or incorrect hands were 0.458 ± 0.208 and 0.112 ± 0.006, respectively.
RESULTS
Cryo-EM reconstruction of AuraB and AuraT.
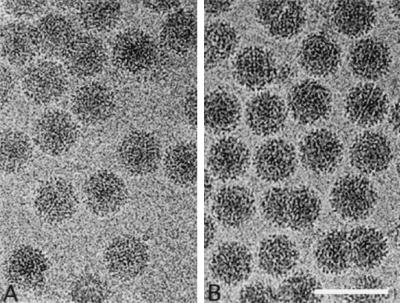
Aura virus propagated in BHK cells is composed of a mixture of virus particles containing both genomic and subgenomic viral RNAs. These particles separate differentially in sucrose gradients into faster-sedimenting particles (AuraB component), which contain both 49S genomic and 26S subgenomic RNA, and slower-sedimenting particles (AuraT component), which contain only the genomic RNA (20). Negative staining and EM revealed two size populations of virions present in the faster-sedimenting band (20; also data not shown). Particles with a diameter of 710 Å were observed, which is consistent with other T=4 alphaviruses. A subset of smaller particles with a diameter of 620 Å was also observed, which was previously hypothesized to be T=3 particles that contain subgenomic RNA. However, this size discrepancy was not apparent in images of the frozen-hydrated Aura virus samples (either the top or bottom bands of the 15 to 30% sucrose gradient) (Fig. 1). Images of AuraT and AuraB particles were visually indistinguishable. In both samples, the viruses exhibit a consistent spherical morphology and a diameter of ∼710 Å. Glycoprotein spikes are discernible at the particle peripheries, and membrane features can be seen on close inspection of close to focus micrographs. The diversity of the projected viral images indicated that virions were randomly orientated in the vitreous ice.
FIG. 1.
Frozen-hydrated samples of AuraT (A) and AuraB (B) viruses. Bar, 1,000 Å.
Image reconstruction maps of AuraT and AuraB virus were computed to 17- and 21-Å resolution (Table 1). Two independent preparations of AuraT virus and three independent preparations of AuraB virus were imaged by cryo-EM and gave consistent results.
TABLE 1.
Data collection and image processing
| Virus | No. of micro- graphs | Under- focus (μm)a | No. of particlesb | Mean correlation coefficientc (SD) | Resolution of the map (Å) |
|---|---|---|---|---|---|
| AuraT | 20 | 1.90-3.52 | 3,506 (2,812) | 0.397 (0.105) | 17.0 |
| AuraB | 21 | 1.46-3.12 | 1,581 (1,263) | 0.454 (0.090) | 21.0 |
Determined from the averaged Fourier transform of boxed images.
Total number of viral particles boxed from all micrographs. The number of particles used for the three-dimensional reconstruction is given in parentheses.
Average real-space correlation coefficient for all particles computed between 165 and 350 Å radii of the final model. The correlation coefficient (CC) was calculated as follows: CC = Σ(ρ1ρ2 − <ρ1><ρ2>)/[Σ(ρ12 − <ρ1>2) Σ(ρ22 − ρ2>2)]0.5. ρ1 and ρ2 refer to the density values of one particle image and its corresponding projection of the three-dimensional reconstruction, respectively. The sum is take over all pixels within the defined radii and <ρ1> and ρ2> refer to the mean values of the image and the corresponding projection of the reconstruction. The standard deviation (SD) of each correlation coefficient is given in parentheses.
Organization of Aura virus.
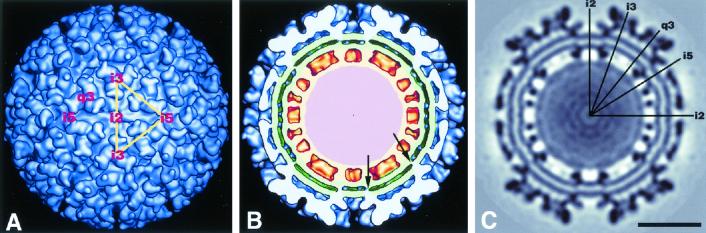
The Aura virus structure appears similar to that of other alphaviruses examined by cryo-EM methods (Fig. 2 and 3). The AuraT structure shows clear T=4 quasi-symmetry and a multilayered organization, which includes a glycoprotein shell (263 to 355 Å), a lipid bilayer (215 to 263 Å), a nucleocapsid protein shell (170 to 215 Å), and viral RNA. The outer glycoprotein shell is subdivided into three distinct layers: the spike projections (53 Å in height) that form the predominant feature on the virus surface, the thin shell of density or skirt (22 Å thick) that, with the exception of holes at the icosahedral twofold axes, covers most of the virus surface and connects the projecting spikes, and the subskirt region (18 Å thick) that connects the skirt to the top of the lipid bilayer. Previous studies of SINV deglycosylation mutants (17) and fitting the atomic structure of SFV E1 into the cryo-EM reconstruction map (12) indicate that the spikes consist primarily of glycoprotein E2, whereas the skirt is attributed to a tangential arrangement of E1 glycoproteins. The nucleocapsid core proteins are organized as 42 capsomers, of which 30 hexamers are situated at the icosahedral twofold axes and 12 pentamers occupy the fivefold axes. Both capsomer types project radially outward from the base of a 15-Å-thick shell. A lipid bilayer membrane tightly envelopes the core, and the inside layer of phosphate head groups bends by approximately 7 Å toward the center of each capsomer. Putative genome-capsid connections are not observed in the reconstructions, though this may be a consequence of enforcing icosahedral symmetry on the entire reconstruction.
FIG. 2.
Image reconstruction of AuraT virus at 17-Å resolution, viewed along a twofold axis. (A) Surface-shaded representation. The yellow triangle demarks one icosahedral asymmetric unit. (B) Surface-shaded representation as in panel A but with the front half removed to reveal interior features. Arrows identify putative transmembrane connections. Aura virus exhibits a multilayered structure similar to that of SINV, which consists of glycoproteins (blue), a lipid bilayer (green), nucleocapsid proteins (orange), and the genome (pink). (C) Electron density map of an equatorial section. Icosahedral twofold (i2), threefold (i3), quasi-threefold (q3), and fivefold (i5) axes are labeled in panels A and C. Bar, 200 Å.
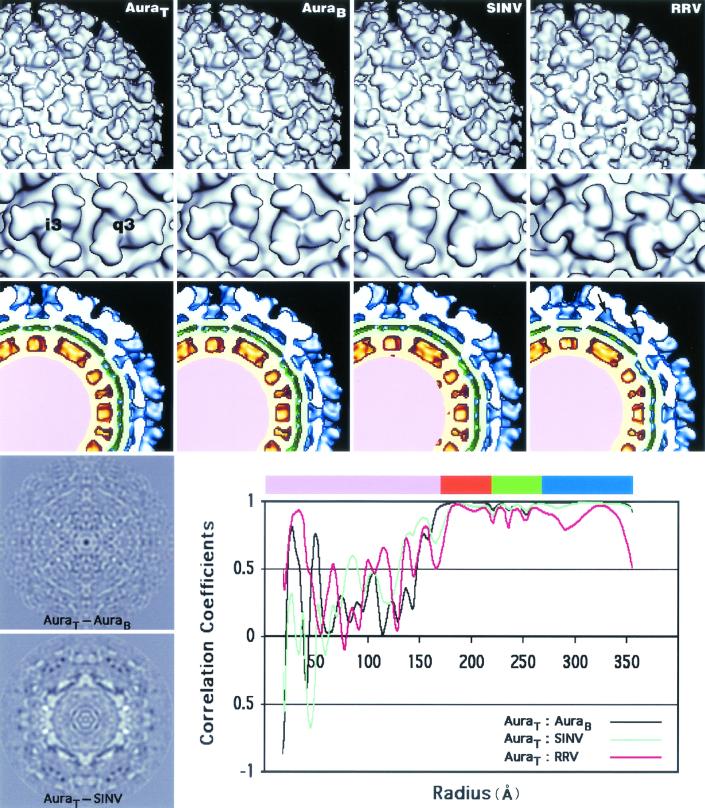
FIG. 3.
Comparison of AuraT, AuraB, SINV, and RRV reconstructions. Surface-shaded representations of the exteriors (top row), spikes (second row) (threefold [i3] and quasi-threefold [q3] axes), and equatorial cross sections (third row) from AuraT and AuraB, SINV, and RRV allow similarities and differences between these reconstruction maps to be compared. Reconstruction maps of AuraT, AuraB, and SINV are remarkably similar, whereas the RRV spike differs both on its exterior and its interior, where a cavity is present (see arrows on RRV cross section). The two panels on the bottom left are the central sections of the AuraT−AuraB and AuraT−SINV difference maps. The bottom right panel plots the correlation coefficients between different pairs of reconstructions (AuraT−AuraB, AuraT−SINV, and AuraT−RRV) as a function of radius. The color bar at the top of the panel coincides with the color scheme used in the equatorial cross sections (third row): viral genome (pink), nucleocapsid core (orange), lipid membrane (green), and envelope glycoproteins (blue).
Structural comparisons of AuraT, AuraB, SINV, and RRV.
The structures of AuraT, AuraB, SINV, and RRV were all rendered at 22-Å resolution to permit qualitative and quantitative comparisons (Fig. 3). Careful comparisons of the sizes and shapes of internal and external features reveal that AuraT and AuraB are virtually identical. The AuraT minus AuraB difference map shows no significant peaks and confirms the similarity of the two reconstructions (Fig. 3). In addition, correlation coefficient analysis, computed as a function of radius, between the AuraT and AuraB reconstructions, indicated that the glycoprotein and nucleocapsid protein layers are almost identical in these Aura viruses (Fig. 3). Therefore, despite differences in RNA content, migration in sucrose gradients, and particle sizes observed using negative staining and EM, AuraT and AuraB are indistinguishable at 22-Å resolution.
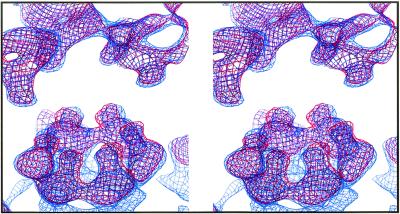
At first glance, the AuraT and SINV structures appear quite similar if not identical (Fig. 3). However, the AuraT minus SINV difference map shows strong positive and negative peaks, which may reflect genuine structural differences. Radial density correlation coefficient analysis shows that SINV is less similar to AuraT than AuraT is to AuraB. Superimposed wire-frame representations of the two maps demonstrate that AuraT hexamers and pentamers in the core and the transmembrane bundles are rotated ∼6° counterclockwise relative to that observed in SINV, whereas densities at the upper surface of the membrane coincide (Fig. 4). Such observations suggest that the CP proteins in Aura and SINV might bind to their respective glycoproteins in slightly different orientations. Similar observations have been made on the basis of recent cryo-EM studies of Venezuelan equine encephalitis virus in which the nucleocapsid capsomeres were found to be rotated counterclockwise with respect to their SINV counterparts (15).
FIG. 4.
Stereo view of the electron density maps of Aura virus (red) and SINV (blue), displayed at a high contour level, with the capsid appearing at the bottom and the glycoprotein at the top. The finger-like projections pointing downwards from the glycoproteins and upward from the capsid, and corresponding to the transmembrane densities of Aura and SINV, are indistinguishable at the upper surface of the membrane but have slightly different orientations at the lower surface.
The spike morphology in RRV differs noticeably from that seen in AuraT, AuraB, and SINV (Fig. 3). The most prominent difference is that RRV lacks a small nodule that occurs on the distal end of the spike in the other alphaviruses. On SINV, this small knob was recently shown to be the sugar moiety at residue 196 of the E2 glycoprotein (17). The RRV E2 glycoprotein has a similar glycosylation site but at residue 200 (E2-200). Consistent with this difference, extra density is found on the inner side of the RRV spike petal, close to the center (threefold or quasi-threefold axis) of each spike. A cavity lies beneath the center of each RRV spike, because in RRV the E1 molecules have no glycosylation site at residue 245, as the E1 glycoproteins of SINV and Aura virus do. Additional densities observed at the skirt level and between adjacent spikes in RRV correspond to the two carbohydrate moieties attached to each of the E2-262 residues that bridge the two spikes. The locations of the RRV E1-141 and E2-200 glycosylation sites were discussed in an earlier study (17).
Glycosylation sites in Aura virus.
The glycoproteins of Aura virus, like SINV, have four glycosylation sites. Two of these sites are located on the same residues as SINV (E1-139 and E1-245). The other two sites occur at E2-197 and E2-319 on Aura virus instead of E2-196 and E2-318 in SINV. All of the glycosylation sites in SINV have recently been mapped through difference map analyses computed between wild-type SINV and each of four, single-site glycosylation mutants (E1-139, E1-245, E2-196, and E2-318) (17). Given the close similarity between the Aura virus and SINV structures, difference maps were calculated by subtracting reconstructions of the individual SINV deglycosylation mutants (17) from the AuraT reconstruction. As expected, on the basis of similarity of the AuraT and SINV structures and conservation in amino acid number for glycosylation sites in E1 and E2, the locations of the sites in Aura virus were comparable to those found in SINV.
The T=4 icosahedral symmetry of the virion means that four unique copies of each glycosylation site are present in each asymmetric unit of the particle. These four, quasi-symmetric positions are listed for AuraT (Table 2) and SINV (17). The HOMOlogy program was used to assess the accuracy with which each carbohydrate difference peak within the icosahedral asymmetric unit obeys the T=4 quasi-symmetry (Table 3). The carbohydrate moiety positions in Aura virus obeyed the T=4 quasi-symmetry to within 1.5 Å for the E2-197 and E2-319 sites and to within 3.1 and 5.4 Å for the E1-245 and E1-139 sites, respectively (Table 3). As the E1-245 sites are located near the threefold and quasi-threefold axes, they should exhibit the smallest deviation from T=4 quasi-symmetry. The large variation detected in the E1-245 sites is mainly attributed to measurement error, because three carbohydrate molecules are too close to be resolved and therefore, their densities merge into one larger feature. The three remaining Aura virus sites show much smaller deviations from T=4 quasi-symmetry than the corresponding SINV sites, whose carbohydrate moieties obey the quasi-symmetry to within 1.6 Å for the E1-245 and E2-318 sites and to within 3.4 and 7.4 Å for the E2-196 and E1-139 sites, respectively.
TABLE 2.
Three-dimensional locations of carbohydrate sites in Aura virusa
| Site | A1
|
B1
|
C1
|
D1
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | x | y | z | x | y | z | |
| E1-139 | −13.5 | 33.5 | 294.5 | 15.4 | 31.4 | 296.0 | 29.5 | 1.8 | 296.1 | 131.9 | 6.6 | 268.0 |
| E1-245 | −9.3 | 95.5 | 270.5 | 75.9 | 57.5 | 273.6 | 84.3 | 42.5 | 273.9 | 93.8 | 58.3 | 268.2 |
| E2-197 | −18.9 | 73.7 | 337.3 | 48.0 | 57.2 | 338.6 | 131.6 | 17.0 | 320.5 | 120.2 | 111.8 | 306.2 |
| E2-319 | −17.4 | 76.5 | 264.6 | 53.1 | 55.1 | 265.5 | 90.4 | 23.1 | 261.5 | 99.1 | 70.3 | 248.9 |
The experimental error of these measurements is estimated to be in the range of ±0.3 to ±1.5 Å. This is based on the deviations from a perfect symmetrical arrangement when the Aura virus glycosylation sites near the icosahedral threefold axes are superimposed after applying threefold symmetry, as shown in the first column in Table 3.
TABLE 3.
Superposition of individual glycosylation sites from Aura virus around threefold (i3) and quasi-threefold (q3) axes
| Site | RMS deviation (Å) from T = 4 quasi-symmetrya
|
||
|---|---|---|---|
| i3 on i3 RMS | q3 on q3 RMS | i3 on q3 RMS | |
| E1-139b | 0.6 | 5.4 | 3.1 |
| E1-245 | 1.5 | 1.3 | 3.1 |
| E2-197 | 0.5 | 1.3 | 1.5 |
| E2-319 | 0.3 | 1.5 | 1.2 |
The i3 rotation axis passes through the particle center. Rotation about the spike symmetry axes (Kappa) was 120.0° for i3 on i3 and q3 on q3 for Aura sites. RMS, root mean square.
Corresponds to the B fit, as reported in reference 17.
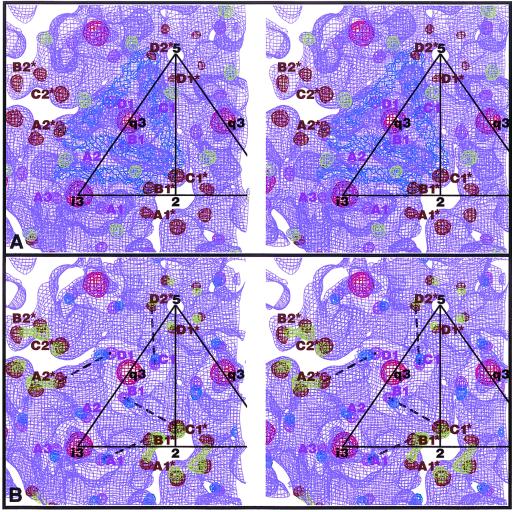
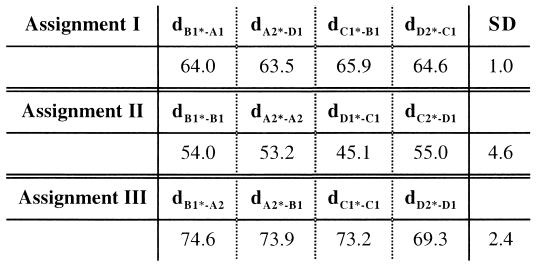
Modeling studies have shown that the long, thin E1 molecules fit nicely onto the skirt region and form an icosahedral scaffold (12, 17) (Fig. 5A). These same studies assigned the E2 molecules to all unoccupied densities, the majority of which occur in the protruding spikes. However, the association between E1 and E2 remains unclear. We have performed statistical analyses to determine which E2-319 sites and E1-139 sites belong to a particular heterodimer based on the locations of the carbohydrate densities. Three E2-319 sites (A1, A2, and B1) surround each B1* site of E1-139 (Fig. 5). Each of these three possible heterodimer assignments (B1*-A1, B1*-A2, or B1*-B1) gives rise to a unique set of four, quasi-symmetric dimers. The distances between E1-139 and E2-319 for each of the four, quasi-symmetric dimers as well as the standard deviations were measured (Fig. 6). Assignment I (Fig. 5B), which links B1* of E1-139 and A1 of E2-319, gives the smallest variation (i.e., ±1.0 Å). The variations found for the other two assignments are ±4.6 Å and ±2.4 Å for B1*-B1 and B1*-A2, respectively. Similar results were found for SINV: ±0.3, ±3.5, and ±3.2 Å for B1*-A1, B1*-B1, and B1*-A2, respectively (17). Therefore, for Aura virus, at a threefold spike, the A1 site of E2-319 and the B1* site of E1-139 probably belong to the same heterodimer.
FIG. 5.
Stereo views of the Aura virus glycosylation sites. (A) Asymmetric unit of the Aura virus glycoproteins. Four SFV E1 molecules (blue) were fitted into the AuraT density map at the lower portion of the spike and in the skirt (violet). The Aura virus carbohydrate densities are shown and were determined by generating difference maps between AuraT and each of the four SINV deglycosylation mutants (17). The AuraT carbohydrate densities are colored (E1-139 in brown, E1-245 in red, E2-197 in light green, and E2-319 in magenta). (B) Electron density of Aura virus and the shift in position between the Aura carbohydrate densities and those of SINV (green for E1-139 and blue for E2-318). The broken lines indicate possible pairs of Aura E1-139 and E2-319 carbohydrate sites which belong to one heterodimer (assignment I in Fig. 6).
FIG. 6.
Distances between E1-139 and E2-319 sites in the asymmetric unit in Aura virus. Distances are given in angstroms. The standard deviation (SD) for each assignment is also given. dB1∗-A1, distance between B1∗ and A1.
DISCUSSION
EM of AuraT and AuraB.
Cryo-EM images of AuraT and AuraB are indistinguishable. As revealed in the three-dimensional reconstruction analyses, particles in both fractions are 710 Å in diameter and they obey T=4 icosahedral symmetry. The surface features of the glycoproteins are essentially indistinguishable from those of SINV virions. Thus, the findings that these particles separate into two components during sucrose gradient sedimentation and that many negatively stained particles in the bottom component appear to be smaller, uniform particles with a diameter of 620 Å are puzzling. These differences presumably arise as a result of differences in the nucleic acid content in the two components. Given the technical difficulties in separating top and bottom components, it appears that the top component consists of particles that contain mostly genomic RNA and that the bottom component consists of particles that contain primarily subgenomic RNA. We suggest that the smaller particles seen by negative staining and EM have a suboptimal content of RNA, perhaps only a single copy of the subgenomic 4.2-kb RNA, and that the structure collapses upon exposure to the negative stain. Perhaps particles that do not collapse have more RNA, or perhaps they survive intact upon addition of stain. The fraction of particles that collapse could depend upon the vagaries of exactly how they are exposed to the stain, and we have found that different preparations for EM observation differ in the fraction of smaller particles observed. If these assumptions are correct, it is still puzzling why particles that contain less RNA would sediment faster. Perhaps the high concentration of sucrose induces a reversible contraction of the particles that contain less RNA, causing them to sediment faster. However, other explanations may be just as plausible.
These results have significant implications for understanding how the structure of the virion forms. They predict that the final T=4 structure is not dependent upon RNA content but is a fundamental property of the proteins that form the structure. Two forces are believed to contribute to the T=4 structure. With most alphaviruses, nucleocapsids form in the cytoplasm independent of interactions with glycoproteins, and nucleocapsids can be assembled in vitro in the absence of glycoproteins (25, 30). Although the precise structure of these nucleocapsids has not been determined, they do possess the requisite number (240) of CP molecules for a T=4 structure. Significantly, when assembled in vitro, the size of the nucleocapsids is independent of the size of the nucleic acid used to initiate assembly (25). Assembly using polynucleotides of length 14 gives rise to the same-size structure as does the viral genomic RNA. The glycoproteins, in particular E1, are also important for forming a stable T=4 structure. The glycoproteins can form a regular lattice in the absence of nucleocapsids (28) and can assist the formation of nucleocapsids in mutants that are otherwise unable to form nucleocapsids (7). Thus, formation of a T=4 structure appears to be an innate property of both the CP protein and the glycoproteins. In this regard, it is noteworthy that RNA molecules that are more than 10 to 20% larger than the wild-type genome cannot be incorporated into virions, which is consistent with the hypothesis that the composition of the structure formed by the viral proteins is predetermined (8).
If this analysis is correct, the assembly pathways of the alphaviruses differ from those used by flaviviruses in a number of fundamental properties. The E1 protein of alphaviruses has been shown recently to have a structure very similar to that of E protein of flaviviruses and to fulfill a similar role in the structure of the mature virion (12, 17). However, the flavivirus virion is a T=1 structure in which the component E proteins are not all present in quasi-equivalent positions (11a), whereas the alphavirus virion is a T=4 structure with all E1 molecules present in quasi-equivalent positions. Of interest is the fact that E and prM of flaviviruses will form a T=1 structure in the absence of nucleocapsids (6), whereas an equivalent structure has never been seen in alphaviruses. Thus, the E protein of flaviviruses can participate in alternative structures, whereas the E protein of alphaviruses appears to be predetermined to a T=4 structure.
Structure of Aura virus at 17-Å resolution.
The structure of the Aura virion is very similar to that of SINV. Based on the difference map analyses, the details of the skirt and subskirt regions resemble those of SINV rather than SFV or RRV. Also the locations of the carbohydrate chains in Aura virus are strikingly similar to those in SINV but differ from those in RRV. The conservation of this feature within the SINV lineage is consistent with the hypothesis that the carbohydrate moieties have a structural role in the assembly or disassembly of the virion and are not present simply to increase the solubility of the proteins during folding. Also consistent with this hypothesis is the fact that of the four carbohydrate chains present on E1 and E2, only one chain (at E2 position 197 in Aura [SINV position 196]) is on the distal surface of the glycoprotein spike where it might interact with the external solvent. The other three chains reside in the skirt region where they appear to interact with domains within the glycoprotein, with each other, or with bound solvent, rather than with the external solvent.
The carbohydrate chains in the SFV/RRV lineage occupy detectably different positions from those in the SINV lineage, suggesting that the structures of the virions differ in ways that may be important for function. In this regard, it is notable that the sequences of the E2 proteins of SINV and Aura virus can be aligned throughout their length without the introduction of gaps, with the exception that Aura virus has one extra amino acid at the N terminus. In contrast, alignment of SINV and SFV or RRV E2 proteins requires the insertion of several gaps. The ectodomains of E1 of Aura virus, SINV, SFV, and RRV can all be aligned without gaps. This may reflect the importance of the structural role of this protein in forming the skirt and imposing a T=4 structure on the virion.
Acknowledgments
We thank Michael Rossmann, Ellen Strauss, and Rushika Perera for useful discussions.
This work was supported in part by NIH grants to T.S.B. (AI45976), R.J.K. (GM56279), and J.H.S. (AI20612) and an NSF shared instrumentation grant (BIR-9112921) to T.S.B. We also thank Purdue University for an instrumentation reinvestment grant to the Purdue Structural Biology Group.
REFERENCES
- 1.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 2.Baker, T. S., J. Drak, and M. Bina. 1988. Reconstruction of the three-dimensional structure of simian virus 40 and visualization of the chromatin core. Proc. Natl. Acad. Sci. USA 85:422-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryoelectron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belnap, D. M., N. H. Olson, and T. S. Baker. 1997. A method for establishing the handedness of biological macromolecules. J. Struct. Biol. 120:44-51. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, R. H., R. J. Kuhn, N. H. Olson, M. G. Rossmann, H. K. Choi, T. J. Smith, and T. S. Baker. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis. Molecular Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 7.Forsell, K., L. Xing, T. Kozlovska, R. H. Cheng, and H. Garoff. 2000. Membrane proteins organize a symmetrical virus. EMBO J. 19:5081-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolov, I., T. A. Hoffman, B. M. Pragai, S. A. Dryga, H. V. Huang, S. Schlesinger, and C. M. Rice. 1996. Alphavirus-based expression vectors: strategies and applications. Proc. Natl. Acad. Sci. USA 93:11371-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller, S. D., S. J. Butcher, R. H. Cheng, and T. S. Baker. 1996. Three-dimensional reconstruction of icosahedral particles-the uncommon line. J. Struct. Biol. 116:48-55. [DOI] [PubMed] [Google Scholar]
- 10.Harrison, S. C., R. K. Strong, S. Schlesinger, and M. T. Schlesinger. 1992. Crystallization of Sindbis virus and its nucleocapsid. J. Mol. Biol. 226:277-280. [DOI] [PubMed] [Google Scholar]
- 11.Kääriäinen, L., and H. Söderlund. 1971. Properties of Semliki Forest virus nucleocapsid. 1. Sensitivity to pancreatic ribonuclease. Virology 43:291-299. [DOI] [PubMed] [Google Scholar]
- 11a.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lescar, J., A. Roussel, M. W. Wein, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 13.Mancini, E. J., M. Clarke, B. E. Gowen, T. Rutten, and S. D. Fuller. 2000. Cryoelectron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5:255-266. [DOI] [PubMed] [Google Scholar]
- 14.Olson, N. H., T. S. Baker, J. E. Johnson, and D. A. Hendry. 1990. The three-dimensional structure of frozen-hydrated Nudaurelia capensis β virus, a T = 4 insect virus. J. Struct. Biol. 105:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paredes, A., K. Alwell-Warda, S. C. Weaver, W. I. Chiu, and S. J. Watowich. 2001. Venezuelan equine encephalomyelitis virus structure and its divergence from Old World alphaviruses. J. Virol. 75:9532-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paredes, A. M., D. T. Brown, R. Rothnagel, W. Chiu, R. J. Schoepp, R. E. Johnston, and B. V. V. Prasad. 1993. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA 90:9095-9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on Sindbis virus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powers, A. M., A. C. Brault, Y. Shirako, E. G. Strauss, W. L. Kang, J. H. Strauss, and S. C. Weaver. 2001. Evolutionary relationships and systematics of alphaviruses. J. Virol. 75:10118-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice, C. M., and J. H. Strauss. 1981. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc. Natl. Acad. Sci. USA 78:2062-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rümenapf, T., D. T. Brown, E. G. Strauss, M. König, R. Rameriz-Mitchel, and J. H. Strauss. 1995. Aura alphavirus subgenomic RNA is packaged into virions of two sizes. J. Virol. 69:1741-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rümenapf, T., E. G. Strauss, and J. H. Strauss. 1995. Aura virus is a New World representative of Sindbis-like viruses. Virology 208:621-633. [DOI] [PubMed] [Google Scholar]
- 22.Rümenapf, T., E. G. Strauss, and J. H. Strauss. 1994. Subgenomic mRNA of Aura alphavirus is packaged into virions. J. Virol. 68:56-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlesinger, S., R. Levis, B. G. Weiss, M. Tsiang, and H. Huang. 1987. Replication and packaging sequences in defective interfering RNAs of Sindbis virus. UCLA Symp. Mol. Cell Biol. New Ser. 54:241-250. [Google Scholar]
- 24.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tellinghuisen, T. L., A. E. Hamburger, B. R. Fisher, R. Ostendorp, and R. J. Kuhn. 1999. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tellinghuisen, T. L., R. Perera, and R. J. Kuhn. 2001. Genetic and biochemical studies of the assembly of an enveloped virus. Genet. Eng. 23:83-112. [DOI] [PubMed] [Google Scholar]
- 27.Vénien-Bryan, C., and S. D. Fuller. 1994. The organization of the spike complex of Semliki Forest virus. J. Mol. Biol. 236:572-583. [DOI] [PubMed] [Google Scholar]
- 28.von Bonsdorff, C.-H., and S. C. Harrison. 1975. Sindbis virus glycoproteins form a regular icosahedral surface lattice. J. Virol. 16:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss, B., H. Nitschko, I. Ghattas, R. Wright, and S. Schlesinger. 1989. Evidence for specificity in the encapsidation of Sindbis virus RNAs. J. Virol. 63:5310-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wengler, G., U. Boege, G. Wengler, H. Bischoff, and K. Wahn. 1982. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology 118:401-410. [DOI] [PubMed] [Google Scholar]