Abstract
Pattern recognition proteins such as lipopolysaccharide and β-1,3-glucan binding protein (LGBP) play an important role in the innate immune response of crustaceans and insects. Random sequencing of cDNA clones from a hepatopancreas cDNA library of white spot virus (WSV)-infected shrimp provided a partial cDNA (PsEST-289) that showed similarity to the LGBP gene of crayfish and insects. Subsequently full-length cDNA was cloned by the 5′-RACE (rapid amplification of cDNA ends) technique and sequenced. The shrimp LGBP gene is 1,352 bases in length and is capable of encoding a polypeptide of 376 amino acids that showed significant similarity to homologous genes from crayfish, insects, earthworms, and sea urchins. Analysis of the shrimp LGBP deduced amino acid sequence identified conserved features of this gene family including a potential recognition motif for β-(1→3) linkage of polysaccharides and putative RGD cell adhesion sites. It is known that LGBP gene expression is upregulated in bacterial and fungal infection and that the binding of lipopolysaccharide and β-1,3-glucan to LGBP activates the prophenoloxidase (proPO) cascade. The temporal expression of LGBP and proPO genes in healthy and WSV-challenged Penaeus stylirostris shrimp was measured by real-time quantitative reverse transcription-PCR, and we showed that LGBP gene expression in shrimp was upregulated as the WSV infection progressed. Interestingly, the proPO expression was upregulated initially after infection followed by a downregulation as the viral infection progressed. The downward trend in the expression of proPO coincided with the detection of WSV in the infected shrimp. Our data suggest that shrimp LGBP is an inducible acute-phase protein that may play a critical role in shrimp-WSV interaction and that the WSV infection regulates the activation and/or activity of the proPO cascade in a novel way.
Invertebrates lack a true adaptive immune system and rely instead on various innate immune responses against invading pathogens (12). Both cellular and humoral immunity play important roles in defense against microorganisms in invertebrates. The cellular immune responses include encapsulation, phagocytosis, and nodule formation (24), whereas the humoral responses include the clotting cascade, the synthesis of a wide array of antimicrobial peptides, and the phenoloxidase (PO)-activating system (prophenoloxidase [proPO] system) (11, 15, 34). Bacteria and fungi have molecules such as lipopolysaccharide (LPS) and β-1,3-glucans on their surfaces that are recognized by nonself recognition molecules and elicit an immune reaction in both vertebrates and invertebrates. Proteins that recognize the LPS and β-1,3-glucans are known as pattern recognition proteins (PRPs) (16). In mammals, the complex of bacterial LPS and the plasma LPS binding protein binds to CD14, which transduces the signal through toll-like receptors and activates the NF-κB signaling cascade (13, 21, 22, 49). In insects and crustaceans, LPS and β-1,3-glucans, upon binding to LPS and glucan binding protein, activate proPO, the coagulation cascade, and the genes for antibacterial effector proteins (11, 15, 33).
Shrimp, like all invertebrates, depend on an innate immune response against pathogenic invasion (1). A number of genes involved in innate immunity in shrimp have been recently characterized. For example, a family of antimicrobial peptides named penaeidins, which have antibacterial and antifungal activities, were isolated from shrimp (Penaeus vannamei) (5). The gene encoding proPO was cloned from shrimp (Penaeus monodon) and found to be very similar to that for crayfish proPO (35). Antiviral substances from shrimp (Penaeus setiferus), blue crab, and crayfish that bind to a variety of DNA and RNA viruses (Sindbis virus, vaccinia virus, vesicular stomatitis virus, mengovirus, Banzi virus, and poliomyelitis virus) have been isolated (29). The inhibitory activity of these antiviral substances is a component of the innate immune response and may serve as an important barrier to delay the onset of viral infection (29). A quasi-immune response in kuruma prawn (Penaeus japonicus) against white spot virus (WSV) has been reported previously (45). When survivors of WSV natural outbreaks or experimental infections were challenged with WSV, a survivability value of 64 to 94% was recorded. Although the survivors of the natural outbreak did not show any WSV-neutralizing activity, it was found in the experimentally infected shrimp 17 days postchallenge. Such an immune response in P. japonicus has been attributed to a quasi-immune response (45).
WSV is the most devastating viral pathogen of cultured penaeid shrimp (Penaeus spp.) worldwide (9, 23). The virus has a wide host range among crustaceans and infects all commercially important species of penaeid shrimp (9). WSV is an enveloped virus with bacilliform morphology, ∼275 by 120 nm in size, and has a tail-like projection at one end of the particle (47). The viral genome contains double-stranded DNA of ∼292 to 305 kb in length (42, 48). WSV is morphologically similar to insect baculovirus. However, phylogenetic analysis of ribonucleotide reductase and protein kinase genes revealed that WSV does not share a common ancestor with baculoviruses (40, 41). Although considerable progress has been made in the molecular characterization of WSV, information on shrimp genes that are involved in WSV pathogenesis remains elusive.
In order to isolate host genes that are differentially expressed due to WSV infection, a cDNA library was constructed from hepatopancreas tissue of WSV-infected shrimp (Penaeus stylirostris). Random sequencing of cDNA clones from the library provided a partial clone that showed significant similarity to a crayfish LPS and β-1,3-glucan binding protein (LGBP) gene (25). The full-length LGBP gene of shrimp was then cloned and sequenced, and its temporal expression was measured by real-time reverse transcription-PCR (RT-PCR) in healthy and WSV-infected shrimp. Since binding of LPS to LGBP is known to activate the proPO cascade, we also measured the temporal expression of proPO in the same healthy and WSV-infected shrimp. The mRNA expression of the LGBP gene was found to be upregulated in WSV-infected animals. The expression of the proPO gene was upregulated initially, followed by a downregulation as the virus infection progressed, suggesting that WSV infection inhibits the activation and/or activity of the proPO gene. The demonstration that the LGBP gene is upregulated in WSV-infected shrimp and the consequent downregulation of proPO with disease progression open a new insight into the involvement of PRPs in viral pathogenesis in invertebrates and in particular in WSV pathogenesis in shrimp.
MATERIALS AND METHODS
WSV challenge experiment.
Specific-pathogen-free (SPF) juvenile (1.5- to 2.0-g) shrimp (P. stylirostris) from Super Shrimp Inc. stocks were used for all WSV challenge work. Animals were maintained in environmentally controlled 40-liter glass salt water aquaria (32- to 36-ppt salinity, 27 to 28°C) and fed a high-protein commercial feed (Aquafauna; Bio-Marine, Inc., Hawthorne, Calif.). A WSV (Chinese isolate, 1995) inoculum was prepared with virus-infected shrimp tail muscle that tested positive by real-time quantitative PCR using SYBR Green chemistry as described previously (6). Frozen infected tissue was homogenized in sterile 2% NaCl (1:10 [wt/vol]) solution and centrifuged in a tabletop centrifuge (Beckman Microfuge Lite model) at 5,000 rpm for 5 min. The supernatant was filtered through an 0.45-μm-pore-size filter and used for injecting the animals. The negative control healthy tissue homogenate was prepared from PCR-confirmed healthy tail tissue by the same method as that used for preparing the WSV inoculum. SPF animals were injected with either ∼30 μl (106 copies) of WSV inoculum or healthy tissue homogenate between the second and third tail tergal plates on the lateral side of the shrimp with a 1-ml tuberculin syringe. Moribund animals and healthy injected animals were collected 36 h postinjection (p.i.) and stored at −80°C. Two independent WSV challenge experiments were performed.
To study the kinetics of expression of LGBP and proPO genes in healthy and WSV-infected shrimp, a time course study was performed. SPF P. stylirostris shrimp were injected either with WSV inoculum or healthy tissue homogenate as described above, and samples were collected at 0, 2, 4, 8, 16, and 32 h p.i. There were seven healthy and seven WSV-injected animals for each time point. Hemolymph (50 to 150 μl) was drawn from the ventral sinuses of each shrimp with a 1-ml syringe containing 10% sodium citrate. The animals and the hemolymph samples were immediately stored at −80°C. Total RNA was isolated from the hepatopancreas and the hemolymph samples with TRI reagent according to the manufacturer's protocol (Molecular Research Center Inc.).
Quantification of WSV load by real-time quantitative PCR using SYBR Green chemistry (SYBR Green PCR).
WSV load was measured in hepatopancreas tissue collected 36 h p.i. Total genomic DNA was extracted from the hepatopancreas of healthy and WSV-infected animals according to the DNAzol protocol (Molecular Research Center Inc.). The sequence of the primers used for WSV quantification and the reaction mixture were the same as described previously (6).
For the WSV time course experiment, DNA was extracted from tail muscle of both healthy and WSV-injected animals at each time point by the DNAzol protocol. WSV load was measured with the primers based on the WSV structural protein gene VP26 (470F, 5′GCAGGAAACATTAAGGGAAATACTATG3′, and 570R, 5′TTGCTGCACACGTCAATGAG3′). The protocol for SYBR Green PCR was the same as described previously (6).
Construction of a cDNA library and isolation of the LGBP gene.
A cDNA library was constructed from the hepatopancreas of WSV-challenged shrimp at 36 h p.i. mRNA was isolated from hepatopancreas total RNA with an oligo(dT) cellulose spin column (Poly A Pure mRNA isolation kit; Ambion Inc., Austin, Tex.). cDNA was synthesized with oligo(dT) primer and directionally cloned into the Uni-Zap XR vector with the ZAP-cDNA synthesis kit and the ZAP-cDNA Gigapack III Gold cloning kit (Stratagene, La Jolla, Calif.). The primary library was amplified to 5.18 × 106 PFU/ml. A small aliquot of the amplified library was taken for mass excision with ExAssist helper phage and Escherichia coli Solar strain according to the manufacturer's protocol (Stratagene). Colony PCR was done on randomly selected phagemid clones with T3 and T7 primers. Eighty recombinant clones containing an insert of >350 bp were taken for sequencing with the T3 primer. Sequencing was done in an automated DNA sequencer (Model ABI 373A) with the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.). An 808-bp clone (PsEST 289) showed similarity to a GenBank database entry for the crayfish LGBP gene. Internal primers were then designed to complete the sequencing of PsEST 289.
Cloning the 5′ end of PsEST 289 by the RACE (rapid amplification of cDNA ends) technique.
The 5′ end of the shrimp LGBP gene was cloned by an RNA ligase-mediated RACE method (RLM 5′-RACE; Ambion Inc.). Two internal primers, 5′-CTGGACTCCAAAATGTCGATCTC-3′ (nucleotide position 604 to 626, Fig. 1) and 5′-TACTCGACGTGGGTCTTCTCGA-3′ (nucleotide position 710 to 731, Fig. 1), were designed based on the sequence of PsEST 289. These two primers were used in conjunction with the RLM 5′-RACE adapter primer to capture the 5′ end of the shrimp LGBP gene. The resulting 627-bp fragment was cloned into pIB/V5-His TOPO vector with the TA cloning kit (Invitrogen Inc., Carlsbad, Calif.). Three recombinant clones were sequenced with an automated DNA sequencer (Model ABI 373A).
FIG. 1.
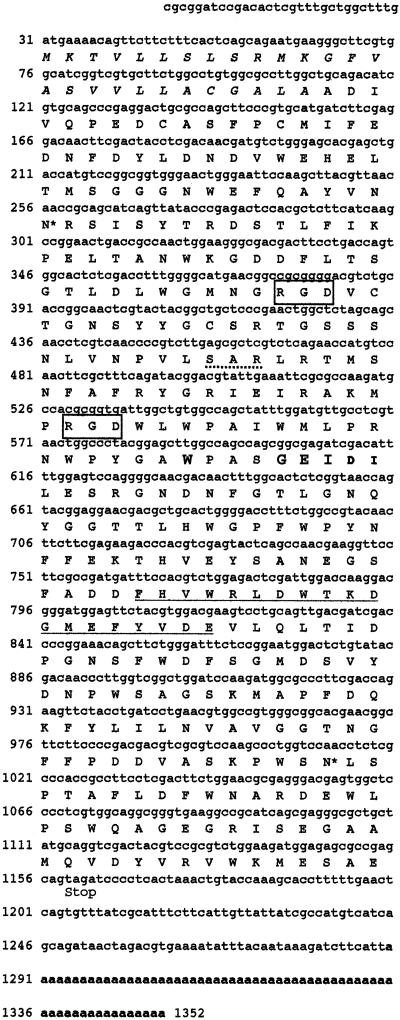
Nucleotide and deduced amino acid sequences of P. stylirostris LPS and LGBP gene. The nucleotide sequence is numbered from the 5′ end, and the single-letter amino acid code is presented below the corresponding codon. The putative signal sequence (amino acid position 1 to 27) is shown in italics. Potential N-glycosylation sites are marked with asterisks. The underlined amino acid sequence represents a potential recognition motif for β-1,3-linkage of polysaccharides. Two RGD (Arg-Gly-Asp) putative cell adhesive sites are boxed. A dotted line marks a potential kinase C phosphorylation site, and the amino acids that compose the functional domain of glucanase are shown in boldface.
Sequence analysis.
The shrimp LGBP gene sequence was used for a GenBank database search with BLASTX and BLASTP searches (http://blast.genome.ad.jp). Multiple alignment of the shrimp LGBP gene was performed with the ClustalW Multiple Alignment program (http://searchlauncher.bcm.tmc.edu/) and BOXSHADE. The protein domain features of the shrimp LGBP gene were determined by using the InterPro website hosted by the European Bioinformatics Institute, Cambridge, United Kingdom (www.ebi.ac.uk/interpro/).
Quantification of LGBP gene expression by real-time RT-PCR.
The mRNA expression of the shrimp LGBP gene in hepatopancreas tissue of healthy and WSV-injected shrimp was measured by real-time RT-PCR in two independent WSV challenge experiments. The cDNA synthesis was carried out in a 20-μl reaction volume containing 1 μg of DNase-treated total RNA, 1× PCR buffer (pH 8.2), 1 mM deoxynucleoside triphosphate (dNTP; PE Applied Biosystems), 0.75 μM oligo(dT), 4 U of RNase inhibitor (PE Applied Biosystems), and 5 U of MutiScribe reverse transcriptase (PE Applied Biosystems). cDNA was diluted to 1:10, and 1 μl of the 1:10 dilution was used for each SYBR Green RT-PCR. The SYBR Green RT-PCR assay was carried out in a GeneAmp 9600 thermocycler coupled with a GeneAmp 5700 Sequence Detection System (PE Applied Biosystems). The amplifications were performed in a 96-well plate in a 25-μl reaction volume containing 7.1 μl of 2× SYBR Green Master Mix (PE Applied Biosystems), 0.24 μM (each) LGBP gene-specific forward (289-170F, 5′GGTAACCAGTACGGAGGAACGA3′) and reverse (289-233R, 5′TACTCGACGTGGGTCTTCTCGA3′) primers, and 1 μl of 1:10-diluted cDNA. The thermal profile for SYBR Green RT-PCR was 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. In a 96-well plate, each sample was run in triplicate along with an internal control gene, the shrimp elongation factor 1α (EF-1α) gene. Recently, we isolated a constitutively expressed gene from P. stylirostris shrimp, the EF-1α gene, while comparing the RNA fingerprints of healthy and WSV-infected animals. Cloning and sequencing of a 382-bp EF-1α cDNA showed that it has significant similarity to yeast EF-1α (score of 157; E = 4e-38; identities = 77 of 97, 79%; positive, 87 of 97, 89%) (A. K. Dhar, K. R. Klimpel, M. M. Roux, K. Astrofsky, and J. G. Fox, Plant and Animal Genome IX, The International Conference on the Status of Plant and Animal Genome Research, 13 to 17 January 2001, San Diego, Calif.). We have also shown previously that the shrimp EF-1α gene is a better internal control for real-time RT-PCR than is the shrimp β-actin gene (7). Based on these data, the shrimp EF-1α gene was used as an internal control for all real-time PCR assays described in this paper. The nucleotide sequences of shrimp EF-1α primers used for SYBR Green RT-PCR assay were as follows: forward, 5′TCGCCGAACTGCTGACCAAGA3′, and reverse, 5′CCGGCTTCCAGTTCCTTACC3′. To determine reproducibility, the SYBR Green assay was repeated three times independently.
In addition to measurement of LGBP gene expression in healthy and WSV-injected shrimp at 36 h p.i., the temporal expression of the LGBP gene in the hepatopancreas of healthy and WSV-injected animals was measured at different time points (0, 2, 4, 8, 16, and 32 h) after virus challenge. The cDNA synthesis, SYBR Green reaction mixture, and the temperature profile for the RT-PCR were the same as described above.
Cloning of the P. stylirostris proPO gene and measurement of the expression of proPO in healthy and WSV-injected shrimp by real-time RT-PCR.
A 563-bp proPO cDNA was amplified by RT-PCR with hemolymph total RNA isolated from WSV-infected shrimp (P. stylirostris). The primers for proPO RT-PCR were designed based on the published sequence of the proPO gene from P. monodon (35). The primer sequences were as follows: 735F, 5′GGAATTGTTTTACTACATGCATCAGC3′, and 1297R, 5′GGAACAAGTCATCCACGAGCTT3′. cDNA synthesis was done with MutiScribe reverse transcriptase (PE Applied Biosystems) as described above. The RT-PCR was carried out in a 20-μl reaction volume containing 1 μl of cDNA reaction mix, 1× AmpliTaq Gold Buffer (Applied Biosystems), 2.5 mM MgCl2, 0.5 μM dNTPs, 1 μM (each) forward and reverse primers, and 2.5 U of AmpliTaq Gold (Applied Biosystems). The temperature profile for PCR was 95°C for 10 min followed by 30 cycles of 95°C for 45 s, 55°C for 1 min, and 72°C for 1 min. The amplified cDNA was run in a 1% agarose gel, gel purified with Qiaquick (Qiagen, Valencia, Calif.) columns, and cloned into a TOPO vector (TOPO TA cloning kit for sequencing; Invitrogen). Three recombinant clones were sequenced, and based on this sequence, primers were designed for measuring the proPO level in the hemolymph of healthy and WSV-injected shrimp by SYBR Green RT-PCR assay. The primers used for proPO assay by real-time PCR were 408F, 5′CGACATCCTGGCCTTCTACAC3′, and 463R, 5′AAAGCCCATTTCCTCCTTGT3′. The hemolymph RNAs isolated from the animals at different time points were dissolved in 25 μl of Tris-EDTA, pH 8.0, and cDNA was synthesized in a 40-μl reaction volume containing 1× PCR buffer (pH 8.2), 1 mM dNTP (PE Applied Biosystems), 0.75 μM oligo(dT), 4 U of RNase inhibitor (PE Applied Biosystems), and 5 U of MutiScribe reverse transcriptase (PE Applied Biosystems). Five out of 40 μl of cDNA was used for each SYBR Green RT-PCR for measuring the proPO level. In a 96-well plate, each sample was run in duplicate along with the internal control gene, the EF-1α gene.
Since proPO expression is known to be upregulated due to infection by gram-negative bacteria, we injected juvenile (1.5- to 2.0-g) shrimp (P. stylirostris) with dead avirulent gram-negative bacteria, Caulobacter crescentus. The bacterial culture was diluted to 1.6 × 107 cells/ml with 2% saline, and 10 μl was injected between the second and third tail tergal plates on the lateral side of the shrimp with a 1-ml tuberculin syringe. Hemolymph collected from bacterially challenged animals served as positive controls for proPO assay by real-time RT-PCR. Animals injected with 2% saline served as negative controls. Hemolymph was drawn from bacterially challenged and control animals at 32 h p.i., and total RNA was isolated with TRI reagent. RNA was treated with DNase I before being used for SYBR Green RT-PCR.
Data analysis for SYBR Green RT-PCR.
At the end of each SYBR Green RT-PCR run, data analysis was performed with the 5700 Sequence Detection System software (SDS version 1.3) and the method described elsewhere (6; Dhar et al., Plant and Animal Genome IX, 2001). Briefly, the threshold PCR cycle (CT) is defined as the cycle number at which a statistically significant increase in fluorescence of SYBR Green against the internal passive dye ROX (ΔRn) is first detected. The copy number of the target gene and CT values are inversely related; thus, a sample containing a higher number of copies of the target gene has a lower CT value than that of a sample with a lower number of copies of the same target. The differences in the CT values of LGBP and proPO genes and the corresponding internal control EF-1α gene, called ΔCT, were calculated to normalize for any difference in the amount of total RNA added to the cDNA reaction mixture and the efficiency of the reverse transcription reaction. The ΔCT for the healthy sample was subtracted from the ΔCT of the WSV-infected sample. The difference was expressed as a ΔΔCT value that allowed measurement of the change in expression of LGBP and proPO genes in the WSV-infected sample relative to the healthy control. A 3.3-CT-value change is considered to be equivalent to 10-fold changes in expression. The CT values were exported into a Microsoft Excel sheet for subsequent data analyses. An unpaired, two-tailed t test was done with the program Instat 1.41 to compare the LGBP and proPO gene expression levels in healthy and WSV-infected shrimp.
Northern blot hybridization.
DNase-treated total RNAs (4 μg) from healthy and WSV-injected shrimp at 32 h p.i. were run in a 1% formaldehyde agarose gel according to the standard protocol (30). The RNA was then blotted onto a nylon membrane (Duralon-UV membranes; Stratagene) and baked at 80°C for 1 h. An 808-bp cDNA representing the shrimp LGBP gene was amplified by PCR with T3 and T7 primers from the cDNA clone PsEST 289. The amplified cDNA was run in a 1% agarose gel and gel purified with Qiaquick columns, and radiolabeled probe was made with [α-32P]dCTP (NEN Life Science Products, Inc.) and the random primer labeling kit Prime It II (Stratagene). Prehybridization and hybridizations were performed according to the SuperHyb Kit protocol (Molecular Research Center Inc.). After hybridization, the membrane was exposed to Kodak film at −80°C. The PsEST 289 probe was then removed, and the membrane was hybridized with an EF-1α probe. The hybridization and washing conditions for the EF-1α probe were same as those used for the LGBP probe. The Northern blots were scanned with the program NIH Image 1.62 (http://rsb.info.nih.gov/nih-image/). First, the density measurements of the EF-1α gene in healthy and WSV-infected animals were analyzed with an unpaired, two-tailed t test to ensure that the shrimp EF-1α gene was constitutively expressed. Then the ratio of LGBP to EF-1α density was calculated for healthy and WSV-infected samples and analyzed by an unpaired, two-tailed t test.
Nucleotide sequence accession number.
The consensus sequence was submitted to the GenBank database under accession no. AF 473579.
RESULTS
cDNA cloning and sequencing of the shrimp LGBP gene.
Eighty cDNA clones obtained from the hepatopancreas cDNA library of a WSV-infected shrimp were sequenced (A. K. Dhar, unpublished data). A GenBank database search showed that an 808-bp clone (PsEST 289) has significant similarity (84% amino acid similarity, score of 363, E = 2e-99) to the 3′ end of the open reading frame (ORF) of a crayfish LGBP gene (25). Northern blot hybridization was then performed with the radiolabeled PsEST 289 probe (see “Northern blot hybridization” above). A single transcript of approximately 1.35 kb was detected. This indicated that the shrimp LGBP partial clone (PsEST 289) was missing approximately 550 bp from the 5′ end of the coding region in the gene. By the RLM 5′-RACE technique, a 627-bp cDNA was amplified. The sequence of 627 bp 5′-RACE clones overlapped with the sequence of clone PsEST 289. The 1,352-bp consensus sequence of PsEST 289 and the RACE clones represented the entire LGBP gene, since it corresponded to the LGBP transcript size detected by Northern blot hybridization (see “Northern blot hybridization” above). The 1,352-bp cDNA sequence contained a 5′ untranslated region (UTR) of 30 bases followed by an ORF of 1,131 bases, a 3′ UTR of 129 bases, and a poly(A) stretch of 62 bases (Fig. 1). The ORF is capable of encoding a polypeptide of 376 amino acids with an estimated molecular mass of 44 kDa. Shrimp LGBP showed significant amino acid similarity with LPS-LGBP of the crayfish Pacifastacus leniusculus (79% similarity, E = e-145 [25]), β-1,3-glucanase of the sea urchin Strongylocentrotus purpuratus (65% similarity, E = 2e-96 [2]), and the coelomic cytolytic factor of the earthworm Eisenia foetida (58% similarity, E = 6e-79 [4]). It also showed similarity to the gram-negative bacterial binding protein of the mosquito Anopheles gambiae (64% similarity, E = 4e-88 [8]), Drosophila melanogaster (50% similarity, E = 7e-38 [20]), and the fall webworm, Hyphantria cunea (46% similarity, E = 1e-30 [32]). Moreover, shrimp LGBP showed amino acid similarity to the LGBP of the tobacco hornworm, Manduca sexta (50% similarity, E = 6e-42 [26]), and the silkworm, Bombyx mori (52% similarity, E = 6e-45 [28]). It is interesting that the shrimp LGBP gene showed similarity to the glucanase domain of the alpha subunit of coagulation factor G in the horseshoe crab (48% similarity, E = 5e-12 [31]) and several other bacterial β-1,3- and β-1,3,4-glucanases.
The shrimp LGBP gene contains a putative signal sequence, two potential glycosylation sites, a potential recognition motif for β-1,3-linkage of polysaccharides, two putative cell adhesion sites (Arg-Gly-Asp), and a protein kinase C phosphorylation site (Fig. 1). Multiple alignment of the shrimp LGBP gene with the homologous genes of insect, crustacean, annelid, and tunicate species revealed that these sequence motifs remained largely conserved across species boundaries, indicating the functional conservation of these sequence motifs (Fig. 2).
FIG. 2.
Multiple alignment of shrimp LGBP gene with the homologous genes of crayfish (AJ250128), silkworm (AB026441), tobacco hornworm (AF177982), purple urchin (U49711), mosquito (AJ001042), fruit fly (AF228472), earthworm (AF030028), and fall webworm (AF023916). The black shading indicates identical amino acids, and the gray shading indicates conservative replacements. The numbers to the left indicate the amino acid position of LGBP in the corresponding species.
Quantification of WSV load by SYBR Green PCR.
WSV load was measured in samples collected at 36 h p.i. from two independent WSV challenge experiments as well as the samples from the temporal regulation study. For all WSV challenge experiments, the virus was detected only in the virus-injected samples, not in the healthy samples. The viral load in shrimp at 36 h p.i. varied from 1.5 × 106 to 2.1 × 108 copies/ng of total cellular DNA. For each sample, the amplification of WSV-specific product was confirmed by examining the amplification plots and the corresponding dissociation curves. A dissociation curve with a single peak at the expected melting temperature (Tm = 76.0°C) indicated the amplification of WSV target only.
For the temporal regulation study, the virus was not detected in healthy injected samples from all six time points studied. Among the WSV-injected animals, the virus was detected only at 16 and 32 h p.i. and not at 0, 2, 4, and 8 h p.i. The average viral loads at 16 and 32 h p.i. were 3.51 × 104 and 3.41 × 104 copies/ng of cellular DNA, respectively.
Expression of LGBP gene in healthy and WSV-injected shrimp.
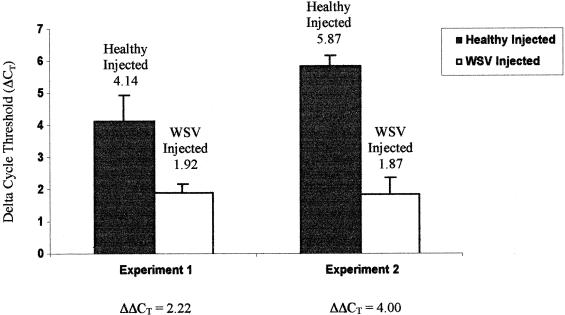
The relative levels of mRNA expression of the LGBP gene in the hepatopancreas tissue of healthy and WSV-challenged animals were measured by SYBR Green RT-PCR at 36 h p.i. In experiment 1, the average levels of the LGBP gene (expressed as ΔCT) in healthy and WSV-infected animals at 36 h p.i. were 4.14 and 1.92, respectively (Fig. 3). Therefore, the difference (ΔΔCT = ΔCT WSV − ΔCT healthy) in the levels of LGBP gene expression between healthy and WSV-infected animals is −2.22. Considering a ΔΔCT of 3.3 equivalent to a 10-fold difference, the WSV-infected animal had an approximately fivefold-higher expression of the LGBP gene than did the healthy animals. In experiment 2, the WSV-infected samples showed 16-fold-higher (ΔΔCT = −4.0) expression of the LGBP gene than did the healthy animals. An unpaired, two-tailed t test showed that a statistically significant difference existed between the two treatment groups for both experiments 1 and 2, with a P value of <0.0001.
FIG. 3.
Measurement of LGBP mRNA expression in healthy and WSV-injected P. stylirostris shrimp by SYBR Green RT-PCR in two independent WSV challenge experiments. Each bar represents the average relative level of LGBP mRNA as measured by SYBR Green RT-PCR. The expression of LGBP and EF-1α genes was measured in healthy and WSV-injected shrimp at 32 h p.i. The ΔCT (CT value of LGBP gene − CT value of EF-1α gene) was measured for each healthy (n = 7) and WSV-infected (n = 7) animal, and the average ΔCT values were used to draw the bars. The numbers above the bars indicate the average ΔCT values. A 3.3-ΔCT-value difference (ΔΔCT) indicates a 10-fold difference between the two treatments. CT values and the copy number of the genes are inversely related, and therefore, a sample with higher CT values contains less of the target gene.
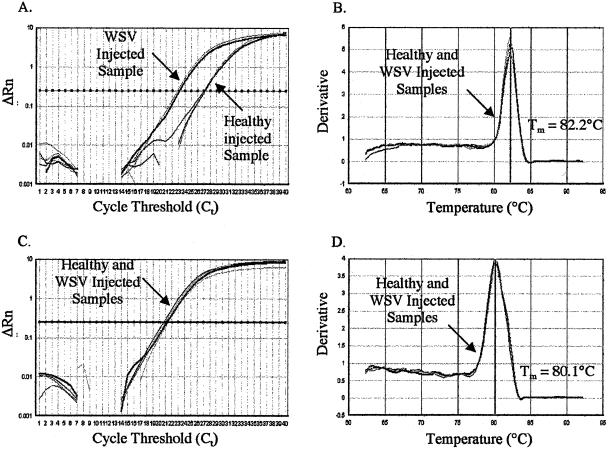
The amplification specificity for LGBP and the corresponding internal control EF-1α was determined for each healthy and WSV-infected animal by analyzing the amplification plots and the dissociation curves. The amplification profiles and dissociation curves of the LGBP and EF-1α genes of representative healthy and WSV-infected samples are shown in Fig. 4. In Fig. 4A, the CT value of the WSV-infected sample was 23, whereas the CT value of the healthy sample was 27. Therefore, the WSV-infected sample had 16-fold-higher expression than the healthy sample. To ensure that the difference in LGBP gene expression observed between healthy and WSV-infected samples was not due to a difference in the amount of input RNA used for the cDNA synthesis, EF-1α was amplified in parallel for both samples (Fig. 4C). Both healthy and WSV-infected samples provided equivalent levels of EF-1α expression (CT value of both healthy and WSV-infected animals was 21.79 [Fig. 4C]). This indicated that the difference in LGBP gene expression observed between healthy and WSV-infected samples was indeed due to the difference in the expression level between the two treatments and not due to variation in input RNA, efficiency of cDNA synthesis, or any other PCR artifact. The specificity of amplification of LGBP and EF-1α genes was determined by analyzing the dissociation curves. A dissociation curve showing a single peak at a melting temperature expected for that amplicon suggests specific amplification. For both LGBP and EF-1α genes, dissociation curves indicated amplification of intended targets with melting temperatures at 82.2 and 80.1, respectively (Fig. 4B and D).
FIG. 4.
Amplification profiles (A and C) and dissociation curves (B and D) of the LGBP gene (A and B) and the internal control EF-1α gene (C and D). The mRNA expression of LGBP and EF-1α genes in healthy and WSV-infected shrimp at 32 h p.i. was quantified by SYBR Green RT-PCR. The melting temperatures (Tm) of the amplicons are indicated alongside their dissociation curves.
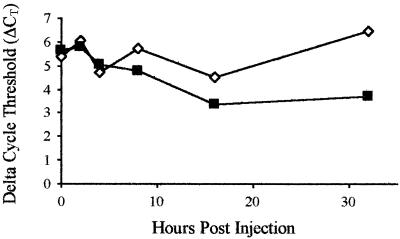
The temporal expression of the LGBP gene in healthy and WSV-infected samples is shown in Fig. 5. At 0, 2, and 4 h p.i, the trend in LGBP gene expression remained the same in healthy and WSV-injected samples. At 8 h p.i., the LGBP gene expression level was about 1.91-fold higher in the virus-injected animals (ΔΔCT = −0.935). At 16 and 32 h p.i., the LGBP gene expression was 2.64-fold (ΔΔCT = −1.402) and 6.97-fold (ΔΔCT = −2.802) higher in the virus-infected animals (Fig. 5). An unpaired, two-tailed t test with healthy and WSV-injected samples showed no significant difference in LGBP gene expression at 0 (P < 0.7570), 2 (P < 0.6514), 4 (P < 0.6631), and 8 (P < 0.1964) h. However, a significant difference at 16 (P < 0.0308) and at 32 (P < 0.0002) h p.i. was observed. Overall, the data indicated that, as the virus infection progressed, the LGBP gene showed an increasingly higher level of expression in the virus-infected animals than in the healthy animals.
FIG. 5.
Temporal expression of the LGBP gene in the hepatopancreas of healthy and WSV-injected P. stylirostris as measured by real-time quantitative RT-PCR. The mRNA expression of the LGBP gene and the internal control EF-1α gene was measured by SYBR Green RT-PCR in each healthy (n = 7) and WSV-injected (n = 7) shrimp at 0, 2, 4, 8, 16, and 32 h p.i. The relative LGBP level as expressed by ΔCT (CT value of LGBP gene − CT value of EF-1α gene) was determined for each sample. The average ΔCT for healthy (⋄) and WSV-injected (▪) samples for each time point was then used to plot the graph.
Northern blot analysis.
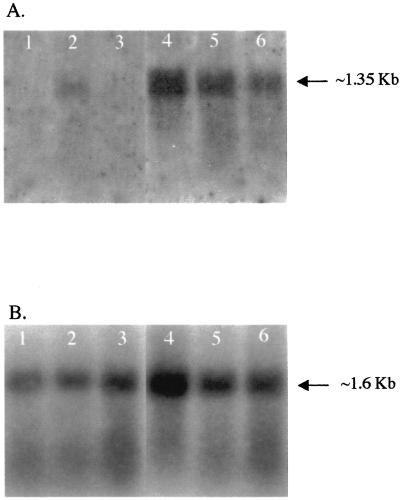
Northern blots containing RNA from healthy and WSV-infected samples were hybridized with a radiolabeled LGBP gene probe. A single transcript of approximately 1.35 kb was detected in both healthy and WSV-infected samples (Fig. 6). The signal intensity was much higher in WSV-infected samples than in healthy samples (Fig. 6). The membrane was then washed to remove the LGBP gene probe and hybridized with the EF-1α probe. A single transcript of approximately 1.6 kb for the EF-1α gene was detected in both healthy and WSV-infected samples. The density of the EF-1α gene in healthy and WSV-infected animals was analyzed by an unpaired, two-tailed t test. The P value was <0.119, indicating that there was no statistically significant difference in the EF-1α gene expression between healthy and WSV-infected shrimp. Then the density ratio of LGBP to EF-1α was calculated for healthy and WSV-infected samples and analyzed by an unpaired, two-tailed t test. The P value was <0.007, which is statistically significant. This indicated that, although there is no statistically significant difference in the mRNA expression level of the constitutively expressed EF-1α gene, the LGBP gene expression showed a significant difference between healthy and WSV-infected animals. The data, therefore, suggested that the LGBP gene was overexpressed in the WSV-infected samples. The Northern blot hybridization data, therefore, supported the real-time RT-PCR findings that the LGBP gene was upregulated in the WSV-infected animals.
FIG. 6.
Autoradiographs of Northern blots containing total RNA from healthy (lanes 1 to 3) and WSV-infected (lanes 4 to 6) shrimp 32 h p.i. (A) Northern blots were hybridized with radiolabeled LGBP probe (PsEST-289). (B) The LGBP probe was removed, and the membrane was hybridized with EF-1α probe. The arrow in each panel indicates the transcript size.
Expression of proPO gene in response to WSV infection.
The mRNA expression of the proPO gene was measured in the hemolymph of healthy and WSV-infected shrimp by SYBR Green RT-PCR at different time points after challenge. In the healthy animals, the proPO level was upregulated at 4 h p.i. followed by a downregulation at 8 h. At 16 h p.i., the proPO level was upregulated again, and at 32 h p.i. the proPO level was comparable to that at 2 h p.i. In the WSV-infected samples, the proPO level showed an upregulation trend until 8 h followed by a downregulation at 16 and 32 h p.i. The proPO level in the WSV-injected animals appeared to correspond to the progression of the virus infection. WSV was detected at 16 and 32 h p.i., with the average virus load being 3.51 × 104 and 3.41 × 104 copies/ng of total DNA, respectively. An unpaired, two-tailed t test showed a significant difference between healthy and WSV-infected samples only at 16 h p.i. (P < 0.0057). Therefore, the data suggest that, in the early hours after infection, the animals are trying to clear the infection and there is an upregulation of proPO expression. As time progresses, the virus multiplies and overwhelms the host defense and proPO expression is significantly downregulated. In contrast, the expression of proPO in the bacterially challenged shrimp remained at a very high level (ΔΔCT = 3.3, 9.85-fold-higher expression) at 32 h p.i. compared to that for the 2% saline-injected animals at the same time point.
DISCUSSION
Crustaceans rely primarily on their innate immune response to protect themselves from a variety of pathogens. While the intricacies of the crustacean immune system have been explored with respect to a number of bacterial and fungal pathogens, little is known about the crustacean response to viral infections. The crustacean's response to viral infection may share some of the same pathways in the innate immune system already explored for defense against bacterial and fungal infections. Indeed, this appears to be the case with respect to WSV infection in shrimp, where a gene normally associated with bacterial infection, the LGBP gene, appears to be upregulated in response to the presence of WSV. The overexpression of the LGBP gene coincided with the activation of the proPO cascade in the initial hours after infection and before the animal was overwhelmed by the viral load and succumbed to the disease.
Toward our overall goal of isolating genes that are differentially expressed in WSV-infected shrimp, we have undertaken a project to randomly sequence clones from a hepatopancreas cDNA library of WSV-infected shrimp. WSV is currently the most economically important viral pathogen of farmed penaeid shrimp. Since the initial report of WSV infection in farmed Marsupenaeus japonicus in Japan in 1993 (14), the virus has spread through much of Asia and Central and South America, causing catastrophic losses to shrimp aquaculture. Therefore, there is an urgent need to understand the molecular basis of WSV pathogenesis in shrimp, which may be helpful in developing strategies for management of the disease and for long-term sustainability of penaeid shrimp farming worldwide.
Random sequencing of expressed sequence tags has accelerated the identification of genes involved in the immune response in many species including shrimp (10). In Pacific white shrimp (P. vannamei) and Atlantic white shrimp (P. setiferus) several immune-function genes have been identified by high-throughput sequencing of expressed sequence tags from a hepatopancreas and hemocyte cDNA library (10). Lectins were the most abundant immune-function genes in the hepatopancreas, and antimicrobial peptides were the most common in the hemocyte (10). Random sequencing of cDNA clones from the hepatopancreas tissue of WSV-infected Pacific blue shrimp (P. stylirostris) provided us with several genes that may potentially be involved in the immune response in shrimp (A. K. Dhar, unpublished data). The hepatopancreas is a key organ involved in the immune response of crustaceans and thus presumed to be a primary site for the production of immune recognition molecules (10, 18). The outer spaces of arterioles in hemal spaces of the hepatopancreas are occupied by the fixed phagocytes that are involved in eliminating the pathogens and other particulate matter from the hemolymph (18, 46).
The organization of the shrimp LGBP gene was found to be very similar to that of the crayfish LGBP gene (25). Unlike the crayfish gene, the shrimp LGBP gene does not contain any repeat element in the 3′ UTR, and the shrimp LGBP gene has two putative cell adhesion motifs (RGD) compared to one in crayfish. In crayfish, the RGD cell adhesion motif was found to serve as a ligand for cell surface integrins and to mediate blood cell adhesion and cellular immunity (17). Many viruses such as foot-and-mouth disease virus and rhinovirus contain a highly conserved RGD domain in the capsid protein VP1 that is involved in recognition of cellular receptors and is dispensable for entry of the virus into the cell (27). It remains to be seen whether WSV binds to LGBP and then whether the complex of LGBP and WSV attaches to the cell surface receptor(s) with the RGD domain.
The amino acid sequence for the shrimp LGBP gene contained a potential recognition motif for β-1,3-linkage of polysaccharides. This motif was conserved in the homologous genes of insects, crustaceans, and other species. It is unknown whether such a motif is involved in the binding of not only LPSs and β-1,3-glucans but also WSV capsid protein(s). WSV encodes four major structural proteins, VP28, VP26, VP24, and VP19 (38). VP26 and VP24 are associated with the nucleocapsid, whereas VP28 and VP19 remain on the virus envelope (38). VP28 has five potential sites for N glycosylation and two sites for O glycosylations (39). An in vitro assay using recombinant LGBP and VP28 proteins will enable us to determine if VP28 could selectively bind to LGBP.
The 3′ end of the shrimp LGBP gene showed similarity with the β-1,3- and β-1,3,4-glucanase of bacteria (Bacillus circulans, Clostridium thermocellum, and Thermotoga maritima). The catalytic residues in the active site of glucanase were found to be conserved in shrimp (W, E, I, and D at amino acid positions 187, 192, 193, and 194, respectively) (Fig. 1). A similar glucanase domain has also been found in the LPS binding protein of insects (8, 28, 32) and earthworm CCF-1 protein (4). Although the CCF-1 protein binds to β-1,3-glucan and bacterial LPS, it lacks glucanase activity, and the glucanase activity has not been demonstrated for the gram-negative bacterial binding protein of other invertebrates. It has been suggested that the LGBP gene of invertebrates might have evolved from bacterial glucanase. During evolution the invertebrate LGBP gene lost the glucanase activity but retained the glucan binding properties and therefore plays a role in innate immunity (25). A β-1,3-glucan binding protein has been isolated from the plasma of both brown shrimp (Penaeus californiensis) and white shrimp (P. vannamei). This 100-kDa protein was shown to enhance the activation of the proPO cascade (43, 44). The N-terminal portion of shrimp β-1,3-glucan binding protein was partially sequenced; however, the gene has not been cloned or characterized.
The shrimp LGBP gene was found to be upregulated in WSV-infected animals. Initially, LGBP gene expression was measured in healthy and WSV-infected shrimp at 36 h p.i. with animals from two independent challenge studies. A considerable difference in LGBP gene expression between experiments 1 and 2 (ΔΔCT of −2.2 and −4.0 [Fig. 5]) was observed. Since no established cell line is available for shrimp, individual animals were used to assess the LGBP gene expression for each experiment. Animal-to-animal variation might have contributed to the difference in LGBP gene expression between the two experiments. Nevertheless, the LGBP gene expression was significantly upregulated (P < 0.0001) in the WSV-injected animals in both experiments. A similar observation was recorded when the temporal expression of the LGBP gene was measured in healthy and WSV-injected shrimp at different hours postinjection. The LGBP gene expression level in healthy and WSV-injected animals did not show any significant difference until 16 h p.i. At 36 h p.i. the difference reached a maximum level, with the WSV-infected animals showing almost 10-fold-higher expression than the healthy animals. The differential expression of the LGBP gene followed the trend in WSV multiplication in the infected animals, since the virus could not be detected until 16 h p.i..
The proPO expression was measured in the hemolymph of healthy and WSV-injected animals from the time course study. In the WSV-infected shrimp, the proPO expression showed an upregulation until 8 h p.i., while at 16 and 32 h p.i. the proPO level was reduced. The proPO expression, therefore, followed a reverse trend compared to LGBP gene expression at 16 and 32 h p.i. In invertebrate blood, the PO remains as an inactive zymogen, proPO, which is activated by serine proteinases to active PO. Inhibitors of serine proteinases that regulate the activation of this enzyme have been cloned from crayfish and insects (34). In shrimp, proPO has been shown to be activated by commercial trypsin proteinase in vitro, although no shrimp serine proteinases have been cloned so far (37). Recently, we have isolated a partial clone of serine protease from a shrimp hepatopancreas cDNA library and measured its temporal expression at 0, 2, 4, 8, 16, and 32 h in the same samples used in the present study. It was observed that serine protease is upregulated in the WSV-infected animals as infection progresses (A. K. Dhar, unpublished data). The fact that proPO expression is not upregulated in the same virus-infected samples leads to the speculation that WSV infection probably activates the expression of a protease inhibitor(s) that blocks the activity of serine protease or the activity of the proPO. In some insect species such as tobacco hornworm (M. sexta) in a parasitoid relationship with the braconid wasp Cotesia congregata and Trichoplusia ni and Heliothes viriscens larvae in a parasitoid relationship with Hyposoter exiguae and Campoletis sonorensis, the parasitoids have evolved mechanisms to inhibit the activation and/or activity of PO in order to suppress or evade the immune mechanisms of the host (3, 36). A similar suppression of PO activity in unparasitized larvae was observed following injection of purified wasp polydnavirus, an enveloped virus with a double-stranded segmented DNA genome. The inactivation of the virus by exposure to psoralen and long-wave UV eliminated the inhibitory activity on PO (3, 36). The female wasp normally injects the virus into the host, suggesting that in parasitized larvae the inhibition of PO is virally mediated (3, 36).
There was a dramatic reduction of hemocyte count as well as PO enzyme activity in the WSV-infected animals compared to the healthy animals at 16 and 32 h p.i. (data not shown). A reduction in hemocyte count in response to infection by nuclear polyhedrovirus, a member of the family Baculoviridae, has been reported for the American bollworm (Heliothes armigera) (19). It is, therefore, tempting to speculate that WSV may employ a strategy similar to the parasitoid wasp to avoid the defense mechanisms of shrimp.
The data presented here clearly indicate a differential expression of the LGBP gene in WSV-infected shrimp. To our knowledge, this is the first report that LGBP gene overexpression has been shown in response to virus infection in an invertebrate host. This suggests that LGBP is an inducible acute-phase protein that may play a critical role not only in bacterial and fungal pathogenesis but also in viral pathogenesis. This also opens up new insights into the role of PRPs in viral pathogenesis in invertebrates in general and WSV pathogenesis in shrimp in particular.
In insects and crustaceans, LGBP has been shown to bind PRPs such as LPS and β-1,3-glucan and thereby activate the proPO cascade. The downregulation of proPO suggests that WSV may employ a novel way(s) to regulate the activation and/or activity of the proPO cascade to evade the host defense.
Acknowledgments
The radioactive work was carried out in the laboratory of Jane C. Burns at the University of California, San Diego (UCSD). Sequencing was conducted at the Molecular Pathology Shared Resources, UCSD Cancer Center, which is funded in part by NCI Cancer Center Support Grant 5POCA23100-16.
We also thank Qing-Gang Xue of Super Shrimp Inc. for his assistance in the WSV time course study.
REFERENCES
- 1.Bachere, E., E. Mialhe, D. Noel, V. Boulo, A. Morvan, and J. Rodriguez. 1995. Knowledge and research prospects in marine mollusk and crustacean immunology. Aquaculture 132:17-32. [Google Scholar]
- 2.Bachman, E. S., and D. R. McClay. 1996. Molecular cloning of the first metazoan β-1,3-glucanase from eggs of the sea urchin Strongylocentrotus purpuratus. Proc. Natl. Acad. Sci. USA 93:6808-6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckage, N. E., J. S. Metcalf, D. J. Nesbit, K. W. Schleifer, S. R. Zetlan, and I. D. Buron. 1990. Host haemolymph monophenoloxidase activity in parasitized Manduca sexta larvae and evidence for inhibition by wasp polydnavirus. Insect Biochem. 20:285-294. [Google Scholar]
- 4.Beschin, A., M. Bilej, F. Hanssens, J. Raymakers, E. Van Dyck, H. Revets, L. Brys, J. Gomez, P. De Baetselier, and M. Timmermans. 1998. Identification and cloning of a glucan- and lipopolysaccharide-binding protein from Eisenia foetida earthworm involved in the activation of prophenoloxidase cascade. J. Biol. Chem. 273:24948-24954. [DOI] [PubMed] [Google Scholar]
- 5.Destoumieux, D., M. Munoz, P. Bulet, and E. Bachere. 2000. Penaeidins, a family of antimicrobial peptides from penaeid shrimp (Crustacea, Decapoda). Cell. Mol. Life Sci. 57:1260-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhar, A. K., M. M. Roux, and K. R. Klimpel. 2001. Detection and quantification of infectious hypodermal and hematopoietic necrosis virus and white spot virus in shrimp using real-time quantitative PCR and SYBR Green chemistry. J. Clin. Microbiol. 39:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhar, A. K., M. M. Roux, and K. R. Klimpel. 2002. Quantitative assay for measuring the Taura syndrome virus (TSV) and yellow head virus (YHV) load in shrimp by real-time RT-PCR using SYBR Green chemistry. J. Virol. Methods 104:69-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimopoulos, G., A. Richman, H. M. Muller, and F. C. Kafatos. 1997. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl. Acad. Sci. USA 94:11508-11513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flegel, T. W. 1997. Major viral diseases of black tiger prawn (Penaeus monodon) in Thailand. World J. Microbiol. Biotechnol. 13:433-442. [Google Scholar]
- 10.Gross, P. S., T. C. Bartlett, C. L. Browdy, R. W. Chapman, and G. W. Warr. 2001. Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific white shrimp, Litopenaeus vannamei, and the Atlantic white shrimp, L. setiferus. Dev. Comp. Immunol. 25:565-577. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann, J. A., J.-M. Reichart, and C. Hetru. 1996. Innate immunity in higher insects. Curr. Opin. Immunol. 8:8-13. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, Jr., and R. A. B. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 13.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Toll-like receptor 4 (TLR 4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the LPS gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 14.Inouye, K., S. Miwa, N. Oseko, H. Makaro, T. Kimura, K. Momoyama, and K. Hiraoka. 1994. Mass mortality of cultured kuruma shrimp Penaeus japonicus in Japan in 1993: electron microscopic evidence of the causative virus. Fish Pathol. 9:149-158. [Google Scholar]
- 15.Iwanaga, S., S. Kawabata, Y. Miura, N. Seki, T. Shigenaga, and T. Muta. 1994. Clotting cascade in the immune response of horseshoe crab, p. 79-96. In J. A. Hoffmann, C. A. Janeway, Jr., and S. Natori (ed.), Phylogenetic perspectives in immunity: the insect host defense. R. G. Landes Company, Austin, Tex.
- 16.Janeway, C. A., Jr. 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symp. Quant. Biol. 54:1-13. [DOI] [PubMed] [Google Scholar]
- 17.Johansson, M. W. 1999. Cell adhesion molecules in invertebrate immunity. Dev. Comp. Immunol. 23:303-315. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, P. T. 1987. A review of fixed phagocytic and pinocytic cells of decapod crustaceans with remark on hemocytes. Dev. Comp. Immunol. 19:679-704. [DOI] [PubMed] [Google Scholar]
- 19.Kalia, V., S. Chaudhari, and G. T. Gujar. 2001. Changes in haemolymph constituents of American bollworm, Helicoverpa armigera (Hubner), infected with nucleopolyhedrovirus. Indian J. Exp. Biol. 39:1123-1129. [PubMed] [Google Scholar]
- 20.Kim, Y. S., J. H. Ryu, S. J. Han, K. H. Choi, K. B. Nam, I. H. Jang, B. Lemaitre, P. T. Brey, and W. J. Lee. 2000. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and β-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J. Biol. Chem. 275:32721-32727. [DOI] [PubMed] [Google Scholar]
- 21.Kopp, E. B., and R. Medzhitov. 1999. The Toll-receptor family and control of innate immunity. Curr. Opin. Microbiol. 11:13-18. [DOI] [PubMed] [Google Scholar]
- 22.Krischning, C. J., H. Wesche, T. M. Ayres, and M. Rothe. 1998. Human Toll-like receptor 2 confers responsiveness to bacterial lipopolysaccharide. J. Exp. Med. 188:2091-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishna, R. R., K. G. Rao, P. Rao, and P. H. Babu. 1997. White spot disease. World Aquac. 12:14-19. [Google Scholar]
- 24.Lackie, A. M. 1988. Hemocyte behavior. Adv. Insect Physiol. 21:85-178. [Google Scholar]
- 25.Lee, S. Y., R. Wang, and K. Soderhall. 2000. A lipopolysaccharide- and β-1,3-glucan-binding protein from hemocytes of the freshwater crayfish Pacifastacus leniusculus. Purification, characterization, and cDNA cloning. J. Biol. Chem. 275:1337-1343. [DOI] [PubMed] [Google Scholar]
- 26.Ma, C., and M. R. Kanost. 2000. A β-1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J. Biol. Chem. 275:7505-7514. [DOI] [PubMed] [Google Scholar]
- 27.Martinez, M. A., N. Verdaguer, M. G. Mateau, and E. Domingo. 1997. Evolution subverting essentiality: dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc. Natl. Acad. Sci. USA 94:6798-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochiai, M., and M. Ashida. 2000. A pattern-recognition protein for β-1,3-glucan. The binding domain and the cDNA cloning of β-1,3-glucan recognition protein from the silkworm, Bombyx mori. J. Biol. Chem. 275:4995-5002. [DOI] [PubMed] [Google Scholar]
- 29.Pan, J., A. Kurisky, B. Xu, A. K. Chopra, D. H. Coppenhaver, I. P. Singh, and S. Baron. 2000. Broad antiviral activity in tissues of crustaceans. Antivir. Res. 48:39-47. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Seki, N., T. Muta, T. Oda, D. Iwaki, K. Kuma, T. Miyata, and S. Iwanaga. 1994. Horseshoe crab (1,3)-β-d-glucan-sensitive coagulation factor G. A serine protease zymogen heterodimer with similarities to β-glucan-binding proteins. J. Biol. Chem. 269:1370-1374. [PubMed] [Google Scholar]
- 32.Shin, S. W., S. S. Park, D. S. Park, M. G. Kim, S. C. Kim, P. T. Brey, and H. Y. Park. 1998. Isolation and characterization of immune-related genes from the fall webworm, Hyphantria cunea, using PCR-based differential display and subtractive cloning. Insect Biochem. Mol. Biol. 28:827-837. [DOI] [PubMed] [Google Scholar]
- 33.Soderhall, K., L. Cerenius, and M. W. Johansson. 1996. The prophenoloxidase activating systems in invertebrates, p. 229-253. In K. Soderhall, S. Iwanaga, and G. R. Vasta (ed.), New directions in invertebrate immunology. SOS Publications, Fair Haven, N.J.
- 34.Sritunyalucksana, K., and K. Soderhall. 2000. The proPO and clotting system in crustaceans. Aquaculture 191:53-59. [Google Scholar]
- 35.Sritunyalucksana, K., L. Cerenius, and K. Soderhall. 1999. Molecular cloning and characterization of prophenoloxidase in the black tiger shrimp, Penaeus monodon. Dev. Comp. Immunol. 23:179-186. [DOI] [PubMed] [Google Scholar]
- 36.Stoltz, D. B., and D. I. Cook. 1983. Inhibition of host phenoloxidase activity by parasitoid hymenoptera. Experientia 39:1022-1024. [Google Scholar]
- 37.Sung, H. H., H. J. Chang, C. H. Her, J. C. Chang, and Y. L. Song. 1998. Phenoloxidase activity of hemocytes derived from Penaeus monodon and Macrobrachium rosenbergii. J. Invertebr. Pathol. 71:26-33. [DOI] [PubMed] [Google Scholar]
- 38.van Hulten, M. C. W., R. W. Goldbach, and J. M. Valk. 2000. Three functionally diverged major structural proteins of white spot syndrome virus evolved by gene duplication. J. Gen. Virol. 81:2525-2529. [DOI] [PubMed] [Google Scholar]
- 39.van Hulten, M. C. W., M. Westenberg, S. D. Goodall, and J. M. Valk. 2000. Identification of two major virion protein genes of white spot syndrome virus. Virology 266:227-236. [DOI] [PubMed] [Google Scholar]
- 40.van Hulten, M. C. W., M.-F. Tsai, C. A. Schipper, C.-F. Lo, G.-H. Kou, and J. M. Valk. 2000. Analysis of a genomic segment of white spot syndrome virus of shrimp containing ribonucleotide reductase genes and repeat regions. J. Gen. Virol. 81:307-316. [DOI] [PubMed] [Google Scholar]
- 41.van Hulten, M. C. W., and J. M. Valk. 2001. Identification and phylogeny of a protein kinase gene of white spot syndrome virus. Virus Genes 22:201-207. [DOI] [PubMed] [Google Scholar]
- 42.van Hulten, M. C. W., J. Witteveldt, S. Peters, N. Kloosterboer, R. Tarchini, M. Fiers, S., Sandbrink, R. K. Lankhorst, and J. M. Valk. 2001. The white spot syndrome virus DNA genome sequence. Virology 286:7-22. [DOI] [PubMed] [Google Scholar]
- 43.Vargas-Albores, F., F. Jimenez-Vega, and K. Soderhall. 1996. A plasma protein isolated from brown shrimp (Penaeus californiensis) which enhances the activation of prophenoloxidase system by β-1,3-glucan. Dev. Comp. Immunol. 20:299-306. [DOI] [PubMed] [Google Scholar]
- 44.Vargas-Albores, F., F. Jimenez-Vega, and G. M. Yepiz-Plascencia. 1997. Purification and comparison of β-1,3-glucan binding protein from white shrimp (Penaeus vannamei). Comp. Biochem. Physiol. 116B:453-458. [DOI] [PubMed] [Google Scholar]
- 45.Venegas, C. A., L. Nonaka, K. Mushiake, T. Nishizawa, and K. Murog. 2000. Quasi-immune response of Penaeus japonicus to penaeid rod-shaped DNA virus (PRDV). Dis. Aquat. Org. 42:83-89. [DOI] [PubMed] [Google Scholar]
- 46.Vogt, G. 1996. Cytopathology of Bay of Piran shrimp virus (BPSV), a new crustacean virus from the Mediterranean Sea. J. Invertebr. Pathol. 68:239-245. [DOI] [PubMed] [Google Scholar]
- 47.Wongteerasupaya, C., J. E. Vickers, S. Sriurairatana, G. L. Nash, A. Akarajamorn, V. Boosaeng, S. Panyim, A. Tassanakajon, B. Withyachumnarnkul, and T. W. Flegel. 1995. A non-occluded, systemic baculovirus that occurs in the cells of ectodermal and mesodermal origin and causes high mortality in the black tiger prawn Penaeus monodon. Dis. Aquat. Org. 21:69-77. [Google Scholar]
- 48.Yang, F., J. He, X. Lin, Q. Li, D. Pan, X. Zhang, and X. Xu. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75:11811-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, R. B., M. R. Mark, A. Gray, A. Huang, M. H. Xie, M. Zhang, A. Goddard, W. I. Wood, A. L. Gurney, and P. J. Godowski. 1998. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signaling. Nature 395:284-286. [DOI] [PubMed] [Google Scholar]