Abstract
Bovine leukemia virus (BLV) is a complex retrovirus that belongs to the Deltaretrovirus genus, which also includes Human T-cell leukemia virus type 1 (HTLV-1). Both viruses contain an X region coding for at least four proteins: Tax and Rex, which are involved in transcriptional and posttranscriptional regulation, respectively, and the accessory proteins R3 and G4 (for BLV) and p12I, p13II, and p30II (for HTLV-1). The present study was aimed at characterizing the subcellular localization of BLV R3 and G4. The results of immunofluorescence experiments on transfected HeLa Tat cells demonstrated that R3 is located in the nucleus and in cellular membranes, as previously reported for HTLV-1 p12I. In contrast, G4, like p13II, is localized both in the nucleus and in mitochondria. In addition, we have shown that G4 harbors a mitochondrial targeting signal consisting of a hydrophobic region and an amphipathic α-helix. Thus, despite a lack of significant primary sequence homology, R3 and p12I and G4 and p13II exhibit similar targeting properties, suggesting possible overlap in their functional properties.
Bovine leukemia virus (BLV) and Human T-cell leukemia virus type 1 (HTLV-1) are members of the Oncovirinae subfamily and belong to the Deltaretrovirus genus. These two viruses share a similar genomic organization and induce lymphoproliferative diseases in their respective host species (reviewed in references 38 and 45). In addition to the gag, pol, and env genes, deltaretroviruses contain a genome segment termed the X region, located between the env sequence and the 3′ long terminal repeat. This X region contains the open reading frames (ORFs) coding for the essential regulatory proteins Tax and Rex as well as a number of so-called accessory ORFs. While considerable attention has been focused on the functional properties of Tax and Rex, very little is known about the other ORFs. Alternative splicing generates at least two mRNAs that have the potential to produce accessory proteins encoded in ORFs located between the env and tax/rex genes: R3 and G4 for BLV and p12I, p13II, and p30II for HTLV-1 (2, 5, 6, 7, 24). HTLV-1 p12I is a highly hydrophobic and poorly immunogenic protein which is localized in cellular membranes (14, 21, 25). It shares distant homology with the bovine papillomavirus E5 oncoprotein and harbors weak oncogenic potential (15). p12I binds to the 16-kDa subunit of a vacuolar ATPase as well as to the β and γ chains of the interleukin 2 receptor and to the major histocompatibility complex class I heavy chain (20, 21, 26, 29). In cell culture, p12I is required for the infection of primary quiescent lymphocytes, suggesting a role in the activation of host cells during early stages of infection (1). Another transcript codes for p30II (also called Tof), a 241-amino-acid nucleolar/nuclear protein containing two arginine-rich domains involved in nuclear targeting (12). Consistent with this nuclear localization, p30II was also shown to regulate transcription (46, 47). The HTLV-1 p13II protein corresponds to the 87 carboxy-terminal amino acids of p30II. p13II lacks p30II's nuclear targeting signal and accumulates mainly in mitochondria, although nuclear accumulation is detected in a subpopulation of cells (9, 25). p13II has been shown to induce changes in mitochondrial architecture (9) and interacts in vitro with proteins of the nucleoside monophosphate kinase superfamily and with the actin-binding protein 280 (18).
The mRNAs coding for the BLV accessory proteins R3 and G4 were originally identified by reverse transcription-PCR in experiments conducted on peripheral blood mononuclear cells from BLV-infected cattle (2). Although the functional role of these proteins still remains unclear, their requirement for viral propagation in vivo was established by the use of recombinant BLV proviruses. Indeed, although the deletion of the R3 and G4 genes does not alter the infectious potential of the virus, long-term studies have revealed a drastic decrease in the proviral loads (22, 23, 33, 42, 43). Although similar discrepancies between conclusions drawn from cell culture and in vivo experiments were also observed for HTLV-1 p12I and p13II/p30II (11, 13, 34), it is now accepted that these proteins are required for HTLV-1 infectivity in rabbits (4, 10). These observations are in fact the best evidence for the biological relevance of the HTLV-1 and BLV accessory proteins.
The 44-amino-acid R3 protein consists of an N-terminal hydrophilic region followed by hydrophobic sequences. The hydrophilic portion corresponds to the first 17 amino acids of Rex, which include the domain that is responsible for the ability of the protein to accumulate in the nucleus and bind to its RNA target, the Rex-responsive element (2). The presence of this shared domain suggests that R3 might modulate or interfere with Rex function, a possibility that has not yet been demonstrated. G4 exhibits oncogenic potential in primary rat embryo fibroblasts (Ref cells) (22). Indeed, cotransfection of G4 and Ha-ras expression vectors into Ref cells generates fully transformed cells able to induce tumors in nude mice. G4 thus appears to belong to the immortalizing class of oncogenes, which also includes the BLV and HTLV Tax proteins (32, 41, 44). The 105-amino-acid sequence of G4 includes an amino-terminal stretch of hydrophobic residues (amino acids 1 to 24) followed by potential proteolytic cleavage sites and an arginine-rich region (amino acids 58 to 72) located in the middle of the protein (2). This latter region was recently demonstrated to be required for the interaction of G4 with its cellular partner, farnesyl pyrophosphate synthetase (27). Furthermore, tumorigenicity assays conducted with Ref cells have underlined the involvement of this domain in the oncogenic potential of G4.
In order to gain further insight into the function of the BLV R3 and G4 proteins, we carried out experiments to determine their subcellular localization. Indeed, the identification of the localization of a protein is often the first step leading to its functional characterization. Preliminary efforts aimed at obtaining specific antisera against these proteins failed, perhaps due to poor immunogenicity of hydrophobic sequences (for R3) or instability (for G4). One way to circumvent the lack of specific antibodies against proteins is to tag them with small epitopes easily recognizable by monoclonal antibodies. Another possibility is to generate hybrids with a protein that lacks a specific intracellular targeting signal, such as the green fluorescent protein (GFP). To minimize artifacts due to the insertion of foreign sequences and avoid misinterpretations of results, we adopted both strategies and designed and tested at least three different R3 and G4 fusion proteins; nucleotide sequencing and Western blot analysis were performed in order to verify the integrity of all constructs. Localization studies were performed with HeLa Tat cells, a HeLa-derived cell line (37) used in several previous studies of the HTLV-1 accessory proteins (9, 12, 25). HeLa Tat cells were cultivated at 37°C in a 5% CO2-air humidified atmosphere in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 0.25 μg of amphotericin B/ml, 100 μg of streptomycin/ml, and 1 mM sodium pyruvate. One day before transfection, the cells were passaged and seeded on coverslips. The cells were then transfected with 1 μg of plasmid DNA by using Lipofectamine reagent (Gibco BRL) as described by the manufacturer. Because of the very low number of positive cells expressing R3 or G4 tagged with the hexahistidine (six-His) or Flag epitope, respectively, a proteasome inhibitor (2 μM β-clasto-Lactacystin; Calbiochem) was added to the medium at least 6 h before the cells were harvested. The addition of proteasome inhibitor did not affect the subcellular localization of the two proteins but drastically increased their stability (data not shown). Cells transfected with GFP-derived vectors were fixed in 3.7% formaldehyde only, whereas cells transfected with epitope-tagged constructs were formaldehyde fixed and then permeabilized with 0.1% Nonidet P-40 and incubated with appropriate antibodies. Coverslips were mounted with a Prolong antifade kit (Molecular Probes) and examined with a confocal microscope (TCS-SP; Leica).
Subcellular localization of R3.
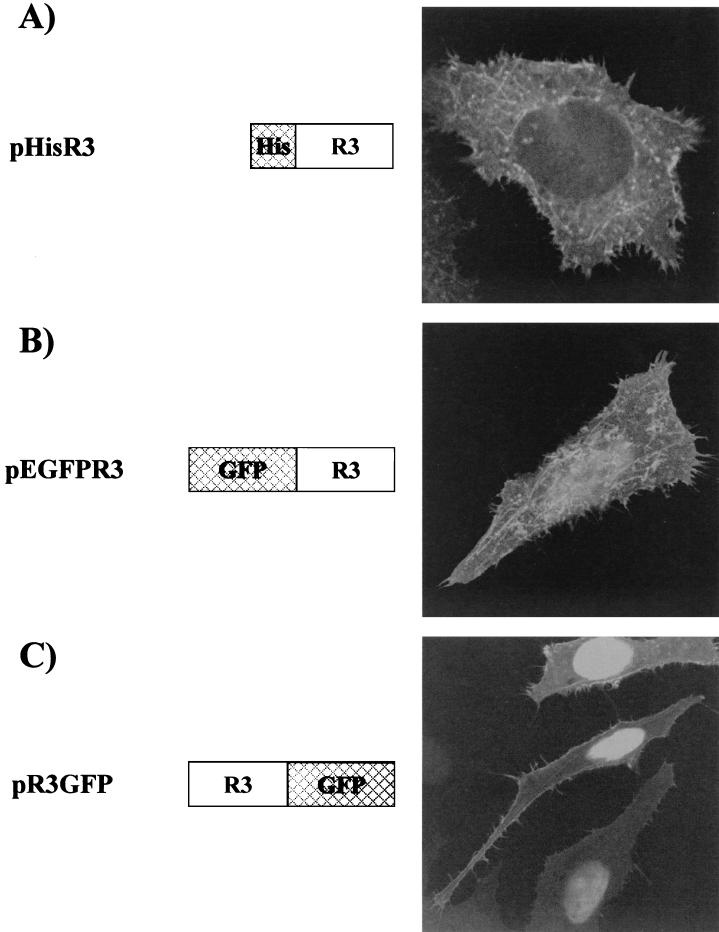
The subcellular localization of R3 was initially studied by using plasmid pHisR3, which codes for R3 modified with a six-His epitope at its amino terminus and is derived from vector pcDNA3.1/HisB (Invitrogen). After fixation and permeabilization steps, pHisR3-transfected cells were incubated with an anti-six-His monoclonal antibody (Sigma) and stained with fluorescein isothiocyanate (FITC)-coupled anti-mouse immunoglobulin. Figure 1A shows a representative cell expressing HisR3, demonstrating that the protein is detected as a diffuse signal superimposed over a cytoplasmic spiderweb-like pattern. The failure to detect substantial amounts of R3 in the nucleus was somewhat unexpected, as the protein contains a nuclear localization signal (NLS) within its first 17 amino acids. This region, which is also present in the BLV and HTLV Rex proteins, has a high content of positively charged residues which are likely to be crucial for its nuclear targeting properties (31, 39). To explain the lack of nuclear accumulation of R3, we hypothesized that the presence of the amino-terminal six-His tag might have interfered with the function of the predicted NLS. To test this hypothesis, we designed two constructs in which the R3 gene was inserted in frame either carboxy terminal or amino terminal of the GFP coding sequence (pEGFPR3 and pR3GFP; Fig. 1B and C, respectively). Confocal analysis conducted on pEGFPR3-transfected cells revealed that approximately 50% of the cells presented mainly the typical diffuse and cytoplasmic spiderweb-like pattern as described for pR3His-transfected cells and that about 50% showed both this mixed distribution and intense nuclear staining (Fig. 1B). This nuclear distribution, which is consistent with the presence of the Rex NLS at the R3 amino terminus, was observed in nearly all of the cells transfected with the pR3GFP vector (Fig. 1C), with some cells also displaying the diffuse and cytoplasmic spiderweb-like pattern (Fig. 1C, upper portion).
FIG. 1.
Subcellular distribution of R3. Plasmids pEGFPR3 and pHisR3 were generated by cloning the R3 gene isolated from pRSR3 (2) into plasmids pEGFP-C1 (Clontech) and pcDNA3.1/HisB (Invitrogen), respectively. To generate pR3GFP, R3 was PCR amplified with primers HindIIIS (5′-TTTAAGCTTGGCTTTAAAATGGCTAAAGAACG-3′) and TAAR3Bam (5′-TTTGGATCCGAGAAAAGGCGGCCCAAATGC-3′), digested with HindIII/BamHI, and inserted in frame upstream of the enhanced GFP gene in plasmid pEGFP-N1 (Clontech). HeLa Tat cells were transfected with pHisR3 (A), pEGFPR3 (B), or pR3GFP (C). Thirty hours posttransfection, cells were fixed and examined directly by confocal microscopy (B and C) or fixed, permeabilized, and incubated with a monoclonal anti-six-His antibody (Sigma) and FITC-coupled anti-mouse immunoglobulin (A).
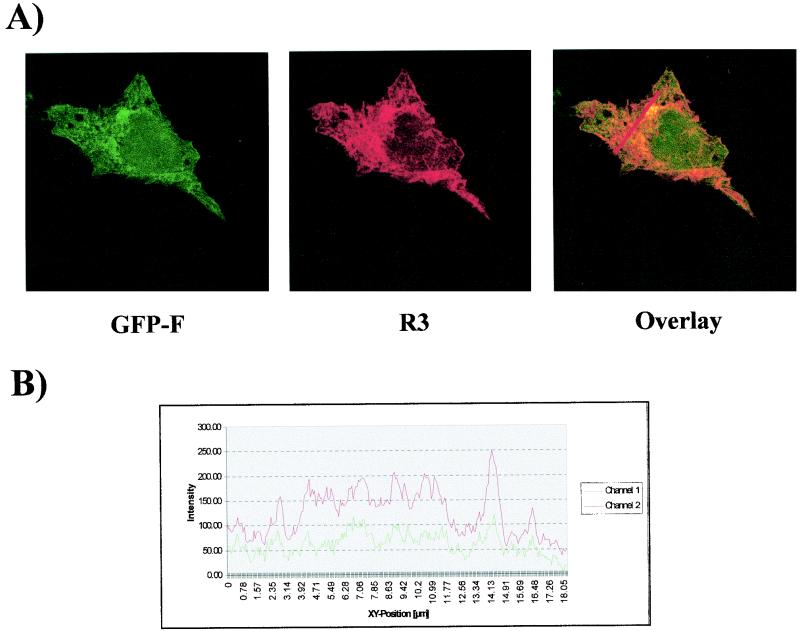
In order to define this cytoplasmic spiderweb-like pattern, immunofluorescence assays were performed with markers for different subcellular structures. Figure 2 shows cells transfected with pHisR3 in combination with the pEGFP-F vector (Clontech), which contains the 20-amino-acid farnesylation signal from c-Ha-Ras fused to the C terminus of enhanced GFP. This farnesylated enhanced GFP (EGFP) is targeted to the plasma and cellular membranes (3). Results showed significant colocalization between this farnesylated form of EGFP (Fig. 2A, green) and R3 (Fig. 2A, red). Besides the appearance of a yellow staining due to the overlapping of both fluorochromes (Fig. 2A, overlay), the colocalization between the two proteins was further demonstrated by the histogram shown in Fig. 2B. In contrast, immunofluorescence assays conducted with Texas Red-conjugated phalloidin, an actin-specific probe, failed to reveal any colocalization between R3 and this cytoskeleton marker (data not shown).
FIG. 2.
R3 is located in cellular membranes and in the nuclear compartment. (A) HeLa Tat cells were cotransfected with pHisR3 and pEGFP-F (Clontech), which contains the 20-amino-acid farnesylation signal from c-Ha-Ras fused to the C terminus of enhanced GFP. Thirty hours posttransfection, cells were fixed, permeabilized, and stained with anti-six-His antibody and Texas Red-conjugated anti-mouse immunoglobulin in order to detect R3. Cells were then analyzed by confocal microscopy. Shown are the signals obtained for GFP-F (green) and R3 (red); the overlay corresponds to the superimposition of the two fluorescent signals. (B) Shown is a quantification of the intensities of both fluorochromes assessed along the line indicated on the overlay in panel A.
Taken together, these data demonstrate that BLV R3 accumulates in two cellular compartments—the nucleus and the cellular membranes.
Subcellular localization of G4.
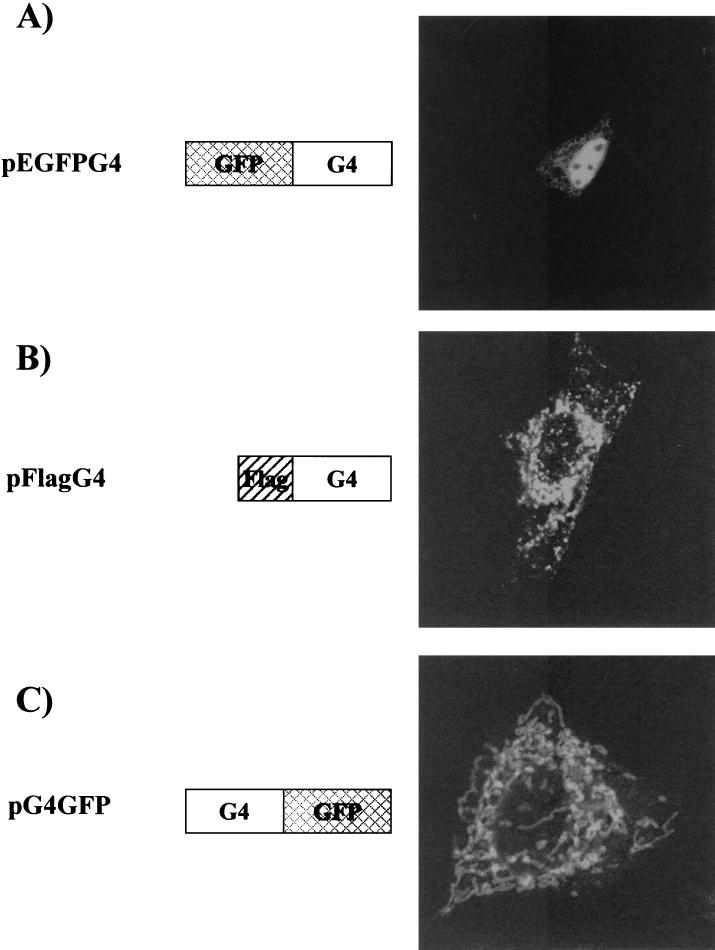
To determine the subcellular distribution of G4, we first fused the G4 gene in frame and carboxy terminal of the EGFP sequence inserted within vector pEGFP-C1 (Clontech), resulting in plasmid pEGFPG4. As illustrated in Fig. 3A, HeLa Tat cells transfected with this construct exhibited a strong nuclear signal. To verify this nuclear staining, we next inserted the G4 sequence into vector pCMV2-Flag (Sigma) in order to generate a G4 fusion protein containing a Flag epitope (DYKDDDDK) at its amino terminus (pFlagG4), and we detected the protein in transfected HeLa Tat cells by using an anti-Flag monoclonal antibody (Sigma) and FITC-conjugated secondary antibody. As shown in Fig. 3B, cells expressing Flag-tagged G4 exhibited a staining pattern consisting of clustered and filamentous structures surrounding the nucleus; unlike in the case of EGFPG4, nuclear staining was not evident.
FIG. 3.
Subcellular localization of G4. pEGFPG4 was constructed by inserting a HindIII/EcoRI fragment obtained from pSGG4 (22) into pEGFP-C1. To generate pFlagG4, the KpnI/XbaI insert of pHisG4 (27) was cloned into the pFlag-CMV-2 vector (Sigma). pG4GFP was constructed by PCR amplification of the G4 gene from pRSG4 (2) with primers HindG4S (5′-TTTAAGCTTCCCGGGAGTATGGCTTGCA-3′) and TGAG4BglII (5′-TTTAGATCTGATTGGACAAAACCAGGGCCG-3′). The resulting DNA fragment was cleaved with HindIII/BglII enzymes and inserted into pEGFP-N1 digested with HindIII/BamHI. HeLa Tat cells were transfected with pEGFPG4 (A), pFlagG4 (B), or pG4GFP (C). Thirty hours after transfection, GFP-fused G4-expressing cells were fixed and examined directly by confocal microscopy (A and C) or fixed, permeabilized, and stained with monoclonal anti-Flag antibody (Sigma) and FITC-coupled anti-mouse immunoglobulin (B).
The amino terminus of G4 includes a stretch of 24 hydrophobic amino acids (2) which might be predicted to be essential for subcellular targeting. To test this possibility, G4 was inserted upstream of the EGFP gene in vector pEGFP-N1 (Clontech) to generate plasmid pG4GFP. Transfection with pG4GFP yielded an interconnected network-like pattern similar to that obtained for FlagG4 (Fig. 3C). Therefore, it is reasonable to conclude that G4 is specifically targeted to a cytoplasmic reticulated compartment, given that two different vectors (pFlagG4 and pG4GFP) yielded similar results.
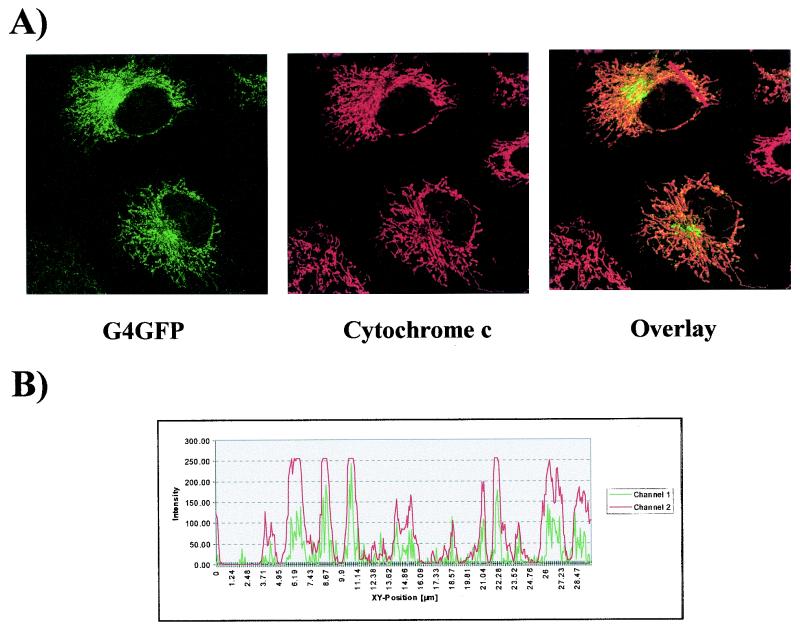
The filamentous structures containing FlagG4 and G4GFP resembled the mitochondrial pattern previously reported for HTLV-1 p13II (9). To positively identify these structures, cells expressing G4GFP were subjected to indirect immunofluorescence by using a monoclonal antibody (Pharmingen) against cytochrome c, a component of the respiratory chain that accumulates in the mitochondrial intermembrane space. As shown in Fig. 4, cytochrome c (red signal) colocalized nearly perfectly with G4GFP (green signal), yielding a yellow signal in the overlay. Results of additional assays demonstrated that G4 also colocalized with the multienzyme complex III of the respiratory chain and with Mitotracker Red CMXRos (Molecular Probes), a probe specific for mitochondria (data not shown). Therefore, we concluded that FlagG4 and G4GFP are targeted to the mitochondria.
FIG. 4.
G4 is targeted to mitochondria. HeLa Tat cells were transfected with vector pG4GFP, and immunofluorescence experiments were performed with an anti-cytochrome c antibody (Pharmingen) and Texas Red-conjugated anti-mouse immunoglobulin G in order to visualize mitochondria. Cells expressing G4 (green) and stained for cytochrome c (red) are shown in the left and middle photographs, respectively, of panel A. The overlay shows the merging of the two fluorochromes. The intensity of fluorescence along the line was quantified and is represented in a profile plot in panel B.
Mapping of the MTS of G4.
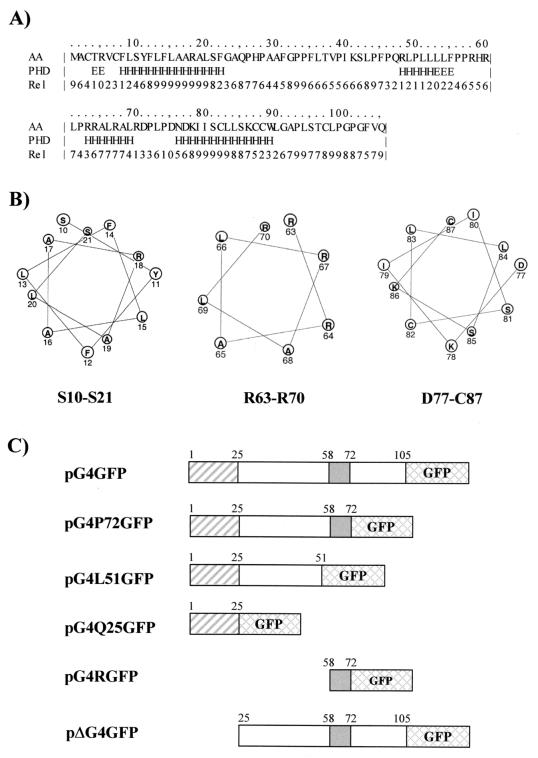
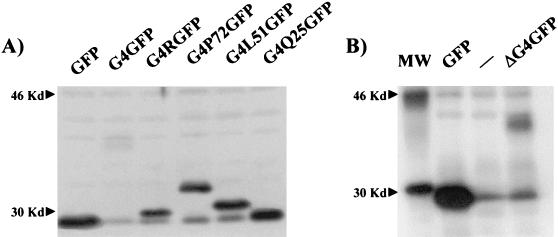
To define the mitochondrial targeting signal (MTS) of G4, we analyzed its amino acid sequence with the PHDsec secondary structure prediction program (35, 36). Results (Fig. 5A) predicted the presence of two long helical segments, the first lying in the hydrophobic amino-proximal portion of the protein (residues 8 to 22) and the second lying near the carboxy terminus (residues 76 to 89), as well as a third shorter α-helix (residues 63 to 69) spanning a region that includes several arginines. An additional α-helix detected between amino acids 48 and 52 displayed very low confidence values, which led us to assume that this region is unlikely to stably display this conformation. In addition, helical wheel models (Fig. 5B) indicate that the arginine-rich α-helix region (R63 to R70) exhibits a clear partitioning of positively charged arginines and hydrophobic leucine and alanine residues on opposite sides of the helical structure. In contrast, this amphipathic property is not evident for the other two α-helices (S10 to S21 and D77 to C87; Fig. 5B). In order to gain insight into the significance of the three α-helices in mitochondrial targeting, we designed a series of constructs coding for different portions of G4 fused in frame to the amino terminus of EGFP. To this end, sequences corresponding to amino acids 1 to 72, 1 to 51, and 1 to 25 of G4 derived from plasmid pRSG4 (2) were inserted into plasmid pEGFP-N1, generating pG4P72GFP, pG4L51GFP, and pG4Q25GFP, respectively (Fig. 5C). To analyze the role of the amphipathic α-helix in mitochondrial targeting, G4 sequences located between amino acids 58 and 72 were amplified by PCR and inserted into pEGFP-N1, yielding pG4RGFP. Furthermore, in order to assess the importance of the amino-terminal α-helix, the first 25 amino acids of G4 were removed from pG4GFP, yielding pΔG4GFP. Given that G4 harbors potential proteolytic cleavage sites located between amino acids 21 and 22 and 23 and 24, one can hypothesize that this truncated G4 protein (ΔG4) corresponds to the processed form of the protein. Expression of the G4-GFP fusion proteins was verified by Western blotting with a rabbit polyclonal anti-GFP antibody (Fig. 6A) and by immunoprecipitation for ΔG4GFP due to its weaker expression (Fig. 6B).
FIG. 5.
Mapping of the MTS of G4. (A) Protein sequence and predicted secondary structure of G4. The secondary structure prediction of G4 was obtained by using the PHDsec method (35, 36). Unstructured (loop) regions, β sheets, and helices are indicated by blank spaces, E's, and H's, respectively. Rel refers to the reliability of the structure predicted for each residue on a scale of 0 to 9 (i.e., lowest to highest reliability). AA, amino acid. (B) Helical wheel models of the three predicted α-helices of G4 corresponding to amino acids 10 to 21, 63 to 70, and 77 to 87. (C) Schematic representation of pEGFP-N1-derived constructs in which the G4 gene (pG4GFP) or its derivatives (pG4P72GFP, pG4L51GFP, pG4Q25GFP, pG4RGFP, and pΔG4GFP) were cloned. The hydrophobic amino-terminal stretch, which is followed by theoretical proteolytic cleavage sites near position Q25, is shown as a hatched box, whereas the arginine-rich region proven to be important for G4 oncogenic potential (amino acids 58 to 72) is depicted as a shaded box. Plasmid pG4P72GFP was derived from pG4GFP by BamHI cleavage and religation and expresses a truncated version of G4 ending at residue P72. Plasmids pG4L51GFP and pG4Q25GFP were obtained by PCR amplification of G4 codons 1 to 51 and 1 to 25, respectively. To generate these mutants, we used oligonucleotide HindG4S as a sense primer (5′-TTTAAGCTTCCCGGGAGTATGGCTTGCA-3′) and antisense oligonucleotide 5′-TTTGGATCCAGAGGCAGCCGTTGTGGA-3′ (for pG4L51GFP) or 5′-TTTGGATCCTGGGCGCCGAAGGAGAGA-3′ (for pG4Q25GFP). To construct pG4RGFP, G4 sequences between amino acids 58 and 72 corresponding to the arginine-rich region were inserted in frame upstream of the GFP gene. This DNA fragment was first PCR amplified by using a primer that provided an AUG codon in the context of a Kozak consensus sequence (5′-TTTAAGCTTACCATGGTGCGGCACCGACTCCCCCGC-3′) and antisense oligonucleotide TGAG4BglII (5′-TTTAGATCTGATTGGACAAAACCAGGGCCG-3′). This PCR fragment was digested with HindIII/BamHI and cloned into pEGFP-N1. Plasmid pΔG4GFP, which lacks the N-terminal part of G4, was designed by PCR amplifying the G4 gene with a primer providing an AUG codon in the context of a Kozak consensus sequence (5′-TTTAAGCTTACCATGGTGCCACATCCAGCAGCATTTG-3′) and antisense oligonucleotide TGAG4BglII. In order to compare the different constructs, all of the resulting PCR fragments were inserted into pEGFP-N1 digested with HindIII/BamHI.
FIG. 6.
Expression of G4-GFP fusion proteins. (A) HeLa Tat cells were transfected with plasmid pEGFP-N1, pG4GFP, pG4RGFP, pG4P72GFP, pG4L51GFP, or pG4Q25GFP. Thirty hours after transfection, the cells were lysed in Laemmli buffer. Aliquots of the lysates were migrated on a denaturing polyacrylamide gel, and the proteins were transferred onto polyvinylidene difluoride membranes. The G4 fusion proteins were revealed with a rabbit polyclonal anti-GFP antibody (Molecular Probes) and by chemiluminescence with horseradish peroxidase-conjugated anti-rabbit immunoglobulin antibody (essentially as described in the BM chemiluminescence Western blot kit [Roche]). (B) To reveal ΔG4GFP, HeLa Tat cells were transfected with pEGFP-N1, with the empty vector pSG5, or with pΔG4GFP. On the day following transfection, the cells were incubated in the presence of 35S-labeled methionine and cysteine. Cleared lysates in radioimmunoprecipitation assay buffer (50 mM Tris-Cl [pH 7.2], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and Complete protease inhibitor cocktail [Roche]) were immunoprecipitated with the rabbit polyclonal anti-GFP antibody coupled with proteine A-Sepharose beads (Amersham-Pharmacia). After several washes in radioimmunoprecipitation assay buffer, the immunoprecipitates were electrophoresed and revealed by autoradiography.
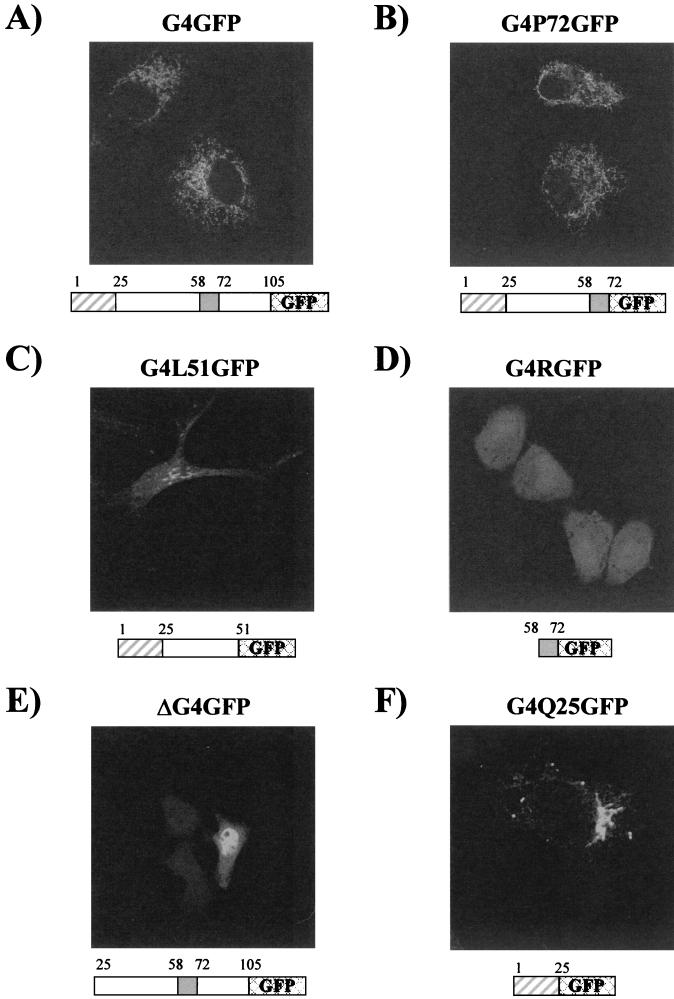
Analysis of cells expressing G4P72GFP revealed that this protein accumulated in mitochondria, indicating that the carboxy-terminal part of G4, which contains the third predicted α-helix, is not involved in mitochondrial targeting. Indeed, the deletion of amino acids 73 to 105 did not impair the typical mitochondrial targeting of the protein (compare G4GFP and G4P72GFP in Fig. 7A and B, respectively). In contrast, results obtained with G4L51GFP indicated that removal of the amphipathic α-helix spanning residues 63 to 70 was sufficient to hamper mitochondrial targeting of G4 (Fig. 7C). Indeed, dual staining assays carried out with an anti-cytochrome c antibody demonstrated that the cytoplasmic structures containing G4L51GFP were not mitochondria, suggesting a key role for this amphipathic α-helix in targeting G4 to this compartment (data not shown). However, this amphipathic sequence was unable by itself to target EGFP to the mitochondria (Fig. 7D). Results obtained for the ΔG4GFP mutant (Fig. 7E) indicated that deletion of the amino-proximal hydrophobic α-helix abolished mitochondrial targeting and instead yielded a nuclear staining pattern superimposed over a diffuse signal. However, as observed for the amphipathic α-helix, this amino-proximal hydrophobic helix was not sufficient to direct mitochondrial targeting of EGFP (Fig. 7F). This lack of colocalization between mutant G4Q25GFP and mitochondria was confirmed by immunofluorescence assays carried out with an anti-cytochrome c antibody (data not shown).
FIG. 7.
The amino-proximal and arginine-rich α-helices of G4 are required for its mitochondrial targeting. HeLa Tat cells were transfected with plasmid pG4GFP (A), pG4P72GFP (B), pG4L51GFP (C), pG4RGFP (D), pΔG4GFP (E), or pG4Q25GFP (F) and were directly visualized by confocal microscopy.
Altogether, these results demonstrate that the amino-proximal and arginine-rich α-helices of G4 are required for its mitochondrial targeting.
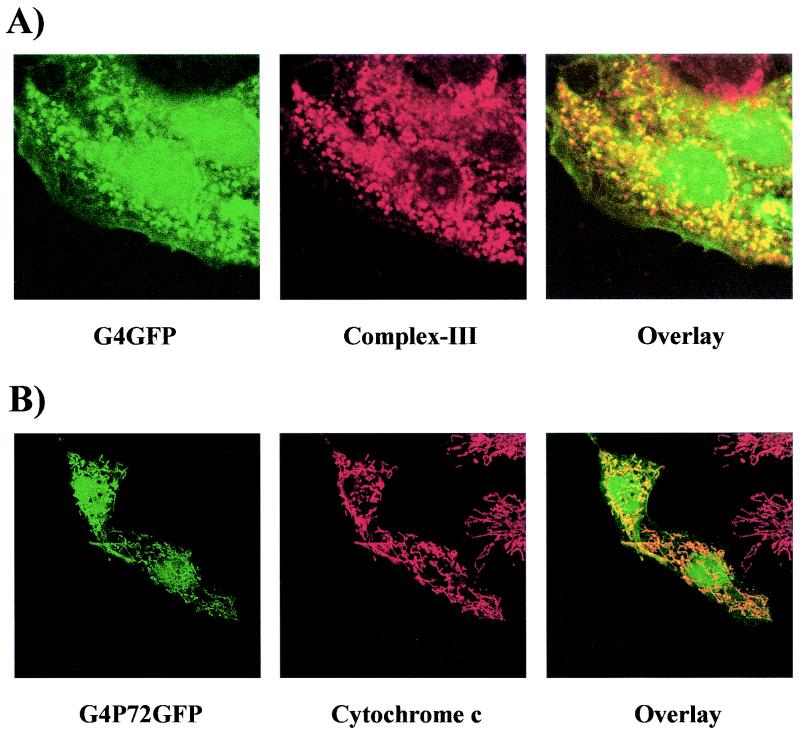
Can we conclude that G4 is exclusively directed to mitochondria and assume that the nuclear distribution observed with the GFPG4 fusion protein is an artifact? An answer to this apparent discrepancy was provided after extensive analysis of cells transfected with G4-derived constructs. These studies revealed that only a minority of cells (less than 5%, depending on the experiment) expressing either the full-length G4 (pG4GFP) or a truncated form (pG4P72GFP, amino acids 1 to 72) fused to the amino terminus exhibited both a nuclear and mitochondrial localization; Fig. 8 shows examples of cells expressing G4GFP (Fig. 8A) and G4P72GFP (Fig. 8B) that exhibit such dual localization. Altogether, these observations demonstrate that G4 is a protein localized in two different subcellular compartments, namely, the nucleus and the mitochondria.
FIG. 8.
G4 accumulates in the nuclear and mitochondrial compartments in a minority of cells. Shown are HeLa Tat cells transfected with pG4GFP (A) or pG4P72GFP (B) after staining with either anti-complex III and Texas Red-conjugated anti-rabbit immunoglobulin G (A) or anti-cytochrome c and Texas Red-conjugated anti-mouse immunoglobulin G (B).
The aim of this study was to cast light on the biological properties of the R3 and G4 accessory proteins by analyzing their subcellular localization. We have demonstrated that R3 accumulates in transfected HeLa Tat cells both in the nucleus and in the cellular membranes. The fact that R3 resides in two different subcellular compartments suggests that this protein may shuttle between these different locations or accumulate in one compartment or the other depending on the status of the cell. Nuclear targeting of R3 was expected, as it contains the first 17 residues of BLV Rex, which are predicted to be an NLS. This nuclear localization was further supported by the fact that these 17 residues were sufficient to direct the EGFP to the nucleus (data not shown). These N-terminal 17 residues can thus be considered to act as a functional NLS. Given that unlike Rex, R3 does not contain a predicted nuclear export signal, which along with the NLS and Rex-responsive element binding sequences is essential for Rex function (2, 31), it is tempting to speculate that R3 could act as a regulator of the posttranscriptional activity of Rex. Besides its nuclear compartmentalization, the association of R3 with membranes, as revealed by three different vectors (Fig. 1), was unexpected and might be conferred by the protein's hydrophobic sequences. A similar localization was previously demonstrated for HTLV-1 p12I (25). However, whether R3 might be considered the homologue of HTLV-1 p12I remains to be established. In this regard, it is noteworthy that HTLV-2, a close relative of HTLV-1, encodes a protein named p10I that consists of the amino terminus of Rex followed by hydrophobic sequences and that accumulates mainly in the nucleus (8).
The present study also provided detailed information regarding the subcellular localization of G4. The data presented here demonstrate that, in addition to being targeted to mitochondria, G4 can also be targeted to the nucleus of transfected HeLa Tat cells. Three different tagging strategies have thus led to a dual localization pattern of G4: in mitochondria and in the nucleus. Evidence for mitochondrial targeting of G4 was obtained by using specific mitochondrial markers (i.e., cytochrome c and multienzyme complex III). Interestingly, a minority of cells exhibited both nuclear and mitochondrial localization. We conclude that G4 is in fact a complex protein that resides both in the nucleus and in the mitochondria. Both nuclear and mitochondrial signals were also reported for HTLV-1 p13II (9, 12, 25); like G4, p13II appears to reside mainly in the mitochondria and is also detected in the nucleus in a minority of cells. These results are in good agreement with our recent data demonstrating that G4 colocalizes perfectly with HTLV-1 p13II (27).
We next asked whether the arginine-rich region of G4 spanning residues 63 to 70 could be involved in mitochondrial targeting, as this has been reported for HTLV-1 p13II (9). Our results revealed that unlike p13II, mitochondrial localization of G4 requires both the arginine-rich region and the hydrophobic NH2-terminus leader. Indeed, the deletion of these regions was sufficient to impair mitochondrial targeting of G4, and neither the amphipathic nor the amino-proximal helix was able to target EGFP to mitochondria. Although the mitochondrial targeting signal of G4 appears to be quite long (72 amino acids instead of 20 to 60 residues), it is consistent with most of the MTSs described to date (30). Indeed, it is located within the N-terminal segment of the protein and harbors a net positive charge (10 residues), five hydroxylated residues, and only one negatively charged amino acid (D71). Finally, since mitochondrial targeting sequences are predicted to form amphipathic α-helices thought to be important for their specific recognition by the protein import machinery, we may reasonably assume that this property is provided by the α-helix situated within the arginine-rich region. The unique feature that distinguishes G4 from other proteins harboring typical MTSs is the apparent lack of MTS cleavage following import into mitochondria. Indeed, G4 was detected at its expected full-length size after analysis of FlagG4 and G4GFP hybrid proteins by Western blotting (data not shown), but we cannot exclude the fact that cleavage of untagged G4 occurs in specific cell types. However, proteolytic processing of MTS sequences does not seem to be a strict prerequisite for mitochondrial localization, since some well-known exceptions exist, including rhodanese, 3-oxo-acyl coenzyme A thiolase, chaperonin 10 (reviewed in reference 30), and p13II (9). These observations suggest that although p13II and G4 possess structurally distinct MTSs, both proteins might utilize an import pathway that differs from that described for most mitochondrial proteins.
From our mapping of the mitochondrial targeting signal of G4, it appears that the arginine-rich region acts as an NLS. Indeed, while attachment of the arginine-rich helix to EGFP led to a diffuse staining pattern in the cytosol and in the nucleus, removal of the hydrophobic helix directed partial nuclear targeting. The fact that the hydrophobic segment is followed by recognition sequences for proteases is of interest and suggests that cleavage events might modulate the targeting of the protein. To confirm this hypothesis, it would be helpful to identify the presumed proteases involved in the recognition of this hydrophobic leader. Alternatively, the dual localization of G4 in the mitochondria and in the nucleus could also be associated with the shuttling of this polypeptide between these compartments, as recently described for a newly identified helicase (40). Such a differential protein distribution could also depend on the physiology of the cells, as described for the human immunodeficiency virus Tat protein (28), or might be controlled by overall expression levels of the transfected gene (16, 17). To address these questions, studies of the G4GFP trafficking and localization in live cells would be helpful. The localization of G4 in the nucleus suggests that it may act as a transcriptional factor, as reported by Alexandersen et al. (2). However, we did not observe any specific activation of the BLV long terminal repeat promoter by G4. It is still possible that G4 activates the transcription of unknown cellular genes in the nucleus, a property possibly related to its transforming potential in primary Ref cells (22). On the other hand, the presence of G4 in mitochondria suggests its possible role in apoptosis, as described for human immunodeficiency virus type 1 Vpr (19) and Tat (28). Alternatively, G4 might affect essential mitochondrial functions such as respiration, ATP synthesis, or lipid metabolism; further investigations are thus required to understand the influence of G4 on mitochondria.
In summary, this report provides additional clues concerning the biological properties of BLV R3 and G4. The analogies between R3 and p12I and G4 and p13II at the level of subcellular localization are likely to be important for future evaluation of their respective functions and could offer new prospects in our understanding of BLV- and HTLV-induced leukemia.
Acknowledgments
L.L. is a Televie fellow, A.V. is a Research Associate, and L.W. and R.K. are Research Directors of the Belgian National Fund for Scientific Research. D.D. was supported by a fellowship from the Fondazione Italiana per la Ricerca sul Cancro. We thank the Belgian Federation against Cancer, the Fortis Bank Assurance, the Fonds national de la Recherche scientifique (FNRS), the UICC, the Service de Programmation pour la Politique scientifique (SSTC P4/30), the Actions de Recherches Concertées du Ministère de la Communauté Française, and the Associazone Italiana per la Ricerca sul Cancro for financial support. The laboratories participated in the concerted action HTLV European Research Network (HERN) of the European Commission Biomed Program.
We are grateful to T. Peremans for excellent technical help.
REFERENCES
- 1.Albrecht, B., N. D. Collins, M. T. Burniston, J. W. Nisbet, L. Ratner, P. L. Green, and M. D. Lairmore. 2000. Human T-lymphotropic virus type 1 open reading frame I p12I is required for efficient viral infectivity in primary lymphocytes. J. Virol. 74:9828-9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandersen, S., S. Carpenter, J. Christensen, T. Storgaard, B. Viuff, Y. Wannemuehler, J. Belousov, and J. A. Roth. 1993. Identification of alternatively spliced mRNAs encoding potential new regulatory proteins in cattle infected with bovine leukemia virus. J. Virol. 67:39-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apolloni, A., I. A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartoe, J., B. Albrecht, N. Collins, M. Robek, L. Ratner, P. Green, and M. D. Lairmore. 2000. Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J. Virol. 74:1094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berneman, Z. N., R. B. Gartenhaus, M. S. Reitz, W. A. Blattner, A. Manns, B. Hanchard, O. Ikehara, R. C. Gallo, and M. E. Klotman. 1992. Expression of alternatively spliced human T-lymphotropic virus type I pX mRNA in infected cell lines and in primary uncultured cells from patients with adult T-cell leukemia/lymphoma and healthy carriers. Proc. Natl. Acad. Sci. USA 89:3005-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cereseto, A., Z. Berneman, I. Koralnik, J. Vaughn, G. Franchini, and M. E. Klotman. 1997. Differential expression of alternatively spliced pX mRNAs in HTLV-I-infected cell lines. Leukemia 11:866-870. [DOI] [PubMed] [Google Scholar]
- 7.Ciminale, V., G. N. Pavlakis, D. Derse, C. P. Cunningham, and B. K. Felber. 1992. Complex splicing in the human T-cell leukemia virus (HTLV) family of retroviruses: novel mRNAs and proteins produced by HTLV type I. J. Virol. 66:1737-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciminale, V., D. M. D'Agostino, L. Zotti, G. Franchini, B. K. Felber, and L. Chieco-Bianchi. 1995. Expression and characterization of proteins produced by mRNAs spliced into the X region of the human T-cell leukemia/lymphotropic virus type II. Virology 209:445-456. [DOI] [PubMed] [Google Scholar]
- 9.Ciminale, V., L. Zotti, D. M. D'Agostino, T. Ferro, L. Casareto, G. Franchini, P. Bernardi, and L. Chieco-Bianchi. 1999. Mitochondrial targeting of the p13II protein coded by the x-II ORF of human T-cell leukemia/lymphotropic virus type I (HTLV-I). Oncogene 18:4505-4514. [DOI] [PubMed] [Google Scholar]
- 10.Collins, N. D., G. C. Newbound, B. Albrecht, J. L. Beard, L. Ratner, and M. D. Lairmore. 1998. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood 91:4701-4707. [PubMed] [Google Scholar]
- 11.Collins, N. D., C. D'Souza, B. Albrecht, M. D. Robek, L. Ratner, W. Ding, P. L. Green, and M. D. Lairmore. 1999. Proliferation response to interleukin-2 and Jak/Stat activation of T cells immortalized by human T-cell lymphotropic virus type 1 is independent of open reading frame I expression. J. Virol. 73:9642-9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Agostino, D. M., V. Ciminale, L. Zotti, A. Rosato, and L. Chieco-Bianchi. 1997. The human T-cell lymphotropic virus type 1 Tof protein contains a bipartite nuclear localization signal that is able to functionally replace the amino-terminal domain of Rex. J. Virol. 71:75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derse, D., J. Mikovits, and F. Ruscetti. 1997. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology 237:123-128. [DOI] [PubMed] [Google Scholar]
- 14.Ding, W., B. Albrecht, R. Luo, W. Zhang, J. R. Stanley, G. C. Newbound, and M. D. Lairmore. 2001. Endoplasmic reticulum and cis-Golgi localization of human T-lymphotropic virus type 1 p12I: association with calreticulin and calnexin. J. Virol. 75:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franchini, G., J. C. Mulloy, I. J. Koralnik, A. Lo Monico, J. J. Sparkowski, T. Andresson, D. J. Goldstein, and R. Schlegel. 1993. The human T-cell leukemia/lymphotropic virus type I p12I protein cooperates with the E5 oncoprotein of bovine papillomavirus in cell transformation and binds the 16-kilodalton subunit of the vacuolar H+ ATPase. J. Virol. 67:7701-7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heger, P., O. Rosorius, J. Hauber, and R. H. Stauber. 1999. Titration of cellular export factors, but not heteromultimerization, is the molecular mechanism of trans-dominant HTLV-1 rex mutants. Oncogene 18:4080-4090. [DOI] [PubMed] [Google Scholar]
- 17.Henkler, F., J. Hoare, N. Waseem, R. D. Goldin, M. J. McGarvey, R. Koshy, and I. A. King. 2001. Intracellular localization of the hepatitis B virus HBx protein. J. Gen. Virol. 82:871-882. [DOI] [PubMed] [Google Scholar]
- 18.Hou, X., S. Foley, M. Cueto, and M. A. Robinson. 2000. The human T-cell leukemia virus type I (HTLV-I) X region encoded protein p13(II) interacts with cellular proteins. Virology 277:127-135. [DOI] [PubMed] [Google Scholar]
- 19.Jacotot, E., K. F. Ferri, C. El Hamel, C. Brenner, S. Druillennec, J. Hoebeke, P. Rustin, D. Metivier, C. Lenoir, M. Geuskens, H. L. Vieira, M. Loeffler, A. S. Belzacq, J. P. Briand, N. Zamzami, L. Edelman, Z. H. Xie, J. C. Reed, B. P. Roques, and G. Kroemer. 2001. Control of mitochondrial membrane permeabilization by adenine nucleotide translocator interacting with HIV-1 viral protein rR and Bcl-2. J. Exp. Med. 193:509-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, J. M., J. C. Mulloy, V. Ciminale, J. Fullen, C. Nicot, and G. Franchini. 2000. The MHC class I heavy chain is a common target of the small proteins encoded by the 3′ end of HTLV type 1 and HTLV type 2. AIDS Res. Hum. Retrovir. 16:1777-1781. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, J. M., C. Nicot, J. Fullen, V. Ciminale, L. Casareto, J. C. Mulloy, S. Jacobson, and G. Franchini. 2001. Free major histocompatibility complex class I heavy chain is preferentially targeted for degradation by human T-cell leukemia/lymphotropic virus type 1 p12I protein. J. Virol. 75:6086-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerkhofs, P., H. Heremans, A. Burny, R. Kettmann, and L. Willems. 1998. In vitro and in vivo oncogenic potential of bovine leukemia virus G4 protein. J. Virol. 72:2554-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerkhofs, P., J. S. Gatot, K. Knapen, M. Mammerickx, A. Burny, D. Portetelle, L. Willems, and R. Kettmann. 2000. Long-term protection against bovine leukaemia virus replication in cattle and sheep. J. Gen. Virol. 81:957-963. [DOI] [PubMed] [Google Scholar]
- 24.Koralnik, I. J., A. Gessain, M. E. Klotman, A. Lo Monico, Z. N. Berneman, and G. Franchini. 1992. Protein isoforms encoded by the pX region of human T-cell leukemia/lymphotropic virus type I. Proc. Natl. Acad. Sci. USA 89:8813-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koralnik, I. J., J. Fullen, and G. Franchini. 1993. The p12I, p13II, and p30II proteins encoded by human T-cell leukemia/lymphotropic virus type I open reading frames I and II are localized in three different cellular compartments. J. Virol. 67:2360-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koralnik, I. J., J. C. Mulloy, T. Andresson, J. Fullen, and G. Franchini. 1995. Mapping of the intermolecular association of human T cell leukaemia/lymphotropic virus type I p12I and the vacuolar H+-ATPase 16 kDa subunit protein. J. Gen. Virol. 76:1909-1916. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre, L., A. Vanderplasschen, V. Ciminale, H. Heremans, O. Dangoisse, J. C. Jauniaux, J. F. Toussaint, V. Zelnik, A. Burny, R. Kettmann, and L. Willems. 2002. Oncoviral bovine leukemia virus G4 and human T-cell leukemia virus type 1 p13II accessory proteins interact with farnesyl pyrophosphate synthetase. J. Virol. 76:1400-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macho, A., M. A. Calzado, L. Jimenez-Reina, E. Ceballos, J. Leon, and E. Munoz. 1999. Susceptibility of HIV-1-TAT transfected cells to undergo apoptosis. Biochemical mechanisms. Oncogene 18:7543-7551. [DOI] [PubMed] [Google Scholar]
- 29.Mulloy, J. C., R. W. Crownley, J. Fullen, W. J. Leonard, and G. Franchini. 1996. The human T-cell leukemia/lymphotropic virus type 1 p12I protein binds the interleukin-2 receptor β and γc chains and affects their expression on the cell surface. J. Virol. 70:3599-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66:863-917. [DOI] [PubMed] [Google Scholar]
- 31.Nosaka, T., H. Siomi, Y. Adachi, M. Ishibashi, S. Kubota, M. Maki, and M. Hatanaka. 1989. Nucleolar targeting signal of human T-cell leukemia virus type I rex-encoded protein is essential for cytoplasmic accumulation of unspliced viral mRNA. Proc. Natl. Acad. Sci. USA 86:9798-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pozzatti, R., J. Vogel, and G. Jay. 1990. The human T-lymphotropic virus type I tax gene can cooperate with the ras oncogene to induce neoplastic transformation of cells. Mol. Cell. Biol. 10:413-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichert, M., G. H. Cantor, L. Willems, and R. Kettmann. 2000. Protective effects of a live attenuated bovine leukaemia virus vaccine with deletion in the R3 and G4 genes. J. Gen. Virol. 81:965-969. [DOI] [PubMed] [Google Scholar]
- 34.Robek, M. D., F. H. Wong, and L. Ratner. 1998. Human T-cell leukemia virus type 1 pX-I and pX-II open reading frames are dispensable for the immortalization of primary lymphocytes. J. Virol. 72:4458-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 36.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fënyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semmes, O. J., and M. L. Hammarskjold. 1999. Molecular pathogenesis of HTLV-I: a current perspective. ABI Professional Publications, Arlington, Va.
- 39.Siomi, H., H. Shida, S. H. Nam, T. Nosaka, M. Maki, and M. Hatanaka. 1988. Sequence requirements for nucleolar localization of human T cell leukemia virus type I pX protein, which regulates viral RNA processing. Cell 55:197-209. [DOI] [PubMed] [Google Scholar]
- 40.Valgardsdottir, R., G. Brede, L. G. Eide, E. Frengen, and H. Prydz. 2001. Cloning and characterization of mddx28, a putative dead-box helicase with mitochondrial and nuclear localization. J. Biol. Chem. 276:32056-32063. [DOI] [PubMed] [Google Scholar]
- 41.Willems, L., H. Heremans, G. Chen, D. Portetelle, A. Billiau, A. Burny, and R. Kettmann. 1990. Cooperation between bovine leukemia virus transactivator protein and Ha-ras oncogene product in cellular transformation. EMBO J. 9:1577-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willems, L., R. Kettmann, F. Dequiedt, D. Portetelle, V. Vonèche, I. Cornil, P. Kerkhofs, A. Burny, and M. Mammerickx. 1993. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 67:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willems, L., P. Kerkhofs, F. Dequiedt, D. Portetelle, M. Mammerickx, A. Burny, and R. Kettmann. 1994. Attenuation of bovine leukemia virus by deletion of R3 and G4 open reading frames. Proc. Natl. Acad. Sci. USA 91:11532-11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willems, L., C. Grimonpont, P. Kerkhofs, C. Capiau, D. Gheysen, K. Conrath, R. Roussef, R. Mamoun, D. Portetelle, A. Burny, E. Adam, L. Lefebvre, J. C. Twizere, H. Heremans, and R. Kettmann. 1998. Phosphorylation of bovine leukemia virus Tax protein is required for in vitro transformation but not for transactivation. Oncogene 16:2165-2176. [DOI] [PubMed] [Google Scholar]
- 45.Willems, L., A. Burny, D. Collete, O. Dangoisse, J. S. Gatot, P. Kerkhofs, L. Lefebvre, C. Merezak, D. Portetelle, J. C. Twizere, and R. Kettmann. 1999. Bovine leukemia virus as a model for human T-cell leukemia virus. Curr. Top. Virol. 1:139-167. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, W., J. W. Nisbet, J. T. Bartoe, W. Ding, and M. D. Lairmore. 2000. Human T-lymphotropic virus type 1 p30II functions as a transcription factor and differentially modulates CREB-responsive promoters. J. Virol. 74:11270-11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, W., J. W. Nisbet, B. Albrecht, W. Ding, F. Kashanchi, J. T. Bartoe, and M. D. Lairmore. 2001. Human T-lymphotropic virus type 1 p30II regulates gene transcription by binding CREB binding protein/p300. J. Virol. 75:9885-9895. [DOI] [PMC free article] [PubMed] [Google Scholar]