Abstract
We report here on the generation of a mouse monoclonal antibody directed against Rous sarcoma virus (RSV) subgroup A Env that will be useful in functional and structural analysis of RSV Env, as well as in approaches employing the RCAS/Tva system for gene targeting. BALB/c mice were primed and given boosters twice with EnvA-expressing NIH 3T3 cells. Resulting hybridomas were tested by enzyme-linked immunosorbent assay against RCANBP virions and SU-A-immunoglobulin G immunoadhesin. One highly reactive hybridoma clone, mc8C5, was subcloned and tested in immunofluorescence, immunoprecipitation (IP), and Western blotting assays. In all three assays, mc8C5-4 subgroup-specifically recognizes SR-A Env, through the SU domain, expressed from different vectors in both avian and mammalian cells. This multifunctionality is notable for a mouse monoclonal. We furthermore observed a preference for binding to terminally glycosylated Env over core-glycosylated Env precursor in IPs, suggesting that the epitope is at least partially conformational and dependent on glycosylation. Most importantly, we found mc8C5-4 inhibited Env function: in vitro, the monoclonal not only interferes with binding of the EnvA receptor, Tva, but it also blocks the Tva-induced conformational change required for activation of the fusion peptide, without inducing that change itself. Infection of Tva-expressing avian or mammalian cells by avian sarcoma and leukosis virus (ASLV) or EnvA-pseudotyped murine leukemia virus, respectively, is efficiently inhibited by mc8C5-4. The apparent interference of the monoclonal with the EnvA-Tva complex formation suggests that the epitope seen by mc8C5 overlaps with the receptor binding site. This is supported by the observation that mutations of basic residues in hr2 or of the downstream glycosylation site, which both impair Tva-binding to EnvA, have similar effects on the binding of mc8C5. Thus, anti-ASLV-SU-A mc8C5-4 proves to be a unique new immunoreagent that targets the receptor-binding site on a prototypical retroviral envelope.
Not only are the avian retroviruses (alpharetroviruses, or avian sarcoma and leukosis viruses [ASLV]) relevant pathogens in poultry with economic impact, but they also have long served as an important model system for studying the biological functions of retroviruses and other enveloped viruses. Attachment to and infection of target cells is mediated by the ASLV envelope (Env) glycoproteins. Env is synthesized as a precursor molecule (Pr95) that is proteolytically processed in the Golgi into two disulfide-linked subunits, SU/gp85 (surface) and TM/gp37 (transmembrane) (18). The TM region is divided into an ectodomain involved in membrane fusion, a membrane-spanning domain anchoring the protein, and a cytoplasmic C terminus containing endocytosis and potentially other trafficking signals (39). The ectodomains of both SU and TM are extensively glycosylated. SU contains two hypervariable regions, hr1 and hr2, that are determinants of host range and thus of the interaction of SU with its respective receptor (4, 5, 14, 15, 35, 36). The receptor for ASLV subgroup A (Tva) (3, 52) has been well characterized, and receptors for subgroups B, D, and E have been recently described (1, 2, 49). A cluster of basic amino acids in hr2 of EnvA has been implicated as important for Tva binding (5, 8, 14, 35, 36, 46), as well as polar residues in hr1 (34). The EnvA-Tva interaction induces conformational changes in the ectodomain of TM, resulting in the activation and exposure of the fusion peptide (9, 26, 30), in the subsequent fusion of host cell and viral membranes, and in infection. Glycosylation of retroviral Env proteins is important for both folding and function (19, 29) and has also been implicated in immune evasion, especially in the case of human immunodeficiency virus (HIV) and simian immunodeficiency virus Env (44).
For our ongoing studies of intracellular targeting of Rous sarcoma virus (RSV) Env molecules (39), as well as of the molecular events involved in entry (11), we are using ASLV subgroup Schmidt-Ruppin A (SR-A). To elucidate steps in entry we are probing the conformational changes in the EnvA ectodomain during the fusion process. To assess intracellular targeting functions we are analyzing transmembrane (TM) cytoplasmic tail mutations and truncations. For these studies we needed a multifunctional immunoreagent(s) directed against the ectodomain of subgroup A Env. Since the generation of broadly applicable polyclonal antisera against SU or the ectodomain of TM has proven difficult in the past, we chose to attempt the generation of monoclonal antibodies. Not only might such reagents recognizing a single epitope be subgroup specific, but they could also be produced in large quantities once stable hybridoma cultures were established. Precedence for the usefulness of anti-SU monoclonal antibodies comes from extensive work on HIV-1 Env (reviewed in references 6, 17, and 38).
Here we report on the successful generation of anti-ASLV SR-A Env mc8C5-4, a unique mouse monoclonal that can be used in a variety of applications including immunofluorescence, flow cytometry, IP, and Western blotting. The mc8C5-4 antibody interferes with Tva receptor binding to EnvA in vitro and in cell culture. It efficiently inhibits infection of Tva-expressing cells with ASLV or EnvA-pseudotyped murine leukemia virus (MLV). Additionally, the ability of mc8C5-4 to bind mutant EnvA proteins that have been described to display reduced receptor binding properties (8, 11, 46) parallels that of Tva.
MATERIALS AND METHODS
DNA expression vectors.
The proviral RSV SR-A expression vector RCANBP was described previously (43). In RCASBP-X-AP, X represents the coding region for the SU domain (between the XhoI and EcoRI sites) of either SR-A, SR-B, or Pr-C Env cloned into the SR-A env gene. The vector furthermore contains an alkaline phosphatase (AP) gene in the place of src, thus allowing virus titer determination by staining for AP activity (21-23).
The expression vectors used to generate MLV pseudotyped with RSV EnvA have been described previously (12).
The recombinant simian virus 40 (SV40) expression vector for RSV EnvC, pSVenvKX, used for Env expression in CV-1 cells has been described previously (10, 39). To generate pCB6-EnvC, the 1.86-kb KpnI/BamHI env fragment from pSVenvKX was inserted into pCB6. We generated pCB6-EnvA, and pCB6-EnvA-S19 by inserting the 2.65-kb KpnI/BamHI env gene fragment from RCANBP or RCANBP-S19 into the pCB6 plasmid vector; the Env cleavage site mutation S19 (RRKR340 changed to SRER) was previously described in the context of EnvC (13, 39), and the mutation was regenerated in the background of the SR-A RCANBP proviral vector by single-strand DNA mutagenesis essentially as described previously (13).
pCB6-EnvA-hr2 mutants sM5 (R223,224,227→I), sM12 (R210,213,223,224,227→I), sM20 (R213,227→A), sM21 (R213,223,224→A), and sM28 (R213→S) were kindly provided by P. Bates and colleagues (8, 46).
The pCB6-based expression vectors for glycosylation site mutants of EnvA are described elsewhere (11).
The plasmid vector for the generation of SU-A-rabbit immunoglobulin G (SU-A-rbIgG) immunoadhesin has been described previously (53); plasmid vectors for Tva-rbIgG immunoadhesin were kindly provided by K. Zingler and J. Young.
Mammalian and avian cell culture.
CV-1 and 293T cells were cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS). The chicken fibroblast cell line Df-1 (32, 48) was kindly provided by D. Foster (University of Minnesota, St. Paul), and was cultivated in DMEM-10%FCS.
NIH 3T3 cell lines stably expressing wild-type SR-A Env (27) or full-length Tva (PG950 cells) (26) have been described previously.
The myeloma line P3X63-Ag8.653 was used as fusion partner for the generation of hybridoma cell clones.
Generation of mouse monoclonal directed against ASLV SR-A SU.
In order to generate antibodies directed against ASLV EnvA in its native form, BALB/c mice were immunized and then twice given boosters with NIH 3T3 cells stably expressing RSV SR-A Env glycoprotein (27). For each injection, each mouse received approximately 107 cells that were removed from confluent culture dishes with 20 mM EDTA in phosphate-buffered saline (PBS), washed twice in PBS, and then resuspended in 100 μl of cold PBS. For the first injection the cells were emulsified in complete Freund's adjuvant. Approximately 3 weeks later the mice received a booster consisting of the same number of cells in incomplete Freund's adjuvant. The cells were injected subcutaneously into the thigh area of the rear legs. Approximately 2 weeks after the booster, the mice were bled and sera were tested for reactivity against gradient-purified RCANBP virus by enzyme-linked immunosorbent assay (ELISA). A virus titer of approximately 1:512 was determined. Approximately 1 month after the first booster, we administered the second booster, which consisted of 107 cells in PBS. The cells were once more injected subcutaneously into the rear thigh. Five days after the final booster, the mice were sacrificed and the popliteal and inguinal lymph nodes were removed. Lymph node cells were fused with cells of the P3X63-Ag8.653 myeloma line using polyethylene glycol. Fused cells were seeded into 24-well plates in HAT medium for selection following standard procedures (28). Fourteen days after fusion, hybridomas were tested for reactivity against SU-A by ELISA, using gradient-purified RCANBP virions, or against chimeric SU-A-rbIgG immunoadhesin (53). Chimeric Tva-rbIgG (expression vector kindly provided by K. Zingler and J. Young) served as a negative control.
Preparation of purified RSV virions and of SU-A-immunoadhesins.
For large-scale production of RCANBP virions, Df-1 cells were infected with high-titer virus stocks and cultured in roller bottles for 6 days with daily medium changes. Virion-containing culture supernatants were kept on ice until a total of 2 liters had been collected. Virions were concentrated by ultracentrifugation. Pellets were resuspended in PBS and subjected to mild sonication, and the nonaggregated portion (∼50%) was further purified by Optiprep gradient centrifugation. The virus fraction was washed, pelleted again, resuspended in PBS, and kept on ice until further use. Protein concentration was quantitated by spectrometry.
Large-scale production of purified SU-A or Tva immunoadhesin over protein A columns was performed essentially as described previously (53).
ELISA screening of hybridoma culture supernatants.
For antibody capture assays, purified RCANBP virus was diluted to 20 μg/ml in borate-buffered saline (BBS) pH 8.5, and wells of a 96-well ELISA plate (Dynex) were coated with 100 μl of the virus dilution overnight at 4°C. Plates were blocked for 1 h with 1% BSA in BBS. After blocking, wells were washed with BBS and incubated with hybridoma supernatants for 4 h at RT. Plates were washed in BBS and incubated with alkaline phosphatase-labeled goat anti-mouse Ig (1:4,000; Jackson ImmunoResearch Laboratories) for 1 h at room temperature. After washing, plates were developed by addition of p-nitrophenyl phosphate (104 phosphatase substrate; Sigma). ELISAs using SU-A-rbIgG to capture antibody were performed essentially the same way, by coating plates with 100 μl of 5-μg/ml immunoadhesin per well.
Purification of mc8C5 from hybridoma supernatants.
Antibody was purified from culture supernatants by affinity chromatography on recombinant protein G columns (Gamma Bind Plus; Pharmacia/LKB) according to the manufacturer's instructions, using a Shimadzu automated preparative/analytical high-performance liquid chromatography system. Briefly, culture supernatants were filter sterilized and loaded directly onto the equilibrated column. The column was washed with equilibration buffer (0.01 M sodium phosphate, pH 7.0; 0.15 M sodium chloride; 0.01 M EDTA), and antibody was eluted with 0.5 M acetic acid, adjusted to pH 3.0 with ammonium hydroxide. The antibody peak was immediately adjusted to pH 7.0 by the addition of 2 M Trizma base. Antibody purity was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining using a Pharmacia/LKB PhastSystem according to the manufacturer's instructions.
Generation of titrated virus stocks and infectivity assay.
Df-1 cells were transfected with proviral RCASBP-A-AP DNA, and the titers of virus supernatants collected 4 days posttransfection were determined on Df-1 cells. For this, the target cells were infected in quadruplicate in 24-well plates (4 × 104 cells per well) with serial 10-fold virus dilutions. After 48 h in culture, cells were fixed in paraformaldehyde (PFA) and stained for exogenous alkaline phosphatase activity. For assays measuring inhibition of infection, a total of 1,000 infectious units were preincubated with increasing concentrations of purified mc8C5-4 for 1 h at 4°C in a total of 200 μl of DMEM-10%FCS. The preincubated virus samples were then used to infect in quadruplicate fresh Df-1 cells grown on coverslips. At 4 h after infection, 400 μl of DMEM-10%FCS was added to a final volume of 600 μl. At 48 h postinfection, cells were fixed and stained for AP activity. Infectivity was determined by counting blue cells on bright-field photographs taken of four to five random fields for each of the four coverslips per sample, at a magnification of ×20 or ×10, using a Zeiss microscope.
Infectivity assays using EnvA-pseudotyped MLV and PG950 cells were performed as previously described (12).
Polyclonal antisera.
Rabbit sera directed against C-terminal peptides of the TM cytoplasmic domain (rb-anti-A-tail and -C-tail) have been described previously (25). rb-Ngp37 is directed against the N-terminal ectodomain of EnvA TM (31).
Secondary antibodies for immunofluorescence and fluorescence-activated cell sorting (FACS) were purchased from Molecular Probes, Eugene, Oreg.
Metabolic labeling, IP, and quantitation of autoradiographs.
Cells were metabolically pulse-labeled with [35S]methionine-cysteine for 30 min (500 μCi/ml) in deficient medium and chased for 2 h in complete medium essentially as previously described (39). Cells were lysed in buffer containing 1% Triton X-100 and 0.5% deoxycholate and subjected to different conditions of IP as specified in the text and figure legends. After SDS-PAGE, autoradiographs were quantitated using a Cyclone scanner and OptiQuant software (Packard Instrument Company, Meriden, Conn.).
Indirect immunofluorescence and Western blotting.
All immunofluorescence analyses were performed essentially as previously described (39, 40). To visualize intracellular steady-state distribution of wild-type and mutant Env, protein-expressing cells grown on glass coverslips were fixed in ice-cold acetone and then probed with rb-anti-A-tail or -C-tail, or with mc8C5-4 ascites, using goat-anti-rb-Alexa488 or rb-anti-mouse Alexa488 as secondary antibody (all primary and secondary antibodies used at 1:200 in PBS-3% BSA). For surface immunofluorescence, unfixed cells on coverslips were incubated with mc8C5-4 antibody on ice for 30 min, washed in PBS, fixed with ice-cold ethanol-acetic acid (95:5), and stained with rb-anti-mouse Alexa488. All samples were observed and photographed with a Zeiss fluorescence microscope.
For analysis of mc8C5-4 in enhanced chemiluminescence (ECL)-Western blotting, EnvA-, RCANBP-, or SU-A-rbIgG-expressing cells were lysed in 2× sample buffer (4% SDS, 0.125 M Tris-HCl [pH 6.8], 10% glycerol, 10% β-mercaptoethanol, 0.002% bromophenol blue) and boiled for 5 min. Samples were subjected to SDS-PAGE, and mini-gels were soaked in blotting buffer (20% methanol, 200 mM glycine, 25 mM Tris) prior to blotting onto nitrocellulose in the semidry apparatus at 12 V for 25 min. Transfer was monitored by reversible staining with Ponceau red, destained in PBS, and then blocked in 5% BSA in PBS-Tween 20 (0.1%) at room temperature for at least 2 h. Antigen was detected after incubation with mc8C5-4 ascites (in 5% BSA-PBS-Tween20; dilutions as indicated), followed by extensive washes and rb-anti-mouse-horseradish peroxidase (-HRP) (in BSA-PBS-Tween) incubation. After washing in PBS-Tween buffer, nitrocellulose membranes were blotted briefly onto tissue before incubation with SuperSignal West Pico chemiluminescence substrate (Pierce, Rockford, Ill.) for 1 min, followed by another brief blotting step and exposure of film for ECL. As a control for SU-A-rbIgG, immunoadhesin was visualized directly with goat-anti-rb-HRP by ECL. As a control for cell-associated SU-A (gp85), the antigen was detected after incubation with rb-anti-A-tail and goat-anti-rb-HRP by ECL.
Liposome flotation assay.
The liposome flotation assay was performed essentially as described previously (30). A soluble form of EnvA (27) was preincubated with or without mc8C5-4 antibody on ice for 30 min, and then s47 (Tva peptide) or PBS was added, and complexes were allowed to form during an additional 10-min incubation on ice. After addition of liposomes, samples were incubated at 37°C for 15 min to induce the fusion-relevant conformational changes. The recooled samples were mixed with 67% sucrose in PBS to a final concentration of 50% and overlaid with equal volumes of 25% and then 10% sucrose. Samples were centrifuged 1 h at 4°C, at 197,000 × g, and six fractions were taken from the top to the bottom of the gradient for Western blot analysis using an antibody that recognizes gp37.
Flow cytometry studies.
To analyze binding of mc8C5-4 to EnvA-hr2 mutants, Df-1 cells were removed from culture plates 48 h posttransfection with 25 mM EDTA in PBS. Cells were washed twice in cold PBS containing calcium, magnesium, and 0.02% azide (PBSA) and then incubated on ice for 30 min in 1:1,000 mc8C5-4 ascites in 5% FCS in PBSA. After washing, cells were counterstained with rb-anti-mouse Alexa488 for 30 min on ice. Washed cells were then fixed in a final concentration of 1.5% PFA and analyzed using a FACScan flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, Calif.).
To test inhibition of s47 binding to surface-expressed EnvA by mc8C5-4, cells were removed from the plates and washed in PBSA. On ice, cells were incubated for 30 min with purified antibody at the indicated concentration in 30% FCS-PSBA and then pelleted at 300 × g in the cold. The supernatant was removed and replaced with biotinylated s47 (1 mg/ml) (30) in the antibody-30% FBS-PBSA solution and incubated 30 min on ice. The cells were washed twice in PBSA and incubated 30 min with avidin-Oregon green 488 in 30% FCS-PBSA, washed twice as above, and fixed in 4% paraformaldehyde, and the binding of s47 was quantitated by flow cytometry.
Binding of mc8C5-4 to EnvAs harboring glycosylation mutations was analyzed after transient transfection of 293T cells. Cells were induced 28 h later and harvested 48 h posttransfection. Cells were treated for FACS analysis as described above, except that no s47 incubation step was performed. The primary antibody was either Ngp37, a polyclonal antibody against the ectodomain of EnvA TM (30), or purified mc8C5-4 (45 μg/ml); the secondary antibody was FITC-conjugated anti-rabbit or anti-mouse, respectively. Mean fluorescence values obtained after incubation with Ngp37 were used to normalize each mc8C5-4 sample for EnvA surface expression.
RESULTS
A mouse monoclonal was generated against ASLV EnvA and reacts with SU-A in ELISA.
In order to generate antibodies directed against ASLV EnvA in its native form, BALB/c mice were immunized and then twice given boosters with NIH 3T3 cells stably expressing RSV SR-A Env glycoprotein (27). Approximately 2 weeks after the first booster, the mice were bled and sera were tested for reactivity against gradient-purified RCANBP virus by ELISA. A virus titer of approximately 1:512 was determined. The mice were sacrificed 5 days after the second boost, and the popliteal and inguinal lymph nodes were removed. Lymph node cells were fused with the P3X63-Ag8.653 myeloma line using polyethylene glycol. Fused cells were seeded into 24-well plates in HAT medium for selection following standard procedures (28).
Fourteen days after fusion, hybridoma supernatants were tested for reactivity against SU-A by ELISA, using gradient-purified RCANBP virions or chimeric SU-A-rbIgG immunoadhesin (53). Chimeric Tva-rbIgG (7) (expression vector kindly provided by K. Zingler and J. Young) served as a negative control. Of the four positive hybridoma clones identified, one clone, mc8C5, which is a mouse IgG1 subtype, was highly reactive in both assays (data not shown) and could be successfully subcloned. mc8C5-4 hybridoma cells were used to generate ascites fluid, and antibody was also purified from hybridoma cell culture supernatants. Subclone mc8C5-4 was used for a majority of the characterizations described below.
mc8C5 reacts with SU-A in immunofluorescence.
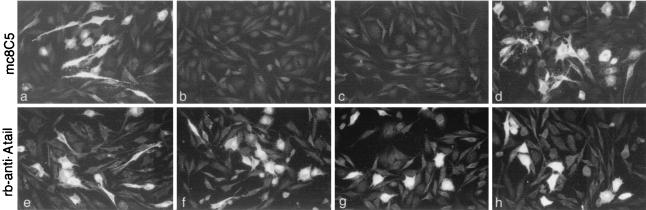
To test whether mc8C5-4 could be used in immunofluorescence applications, EnvA or chimeric molecules containing the SU-A domain were expressed from various expression vectors in either avian Df-1 cells (32, 48) or a variety of mammalian cells. mc8C5-4 was able to recognize the SU-A domain in a subgroup-specific manner using various indirect immunofluorescence approaches: EnvA could be detected after expressing cells were incubated with mc8C5-4 without prior fixation on ice or at 37°C, after fixation with ice-cold acetone or ethanol-acetic acid (95:5) or after fixation with 1 to 4% PFA. Figure 1 shows an example of Df-1 cells which were transfected with proviral vectors RCANBP or RCASBP-X-AP encoding A, B, and C subgroup SU domains (21-23). When acetone-fixed cells were incubated with an anti-cytoplasmic tail peptide antiserum (rb-anti-A-tail; [25]) detecting the C terminus of the SR-A TM protein shared by all constructs, Env expression was seen in all samples (Fig. 1e to h). However, when parallel samples were incubated with mc8C5-4, only those glycoproteins containing the SR-A SU domain were detected (Fig. 1a to d), demonstrating the subgroup A specificity of the monoclonal antibody.
FIG. 1.
ASLV Env glycoprotein immunofluorescence staining by mc8C5-4 is specific for the SU domain and is subgroup A specific. Avian Df-1 cells were transfected with proviral plasmids RCASBP-A-AP (a and e), RCASBP-B-AP (b and f), RCASBP-C-AP (c and g), and RCANBP (d and h), expressing SU of subgroup SR-A, SR-B, Pr-C, and SR-A, respectively. At 48 posttransfection, duplicate samples of cells were acetone fixed for whole-cell immunofluorescence staining with either mc8C5 ascites or with rb-anti-A-tail.
Use of mc8C5-4 for detection of SU-A in Western blot and IP.
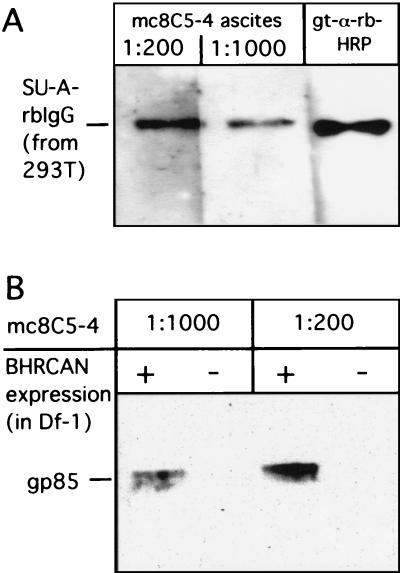
mc8C5-4 also demonstrated activity in ECL-Western blotting against SU-A-rbIgG immunoadhesin (53) expressed in 293T cells (Fig. 2A), as well as the gp85 subunit of EnvA expressed from RCANBP in Df-1 cells (Fig. 2B). Tva-rbIgG, EnvA gp37, and EnvC were not detected (not shown), indicating again specificity for SU-A. When purified SU-A-rbIgG was completely N deglycosylated by PNGase F digestion, subjected to SDS-PAGE, and blotted onto nitrocellulose in SDS-free, but 20% methanol-containing buffer (see Materials and Methods), mc8C5-4 was still able to detect the antigen (not shown). Thus, the monoclonal reagent is not directed against a carbohydrate epitope. However, after purified, PNGase F-treated SU-immunoadhesin was boiled in 1% SDS and then applied directly (dot blotted) onto nitrocellulose presoaked in blotting buffer, mc8C5-4 did not bind to the N-deglycosylated SU-A domain, while glycosylated antigen was still detected under such conditions (not shown). These observations suggest that the presence of carbohydrate side chains and/or electroblotting in the presence of methanol allows the partial conservation and/or refolding of epitope conformation, while such conformation is completely lost in the presence of 1% SDS when N glycosylation is absent.
FIG. 2.
Detection of SU-A in Western blot by mc8C5-4. (A) The immunoadhesin SU-A-rbIgG, which was transiently expressed in 293T cells, was detected after incubation with mc8C5-4 ascites and rb-anti-mouse-HRP or directly with goat-anti-rb-HRP by ECL. (B) Df-1 cells were infected with RCANBP, and cell-associated SU-A (gp85) was detected by ECL-Western blotting after incubation with mc8C5-4 ascites and rb-anti-mouse-HRP.
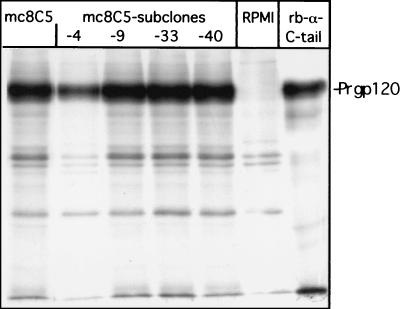
We next tested hybridoma supernatants of mc8C5 and its subclones mc8C5-4, -9, -33, and -40 in an immunoprecipitation (IP) assay (Fig. 3). In the initial experiment, we employed a chimeric ASLV Env molecule, similar to those expressed from RCASBP-X-AP, in which the coding region for SU-A between the XhoI and EcoRI site was cloned into the background of the Pr-C Env gene (designated SU-A/TM-C). This molecule also contains a cleavage site mutation (S19) (39, 42) and is therefore expressed on the cell surface as a terminally glycosylated precursor molecule, Prgp120. CV-1 cells expressing the chimeric protein Env-SU-A/TM-C-S19 were metabolically labeled and lysed in nondenaturing detergent. Aliquots of the lysate were then incubated with hybridoma supernatants, RPMI, or rb-anti-C-tail (25), respectively. All hybridoma supernatants immunoprecipitated Prgp120, as did the polyclonal serum control under these same IP conditions. mc8C5-4 was also successfully used for IP of nonchimeric EnvA and EnvA-S19 from Df-1, CV-1, 293T, NIH 3T3, and MDCK cell lysates, as well as of soluble monomeric EnvA ectodomain fragments (see Fig. 7, and data not shown), but did not react with subgroup C Env (not shown). It is important to note that the selection of protein G-Sepharose beads (protein G-agarose from Roche Diagnostics, Mannheim, Germany, or Protein G Gamma Bind Plus from Amersham Pharmacia Biotech, Piscataway, N.J.) used in IP was found to be critical for successful pull down of the monoclonal antibody.
FIG. 3.
mc8C5-4 antibody efficiently immunoprecipitates EnvA. The chimeric glycoprotein Env-SU-A/TM-C-S19, which contains a cleavage site mutation (S19) and is processed to a fully glycosylated precursor molecule, Prgp120, was expressed in CV-1 cells. Cells were metabolically labeled with [35S]methionine-cysteine and lysed in nondenaturating buffer (1% Triton X-100, 0.5% deoxycholate). Lysate (100 μl) was incubated with 100 μl of hybridoma supernatant, and antigen-antibody complexes were precipitated with 30 μl of protein A plus protein G-Sepharose beads (1:1) plus 1 μl of rb-anti-mouse-IgG. IP with rb-anti-C-tail served as a positive control. After SDS-PAGE, bands were visualized by autoradiography.
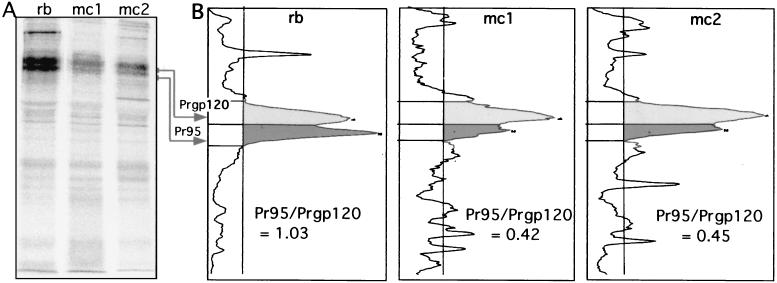
FIG. 7.
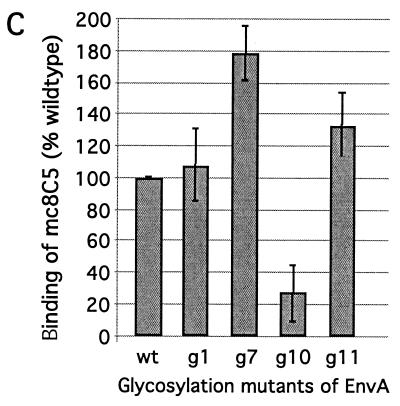
Recognition and binding of EnvA by mc8C5-4 is modulated by glycosylation. (A and B) IP. Df-1 cells expressing the cleavage mutant EnvA-S19 were metabolically labeled, lysed in nondenaturing buffer, and divided into three samples. One was immunoprecipitated with 1 μl of rb-anti-A-tail serum plus staphylococcus A (rb), and two were immunoprecipitated with 5 μl of mc8C5-4 ascites and either protein G (mc1) or 2 μl of rb-anti-mouse IgG plus staphylococcus A (mc2). (A) Samples were analyzed by autoradiography following SDS-PAGE; quantitation of the specific bands for Pr95 precursor and Prgp120 fully glycosylated precursor was performed with an OptiQuant scanner and software. (B) Profiles are shown, and the ratios of Pr95 to Prgp120 per sample are indicated. (C) FACS assay. Binding of mc8C5-4 to selected EnvA glycoproteins harboring glycosylation deletion mutations in the SU subunit (11) was analyzed after transient transfection of 293T cells with cDNA. Cells were treated for FACS analysis as described in Materials and Methods, using either Ngp37, a polyclonal antibody against the EnvA TM domain (31), or mc8C5-4, directed against SU-A as primary antibody. Mean fluorescence values as a measure of mc8C5-4 binding to EnvA wild-type and mutants were corrected for potential differences in EnvA surface expression by normalizing with the values obtained from the Ngp37-incubated duplicate samples.
The formation of EnvA-Tva complexes, and the Tva-induced activation of EnvA to fusion competence are inhibited by mc8C5-4.
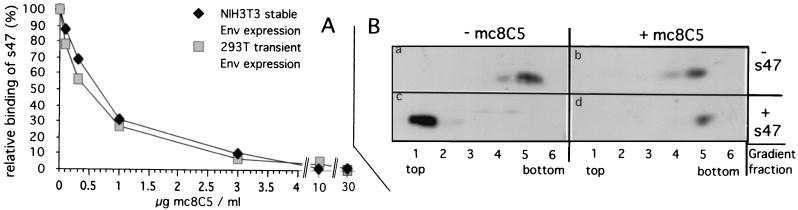
We next asked whether the SU-A specific monoclonal antibody could interfere with functions of EnvA, such as Tva binding. This was first investigated using a flow cytometry assay (Fig. 4A). NIH 3T3 cells and 293T cells expressing EnvA, either stably or transiently, were incubated in suspension with different concentrations of purified mc8C5-4 on ice. This was followed by addition of biotinylated s47, a peptide fragment of the Tva receptor that has previously been shown to bind to EnvA and to induce the conformational changes necessary for fusion peptide activation (9, 30). s47 bound to cells was detected after incubation with avidin-Oregon green 488 and quantitated by FACS. Mean fluorescence values indicative of s47 binding to EnvA were expressed as a percentage of s47-only fluorescence and plotted over the concentration of mc8C5-4 present during incubation. The resulting exponential inhibition curves for both cell types are nearly identical, even though EnvA was expressed to different levels on the cell surface of the different cell lines (mean fluorescent values differed by more than sixfold between the two cell lines; data not shown). Nevertheless, inhibition of s47 binding by mc8C5-4 was reproducibly concentration dependent. This suggests that the epitope recognized by mc8C5-4 is adjacent to or partially overlaps with the Tva binding site, resulting in steric hindrance of receptor binding.
FIG. 4.
Binding of the Tva receptor peptide, s47, to EnvA on the cell surface (A), as well as s47-induced liposome binding of a soluble EnvA (B), is inhibited by mc8C5-4. (A) For flow cytometry analysis of s47 binding, cells expressing EnvA either stably (NIH 3T3) or transiently from plasmid pCB6-EnvA (293T) were suspended and preincubated on ice with mc8C5-4 antibody at the indicated concentrations. Biotinylated s47 was added, and after incubation on ice and washes, Tva peptide that had bound to EnvA was reacted with avidin-Oregon green 488. Cells were then fixed in PFA, and the fluorescence intensity of Oregon green 488 as a measure of the (relative) amount of s47 bound to cells was quantitated. (B) Liposome flotation assay. A soluble form of EnvA (which includes the fusion subunit ectodomain of TM [gp37]) was incubated in PBS with or without mc8C5-4 prior to addition of s47 or PBS. The formation of EnvA-s47 complexes able to interact with liposomes was probed by sucrose gradient centrifugation as described in Materials and Methods. Six fractions were taken from the top (fraction 1) to the bottom (fraction 6) of the gradient and probed by Western blotting with an antibody which recognizes gp37. Env proteins associated with liposomes are expected to float to the top of the gradient.
Inhibition of Env function by the monoclonal antibody was next analyzed in a liposome flotation assay (30) (Fig. 4B). A soluble form of EnvA (which includes the fusion subunit ectodomain of TM [gp37] [27]) was preincubated with or without antibody on ice, and then s47 or PBS was added and complexes were allowed to form on ice. The samples were then incubated at 37°C for 15 min to allow for the fusion-relevant conformational changes to occur in the presence of liposomes (activation of the fusion peptide by s47 mediates the association of the soluble EnvA molecules with liposomes [30]). This association results in coflotation of Env with liposomes as the liposomes rise through a sucrose gradient. Fractions from each sample were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with an antibody that recognizes gp37 (Fig 4B). In the absence of soluble receptor, EnvA does not bind liposomes and is recovered from near the bottom of the gradient (panel a). s47 induces liposome binding, consistent with the induction of conformational changes which initiate fusion (panel c). These conformational changes are inhibited by mc8C5-4 (panel d), and binding of mc8C5-4 to EnvA does not induce these conformational changes on its own (panel b).
Infection of Tva-expressing cells with ASLV-A or EnvA-pseudotyped MLV is blocked by mc8C5-4.
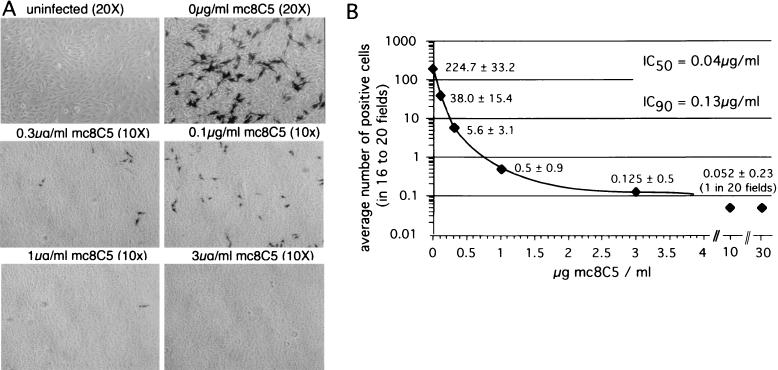
The results described above prompted us to investigate whether the monoclonal antibody was able to neutralize ASLV-A virus infection. Titrated RCASBP-A-AP virus supernatant was preincubated with different concentrations of purified mc8C5-4 and was used to infect Df-1 cells grown on coverslips. At 48 h postinfection, cells were fixed and stained for AP activity, and the blue, i.e., infected, cells per field were counted and plotted (Fig. 5). The 50% inhibitory concentration (IC50) and IC90 were calculated from the resulting exponential inhibition curve and were determined to be 0.04 and 0.13 μg/ml, respectively. These results demonstrate that mc8C5-4 efficiently inhibits infection by ASLV-A virions even at low concentrations.
FIG. 5.
Infection of Df-1 cells by ASLV-A is inhibited by mc8C5-4. Df-1 cells were seeded onto glass coverslips in eight quadruplets. One day postseeding, cells were infected with 1,000 infectious units of RCASBP(A)-AP virus supernatant or were mock infected. Prior to infection, virions had been preincubated with seven different concentrations of mc8C5-4. Cells were cultured for 48 h before being fixed and stained for AP activity. (A) Bright-light photographs were taken of four to five random fields per each of the four coverslips per sample, at a magnification of ×20 or ×10, as indicated. (B) The blue, i.e., infected, cells per field were counted (corrected for magnification) and plotted. IC50 and IC90 were calculated from the resulting exponential decay curve.
Similar results were obtained when PG950 cells (NIH 3T3 cells stably expressing Tva [26]) were infected with ASLV EnvA-pseudotyped MLV (12) in the presence of different concentrations of mc8C5-4 (data not shown).
Mutations in the hr2 domain of SU-A that interfere with Tva binding also reduce binding of mc8C5-4.
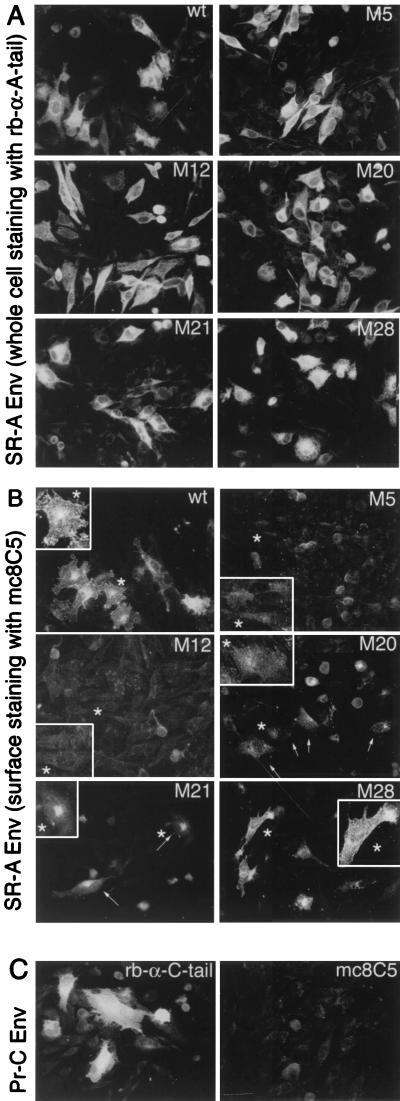
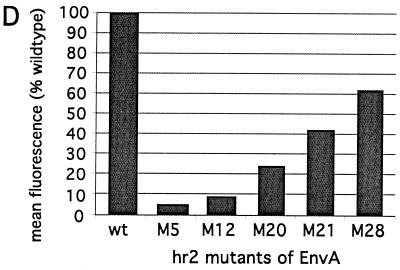
Since the anti-SU-A monoclonal antibody mc8C5-4 interferes with binding of Tva to SU-A and with virus infection, we were interested in determining whether mutations within the hypervariable, host range-determining region hr2 affected recognition by mc8C5-4. hr2 constitutes part of the receptor-binding domain for EnvA, and a cluster of basic residues within hr2 (R210, R213, R223, R224, and R227), has been implicated in receptor recognition and efficient entry. We chose a set of five mutants (sM5, sM12, sM20, sM21, and sM28) with alterations in those residues, which have been characterized in detail previously (8, 46). sM5, sM12, sM20, and sM21 were previously found to be greatly reduced both in their Tva binding capability and infectivity. M28 (R213S) differed since here Tva binding nearly reached wild-type levels, while infectivity was still impaired by 95%. In order to assay mc8C5 binding, we expressed the mutant EnvA proteins, and wild-type EnvA, from pCB6-EnvA plasmids in Df-1 cells, which were then either prepared for immunofluorescence (Fig. 6A to C), or flow cytometry (Fig. 6D). Immunostaining of acetone-fixed cells with rb-anti-A-tail (Fig. 6A) revealed comparable transfection efficiencies and a similar overall intracellular and surface staining pattern and intensity for each of the constructs. This is consistent with the previous observation regarding overall and surface expression levels compared to wild type EnvA. To analyze, by immunofluorescence, the ability of mc8C5-4 to bind surface-resident EnvA, cells were incubated with mc8C5-4 on ice prior to fixation (Fig. 6B). Clear differences in the ability of the mutants to bind mc8C5-4 became apparent: While M28 showed strong surface fluorescence similar to wild type, M5 and M12 showed near-background levels, M20 showed weak but discernible levels, and M21 showed intermediate levels of fluorescence. In order to quantitate these differences, transfected Df-1 cells were prepared for flow cytometry analysis (Fig. 6D); incubation with mc8C5-4 and secondary antibody were performed on ice prior to PFA fixation. When background-corrected mean fluorescence levels of the gated populations were compared to the wild-type EnvA antibody binding level, the values reflected the results of the immunofluorescence analysis. Thus, the mutations that affected Tva binding in the previous studies (with somewhat-pleiotropic effects) also affect mc8C5-4 binding in a similar manner. Our results presented in Fig. 6 indicate that the structural requirements for efficient binding of mc8C5-4 to SU-A are related to those necessary for efficient receptor binding. As shown below, this interpretation can be extended to a critical role of glycosylation as well.
FIG. 6.
Mutations within the hr2 of SU-A that result in reduced Tva binding also result in reduced binding of mc8C5-4. (A to C) Df-1 cells were transfected in duplicate with pCB6-EnvA DNA coding for either wild-type or hr2 mutant glycoprotein (sM5, sM12, sM20, sM21, and sM28 [8, 46]) or with pCB6-EnvC. At 24 h posttransfection, cells were prepared for immunofluorescence. For whole-cell staining and control of transfection efficiency (A and C), Env in fixed cells was detected with rb-anti-A-tail or rb-anti-C-tail, respectively. Binding of mc8C5-4 to cell surface-expressed Env (B and C), was performed with unfixed cells on ice. (B) Individual cells from each field have been enlarged for better viewing. (D) Df-1 cells mock transfected or transfected with pCB6-EnvA wild type (wt) or hr2 mutants were analyzed by FACS after incubation of unfixed cells on ice with mc8C5-4 ascites, followed by rb-anti-mouse Alexa488, and PFA fixation. Binding affinity of mc8C5-4 is reflected in the mean fluorescence values of the gated populations, which were corrected for background fluorescence of nontransfected cells, and are plotted as relative mean fluorescence values compared to wild-type EnvA.
The effect of the SU-A glycosylation state on mc8C5-4 binding reflects the effect on Tva binding.
When performing IPs of cleavage mutant EnvA-S19 with mc8C5-4 and rb-anti-A-tail in parallel (Fig. 7A), we repeatedly observed (Fig. 7B) that the ratio of core-glycosylated precursor, Pr95, to fully glycosylated precursor, Prgp120, was different for the different antibodies. mc8C5-4 preferentially precipitated Prgp120, not only when EnvA-S19 was expressed from Df-1 cells (shown) but also when it was expressed from CV-1 cells (not shown).
This suggested that the glycosylation of EnvA influenced recognition by mc8C5-4, which is supported by our findings regarding detection of SU-A by mc8C5-4 in Western blotting (see above).
To further investigate this matter, we analyzed, by flow cytometry, a set of SU-A glycosylation mutants that have recently been characterized with regard to s47 (Tva) binding and virus infectivity (11). The mutants are designated EnvAΔN-g1, -g7, -g10, and -g11 and harbor mutations T19A, S232A, S256A, and S333A, respectively. The N-glycosylation sites (NXS/T) altered in the chosen mutants, with the exception of the g1 mutant, are conserved among all ASLV Env sequences examined. While EnvAΔN-g1, -g7, and -g11 are proteolytically cleaved, incorporated into virions, and result in virus infectivity similar to wild-type levels, EnvAΔN-g10 was found to be very poorly processed, and though cleaved Env was found in virions, infectivity was reduced approximately 1,000-fold. All mutants but EnvAΔN-g10 were found to bind s47 near wild-type levels in an in vitro assay, while EnvAΔN-g10 did not interact with s47 at all.
For analysis of mc8C5-4 binding to the glycosylation mutants by flow cytometry (Fig. 7C), the respective mutants were transiently expressed in 293T cells in duplicate. Cells were fixed with PFA and stained with either rb-anti-Ngp37 or mc8C5-4 and the appropriate secondary antibody. Since the N terminus of gp37 is identical in all cases, the staining with the rb-anti-Ngp37 serum (31) could be used to adjust for differences in surface expression levels. When corrected mean fluorescence values for mc8C5-4 binding were compared to wild-type levels, EnvAΔN-g10 showed the most dramatic reduction (∼55%), while, interestingly EnvAΔN-g7 and -g11 displayed an increased mc8C5-4 affinity.
DISCUSSION
Here we report on the generation of a monoclonal antibody, mc8C5-4, directed against the SU domain of the ASLV subgroup A glycoprotein. The antibody is specific for subgroup A Env expressed in avian as well mammalian cells in immunofluorescence and IP assays. Our results indicate that it recognizes an epitope that is adjacent to or overlaps with the binding site for the EnvA receptor, Tva, since it can interfere with Tva-EnvA complex formation in vitro and in cell culture. Consequently, mc8C5-4 also prevents Tva-induced conformational changes necessary to activate the fusion peptide located near the N terminus of the EnvA TM subunit. While binding sites for Tva and mc8C5-4 may overlap, the monoclonal antibody does not induce such conformational changes on its own.
With all observations taken into account, it is plausible that the structural requirements for efficient binding of mc8C5-4 to SU-A are related to those necessary for efficient receptor binding. It appears that, even though mc8C5-4, by Western blotting, recognizes Env that was denatured during SDS-PAGE, the epitope is partially conformational and not merely linear. While it was determined that mc8C5-4 does not bind to carbohydrate moieties, glycosylation does influence antibody recognition, most likely by affecting overall conformation of the protein, and thus epitope structure. A single glycosylation site mutation (S256A), approximately 25 amino acids downstream of the host range-determining region hr2, significantly reduces EnvA interaction with mc8C5-4. The same mutation, we recently reported, abrogates Tva binding in an in vitro assay, suggesting that removal of a distal, but crucial glycosylation site might result in a structurally significant rearrangement of protein domains (11). Furthermore, mutations of basic residues in the host range-determining region hr2 itself that impair Tva binding to EnvA (8, 46) also affect mc8C5-4 binding. Tva is related to the low density lipoprotein (LDL) receptor, and basic amino acids in the receptor binding site of LDL have been implicated in interacting with acidic residues in the ligand binding site of LDL receptor. Similar interactions have been suggested for SU-A and Tva (for an overview, see reference 46), and basic residues have also been implicated as structurally important in interactions between antibodies and viral antigens (50, 51).
The observation that glycosylation affects the binding of the monoclonal anti-SU-A mc8C5-4 is further supported by the observation that terminally glycosylated EnvA is immunoprecipitated more efficiently than the core-glycosylated precursor, Pr95. While deglycosylated SU-A-rbIgG is not detected after dot blotting in the presence of SDS, it is detected after electroblotting conditions that allow partial removal of SDS and thus limited refolding. In contrast, mc8C5-4 can bind to fully glycosylated SU-A-rbIgG under both conditions, raising the possibility that the sugar side chains limit the access of SDS to the peptide backbone and thus prevent complete denaturation.
The generation of antibodies in response to retroviral infection often fails to yield high-titer neutralizing antibodies. However, mc8C5-4 has the ability to potently inhibit infection of avian or Tva-expressing mammalian cells by ASLV- or EnvA-pseudotyped MLV. Inhibition of RCASBP-A-AP infection of avian Df-1 cells is very efficient, with an IC50 of 0.04 μg/ml, consistent with the antibody blocking the binding of receptor by the Env protein.
The EnvA function-inhibiting characteristics and the fact that mc8C5-4 seems to interact with the receptor binding site without triggering the activation cascade of conformational changes induced by Tva make this novel monoclonal antibody a unique reagent for structural and functional studies of the ASLV glycoprotein. A similar monoclonal antibody, monoclonal antibody (MAb) b12, recognizes a conformational epitope that overlaps the CD-4-binding site of the human immunodeficiency virus type 1 (HIV-1) envelope protein. MAb b12 neutralizes a broad range of HIV-1 primary isolates and protects against primary virus challenge in animal models (37, 41, 45). The crystal structure of this monoclonal antibody has recently been determined, and together with peptides selected by phage display for binding to MAb b12, is providing new insights into the binding of HIV-1 to its receptor (47, 54). Thus, mc8C5 has the potential to facilitate functional studies of EnvA, especially with regard to receptor binding, and offers a reagent potentially useful in efforts to solve the structure of ASLV-A Env. In addition, mc8C5-4, and monoclonal antibodies directed against Tva which we recently generated and will describe elsewhere, may prove useful in the refinement of gene-targeting strategies based on the ASLV retroviral vector RCAS and its receptor, Tva (16, 20, 21, 24, 33).
Acknowledgments
We thank J. Young for the kind gift of SU-A-rbIgG and Tva-rbIgG expression plasmids. The chicken cell line Df-1 was kindly provided by D. Foster. M. Federspiel kindly provided the RCASBP-X-AP plasmids and valuable technical advice. We also thank P. Bates for the generous gift of the hr2 mutant EnvA expression vectors.
This work was supported by grant CA29884-22 from the National Institutes of Health to E.H. and by NIH grant AI22470 to J.M.W. C.O.-J. received a research fellowship from the Deutsche Forschungsgemeinschaft (Germany). FACS analyses were performed at the Flow Cytometry Core Facilities of the University of Alabama at Birmingham Center for AIDS Research, which is supported by NIH grant P30-AI-27767, and of the University of Virginia Health System, supported by Cancer Center Support Grant P30-CA44579.
REFERENCES
- 1.Adkins, H. B., J. Brojatsch, J. Naughton, M. M. Rolls, J. M. Pesola, and J. A. Young. 1997. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc. Natl. Acad. Sci. USA 94:11617-11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, H. B., J. Brojatsch, and J. A. Young. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J. Virol. 74:3572-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, P., J. A. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 4.Bova, C. A., J. P. Manfredi, and R. Swanstrom. 1986. env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology 152:343-354. [DOI] [PubMed] [Google Scholar]
- 5.Bova, C. A., J. C. Olsen, and R. Swanstrom. 1988. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J. Virol. 62:75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton, D. R., and P. W. Parren. 2000. Vaccines and the induction of functional antibodies: time to look beyond the molecules of natural infection? Nat. Med. 6:123-125. [DOI] [PubMed] [Google Scholar]
- 7.Connolly, L., K. Zingler, and J. A. Young. 1994. A soluble form of a receptor for subgroup A avian leukosis and sarcoma viruses (ALSV-A) blocks infection and binds directly to ALSV-A. J. Virol. 68:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damico, R., L. Rong, and P. Bates. 1999. Substitutions in the receptor-binding domain of the avian sarcoma and leukosis virus envelope uncouple receptor-triggered structural rearrangements in the surface and transmembrane subunits. J. Virol. 73:3087-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damico, R. L., J. Crane, and P. Bates. 1998. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc. Natl. Acad. Sci. USA 95:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, G. L., and E. Hunter. 1987. A charged amino acid substitution within the transmembrane anchor of the Rous sarcoma virus envelope glycoprotein affects surface expression but not intracellular transport. J. Cell Biol. 105:1191-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delos, S. E., M. J. Burdick, and J. M. White. 2002. A single glycosylation site within the receptor binding domain of the avian sarcoma/leukosis virus glycoprotein is critical for receptor binding. Virology 294:354-363. [DOI] [PubMed] [Google Scholar]
- 12.Delos, S. E., J. M. Gilbert, and J. M. White. 2000. The central proline of an internal viral fusion peptide serves two important roles. J. Virol. 74:1686-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, J. Y., J. W. Dubay, L. G. Perez, and E. Hunter. 1992. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J. Virol. 66:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorner, A. J., and J. M. Coffin. 1986. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell 45:365-374. [DOI] [PubMed] [Google Scholar]
- 15.Dorner, A. J., J. P. Stoye, and J. M. Coffin. 1985. Molecular basis of host range variation in avian retroviruses. J. Virol. 53:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn, K. J., B. O. Williams, Y. Li, and W. J. Pavan. 2000. Neural crest-directed gene transfer demonstrates Wnt1 role in melanocyte expansion and differentiation during mouse development. Proc. Natl. Acad. Sci. USA 97:10050-10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edinger, A. L., M. Ahuja, T. Sung, K. C. Baxter, B. Haggarty, R. W. Doms, and J. A. Hoxie. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Einfeld, D., and E. Hunter. 1988. Oligomeric structure of a prototype retrovirus glycoprotein. Proc. Natl. Acad. Sci. USA 85:8688-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellgaard, L., M. Molinari, and A. Helenius. 1999. Setting the standards: quality control in the secretory pathway. Science 286:1882-1888. [DOI] [PubMed] [Google Scholar]
- 20.Federspiel, M. J., P. Bates, J. A. Young, H. E. Varmus, and S. H. Hughes. 1994. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup A avian leukosis virus-based retroviral vectors. Proc. Natl. Acad. Sci. USA 91:11241-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Federspiel, M. J., and S. H. Hughes. 1997. Retroviral gene delivery. Methods Cell Biol. 52:179-214. [PubMed] [Google Scholar]
- 22.Fekete, D. M., and C. L. Cepko. 1993. Retroviral infection coupled with tissue transplantation limits gene transfer in the chicken embryo. Proc. Natl. Acad. Sci. USA 90:2350-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields-Berry, S. C., A. L. Halliday, and C. L. Cepko. 1992. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc. Natl. Acad. Sci. USA 89:693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisher, G. H., S. Orsulic, E. Holland, W. P. Hively, Y. Li, B. C. Lewis, B. O. Williams, and H. E. Varmus. 1999. Development of a flexible and specific gene delivery system for production of murine tumor models. Oncogene 18:5253-5260. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert, J. M., P. Bates, H. E. Varmus, and J. M. White. 1994. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J. Virol. 68:5623-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert, J. M., L. D. Hernandez, J. W. Balliet, P. Bates, and J. M. White. 1995. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J. Virol. 69:7410-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert, J. M., L. D. Hernandez, T. Chernov-Rogan, and J. M. White. 1993. Generation of a water-soluble oligomeric ectodomain of the Rous sarcoma virus envelope glycoprotein. J. Virol. 67:6889-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 291:2364-2369. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez, L. D., and J. M. White. 1998. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J. Virol. 72:3259-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 33.Holland, E. C., W. P. Hively, R. A. DePinho, and H. E. Varmus. 1998. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev 12:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmen, S. L., D. C. Melder, and M. J. Federspiel. 2001. Identification of key residues in subgroup A avian leukosis virus envelope determining receptor binding affinity and infectivity of cells expressing chicken or quail Tva receptor. J. Virol. 75:726-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter, E. 1997. Receptors and entry, p. 71-119. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses, 1st ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Hunter, E., E. Hill, M. Hardwick, A. Bhown, D. E. Schwartz, and R. Tizard. 1983. Complete sequence of the Rous sarcoma virus env gene: identification of structural and functional regions of its product. J. Virol. 46:920-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kessler, J. A., II, P. M. McKenna, E. A. Emini, C. P. Chan, M. D. Patel, S. K. Gupta, G. E. Mark III, C. F. Barbas III, D. R. Burton, and A. J. Conley. 1997. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 13:575-582. [DOI] [PubMed] [Google Scholar]
- 38.Nyambi, P. N., H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, A. Nadas, and S. Zolla-Pazner. 2000. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J. Virol. 74:7096-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochsenbauer, C., S. R. Dubay, and E. Hunter. 2000. The Rous sarcoma virus Env glycoprotein contains a highly conserved motif homologous to tyrosine-based endocytosis signals and displays an unusual internalization phenotype. Mol. Cell Biol. 20:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochsenbauer-Jambor, C., D. C. Miller, C. R. Roberts, S. S. Rhee, and E. Hunter. 2001. Palmitoylation of the Rous sarcoma virus transmembrane glycoprotein is required for protein stability and virus infectivity. J. Virol. 75:11544-11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez, L. G., and E. Hunter. 1987. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein that block processing to gp85 and gp37. J. Virol. 61:1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petropoulos, C. J., and S. H. Hughes. 1991. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J. Virol. 65:3728-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 45.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rong, L., A. Edinger, and P. Bates. 1997. Role of basic residues in the subgroup-determining region of the subgroup A avian sarcoma and leukosis virus envelope in receptor binding and infection. J. Virol. 71:3458-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 48.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 49.Smith, E. J., J. Brojatsch, J. Naughton, and J. A. Young. 1998. The CAR1 gene encoding a cellular receptor specific for subgroup B and D avian leukosis viruses maps to the chicken tvb locus. J. Virol. 72:3501-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda, K., M. Haque, T. Sunagawa, T. Okuno, Y. Isegawa, and K. Yamanishi. 1997. Identification of a variant B-specific neutralizing epitope on glycoprotein H of human herpesvirus-6. J. Gen. Virol. 78:2171-2178. [DOI] [PubMed] [Google Scholar]
- 51.Tourdot, S., S. Herath, and K. G. Gould. 2001. Characterization of a new H-2Dk-restricted epitope prominent in primary influenza A virus infection. J. Gen. Virol. 82:1749-1755. [DOI] [PubMed] [Google Scholar]
- 52.Young, J. A., P. Bates, and H. E. Varmus. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J. Virol. 67:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zingler, K., and J. A. Young. 1996. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J. Virol. 70:7510-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zwick, M. B., L. L. Bonnycastle, A. Menendez, M. B. Irving, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. K. Scott. 2001. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 75:6692-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]