Abstract
Human papillomavirus (HPV) vaccines have the potential to prevent cervical cancer by preventing HPV infection or treating premalignant disease. We previously showed that DNA vaccination with the cottontail rabbit papillomavirus (CRPV) E6 gene induced partial protection against CRPV challenge and that the vaccine's effects were greatly enhanced by priming with granulocyte-macrophage colony-stimulating factor (GM-CSF). In the present study, two additional strategies for augmenting the clinical efficacy of CRPV E6 vaccination were evaluated. The first was to fuse a ubiquitin monomer to the CRPV E6 protein to enhance antigen processing and presentation through the major histocompatibility complex class I pathway. Rabbits vaccinated with the wild-type E6 gene plus GM-CSF or with the ubiquitin-fused E6 gene formed significantly fewer papillomas than the controls. The papillomas also required a longer time to appear and grew more slowly. Finally, a significant proportion of the papillomas subsequently regressed. The ubiquitin-fused E6 vaccine was significantly more effective than the wild-type E6 vaccine plus GM-CSF priming. The second strategy was to vaccinate with multiple CRPV early genes to increase the breadth of the CRPV-specific response. DNA vaccines encoding the wild-type CRPV E1-E2, E6, or E7 protein were tested alone and in all possible combinations. All vaccines and combinations suppressed papilloma formation, slowed papilloma growth, and stimulated subsequent papilloma regression. Finally, the two strategies were merged and a combination DNA vaccine containing ubiquitin-fused versions of the CRPV E1, E2, and E7 genes was tested. This last vaccine prevented papilloma formation at all challenge sites in all rabbits, demonstrating complete protection.
Cervical human papillomavirus (HPV) infection is an extremely common sexually transmitted disease that affects an estimated 15% of women in the United States (18). Persistent lesions can be treated, but available treatments do not necessarily prevent recurrence (from latently infected tissues) and reinfection (with the same or other HPV types). Cervical carcinogenesis is initiated by infection with high-risk types of HPV (3, 28, 39), the most common of which are HPV type 16 (HPV-16) and HPV-18. Worldwide, cervical cancer is the second or third most common cancer in women (23).
Vaccination against HPV to prevent infection and to treat premalignant disease could substantially decrease morbidity and mortality from cervical cancer. The ideal HPV vaccine would not only prevent primary lesions from forming but also provide therapy for established lesions. While humoral antibodies can prevent infection, only cellular immune responses against the early (intracellular) papillomavirus proteins can mediate both functions, by providing the helper cell activities necessary for proliferation and differentiation of B cells and/or differentiation into cytotoxic T cells (CTLs) as well as by elaborating inhibitory cytokines, thereby participating in the responses that occur during the preclinical stage of infection as well as those that occur after the establishment of lesions.
The only small-animal model of papillomavirus infection with long-term persistence and malignant progression of lesions is the cottontail rabbit papillomavirus (CRPV)-domestic rabbit model (4). We previously reported that a DNA vaccine encoding the CRPV early protein E6 induced significant protection against CRPV challenge (34). The E6 vaccine's efficacy was dramatically improved by priming the sites of vaccination with granulocyte-macrophage colony-stimulating factor (GM-CSF), a cytokine that stimulates the local recruitment and maturation of professional antigen-presenting cells (20), which initiate DNA vaccine-induced immune responses (6, 19).
Another strategy for enhancing immune responses to vaccination is to increase the efficiency of antigen processing—for example, by using a gene that encodes a ubiquitin-fused version of the target protein (13). Ubiquitinated proteins enter the proteasomal pathway, where they are processed and presented through the major histocompatibility complex (MHC) class I pathway to stimulate differentiation and clonal expansion of MHC class I-restricted, typically CD8+, cytotoxic T cells (16, 29, 42). Rodriguez et al. showed that the fusion of a ubiquitin monomer to the nucleoprotein of lymphocytic choriomeningitis virus led to more rapid and complete degradation (25). When incorporated into a prophylactic vaccine for mice, the ubiquitin-fused nucleoprotein gene dramatically reduced the viral titer and the severity of lymphocytic choriomeningitis virus-induced disease. Similar ubiquitin-based strategies for other pathogens also have been effective (22, 36, 37).
A different approach for generating broad immune responses is to vaccinate against multiple target proteins (2), thereby increasing the chances of inducing population-wide immunity in individuals, whose immune responses to different epitopes within a given protein may differ widely. The early papillomavirus genes most likely to be useful for prevention of disease and treatment of low-grade lesions are E1, E2, E6, and E7. Each of these genes performs critical functions for the virus life cycle (30, 38), is expressed in virtually all premalignant lesions (7, 31), and is required for papilloma formation (5, 21, 41). Another potential advantage of targeting the early proteins, particularly E1 and E2, is that they contain highly conserved regions (1), so immunity to an early protein of one HPV type might cross-react with other HPV types. Vaccines targeting E6 and E7 oncoproteins could additionally be used to treat high-grade lesions and cancer (7, 31).
The first goal of the present study was to determine whether DNA vaccination with a ubiquitin-fused E6 gene could protect rabbits against papilloma formation following CRPV challenge and whether this strategy would be superior to GM-CSF priming. The second goal was to determine whether DNA vaccines encoding the E1, E2, E6, or E7 protein were capable of inducing prophylactic immunity and whether any combinations of the vaccines would help or hinder the development of protective immunity. Finally, we combined both strategies and tested the efficacy of a combination DNA vaccine encoding ubiquitin-fused versions of the E1, E2, and E7 proteins.
MATERIALS AND METHODS
DNA expression vectors.
Four plasmids used in this study were generated previously: pdCMV-E6 encodes the full-length CRPV E6 protein (34), pCMV-β (Clontech, Palo Alto, Calif.) encodes β-galactosidase (β-gal), pPJV3226 (a gift from PowderJect Vaccines, Madison, Wis.) (20) encodes mouse GM-CSF, and pcDNA3.0 (Invitrogen, Carlsbad, Calif.) is a cloning vector without an insert.
Ubiquitin-fused DNA vaccine encoding the CRPV E6 protein.
A DNA construct encoding a ubiquitin-fused CRPV E6 protein was constructed using a PCR fragment synthesized from CRPV-pLAII template DNA (40) and primers CR153C (5′ aac tta gat ctg ATG GAG AAC TGC) and CR981N (5′g ata ctc gag ATA CTA TCA TCT) (CRPV sequences are in uppercase, start and stop codons are in bold, and restriction sites are underlined). The E6-containing PCR product was digested with BglII and XhoI and cloned into pCMVi(H3)Ubs (2) between its BglII and SalI sites. pCMVi(H3)Ubs was a gift from Michael A. Barry (Center for Cell and Gene Therapy, Baylor College of Medicine, Houston, Tex.) (17). The ubiquitin coding sequence in pCMVi(H3)Ubs has a mutation at residue 76 of ubiquitin, changing a glycine codon to alanine. This mutation in other ubiquitin constructs generates a protein that resists ubiquitin cleavage, augments antigen processing, and increases CTL responses (2, 9, 25). The ubiquitin-E6 gene (Ub-E6) was excised from the new clone by EcoRI and XbaI digestion and then subcloned into pcDNA3.0 between its EcoRI and XbaI sites. DNA sequencing demonstrated that the new construct, named pdCMV-Ub-E6, was identical to the intended sequence.
Ubiquitin-fused vaccine encoding β-gal.
A DNA construct encoding a ubiquitin-fused β-gal gene was constructed with a PCR fragment synthesized using pCMV-β as template DNA and primers B973C (5′ AAA GAA AGA TCT ATG TCG TTT ACT TTG) and B4026N (5′ CGA AAC TCG AGC AGA CAT GGC CTG CC). The Ub-β-gal fused gene was excised with BglII and XhoI and cloned into pCMVi(H3)Ubs between its BglII and SalI sites. The Ub-β-gal gene was excised from the new clone by EcoRI and XbaI digestion and then subcloned into pcDNA3 between its EcoRI and XbaI sites. DNA sequencing demonstrated that the new construct, named pcDNA3-Ub-β, was identical to the intended sequence.
Ubiquitin-fused DNA vaccines encoding the CRPV E1, E2, or E7 protein.
To construct ubiquitin-fused clones for the CRPV E1, E2, and E7 genes, pcDNA3.0 was first modified to destroy its BamHI site, using a Klenow fill-in reaction. The ubiquitin-encoding sequence between the EcoRI and XbaI sites of pCMVi(H3)Ubs was then subcloned into corresponding sites of the modified pcDNA3.0 vector to generate a new plasmid named pcDNA3-Ub. PCR fragments containing the E1, E2, or E7 gene were amplified using CRPV-pLAII template DNA and then cloned into pcDNA3-Ub such that a contiguous open reading frame was maintained between the ubiquitin sequence and each CRPV gene. For the E1 gene, primers 1362Ub-E1C (5′c gct agc tag gga tcc ATG GCT GAA GGT ACA) and 3171Ub-E1N (5′ tca cgt cag tct aga C TCA TAG AGA CTG AGA) were used; for the E2 gene, primers 3112Ub-E2C (5′ c gct agc tag gga tcc ATG GAG GCT CTC AGC) and 4570Ub-E2N (5′ gt cac gtc agtctagaA CTA AAG CCC ATA AAA) were used; and for the E7 gene, primers 1075Ub-E7C (5′ c gct agc tag gga tcc ATG ATA GGC AGA ACT) and 1360Ub-E7N (5′ gtc acg tca gtc tag aTT CAG TTA CAA CAC TC) were used. DNA sequencing demonstrated that each construct was identical to the intended sequence except that the E1 gene in pcDNA3-Ub-E1 contained a transition at nucleotide (nt) 2638 (TGC → TAC), resulting in an amino acid change from cysteine to tyrosine at codon 426. The E7 gene in pcDNA3-Ub-E7 contained a silent transition at nt 1111 (CTA → TTA) in codon 13 (Leu).
DNA vaccines encoding the wild-type CRPV E1, E2, or E7 protein.
To construct DNA vaccines encoding the wild-type CRPV E1, E2, or E7 protein, a PCR fragment of each gene was synthesized, cleaved with BamHI and XbaI, and cloned individually into pcDNA3.0 between the BamHI and XbaI sites. Each PCR fragment contained a Kozak sequence upstream of the CRPV ATG. PCR fragments were synthesized using CRPV-pLAII template DNA and primers 1362 E1Ck (5′ c gct agc tag gga tcc acc ATG GCT GAA GGT ACA) and 3171Ub-E1N (5′ t cac gtc agt cta gaC TCA TAG AGA CTG AGA) for the E1 gene, primers 3112E2Ck (5′ cgc tagc tag gga tcc acc ATG GAG GCT CTC AGC) and 4285Ub-E2N (5′ gt cac gtc agt cta gaA CTA AAG CCC ATA AAA) for the E2 gene, and primers 1075 E7Ck (5′c gct agc tag gga tcc acC ATG ATA GGC AGA ACT) and 1360Ub-E7N (5′ gtc acg tca gtc taga T TCA GTT ACA ACA CTC) for the E7 gene. DNA sequencing demonstrated that each construct was identical to the intended sequence. The constructs were named pcDNA3-E1K, pcDNA3-E2K, and pcDNA3-E7K, respectively.
In vitro translation.
Genes on plasmid DNAs were transcribed from a T7 promoter and translated in rabbit reticulocyte lysates in the presence of [35S]methionine by coupled in vitro transcription-translation (TNT system) using the protocol supplied by the manufacturer (Promega Corp., Madison, Wis.). One microliter of the in vitro-translated lysate was fractionated on a sodium dodecyl sulfate-12% polyacrylamide gel and processed for fluorography as described previously (32).
Rabbits.
Two-kilogram female Pasteurella-free New Zealand White rabbits (Oryctolagus cuniculus) were purchased from Millbrook Farms (Amherst, Mass.). They were maintained in the animal facilities at Yale University School of Medicine. All experiments were performed in accordance with procedures approved by the Yale Institutional Animal Care and Use Committee.
DNA vaccination.
DNA-coated gold beads were prepared for in vivo inoculation as described previously (33), except that the gold beads were 1.9 μm in diameter. DNAs were delivered intracutaneously to the left side of the back at a concentration of 1 μg of DNA per site, using a helium-driven gene gun (XR 1 device; PowderJect Vaccines) at 350 lb/in2. One microgram of DNA was delivered twice to each of 10 sites per rabbit. In the first experiment, priming DNAs were delivered on days 0 and 21 and vaccine DNAs were delivered on days 3 and 24. In the second experiment, vaccine DNAs were delivered on days 0 and 21. Rabbits were challenged 2 weeks after the booster.
CRPV.
Two virus stocks were used. The K154 stock of CRPV was extracted from papillomas experimentally induced in a cottontail rabbit (O. sylvilagus). A preliminary experiment showed that the K154 stock was infectious, but papilloma formation was not quantified. The K216 stock of CRPV was generated from K154 in scid mice (20, 33-35). Previous experiments showed that 1:50 to 1:450 dilutions of the K216 virus induced papillomas at 96 to 100% of challenge sites in control rabbits (20).
CRPV challenge.
Two weeks after the booster immunization, rabbits were challenged on the right flank (contralateral to the vaccination sites) (33). After the fur was clipped, the site for each challenge was marked as a circle with a diameter of 1 cm and then meticulously prepared by superficial cross-hatching of the skin with a razor blade in each of four directions (seven strokes per direction).
Monitoring papilloma formation, growth, and regression.
CRPV challenge sites were examined weekly; for each papilloma, the location was recorded and measurements in three dimensions were taken (20, 34). Papilloma volumes were calculated (4/3π × [length/2] × [width/2] × [height/2]) and plotted using the natural logarithm. Papilloma regression was measured with respect to frequency and duration. No regressed papillomas reformed. The data were analyzed with respect to incidence of papilloma formation, time to papilloma onset, frequency (prevalence) of papilloma-positive sites over time, rate of papilloma growth over time, and final papilloma burden per rabbit. Papilloma burden was calculated as the sum of the volumes of all papillomas per rabbit. The time to papilloma onset and the rate of papilloma growth were analyzed by counting only the sites that formed continuously growing papillomas and excluding all negative sites.
Statistical analysis.
The log-rank test was used to compare differences in the length of time required for the first papilloma to appear. The appearance of lesions, or lack thereof, was modeled using generalized estimating equations that enabled us to estimate the correlation between these binary-valued sites within each rabbit at every longitudinal time measurement. These estimated correlations were very weak, and we performed subsequent analyses using logistic regression and individual growth curves, treating each lesion within each rabbit independently. Models for growth curves of papilloma volumes were log linear in time, and standard deviations were proportional to their means. Statistical differences in mean growth rates were analyzed utilizing the unpaired Student t test. Regression frequency was analyzed utilizing the chi-square approximation except where noted.
RESULTS
Expression of the ubiquitin-fused E6 and β-gal proteins in vitro.
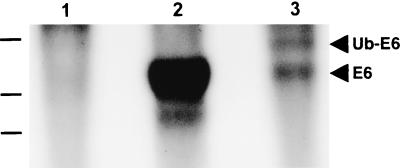
In vitro transcription-translation reactions showed that the Ub-E6 vaccine expressed a protein of about 40 kDa, the molecular weight predicted for the 351-amino-acid Ub-E6 fusion protein (Fig. 1, lane 3). The vaccine also produced a smaller band that comigrated with the unfused E6 protein (lane 2).
FIG. 1.
Expression of the Ub-E6 protein in vitro. Protein products of in vitro transcription-translation reactions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Products of the pCMV-UbE6 construct are shown in lane 3. Control reactions were performed with pcDNA3 (lane 1) and the pdCMV-E6 construct (lane 2).
Vaccination with the ubiquitin-fused E6 gene.
The first rabbit experiment compared the prophylactic efficacy of E6 DNA vaccination using no augmentation, GM-CSF priming, and ubiquitin fusion. Three corresponding control groups received an irrelevant plasmid instead of an E6 DNA vaccine but were otherwise treated identically to the experimental groups. Rabbits were vaccinated and boosted once. Two weeks later, they were challenged at five sites each with a 1:100 dilution of the K216 stock of CRPV. Analysis of the time to papilloma onset showed no differences among the rabbit groups. Regardless of the treatment, papillomas first appeared 19.2 ± 0.3 days (mean ± standard error of the mean [SEM]) after CRPV challenge.
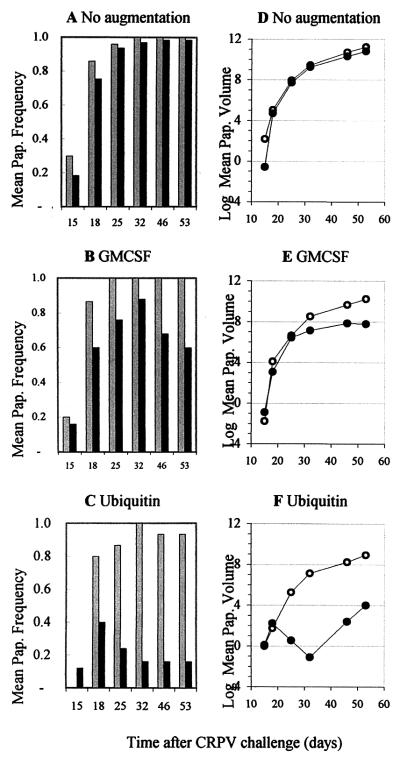
The time course of papilloma formation and regression is plotted in Fig. 2A to C. At each time point, the frequencies of papilloma-positive sites in E6-vaccinated groups were lower than those in corresponding control groups. By 32 days after CRPV challenge, all sites in control rabbits had formed papillomas. In E6 groups vaccinated without augmentation or with GM-CSF priming, 1 to 2 additional weeks were required to reach peak papilloma frequency. Peak frequencies in the groups vaccinated with GM-CSF plus E6 (GM-CSF+E6) or Ub-E6 were not maintained but were followed by a 2- to 3-week period of rapid rates of regression.
FIG. 2.
Time course of papilloma formation, regression, and growth in rabbits vaccinated with the E6 gene without augmentation (A and D), after GM-CSF priming (B and E), and after ubiquitin fusion (C and F). (A to C) Mean proportions of challenge sites with papillomas. The control groups are represented by gray-shaded bars; the E6-vaccinated groups are represented by black bars. Note that papilloma frequencies may not reach the maximum number of positive sites (which are given in Table 1) if some papillomas regress before others form. (D to F) Changes in mean papilloma volumes (in cubic millimeters) plotted on a natural logarithmic scale. Symbols represent the control groups (open circles) and the E6-vaccinated groups (closed circles).
The time course of papilloma growth is shown in Fig. 2D to F. Slope analysis of lines representing rates of growth in each group revealed that, relative to controls, E6 DNA vaccination reduced the growth rate by 42.6% with no augmentation, by 93.4% with GM-CSF priming, and by 99.4% with ubiquitin fusion. (Each effect was significant at P < 0.002.) Note that papilloma volumes are plotted logarithmically. Among the E6-vaccinated groups, papilloma growth was more strongly suppressed in the GM-CSF-primed group than in the nonaugmented group (P < 0.001) and in the Ub-E6-vaccinated group than in the GM-CSF+E6 group (P = 0.02). The large reduction in volume observed in the Ub-E6 group between days 18 and 32 (Fig. 2F) was due to regression of nine papillomas (Fig. 2C). The subsequent rise in volume reflected the continued growth of three small papillomas plus a new papilloma that formed on day 32.
The overall effects of E6 vaccination on papilloma formation, regression, and final frequency for E6-vaccinated and control groups are tabulated in Table 1. The data show that E6 vaccination reduced the percentage of sites that formed a papilloma from 100% to 88% when GM-CSF priming was used and from 100% to 52% when ubiquitin fusion was employed. Papilloma frequency was further reduced by regression, which occurred in E6-vaccinated rabbits at rates of 2% without augmentation, 32% with GM-CSF priming, and 69% with ubiquitin fusion. Stronger effects on each outcome were induced after GM-CSF priming than without augmentation (P < 0.001) and after ubiquitin fusion than with GM-CSF priming (P = 0.02).
TABLE 1.
Frequencies of papilloma formation and regression in rabbits vaccinated with the E6 gene, using no augmentation, GM-CSF priming, or ubiquitin fusion
| Augmentation | Antigen | Nd | Frequency off:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Formationa
|
Regressionb
|
Final frequencyc
|
||||||
| No. | Percentage | No. | Percentage | No. | Percentage | |||
| Nonee | β-gal | 10 | 50/50 | 100 | 0/50 | 0 | 50/50 | 100 |
| E6 | 13 | 65/65 | 100 | 1/65 | 2 | 64/65 | 98 | |
| GM-CSF | β-gal | 3 | 15/15 | 100 | 0/15 | 0 | 15/15 | 100 |
| E6 | 5 | 22/25 | 88 | 7/22 | 32 | 15/25 | 60* | |
| Ubiquitin | β-gal | 3 | 15/15 | 100 | 1/15 | 7 | 14/15 | 93 |
| E6 | 5 | 13/25 | 52* | 9/13 | 69* | 4/25 | 16** | |
Maximum number of sites that ever formed a papilloma/total number of sites.
Number of papillomas that completely regressed/number that formed.
Final number of papillomas/total number of sites.
N, number of rabbits per group.
The nonaugmented control group consisted of rabbits vaccinated with the β-gal gene after priming with pcDNA3 (N = 2) or pCMV-β (N = 3) and rabbits that were both primed and vaccinated with pcDNA3 (N = 2).
∗, significant at P ≤ 0.05; ∗∗, significant at P = 0.001.
In summary, E6 gene vaccination protected rabbits against papilloma formation, suppressed papilloma growth, and stimulated papilloma regression to differing degrees depending on the vaccination strategy. The most effective strategy was ubiquitin fusion, followed by GM-CSF priming and, finally, vaccination with the E6 gene alone.
Vaccination with the E1+E2, E6, and/or E7 gene.
We next tested the clinical efficacy of DNA vaccines encoding CRPV E1+E2, E6, and/or E7 when administered (without augmentation) prior to CRPV challenge. To test for potential synergy or incompatibility among the vaccine components, groups of four rabbits each were immunized with each component individually and in all possible combinations (see Table 2). Control rabbits received only vector DNA. After a single boost, each rabbit was challenged at nine sites, with each of three sites receiving a 1:50, 1:150, or 1:450 dilution of the K154 stock of CRPV, i.e., high, moderate, and low doses of virus. All clinical outcomes were affected by CRPV dose. As the challenge dose increased, the time to papilloma onset decreased, the frequency of papilloma formation increased, the rate of growth increased, and the frequency of regression decreased. Each of these dose-response relationships was significant at P < 0.0001. Analysis of the time to papilloma onset showed no differences among the rabbit groups: papillomas formed 24.2 ± 0.5 days (mean ± SEM), 26.9 ± 0.9 days, and 27.0 ± 1.5 days after high-, moderate-, and low-dose CRPV challenge, respectively.
TABLE 2.
Papilloma formation and regression in rabbits vaccinated with the E1+E2, E6, and/or E7 gene
| Variable measured | Vaccine | No. of papillomas at CRPV dosea:
|
Total/no. of sitesc | Normb | Pd | ||
|---|---|---|---|---|---|---|---|
| H | M | L | |||||
| No. of papillomase formed | Vector | 11 | 10 | 4 | 25/36 | 1 | 1 |
| E6 | 10 | 4 | 4 | 18/36 | 0.72 | 0.09 | |
| E7 | 6 | 3 | 0 | 9/36 | 0.36 | ∗ | |
| E6+E7 | 9 | 4 | 1 | 14/36 | 0.56 | 0.01 | |
| E1+E2 | 8 | 8 | 2 | 18/36 | 0.72 | 0.09 | |
| E1+E2+E6 | 4 | 3 | 1 | 8/36 | 0.32 | ∗ | |
| E1+E2+E7 | 9 | 4 | 0 | 13/36 | 0.52 | ∗ | |
| E1+E2+E6+E7 | 5 | 4 | 1 | 10/36 | 0.40 | ∗ | |
| No. of papillomase regressed | Vector | 4 | 4 | 1 | 9/25d | 1 | 1 |
| E6 | 7 | 4 | 4 | 15/18 | 2.31 | ∗ | |
| E7 | 4 | 2 | 0 | 6/9 | 1.85 | 0.11 | |
| E6+E7 | 4 | 3 | 1 | 8/14 | 1.59 | NS | |
| E1+E2 | 8 | 8 | 2 | 18/18 | 2.78 | ∗ | |
| E1+E2+E6 | 4 | 3 | 1 | 8/8 | 2.78 | ∗ | |
| E1+E2+E7 | 9 | 4 | 0 | 13/13 | 2.78 | ∗ | |
| E1+E2+E6+E7 | 2 | 1 | 0 | 3/10 | 0.83 | NS | |
| Final frequency of papillomasf | Vector | 7 | 6 | 3 | 16/36e | 1 | 1 |
| E6 | 3 | 0 | 0 | 3/36 | 0.19 | ∗ | |
| E7 | 2 | 1 | 0 | 3/36 | 0.19 | ∗ | |
| E6+E7 | 5 | 1 | 0 | 6/36 | 0.38 | 0.01 | |
| E1+E2 | 0 | 0 | 0 | 0/36 | 0 | ∗ | |
| E1+E2+E6 | 0 | 0 | 0 | 0/36 | 0 | ∗ | |
| E1+E2+E7 | 0 | 0 | 0 | 0/36 | 0 | ∗ | |
| E1+E2+E6+E7 | 3 | 3 | 1 | 7/36 | 0.44 | 0.02 | |
Dose of CRPV used for challenge: H, high; M, moderate; L, low.
Value normalized to that of the control group (proportion of affected sites).
For papilloma formation, values are number of sites that ever formed a papilloma/total number of sites; for papilloma regression, values are number of papillomas that completely regressed/number that formed; and for final frequency, values are final number of papillomas/total number of sites.
∗, P < 0.005; NS, not significant.
Analyzed by chi-square test.
Analyzed by Fisher's exact test.
Control rabbits that received vector DNA formed papillomas at 11 of 12, 10 of 12, and 4 of 12 sites challenged with high, moderate, and low doses of CRPV, respectively, for a total of 25 of 36. Relative to the control group, each group vaccinated with a CRPV early-gene or combination vaccine formed papillomas at 28 to 64% fewer sites, with P values ranging from <0.005 to 0.09 (Table 2).
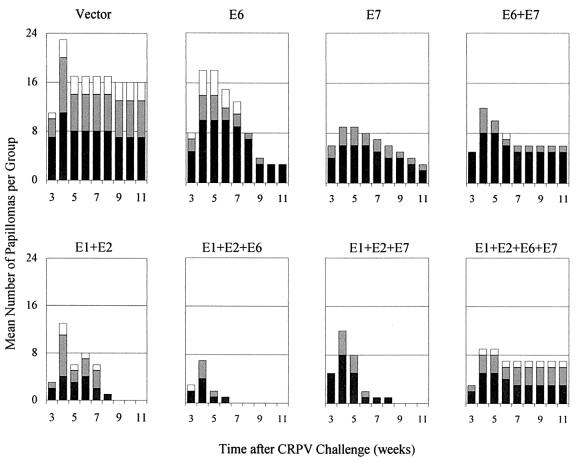
The time course of papilloma formation and regression is plotted in Fig. 3 and quantified in Table 2. In all groups, papilloma frequency increased until week 4, after which a high percentage of papillomas completely regressed. In the E6-only and E7-only groups, regression occurred at about twice the rate of the controls and was still ongoing at the end of the experiment (Fig. 3). All papillomas that formed in the groups vaccinated with the early genes E1+E2, E1+E2+E6, or E1+E2+E7 completely regressed. The rates of regression in the groups vaccinated with the E6+E7 combination or the full combination of E1+E2+E6+E7, however, were not significantly different from the control group, and further analysis showed that adding the E7 component to any vaccine containing the E6 component (E6 alone or E1+E2+E6) significantly inhibited regression (P < 0.001, Fisher's exact test).
FIG. 3.
Time course of papilloma formation and regression in rabbits vaccinated with the CRPV E1, E2, E6, and/or E7 gene. Panels are labeled according to the vaccine gene(s) that was used. The doses of CRPV used for challenge are represented as black bars (high dose), dark-gray-shaded bars (moderate dose), and light-gray-shaded bars (low dose).
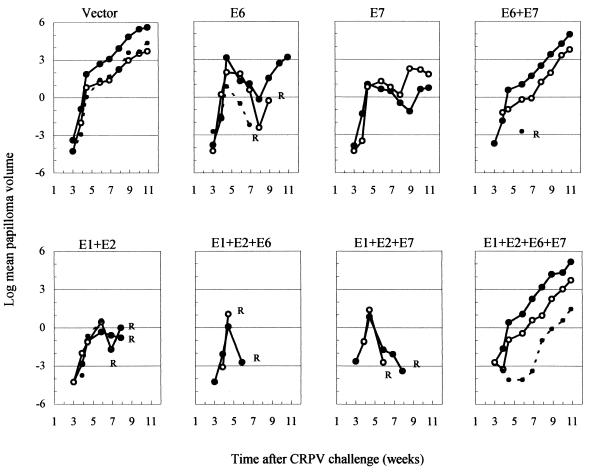
Changes in papilloma volumes (Fig. 4) reflected the course of papilloma formation and regression (Fig. 3). Initially, volumes increased rapidly in all groups (Fig. 4). Thereafter, papilloma growth in the vaccinated groups decreased sharply, particularly in groups that underwent complete regression, i.e., those vaccinated with E1+E2, E1+E2+E6, or E1+E2+E7. In the groups vaccinated with E6 or E7 alone, papilloma volumes fell but resumed positive growth after the number of papillomas stabilized (Fig. 3). At the end of the experiment, papilloma volumes in the E6-only and E7-only groups were more than 95% smaller than that of the control group. Volumes in the groups vaccinated with both E6 and E7 (E6+E7 and E1+E2+E6+E7) were about 70% smaller. Thus, adding the E7 component to either of the vaccines containing the E6 component impeded the reversal of papilloma growth. This finding did not achieve significance, in all likelihood because of the small numbers of papillomas per group and the innate variability in volumes due to the different doses of CRPV used for challenge.
FIG. 4.
Time course of papilloma growth in rabbits vaccinated with the CRPV E1, E2, E6, and/or E7 gene. Panels are labeled according to the vaccine genes that were used. The graphs show the changes in mean papilloma volumes (in cubic millimeters) plotted on a natural logarithmic scale. The doses of CRPV used for challenge are represented as large closed circles (high dose) connected by continuous lines, large open circles (moderate dose) connected by continuous lines, and small closed circles connected by dashed lines (low dose). R, all papillomas at a given CRPV dose regressed.
In summary, each of the E1+E2, E6, and E7 DNA vaccine components and all combinations thereof significantly reduced the rates of papilloma formation and growth. In addition, all vaccines except the two containing both E6 and E7 induced significant papilloma regression. Ultimately, the probability of having persistent clinical disease (papillomas) 11 weeks after CRPV challenge was reduced in each of the early-gene-vaccinated groups by 56 to 81% compared to the control group (Table 2).
Vaccination with ubiquitin-fused CRPV E1, E2, and E7 genes.
Having shown in the first experiment that ubiquitin fusion is an effective strategy for improving the efficacy of an E6-targeting DNA vaccine, and presuming that DNA vaccines targeting the E1, E2, and E7 proteins also would be prophylactic (which they were), we then generated additional DNA constructs individually encoding Ub-E1, Ub-E2, and Ub-E7. These three constructs were combined as a multivalent vaccine and tested in a ninth rabbit group included in the second experiment. The Ub-E6 vaccine was omitted from the combination because it was protective per se and would obscure the prophylactic effects of the other ubiquitin-fused vaccine genes. The effect of the Ub-E1+Ub-E2+Ub-E7 vaccine was obvious: whereas in the control group multiple papillomas formed, grew, and persisted (shown in Fig. 3 and 4), all rabbits in the vaccinated group showed 100% protection, and none of the CRPV challenge sites in any rabbit formed even a transient papilloma. Since only one of four rabbits immunized with the unfused E1+E2+E7 vaccine combination (in the same experiment) was completely protected, these results expand the findings of the first experiment by demonstrating that ubiquitin fusion is a good strategy for augmenting the efficacy of multiple CRPV early-gene vaccines.
DISCUSSION
This study provided several new findings. First, ubiquitin fusion was found to be a highly effective means of augmenting the effects of CRPV E6 DNA vaccination. Ubiquitin fusion most likely increased the rate of E6 polyubiquitination, entry into the proteasome, and antigen processing through the MHC class I pathway (16, 24, 29), events expected to enhance production of E6-specific CTLs. Although the E6 (and other CRPV) early proteins in infected cells in vivo generally do not elicit effective immunity (i.e., rabbit papillomas rarely regress spontaneously), their passage through the ubiquitin-proteasome pathway must not be totally blocked because CRPV-infected cells can be eliminated once a cell-mediated immune response is induced by vaccination. The Ub-E6 vaccine also expressed some apparently unfused E6 protein, but its greatly increased efficacy relative to the unfused E6 vaccine indicates that the fusion protein was the critical component.
This ubiquitin fusion strategy was significantly more effective than GM-CSF priming, which we had previously reported to be beneficial (20). Comparing the two studies, the effects of GM-CSF priming may appear to be less potent in the present study than in the earlier one, primarily because more stringent challenge conditions were used; e.g., papillomas in all groups formed earlier and grew more rapidly than in the earlier study. Both studies used similar dilutions of the K216 stock of CRPV; however, more meticulous preparation of CRPV challenge sites (see Materials and Methods) probably resulted in a larger number of infected cells with a greater number of CRPV copies per cell.
Proteins destined for degradation via the ubiquitin-proteasomal pathway contain one of a large number of peptide sequences that mark them as substrates for E3-mediated ubiquitination, the rate-limiting step in the process of polyubiquitination (16, 24, 29). Few of these sequences have been identified, and it is not known whether the E6 protein (or other papillomavirus proteins) contains such a sequence. A major function of the high-risk-HPV E6 proteins is to cause degradation of the tumor suppressor protein p53, which it achieves through direct interaction with E6AP, a member of the E3 family of ubiquitin ligases (27). Ubiquitin-mediated degradation of p53 by E6 contributes to cellular transformation and is believed to promote malignant progression (26). It therefore seems unlikely that the E6 protein itself would contain a strong ubiquitination signal.
The CRPV and HPV genomes have the same organization, and their proteins have sequence homology (11). For example, the CRPV E1, E2, E6, and E7 proteins are 61, 49, 47, and 47% similar to the HPV-16 early proteins, respectively. CRPV- and HPV-encoded proteins have conserved functions as well as conserved domains (8, 10-12, 14, 21), suggesting that multigene vaccination against CRPV may provide information of relevance to multigene vaccination against HPV. A third finding of this study was that vaccination with DNAs encoding the CRPV protein E1+E2, E6, and/or E7 suppressed papilloma formation and increased papilloma regression. Prior to regression, the CRPV infection itself most likely provided antigenic stimulation for the activation and proliferation of memory T cells induced by previous vaccination, as well as for the generation of new T-cell responses against the E6 proteins (and other viral or cellular antigens) in the lesions. Multigene vaccination provides much of the antigenicity of a true infection without the associated pathogenicity (2) and favors the induction of a broad spectrum of immune responses in both individuals and populations. It also could permit a host to respond to a tumor, even if some of the antigenic epitopes were lost.
Finally, we demonstrated that complete protection against CRPV challenge was obtained with a DNA vaccine that combined the ubiquitin fusion strategy with multigene vaccination, using the Ub-E1+Ub-E2+Ub-E7 gene combination. Since this last vaccine was tested side by side with the unfused E1+E2+E7 DNA vaccine, the data show that ubiquitin fusion is an effective vaccination strategy not only for the E6 protein but also for the E1+E2 and E7 proteins. Although the Ub-E1+E2+E7 vaccine induced complete protection and the Ub-E6 vaccine induced only partial protection, caution must be exercised when comparing the results of the two experiments because the challenge stringencies were different. Thus, it is entirely possible that the Ub-E6 vaccine, if retested under less stringent conditions, would also induce complete protection.
DNA vaccines encoding CRPV E1, E2, E6, and E7 were also tested by Han et al. (15). Their study differed from ours in that their vaccines were constructed with a different vector backbone and in that their rabbits were boosted twice instead of once and were challenged by inoculation of CRPV DNA rather than by infection with CRPV virus. In addition, their challenge conditions appear to have been more stringent than ours, because papillomas formed at all sites in their control group and because none of the papillomas in any of their CRPV E1-, E2-, E6-, or E7-vaccinated groups regressed. We believe that our lower-stringency conditions were due to the CRPV stock used in this experiment, K154, which also induced relatively mild disease in subsequent experiments (data not shown).
After normalization of our data to that of the control group to account for differences in challenge stringency, a reasonable comparison of the outcomes can be made. Both studies found that the E1, E2, and E6 vaccines provided partial protection, at 12 to 62% of sites. Our E7 vaccine protected 64% of the sites, while theirs protected only 4%. The reasons for this discrepancy are unknown but may involve different in vivo levels of E7 expression from the different vector backbones. Rabbit-to-rabbit variability may also be a factor, and both studies used only four rabbits per group. An E1+E2+E6+E7 combination vaccine administered with a single booster, tested in both studies, was found to protect 60% of sites in their study and 62% in ours. All papillomas in their E1+E2+E6+E7-vaccinated group regressed, indicating that their combination vaccine was more effective than their individual gene vaccines since none of the latter induced regression, even after two boosters.
Han et al. retested their E1+E2+E6+E7 combination vaccine using two boosters, which gave complete protection. In our study, regression brought the final papilloma frequency in the E1+E2+E6+E7 group to 44% (P = 0.02); however, in contrast to their study, this outcome was poor compared to those of our other CRPV-vaccinated groups, except for the E6+E7 group, whose outcome was nearly as poor. The papillomas in our groups that received both E6 and E7 became both highest in number and largest in volume, exceeded only by the control group. Hypothetically, the coadministration of the E6+E7 combination vaccines might have stimulated a type of immunity that was protective but not therapeutic or that was detrimental to the elimination of established lesions. Since the other E7 vaccine was only minimally effective (15), the level of immunity it induced may have been below the threshold needed to observe an inhibitory effect, which might help explain why our combination vaccine was less effective than theirs.
In conclusion, this study shows that highly significant protection against papilloma formation was obtained by immunization with the unfused E1+E2, E6, and E7 vaccines, administered individually and in combinations. The Ub-E6 vaccine also induced significant protection against papilloma formation, and the UbE1+UbE2+UbE7 combination vaccine induced complete protection. Moreover, a high proportion of papillomas in the partially protected rabbits subsequently underwent complete regression. Since regression in an immunotherapeutic response, these results suggest a potential for these vaccines to be useful for the treatment of rabbits with established papillomas.
Acknowledgments
This work was supported by grants from the Robert Leet and Clara Guthrie Patterson Trust (R01622), the Yale Skin Disease Research Center (7 P30 041942), and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (T32-AR000716-28). It was also supported by generous gifts from the Ted Mann Foundation and from Merck Research Laboratories, West Point, Pa.
REFERENCES
- 1.Baker, C. 1987. Sequence analysis of papillomavirus genomes, p. 321-385. In P. Salzman and P. M. Howley (ed.), The Papovaviridae, vol. 2. Plenum Press, New York, N.Y.
- 2.Barry, M. A., W. C. Lai, and S. A. Johnston. 1995. Protection against mycoplasma infection using expression-library immunization. Nature 377:632-635. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, and K. V. Shah for the International Biological Study on Cervical Cancer (IBSCC) Study Group. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 4.Brandsma, J. L. 1996. Animal models for human papillomavirus vaccine development, p. 69-78. In C. Lacey (ed.), Papillomavirus review: current research on papillomaviruses. Leeds University Press, Leeds, United Kingdom.
- 5.Brandsma, J. L., Z. H. Yang, S. W. Barthold, and E. A. Johnson. 1991. Use of a rapid, efficient inoculation method to induce papillomas by cottontail rabbit papillomavirus DNA shows that the E7 gene is required. Proc. Natl. Acad. Sci. USA 88:4816-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condon, C., S. C. Watkins, C. M. Celluzzi, K. Thompson, and L. D. Falo, Jr. 1996. DNA-based immunization by in vivo transfection of dendritic cells. Nat. Med. 2:1122-1128. [DOI] [PubMed] [Google Scholar]
- 7.Crum, C. P., M. Symbula, and B. E. Ward. 1989. Topography of early HPV 16 transcription in high-grade genital precancers. Am. J. Pathol. 134:1183-1188. [PMC free article] [PubMed] [Google Scholar]
- 8.Defeo-Jones, D., G. A. Vuocolo, K. M. Haskell, M. G. Hanobik, D. M. Kiefer, E. M. McAvoy, M. Ivey-Hoyle, J. L. Brandsma, A. Oliff, and R. E. Jones. 1993. Papillomavirus E7 protein binding to the retinoblastoma protein is not required for viral induction of warts. J. Virol. 67:716-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ecker, D. J., J. M. Stadel, T. R. Butt, J. A. Marsh, B. P. Monia, D. A. Powers, J. A. Gorman, P. E. Clark, F. Warren, A. Shatzman, et al. 1989. Increasing gene expression in yeast by fusion to ubiquitin. J. Biol. Chem. 264:7715-7719. [PubMed] [Google Scholar]
- 10.Fujii, T., J. L. Brandsma, X. Peng, S. Srimatkandada, L. Li, A. Canaan, and A. Deisseroth. 2001. High and low levels of cottontail rabbit papillomavirus E2 protein generate opposite effects on gene expression. J. Biol. Chem. 276:867-874. [DOI] [PubMed] [Google Scholar]
- 11.Giri, I., O. Danos, and M. Yaniv. 1985. Genomic structure of the cottontail rabbit (Shope) papillomavirus. Proc. Natl. Acad. Sci. USA 82:1580-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giri, I., and M. Yaniv. 1988. Study of the E2 gene product of the cottontail rabbit papillomavirus reveals a common mechanism of transactivation among papillomaviruses. J. Virol. 62:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant, E. P., M. T. Michalek, A. L. Goldberg, and K. L. Rock. 1995. Rate of antigen degradation by the ubiquitin-proteasome pathway influences MHC class I presentation. J. Immunol. 155:3750-3758. [PubMed] [Google Scholar]
- 14.Han, R., N. M. Cladel, C. A. Reed, and N. D. Christensen. 1998. Characterization of transformation function of cottontail rabbit papillomavirus E5 and E8 genes. Virology 251:253-263. [DOI] [PubMed] [Google Scholar]
- 15.Han, R., N. M. Cladel, C. A. Reed, X. Peng, and N. D. Christensen. 1999. Protection of rabbits from viral challenge by gene gun-based intracutaneous vaccination with a combination of cottontail rabbit papillomavirus E1, E2, E6, and E7 genes. J. Virol. 73:7039-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 17.Johnston, S. A., and M. A. Barry. 1997. Genetic to genomic vaccination. Vaccine 15:808-809. [DOI] [PubMed] [Google Scholar]
- 18.Koutsky, L. 1997. Epidemiology of genital human papillomavirus infection. Am. J. Med. 102:3-8. [DOI] [PubMed] [Google Scholar]
- 19.Leachman, S. A., and J. L. Brandsma. 1999. DNA vaccines for papillomaviruses, p. 105-148. In R. W. Tindle (ed.), Human papillomavirus vaccines, vol. 14. R. G. Landes Company, Austin, Tex.
- 20.Leachman, S. A., R. E. Tigelaar, M. Shlyankevich, M. D. Slade, M. Irwin, E. Chang, T. C. Wu, W. Xiao, S. Pazhani, D. Zelterman, and J. L. Brandsma. 2000. Granulocyte-macrophage colony-stimulating factor priming plus papillomavirus E6 DNA vaccination: effects on papilloma formation and regression in the cottontail rabbit papillomavirus-rabbit model. J. Virol. 74:8700-8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers, C., J. Harry, Y. L. Lin, and F. O. Wettstein. 1992. Identification of three transforming proteins encoded by cottontail rabbit papillomavirus. J. Virol. 66:1655-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niethammer, A. G., R. Xiang, J. M. Ruehlmann, H. N. Lode, C. S. Dolman, S. D. Gillies, and R. A. Reisfeld. 2001. Targeted interleukin 2 therapy enhances protective immunity induced by an autologous oral DNA vaccine against murine melanoma. Cancer Res. 61:6178-6184. [PubMed] [Google Scholar]
- 23.Parkin, D. M., P. Pisani, and J. Ferlay. 1999. Estimates of the worldwide incidence of 25 major cancers in 1990. Int. J. Cancer 80:827-841. [DOI] [PubMed] [Google Scholar]
- 24.Rock, K. L., and A. L. Goldberg. 1999. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17:739-779. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez, F., J. Zhang, and J. L. Whitton. 1997. DNA immunization: ubiquitination of a viral protein enhances cytotoxic T-lymphocyte induction and antiviral protection but abrogates antibody induction. J. Virol. 71:8497-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheffner, M. 1998. Ubiquitin, E6-AP, and their role in p53 inactivation. Pharmacol. Ther. 78:129-139. [DOI] [PubMed] [Google Scholar]
- 27.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 28.Schiffman, M. H., H. M. Bauer, R. N. Hoover, A. G. Glass, D. M. Cadell, B. B. Rush, D. R. Scott, M. E. Sherman, R. J. Kurman, S. Wacholder, et al. 1993. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 85:958-964. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz, A. L., and A. Ciechanover. 1999. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu. Rev. Med. 50:57-74. [DOI] [PubMed] [Google Scholar]
- 30.Shah, K. V., and P. M. Howley. 1996. Papillomaviruses, p. 2077-2109. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology. Lippincott-Raven Press, Philadelphia, Pa.
- 31.Stoler, M. H., C. R. Rhodes, A. Whitbeck, S. M. Wolinsky, L. T. Chow, and T. R. Broker. 1992. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum. Pathol. 23:117-128. [DOI] [PubMed] [Google Scholar]
- 32.Sundaram, P., and J. L. Brandsma. 1996. Rapid, efficient, large-scale purification of unfused, non-denatured E7 protein of cottontail rabbit papillomavirus. J. Virol. Methods 57:61-70. [DOI] [PubMed] [Google Scholar]
- 33.Sundaram, P., R. E. Tigelaar, and J. L. Brandsma. 1997. Intracutaneous vaccination of rabbits with the cottontail rabbit papillomavirus (CRPV) L1 gene protects against virus challenge. Vaccine 15:664-671. (Erratum, 16:655, 1998.) [DOI] [PubMed] [Google Scholar]
- 34.Sundaram, P., R. E. Tigelaar, W. Xiao, and J. L. Brandsma. 1998. Intracutaneous vaccination of rabbits with the E6 gene of cottontail rabbit papillomavirus provides partial protection against virus challenge. Vaccine 16:613-623. [DOI] [PubMed] [Google Scholar]
- 35.Sundaram, P., W. Xiao, and J. L. Brandsma. 1996. Particle-mediated delivery of recombinant expression vectors to rabbit skin induces high-titered polyclonal antisera (and circumvents purification of a protein immunogen). Nucleic Acids Res. 24:1375-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sykes, K. F., and S. A. Johnston. 1999. Genetic live vaccines mimic the antigenicity but not pathogenicity of live viruses. DNA Cell Biol. 18:521-531. [DOI] [PubMed] [Google Scholar]
- 37.Tellam, J., M. Sherritt, S. Thomson, R. Tellam, D. J. Moss, S. R. Burrows, E. Wiertz, and R. Khanna. 2001. Targeting of EBNA1 for rapid intracellular degradation overrides the inhibitory effects of the Gly-Ala repeat domain and restores CD8+ T cell recognition. J. Biol. Chem. 276:33353-33360. [DOI] [PubMed] [Google Scholar]
- 38.Turek, L. P., and E. M. Smith. 1996. The genetic program of genital human papillomaviruses in infection and cancer. Obstet. Gynecol. Clin. N. Am. 23:735-758. [DOI] [PubMed] [Google Scholar]
- 39.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 40.Wettstein, F. O., and J. G. Stevens. 1980. Distribution and state of viral nucleic acid in tumors induced by Shope papilloma virus. Cold Spring Harbor Conf. Cell Proliferation 7:301-307. [Google Scholar]
- 41.Wu, X., W. Xiao, and J. L. Brandsma. 1994. Papilloma formation by cottontail rabbit papillomavirus requires E1 and E2 regulatory genes in addition to E6 and E7 transforming genes. J. Virol. 68:6097-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.York, I. A., A. L. Goldberg, X. Y. Mo, and K. L. Rock. 1999. Proteolysis and class I major histocompatibility complex antigen presentation. Immunol. Rev. 172:49-66. [DOI] [PubMed] [Google Scholar]