Abstract
Wild waterfowl captured between 1915 and 1919 were tested for influenza A virus RNA. One bird, captured in 1917, was infected with a virus of the same hemagglutinin (HA) subtype as that of the 1918 pandemic virus. The 1917 HA is more closely related to that of modern avian viruses than it is to that of the pandemic virus, suggesting (i) that there was little drift in avian sequences over the past 85 years and (ii) that the 1918 pandemic virus did not acquire its HA directly from a bird.
The influenza virus pandemic of 1918 was exceptionally severe, killing 20 to 40 million people worldwide, with unusually high death rates among young, healthy adults. The disease swept the globe in 6 months, killing more than 10,000 per week in some U.S. cities at the height of the second wave (3). Pandemic influenza results when a type A influenza virus strain emerges with a hemagglutinin (HA) subtype to which few people have prior immunity (4). The source of HA genes new to humans appears to be the extensive pool of influenza A viruses maintained in wild birds (14). The pandemic influenza virus strains of 1957 and 1968 had new HA genes that were very similar to those in avian strains (10). We have isolated and sequenced a portion of the HA gene from a bird captured in 1917. Comparisons of this sequence with that of the 1918 pandemic virus suggest that the pandemic viral HA gene was not derived directly from an avian source.
Recently, we analyzed samples of 1918 pandemic virus RNA isolated from the lungs of three individuals: two U.S. Army soldiers and a woman who had been interred in the permafrost in Alaska (1, 8, 9). The HA gene sequences were virtually identical in all three cases. The genetic uniformity, along with the rapid spread and high morbidity experienced in 1918, suggest that the pandemic virus was mammal-adapted but had not previously circulated widely in humans. More than 90% of the 1918 viral HA amino acids subject to antigenic drift pressure in H1 subtype HAs in humans matched that of the avian consensus sequence, suggesting that there was little immunologic pressure on the HA protein before the autumn of 1918. Phylogenetic analyses, however, placed the 1918 HA gene within the mammalian clade, suggesting that the 1918 HA had acquired many changes that distinguished it from avian sequences (8). However, since only modern avian strains were available for comparison, the possibility remained that if avian strains had drifted substantially over the last century, the 1918 pandemic virus HA might more closely resemble an avian viral HA from around 1918.
The Smithsonian Institution in Washington, D.C., has several thousand preserved bird specimens from the early part of the 20th century. Wild waterfowl collected by Smithsonian Institution researchers between 1915 and 1920, cataloged and preserved in ethanol, were used for the study. These specimens were originally fixed in formalin and then preserved in 70% ethyl alcohol. We identified those birds from the 1918 time period that had the highest probability of containing influenza virus, especially juvenile wild waterfowl. Knowing that natural and experimental influenza virus exposure in ducks results in enteric infection, birds were sampled by cloacal lavage and cloacal and intestinal biopsy (11, 12). Forty-five samples were obtained from 25 birds representing 16 species. Birds were externally examined, and a cloacal lavage was performed by using a 1-ml syringe, an 18-gauge needle, and 0.2 ml of extraction buffer (20 mM Tris-HCl, pH 7.4, 20 mM EDTA, 1% [vol/vol] sodium dodecyl sulfate in diethyl pyrocarbonate-treated water). Biopsy samples of cloacal and large intestinal tissue were placed in 0.5 ml of extraction buffer.
After extraction and reverse transcription-PCR (RT-PCR) of a conserved region of the influenza virus matrix gene (5, 13), samples from six birds proved positive for influenza virus RNA: five of the positive samples were obtained by cloacal biopsy and one by cloacal lavage. Four of the birds were collected by A. Wetmore (later Secretary of the Smithsonian Institution) from the Bear River in Utah: a cinnamon teal (Anas cyanoptera) on 11 Aug. 1916, a ruddy duck (Oxyura jamaicensis) on 11 August 1916, a greenwing teal (Anas crecca) on 24 Sept. 1916, and a bufflehead duck (Bucephala albeola) on 15 Oct. 1916. One bird was collected by C. Johnson from Dry Tortugas, Fla.: a bluewing teal (Anas discors) on 25 Feb. 1919. A brant goose (Branta bernicla) was collected by G. D. Hanna from St. Paul Island, Pribilof Islands, Alaska, on 9 Sept. 1917. RT-PCR with primers specific for the H1 subtype gave a positive signal from the 1917 brant goose. A complete description of the lavage and/or biopsy results, as well as a listing of birds that proved negative for influenza virus RNA, is available upon request.
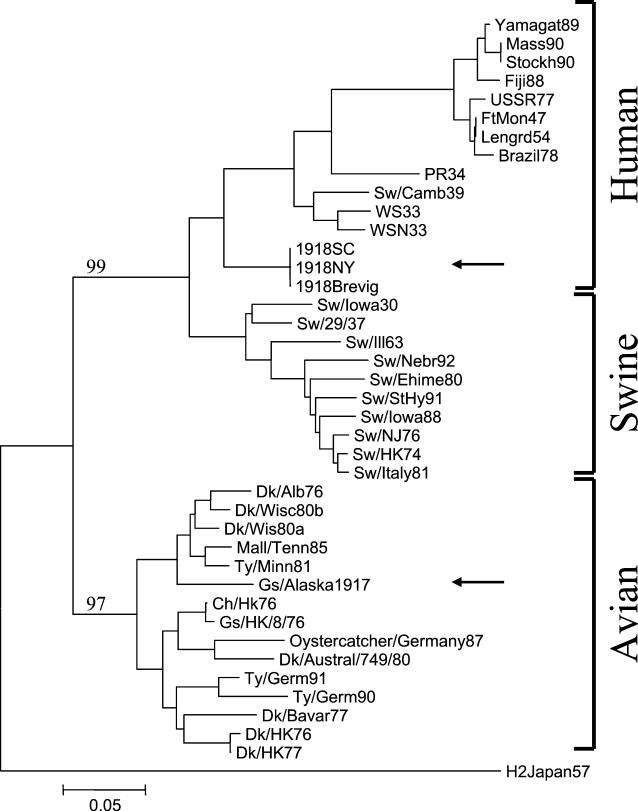
We sequenced a 166-base fragment of the HA1 domain of the HA gene of the virus we designate as A/Brant goose/Alaska/1/1917 (H1N?). Primer sequences used are available upon request. The 166-base fragment spans a region coding for 24 antigenic site amino acids known to be under significant positive selective pressure in humans (2, 7). The 1917 goose sequence matches the avian consensus at all 24 antigenic sites, whereas the human 1918 sequences differ at 3 of the 24 sites (8). A phylogenetic analysis of this region is shown in Fig. 1. Analysis of a 288-base HA2 fragment, as well as a 151-base segment of the nucleoprotein gene (not shown), supports the topology shown in Fig. 1.
FIG. 1.
Phylogenetic analysis of a portion (bases 494 to 659 as aligned to the sequence of A/Puerto Rico/8/34) of the influenza A virus HA gene. The bootstrap analysis (500 replications) used neighbor-joining and the proportion of differences as the distance measures (6). The arrows identify A/Brant goose/Alaska/1/1917 (H1N?) (Gs/Alaska1917) and the 1918 pandemic viral sequences (1918SC, 1918NY, and 1918Brevig). Bootstrap values are given for selected nodes, and a distance bar is shown under the tree. Viral strain abbreviations are as described previously (8). Abbreviations: Sw, swine; Dk, duck; Gs, goose; Ty, turkey; Mall, mallard; Ch, chicken.
Phylogenetic analyses of the partial 1917 goose HA sequence demonstrate that it falls within the avian branch of the tree (Fig. 1), while the 1918 pandemic viral sequences are clearly mammalian, as shown by the high bootstrap values (Fig. 1). The topology of the tree shown in Fig. 1 is virtually identical to that of the complete HA gene, demonstrating that the short region of the gene used to generate the tree contains significant phylogenetic information (8). The 1917 goose sequence is within and near the root of the North American clade of avian viral sequences. This result is in agreement with the suggestion by Webster et al. that avian HAs exhibit little drift over time (15).
It is formally possible that the goose HA sequence is aberrant and not representative of avian HA genes from around 1918. We think this unlikely for two reasons. (i) When a sequence is picked randomly from a (reasonable) distribution of sequences around a consensus, one should pick a sequence near the consensus with a much higher probability than a sequence at the extreme(s) of the distribution. (ii) As noted above, the sequence is placed within and near the root of the North American avian clade, exactly as expected.
Could the HA gene from a contemporaneous Eurasian bird more closely resemble the 1918 pandemic virus HA gene? This seems unlikely for several reasons. First, since North American avian HA sequences do not appear to drift, it is unlikely that Eurasian avian HA sequences would do so. Second, we have previously shown that the pandemic viral HA gene is genetically equidistant from both North American and Eurasian lineages (8).
The 1918 pandemic viral HA sequences had more changes from a typical avian sequence than was true for the 1957 and 1968 HA genes. Whether these changes accumulated during an indefinite amount of time undetected in another mammalian host cannot be determined from the present data. However, the 1917 avian HA sequence strongly supports the hypothesis that the 1918 influenza virus strain did not acquire its HA segment from a bird immediately before the pandemic.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 1917 goose sequence fragments are AY095226 (HA1), AY095227 (HA2), and AY095228 (NP).
Acknowledgments
This work was supported by grants from the American Registry of Pathology and by intramural funds of the Armed Forces Institute of Pathology.
We thank Storrs Olson and Gary Graves, curators of the Division of Birds, Smithsonian Institution, for permission to sample birds in their care.
The opinions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense.
REFERENCES
- 1.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caton, A. J., G. G. Brownlee, J. W. Yewdell, and W. Gerhard. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417-427. [DOI] [PubMed] [Google Scholar]
- 3.Crosby, A. 1989. America's forgotten pandemic. Cambridge University Press, Cambridge, United Kingdom.
- 4.Kilbourne, E. 1977. Influenza pandemics in perspective. JAMA 237:1225-1228. [PubMed] [Google Scholar]
- 5.Krafft, A. E., B. W. Duncan, K. E. Bijwaard, J. K. Taubenberger, and J. H. Lichy. 1997. Optimization of the isolation and amplification of RNA from formalin-fixed, paraffin-embedded tissue: the Armed Forces Institute of Pathology experience and literature review. Mol. Diagn. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 6.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software, 2.1 ed. Arizona State University, Tempe, Ariz. [DOI] [PubMed]
- 7.Raymond, F., A. Caton, N. Cox, A. P. Kendal, and G. G. Brownlee. 1986. The antigenicity and evolution of influenza H1 haemagglutinin, from 1950-57 and 1977-1983: two pathways from one gene. Virology 148:275-287. [DOI] [PubMed] [Google Scholar]
- 8.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 96:1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid, A. H., T. G. Fanning, T. A. Janczewski, and J. K. Taubenberger. 2000. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc. Natl. Acad. Sci. USA 97:6785-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholtissek, C., W. Rohde, V. von Hoyningen, and R. Rott. 1978. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87:13-20. [DOI] [PubMed] [Google Scholar]
- 11.Slemons, R. D., and B. C. Easterday. 1977. Type-A influenza viruses in the feces of migratory waterfowl. J. Am. Vet. Med. Assoc. 171:947-948. [PubMed] [Google Scholar]
- 12.Slemons, R. D., D. C. Johnson, J. S. Osborn, and F. Hayes. 1974. Type-A influenza viruses isolated from wild free-flying ducks in California. Avian Dis. 18:119-124. [PubMed] [Google Scholar]
- 13.Taubenberger, J. K., A. H. Reid, A. E. Krafft, K. E. Bijwaard, and T. G. Fanning. 1997. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science 275:1793-1796. [DOI] [PubMed] [Google Scholar]
- 14.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webster, R. G., G. B. Sharp, and E. C. Claas. 1995. Interspecies transmission of influenza viruses. Am. J. Respir. Crit. Care Med. 152:S25-30. [DOI] [PubMed] [Google Scholar]