Abstract
Boid inclusion body disease (BIBD) is a fatal disorder of boid snakes that is suspected to be caused by a retrovirus. In order to identify this agent, leukocyte cultures (established from Python molurus specimens with symptoms of BIBD or kept together with such diseased animals) were assessed for reverse transcriptase (RT) activity. Virus from cultures exhibiting high RT activity was banded on sucrose density gradients, and the RT peak fraction was subjected to highly efficient procedures for the identification of unknown particle-associated retroviral RNA. A 7-kb full retroviral sequence was identified, cloned, and sequenced. This virus contained intact open reading frames (ORFs) for gag, pro, pol, and env, as well as another ORF of unknown function within pol. Phylogenetic analysis showed that the virus is distantly related to viruses from both the B and D types and the mammalian C type but cannot be classified. It is present as a highly expressed endogenous retrovirus in all P. molurus individuals; a closely related, but much less expressed virus was found in all tested Python curtus individuals. All other boid snakes tested, including Python regius, Python reticulatus, Boa constrictor, Eunectes notaeus, and Morelia spilota, were virus negative, independent of whether they had BIBD or not. Virus isolated from P. molurus could not be transmitted to the peripheral blood mononuclear cells of B. constrictor or P. regius. Thus, there is no indication that this novel virus, which we propose to name python endogenous retrovirus (PyERV), is causally linked with BIBD.
Retroviruses were identified in snakes more than 30 years ago, when C-type-like particles were observed by electron microscopy in the tissues of a tumor-bearing viper (25-27) and were later shown to be related to primate D-type retroviruses (1). Retrovirus-like particles subsequently detected in snakes also involved mostly animals affected with proliferative disorders (12, 14, 17) but were also found in normal tissues (6). Recently, a systematic PCR-based search for endogenous retroviruses related to murine leukemia virus (MLV) showed a widespread distribution of such C-type sequences among terrestrial animals, including amphibians, reptiles, birds, and mammals (15).
In 1994, a retrovirus responsible for boid inclusion body disease (BIBD) in snakes, a disease known for more than 20 years to occur in snakes of both the Boinae and Pythoninae subfamilies of Boidae, was described in private and zoological snake collections all over the world (5, 16, 19). Clinical signs of BIBD include chronic regurgitation and central nervous system (CNS) disease manifested in head tremors, disorientation, lack of motor coordination, paresis, and/or paralysis. Histologic examination revealed numerous eosinophilic intracytoplasmic inclusion bodies in the epithelial cells of all major organs, including neurons within the CNS. The inclusion bodies contain an antigenically distinct 68-kDa protein against which a specific monoclonal antibody has been developed (24). In all snakes with CNS disease, a nonsuppurative meningoencephalitis with neuronal degeneration and perivascular cuffing was present and viral particles resembling C-type retroviruses were detected in the brain, pancreas, and kidney, as well as in cultured primary kidney cells. The disease was shown to be transmissible by cell-free primary kidney cell culture supernatants from infected Boa constrictor snakes to young Burmese pythons (Python molurus bivittatus) (19) or by liver homogenates from B. constrictor (24). A polyclonal antibody raised against sucrose density gradient-purified BIBD virus produced by in vitro cocultivation of leukocytes from a BIBD-positive B. constrictor snake and VH2 viper heart cells reacted with specific proteins associated with virus particles of C-type morphology that were purified from the primary kidney cell cultures established from another BIBD-positive B. constrictor snake (13). These data highly suggest that the implied C-type virus may indeed be the causative agent of BIBD. However, since the VH2 cells are known to harbor an endogenous retrovirus which may sometimes be expressed (8) and because they were indeed positive for reverse transcriptase (RT) activity (13), the specificity of this antiserum is somewhat uncertain. It is thus conceivable that the virus isolated from BIBD-positive boas, which was recognized by this antibody, may be a widely distributed endogenous retrovirus present in both vipers and boas. The situation is further complicated by the recent detection of C-type particles in malignant tumors from four P. molurus bivittatus snakes (7).
In an attempt to identify the retroviral agent associated with BIBD, we investigated RT-positive peripheral blood mononuclear cell (PBMC) cultures from several P. molurus snakes from a privately owned population in which some animals exhibited symptoms of BIBD. Using highly effective methods for the identification and characterization of unknown retroviruses, we here report our identification of a novel group of endogenous retroviruses present in P. molurus and Python curtus.
MATERIALS AND METHODS
Samples.
EDTA or heparin blood samples were collected from the tail vein or by cardial puncture, and PBMC were purified by Lymphodex (Fresenius, Stans, Switzerland) centrifugation.
Cell cultures and in vitro virus transmission.
Purified PBMC were stimulated with 0.2% phytohemagglutinin (PHA; Sigma) in RPMI 1640 medium with Glutamax-I and 20% fetal calf serum (Gibco Life Technologies) for 24 h. After two washing steps, the cells were incubated in RPMI 1640 medium with 10% fetal calf serum and 2% natural human T-cell growth factor (interleukin 2; ZeptoMetrix, Buffalo, N.Y.) at 30°C in 5% CO2. Ninety percent of the culture medium was exchanged twice per week. For infection experiments, PHA-stimulated PBMC were incubated with 0.8 ml of a virus pool that contained an activity equivalent of 0.54 mU of human immunodeficiency virus type 1 RT or medium control for 4 h at 30°C, washed twice, transferred to culture flasks, and grown for 25 days under the same conditions as described above.
RT assays.
RT activity was measured by a TaqMan real-time PCR modification of the product-enhanced RT (PERT) assay (18) as described previously (4). Prior to testing, plasma and serum samples were diluted 1/100 with phosphate-buffered saline (PBS) and filtered through 0.2-μm-diameter filters. Sucrose gradient fractions and culture supernatants were tested undiluted and unfiltered.
Sucrose gradient density banding.
Culture supernatant was precleared at 12,000 rpm for 3 min and pelleted at 70,000 × g for 45 min. The pellet was resuspended in 200 μl of PBS, layered on a 7.5% to 60% sucrose gradient, and ultracentrifugated at 70,000 × g for 16 h at 4°C in a Kontron TFT32.13 rotor. Fractions of 400 μl were manually collected from the top, and their densities were determined in a Zeiss refractometer. Of each fraction, 10 μl was analyzed for RT activity.
Particle-associated retroviral RNA amplification.
Retroviral sequences from peak RT gradient fractions were identified by using a modification of a previously published procedure (9, 22, 23) as follows.
(i) Poly(A) polymerase.
To increase binding of partially degradated RNA to oligo(dT)25 Dynabeads (Dynal), nucleic acids from 20 μl of sucrose peak fraction were polyadenylated for 20 min at 37°C in 40 μl of reaction mixture containing 50 mM Tris-HCl (pH 7.9), 250 mM NaCl, 10 mM MgCl2, 2.5 mM MnCl2, 10 mM dithiothreitol (DTT), 40 U of RNasin (Promega), 0.15% Nonidet P-40 (NP-40), 100 μM ATP, 4 U of poly(A) polymerase (Gibco BRL), and 50 μg of oligo(dT)25 Dynabeads.
(ii) cDNA synthesis.
cDNA synthesis was performed in a final volume of 20 μl containing Dynabeads washed in RT buffer, 10 pmol (each) of L-mix primers, 0.5 mM each deoxynucleoside triphosphate (dNTP), 10 mM DTT, 0.2% NP-40, 30 U of RNasin, 50 U of Expand (Roche Molecular Biochemicals) RT, and 1× RT reaction buffer (Roche Molecular Biochemicals). The reaction was carried out at 37°C for 30 min, at 42°C for 20 min, and at 45°C for 10 min and stopped at 75°C for 15 min.
(iii) Tailing of cDNA.
Prior to tailing, the beads were washed with wash buffer (20 mM Tris-HCl [pH 8], 50 mM KCl) and then the cDNA-RNA hybrids were subjected to RNase H degradation in a volume of 7.5 μl containing 10 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 50 mM KCl, 10 mM DTT, 0.005% NP-40, and 0.3 U of RNase H (Boehringer Mannheim). The reaction was carried out at 37°C for 15 min, followed by 65°C for 5 min. A volume of 7.5 μl containing 10 mM Tris-HCl (pH 9), 50 mM KCl, 200 mM dCTP, and 15 U of terminal deoxynucleotidyl transferase (Gibco BRL) was added. The mixture was incubated at 37°C for 20 min, followed by inactivation of the enzyme at 65°C for 5 min.
(iv) Anchored PCR of the 5′ terminal region.
The first of two successive rounds of PCR was performed with the Expand Long Template PCR System kit (Boehringer Mannheim). A total volume of 20 μl containing 5 μl of tailing reaction mixture and 0.5 pmol of anchor primer flu37 was subjected to three initial cycles at 92°C for 30 s, 55°C for 2 min, and 68°C for 1.5 min (Touchgene cycler; Techne). Then, a PCR mixture containing 16 pmol of primer flu19 and 10 pmol of each L-mix was added to a final volume of 50 μl, followed by 5 cycles at 92°C for 30 s, 50°C for 1 min with a temperature decrement of 1°C per cycle, and 68°C for 1 min and finally 15 cycles at 92°C for 20 s, 45°C for 30 s, and 68°C for 45 s. One microliter from the first amplification mixture was subjected to a second seminested PCR. After heat activation of the polymerase AmpliTaq Gold (Applied Biosystems) for 10 min at 95°C, 40 cycles of thermal cycling at 95°C for 30 s, 60°C for 30 s with primers pK1, pK2, pP, pW, pM1, and pG1, or 50°C for 30 s with primers pH and pM2 for 30 s, and 72°C for 1 min were performed. The second primer in all reactions was flu19, and dNTP concentration in the buffer was 200 μM. PCR products were separated on low-melting-point agarose gels. DNA bands were excised and processed with the QIAquick gel extraction kit (Qiagen) according to the manufacturer's instructions and sequenced on an ABI model 310 DNA sequencer.
cDNA synthesis and PCR of viral genome RNA.
Synthesis of cDNA from primed RNA present in sucrose gradient fractions was performed for 1 h at 37°C in a reaction volume of 25 μl containing 5 μl of sample, 50 mM KCl, 50 mM Tris-HCl (pH 8.3), 8 mM MgCl2, 3 μg of bovine serum albumin, 10 mM DTT, 1 mM dNTP, 0.3% Triton X-100, 0.4 μM random d(N)6, 20 U of MLV RT (Gibco BRL), and 25 U of RNasin (Promega). Fifteen percent of the cDNA-RNA hybrids was used for long-distance PCR with the Expand 20 kbPLUS PCR System kit (Roche Molecular Biochemicals) and a volume of 50 μl containing primers Rf and BES40TG at a concentration of 0.4 μM each and dNTPs at 500 μM. After five cycles at 92°C for 15 s, 55°C for 30 s, and 68°C for 8 min, 30 cycles with a time increment of 10 s at 68°C were performed.
Isolation of chromosomal DNA and long-distance PCR.
Erythrocytes sedimented from 0.5 ml of blood were resuspended in 2.5 ml of PBS and mixed with 2.5 ml of 2× lysis buffer containing 2 mg of proteinase K (Roche Molecular Biochemicals)/ml. Protein was digested at 37°C overnight, and DNA was purified by standard phenol-chloroform extraction and ethanol precipitation. Precipitated DNA was dissolved in Tris-EDTA buffer. DNA from paraffin-embedded tissue was isolated with Qiagen's QIAamp DNA Mini Kit and protocol. DNA from PBMC was extracted with the QIAamp Blood Kit (Qiagen) following the manufacturer's protocol. For long-distance PCR, the buffer of the Expand 20 kbPLUS PCR System kit (Roche Molecular Biochemicals) was supplemented with 1% dimethyl sulfoxide and the primers and dNTPs at the same concentrations as described above. Amplification was carried out for 5 cycles at 92°C for 15 s, 60°C with a decrement of 0.5°C for 30 s, and 68°C for 9 min; 5 more cycles at 92°C for 15 s, 58°C for 30 s, and 68°C for 9 min; and finally 20 cycles at 92°C for 15 s, 58°C for 30 s, and 68°C for 9 min with a time increment of 12 s.
Cloning and sequencing.
PCR products were isolated and purified with the QIAquick Gel Extraction Kit (Qiagen). Cloning was performed by use of the pGEM-T Easy Vector System kit (Promega) and competent Escherichia coli JM109 cells (Promega). Plasmids were purified with the Plasmid Midi Kit (Qiagen). Primer walking sequencing was done on an ABI model 310 Sequence Detector. Primers were selected by means of Oligo 6.1 primer analysis software (Molecular Biology Insights) and synthesized by GmbH (Microsynth, Balgach, Switzerland). Nucleic acid sequences were analyzed with AutoAssembler version 2.1 (Applied Biosystems) and MacVector version 7.0 (GCG Inc., Oxford Molecular Group).
Primers
The primers used in this work are listed in Table 1.
TABLE 1.
List of primers with orientation, localization, and application
| Primera | Sequence (5′-3′)b | Sense or antisensec | Localizationd | Usee |
|---|---|---|---|---|
| flu37 | CAGCACGACAGACAGAGGACAGGGGIIGGIGIGGGIG | S | Poly(dC) tail | 5′ RACE |
| flu19 | GCACGACAGACAGAGGACA | S | flu19 | 5′ RACE |
| LG1 | ACCACCTTCCCGGCCA | A | tRNAGly PBS | 5′ RACE |
| LH1 | ACCACCATCCGAGTCACG | A | tRNAHis PBS | 5′ RACE |
| LK1 | ACCACCGCCCCACGTT | A | tRNA1,2Lys PBS | 5′ RACE |
| LK2 | ACCACCGTCCCTGTTCG | A | tRNA3Lys PBS | 5′ RACE |
| LM1 | ACCACCTCCTCACACGG | A | tRNAMet PBS | 5′ RACE |
| LM2 | ACCACCAACCATCCTCTGC | A | tRNAMet PBS | 5′ RACE |
| LP | ACCACCATCCCGGACGA | A | tRNAPro PBS | 5′ RACE |
| LW | ACCACCATCACGTCGGG | A | tRNATrp PBS | 5′ RACE |
| pG1 | CCGGCCAATGCACCA | A | tRNAGly PBS | 5′ RACE |
| pH | CGAGTCACGGCACCA | A | tRNAHis PBS | 5′ RACE |
| pK1 | CCACGTTGGGCGCCA | A | tRNA1,2Lys PBS | 5′ RACE |
| pK2 | CCTGTTCGGGCGCCA | A | tRNA3Lys PBS | 5′ RACE |
| pM1 | CCTCACACGGGGCACCA | A | tRNAMet PBS | 5′ RACE |
| pM2 | CAACCATCCTCTCTGCTACCA | A | tRNAMet PBS | 5′ RACE |
| pP | CCGGACGAGCCCCCA | A | tRNAPro PBS | 5′ RACE |
| pW | ATCACGTCGGGGTCACCA | A | tRNAPro PBS | 5′ RACE |
| BES40TG | GAATTCGTCGACAGG(T)24G | A | Poly (dA) | PCR clones |
| Rf | CTCACTCACCTCGAGGAAA | S | U5 | PCR clones |
| U5-1 | CCTGCGTGTTTGTCTTCACT | S | U5 | PCR clones |
| U3r-1 | GCAGAAGCCTGAATCAAGT | A | U3 | PCR clones, Envamp |
| U3-2 | GCTTAAAGCTGTTGCATGACT | S | U3 | LTRamp |
| R-2 | AGGTGAGTGAGCCCAGAG | A | U5 | Sequencing, inverse PCR |
| G-4 | GCGCTGTCTGTTGTTGAG | A | gag | Inverse PCR, LTRamp |
| G-1 | CAATGGTCCCACGGCTC | S | gag | Inverse PCR |
| P-3 | TACCGCTCAAGGAGTCATAG | S | pro | Inverse PCR |
| endo-1 | CTTAACGACCATGGTGATTC | S | Chromosomal | Integration site |
| E-4 | GGCCGAGTTCATGATAAAAC | A | env | Envamp |
| P-2 | CTGGTGGAAATAGGAATGAC | A | pol | pro-polamp |
| Q-1 | CGGACTGTCAAAAGGTGCTC | S | gag | Quant. CN, pro-polamp |
| Q-2 | CATTTTGTGGCTGGTGGTT | A | gag | Quant. CN |
| Q-4 | TCTAAGGCCCTAGCTGGTGTTG | S | pol | Quant. CN |
| Q-5 | CAAACTGAAGGTGGTCGCTGAC | A | pol | Quant. CN |
| mos-1 | AGTAAGAACCATTTGGCATCAC | S | c-mos | Quant. CN |
| mos-2 | CTTGCAAACATTAYGTTCTGTG | A | c-mos | Quant. CN |
| mos-3 | GGAATATGTAGGTAATTGCACTC | S | c-mos | Quant. CN |
| mos-4 | GGTTTTAAATCCAGATGCAC | A | c-mos | Quant. CN |
| mos-7 | CTGGGCAGAACTAAATGTAGC | S | c-mos | Quant. CN |
| mos-8 | GAGTGCAATTACCTACATATTCCA | A | c-mos | Quant. CN |
L-mix primers, all primer designations beginning with L.
I, inosine.
S, sense; A, antisense.
PBS, primer binding site.
quant. CN, quantification of copy number; amp, amplification.
Quantitative PCRs.
All quantitative PCRs were performed on an ABI model 7700 Prism Sequence Detector. All reactions were performed in a volume of 50 μl with 1.25 U of AmpliTaq Gold, manufacturer's buffer, 0.0008 μl of SYBR green I nucleic acid gel stain (10,000×; FMC Bio Products), and 0.375 μM tetramethylrhodamine (TAMRA) 5′-labeled (dT)6 (Microsynth) as an internal reference. Reaction conditions were as follows. gag was amplified with primers Q-1 and Q-2 (20 pmol each) in 150 μM dNTP for 43 cycles at 95°C for 15 s, 62.5°C for 30 s, and 72°C for 30 s. pol was amplified with primers Q-4 and Q-5 (18 pmol each) in 180 μM dNTP for 43 cycles at 95°C for 15 s, 64°C for 30 s, and 72°C for 30 s. Quantitative PCR in reference gene c-mos was carried out by amplification with primers mos-3 and mos-4 (20 pmol each) in 150 μM dNTP and additional MgCl2 (0.25 mM) for 43 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Amplification with primers mos-5 and mos-6 (16.7 pmol each) was carried out in 167 μM dNTP and additional MgCl2 (1 mM) for 40 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Amplification with primers mos-7 and mos-8 was done at the same concentrations and under the same conditions as those with primers Q-1 and Q-2.
Standardization of quantitative PCRs.
All quantitative PCRs were standardized in relation to 1 genome equivalent of the single gene sequence of c-mos (15). Comparison of the published c-mos sequences from Python reticulatus (AF039472), Candoia carinata (AF039473), and Ramphotyphlops australicus (AF039474) was used for designing primers mos-1 and mos-2. Amplification with these primers yielded a 369-bp amplicon with DNA samples from Python regius, P. molurus, P. curtus, Morelia spilota, Eunectes notaeus, and B. constrictor. Multiple alignments of these amplicons were used to design primers in conserved sequences for quantitative real-time fluorogenic PCR. Primer combinations mos-3 and mos-4 yielded a 190-bp amplicon, while those of mos-7 and mos-8 yielded a 131-bp amplicon. A serial dilution (107 to 100 copies/μl) of the P. molurus mos-1/mos-2 amplicon in herring sperm was used as an exogenous standard.
Inverse PCR.
Two hundred nanograms of chromosomal DNA was incubated at 37°C for 2 h with 10 U of HindIII endonuclease (Roche Molecular Biochemicals) in manufacturer's buffer. The enzyme was inactivated at 65°C for 20 min. Ten nanograms of this DNA was ligated with 1 U of T4 DNA ligase (Promega) in 10 μl of the recommended buffer at 20°C for 4 h. Ligated DNA (0.5 ng) was amplified in a first round of PCR with primers G-1 and G-4 (20 pmol each), 200 μM dNTP, and 1% dimethyl sulfoxide with the Expand 20 kbPLUS PCR System kit (Roche Molecular Biochemicals), and 13 cycles at 92°C for 20 s, 55°C for 30 s, and 68°C for 8 min were performed. One microliter of the first-round amplification product was entered into a second round of seminested PCR in which the Expand High Fidelity PCR System kit (Roche Molecular Biochemicals) was used with primers G-4 and P-3 (0.4 μM) and dNTP (200 μM), and amplification was carried out for 40 cycles at 92°C for 20 s, 55°C for 30 s, and 68°C for 2 min. PCR products were separated on agarose gel, and the DNA of interesting bands was isolated and sequenced as described with primer R-2.
Phylogenetic analysis.
Protein sequences were aligned using the ClustalW Alignment with Blosum30 matrix in MacVector version 7.0. Aligned sequences were used gap-free to construct neighbor-joining trees by using the Phylogenetic Analysis Using Parsimony software package (PAUP; L. Swofford, version 4.0b4a for power PC, 2001; Sinauer Associates, Sunderland, Mass.).
Nucleotide sequence accession numbers.
The complete nucleotide sequence of the PyT2RV genome and the fragment of the PyB1RV genome are available in the GenBank database under accession no. AF500295 to AF500299.
RESULTS
Detection of RT activity in PBMC cultures.
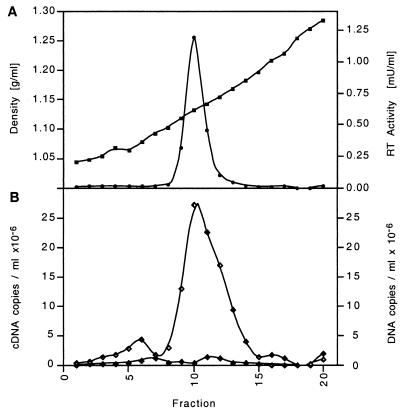
In order to identify an infectious retrovirus as the likely etiologic agent of BIBD, blood samples were obtained from 7 animals with clinical symptoms consistent with BIBD, from 18 asymptomatic animals housed together with symptomatic animals, and from 14 animals epidemiologically unlinked to the disease. The snakes originated from four different collections and included 12 P. molurus (Burmese python), 3 P. reticulatus (reticulated python), 3 P. regius (royal python), 4 P. curtus (blood python), 1 Liasis childreni (Children's python), 5 M. spilota (carpet python), 1 E. notaeus (yellow anaconda), 3 Epicrates cenchria (rainbow boa), 3 Corallus enydris (tree boa), and 4 B. constrictor (boa constrictor) snakes. PHA-stimulated PBMC cultures established in the first phase of this project were analyzed after 4 weeks of culture for RT activity in supernatants by PERT assay, as described in Materials and Methods. Three cultures, all from Burmese pythons, exhibited remarkably high RT activity (13 ± 7 μU/ml) compared with all others, which remained below the cutoff of 0.15 μU/ml. The snake with the highest RT activity, a healthy Burmese python housed together with animals suspected to have BIBD, was selected for further analysis. All collected supernatants from animal T2 were pooled, presumptive virus was pelleted by ultracentrifugation, and the resuspended pellet was subjected to sucrose density gradient analysis. RT analysis of all gradient fractions yielded a single peak of activity slightly below 1.15 g/ml, which was compatible with the RT contained in retroviral particles (Fig. 1A).
FIG. 1.
Colocalization of RT activity and PyT2RV RNA on the sucrose density gradient of PyT2 culture supernatant. (A) RT activity (circles) peaked at a sucrose density (squares) of 1.15 g/ml. (B) PCR of PyT2RV cDNA (open diamonds) and of PyT2RV DNA (black-and-white diamonds) with primers Q-1 and Q-2.
Amplification and sequencing of particle-associated retroviral RNA.
RNA from the peak RT fraction of the gradient from animal T2 was subjected to particle-associated retroviral RNA amplification (see Materials and Methods), an efficient method for amplification of the retroviral RNA contained in a small number of unknown particles for which no specific sequence information other than that of the various 18-nucleotide (nt) tRNA primers is available (23). Priming of the viral RNA with a Trp-like primer, followed by cDNA synthesis and tailing of the cDNA (5′ rapid amplification of cDNA ends [RACE]) consistently yielded a 189-bp product which exhibited no detectable sequence homology to any known retrovirus but contained a possible polyadenylation signal and a CA dinucleotide located 3 bp upstream of the primer binding site, both of which had features characteristic of retroviral R-U5 sequences. Long-distance PCR of the cDNA with primer Rf specific for this amplicon and the poly(T)-tailed primer BES40TG generated a 5-kb amplicon which revealed significant, though distant, homology to the Gag proteins of various retroviruses. As further evidence for a retroviral origin, the concentration of the 5-kb amplicon correlated with the RT activity on the sucrose gradient, as demonstrated by a semiquantitative RT-PCR using primers Q-1 and Q-2 located in gag (Fig. 1B). Since this 5-kb sequence was rather short for a full-length viral RNA, long-distance PCR of the cDNA was repeated with two specific primers, U5-1 and U3r-1, which hybridize to the U5 or U3 region of the viral RNA, respectively. This amplification yielded, in addition to the 5-kb sequence, a second product of approximately 7 kb in length. Both amplicons were cloned and sequenced, and the sequence was combined with that obtained from the initial 5′ RACE. This resulted in a 7,439-nt-long complete viral genome provisionally named python retrovirus (PyT2RV). nt 1 to 76 of this genome were derived from sequencing of the amplified, uncloned cDNA product, nt 77 to 7126 were derived from sequencing of the 7-kb clone, and nt 7127 to 7439 were derived from sequencing of the 5-kb clone.
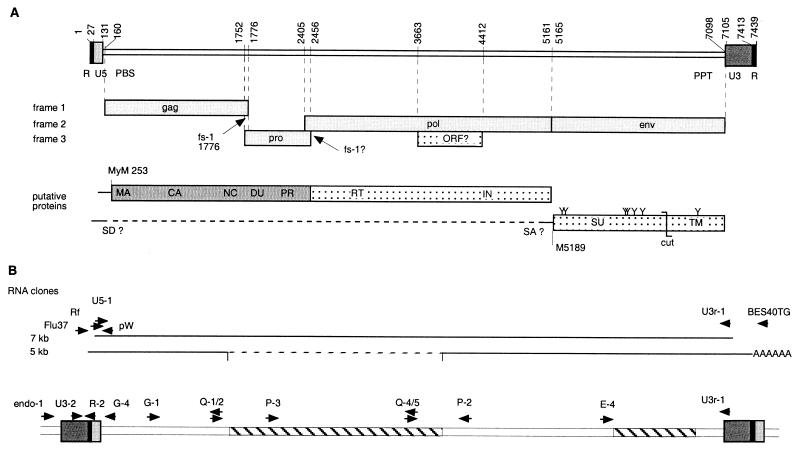
Genomic organization.
The genome of PyT2RV contained open reading frames (ORFs) for gag, pro, pol, env, and a fifth ORF overlapping pol (Fig. 2). The reading frames for gag, pro, and pol were each shifted by a −1 position, as also seen in type B and D retroviruses and the human T-cell leukemia virus (HTLV)/bovine leukemia virus group. As in type D simian retrovirus type 2 (SRV-2), pol and env were in frame and separated by only a stop codon. Additional ORFs between pol and env or, respectively, env and U3R, both typical localizations of accessory genes or viral oncogenes, were not detected.
FIG. 2.
Genomic organization of PyT2RV. (A) Localization of ORFs, frameshifts (fs), deduced proteins, glycosylation sites (Y), putative splice donor (SD), and acceptor sites (SA). MyM, myristoylated methionine. (B) Localization of oligonucleotide primers for 5′ RACE and cDNA clones and for inverse PCR, quantitative PCR, and location of the deletions (hatching).
Long terminal repeats (LTR).
A repetitive R sequence was identified at both the 5′ (nt 1 to 27 of the virion genome) and the 3′ ends (nt 7413 to 7439) and included a typical AATAAA polyadenylation signal. U5 starts at nt 28 and ends with the mandatory CA dinucleotide (nt 130 to 131) 3 nt upstream of the primer binding site for tRNATrp (nt 135 to 152). Inverse PCR of digested and ligated chromosomal DNAs with primers G-1 and G-4 and seminested PCR primers G-4 and P-3, followed by sequencing of the amplicon (with primer R-2 [see Materials and Methods]), identified an integration site at the 5′ end and made it possible to determine the length of U3 (315 bp, nt 7098 to 7412). U3 contained an overlapping 16-nt direct repeat (nt 7370 to 7385 and 7383 to 7398), a possible CAT box, CCAAT (7325 to 7329), and a possible TATA box, TATAAAA (nt 7356 to 7362). The region between RU5 and U3 contained a purine-rich sequence (nt 7079 to 7097; polypurine tract) directly upstream of U3.
gag.
The gag ORF extended from nt 160 to 1776. It had a possible start codon between positions 253 and 255. A BLASTP search with the deduced protein sequence showed the highest score for Jaagsiekte sheep retrovirus (JSRV) Gag protein (24% identity; 136 of 556 aa) and identified this protein as one with a retroviral origin. Consistent with the presence of an N-terminal matrix (MA) protein, the ORF showed a conserved myristoylation site, MetGly X3Ser/Thr (21), from nt positions 253 to 270. In a BLASTP search, the deduced protein of the MA showed homology to the MA of D-type retroviruses, with the highest sequence identities of 34% (21 of 61 aa) with the JSRV (accession no. M80216) and 32% (21 of 64 aa) with human endogenous virus (HERV)-K(I) (accession no. AB047209) within nt 358 to 537. An L-domain, a PPPY motif frequently found between the MA and CA proteins, was identified at nt 583 to 594. In the putative CA protein, a possible major homology region, IIQSPTEDYSSFISRLQVALERQ, was located at nt positions 1261 to 1329. Finally, two CX2CX4HX4C zinc finger motifs of a putative nucleocapsid protein were identified at the C-terminal region at nt 1594 to 1635 and 1699 to 1740, similar to those for other retroviruses.
pro.
A second ORF extended from nt 1752 to 2456. A classical XXXAAAC −1 frameshift signal was located at nt 1770 to 1776 and was followed by a stable palindrome, GTGGGGCX9GCCCCAC (nt 1782 to 1804), and presumably permitted translation of a Gag-Pro precursor protein. A BLASTP search of the deduced 233-aa-long full-length protein showed a 30% (57 of 188 aa) sequence identity at the C terminus to the proteases of SRV-2 (accession no. AF126468) from nt 1935 to 2438 and a 36% (38 of 104 aa) identity to HERV K-109 (accession no. AF164615) between nt 2148 and 2453. Two conserved protein sequence motifs of retroviral proteases, DTG at nt 2211 to 2219 and W/LGRD or GRD at nt 2409 to 2420, were found in the same order and in similar distances and positions as they were found in other retroviral proteases. The upstream region of the second ORF presumably encoded a dUTPase, as was suggested by low sequence homologies with nonprimate lentiviruses and B-type (mouse mammary tumor virus [MMTV]) or D-type (Mason-Pfizer monkey virus, JSRV, SRV) retroviruses (data not shown).
pol.
A third ORF extended from nt 2405 to 5161. The deduced protein showed sequence homology and identity to the polymerase protein of several retroviruses in a BLASTP search. The highest sequence identities found were 41% (348 of 848 aa) for SRV-2 (accession no. M1605) and 40% (350 of 862 aa) for HERV-K(HML-2.HOM) (accession no. AF298588). The polymerase protein deduced from the 7-kb clone contained the classical LPQG motif (nt 2888 to 2899) in the active center, but the usual YMDD motif was replaced by YMDG (nt 2990 to 3001). A YMDD motif was found, however, by population sequencing of the amplified viral cDNA and also of proviral DNA in three investigated Burmese pythons and one blood python. The putative integrase region contained two classical protein motifs. An HX3HX23CX2C motif was identified at nt positions 4247 to 4342, and DDX35E was encoded as N(4415 to 4417)D(4586 to 4588)X35G(4694 to 4696) on the 7-kb clone and as DDX35E at the analogous position in the 5-kb clone.
env.
A fourth ORF (nt 5165 to 7105) codes likely for surface (SU) and transmembrane (TM) proteins. The highest sequence identity found by BLASTP was 40% (240 of 596 aa) with a Sus scrofa porcine endogenous retrovirus isolate, endogenous retrovirus PK15 (accession no. AF038601), and 39% (265 of 675 aa) with the Mus dunni MLV isolate, Mus dunni endogenous virus (accession no. AF053745). As in SRV-2, pol and env were separated by a single stop codon (TAG, nt 5162 to 5164). The deduced Env protein contained a putative protease cleavage site, RHKR, embedded in a conserved region from nt positions 6425 to 6436, indicating the probable transition from the SU protein to the TM protein (11). A likely hydrophobic fusion domain (VALLVGLGLTGAGTGIASFANQYV) of the TM protein was at nt 6458 to 6529, and a putative hydrophobic membrane anchor (LIGTIAGPLILLLLACTLGPCIL) was at nt 6848 to 6916. Six possible glycosylation sites NXS or NXT were found in the SU protein (nt 5303 to 5311, 5345 to 5353, 6008 to 6016, 6023 to 6031, 6101 to 6109, and 6218 to 6226), and one was found in the TM protein (nt 6824 to 6832).
ORF 5.
A remarkably long additional ORF (nt 3663 to 4412) with a possible start codon at position 3801 was localized in frame 3 and overlapping with pol. The deduced protein sequence of 250 aa did not show any homology to known proteins.
Phylogenetic analysis.
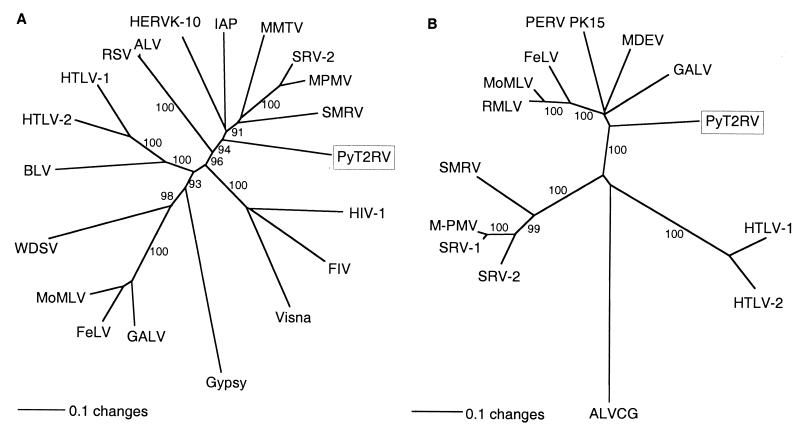
A BLASTN search with the PyT2RV nucleic acid sequence showed poor homology to other known retroviruses, including those recently reported, widely distributed endogenous MLV-related C-type sequences found among terrestrial vertebrates (15). Sequence comparison at the protein level confirmed the dissimilarity of PyT2RV and these C-type viruses. The protein deduced from pol showed, however, good homology to those of other known retroviruses. In a phylogenetic analysis, the best-conserved region (nt 2609 to 3241), a 211-aa peptide containing the YMDD active site of the polymerase, branched closely to the B- and D-type viruses by both the neighbor-joining and unweighted pair-group method with arithmetic mean (UPGMA) algorithms (Fig. 3A). Analysis based on parsimony confirmed these findings, although the bootstrap values were generally somewhat lower (data not shown). The protein deduced from env showed homology to both the SU and TM of various other retroviruses. In particular, a 141-aa peptide (nt 6531 to 6948) in the TM protein was sufficiently well conserved to perform a phylogenetic analysis. In contrast to the peptide in pol, PyT2RV in env showed a clear affiliation with the origin of mammalian C-type viruses by both the neighbor-joining and UPGMA algorithms (Fig. 3B). Phylogenetic analyses with other peptides of PytT2RV, e.g., in gag, could not be carried out because the high degree of protein sequence heterogeneity, combined with extensive length variations in the various retroviral reference sequences, prevented satisfactory multiple sequence alignments.
FIG. 3.
Phylogenetic analysis of PyT2RV. (A) Analysis of polymerase protein (211 aa, nt 2609 to 3241). (B) Analysis of Env protein (140 aa, nt 6530 to 6949). Aligned protein sequences were used gap-free to construct neighbor-joining trees. Bootstrap values for 1,000 replicated trees (over 90%) are indicated as percent values. The polymerase tree was rooted using Gypsy, and the Env tree was rooted using avian leukosis virus (ALV) as the outgroup. Refer to Materials and Methods for the identities of the compared sequences. Retrovirus sequences (with GenBank accession numbers in parentheses) used in the comparisons included ALV (M37980); BLV, bovine leukemia virus (AF033818); FeLV, feline leukemia virus, subgroup A (M18247); FIV, feline immunodeficiency virus (M25381); GALV, gibbon ape leukemia virus (M26927); Gypsy, Drosophila melanogaster gypsy transposable element (AF033821); HERV K-10, human endogenous retrovirus (M14123); HIV-1, human immunodeficiency virus type 1 (AF033819); HTLV-1, HTLV type 1 (AF033817); HTLV-2, HTLV type 2 (M10060); IAP, Syrian hamster intracisternal A particle (K02288); MDEV, Mus dunni endogenous virus (AF053745); MMTV (AF033807); MoMLV, Moloney MLV (AF033811); MPMV, Mason-Pfizer monkey virus (AF033815); PERV PK15, Sus scrofa porcine endogenous retrovirus PK15 (AF038601); RMLV, Rauscher murine leukemia virus (U94692); RSV, Rous sarcoma virus (AF033808); SMRV, simian sarcoma virus (M23385; squirrel monkey retro); SRV-1, simian retrovirus type 1 (M11841); SRV-2 (M16605); Visna lentivirus (M10608); and WDSV, walleye dermal sarcoma virus (AF033822).
Molecular epidemiology. (i) Prevalence among different species of Boidae.
In order to determine whether PyT2RV was associated with BIBD, PBMC samples from additional Burmese pythons and other species of the Boidae family were analyzed by PCR for PyT2RV gag sequences with primers Q-1 and Q-2. These investigations showed that all of the tested Burmese and blood pythons but none of the royal pythons, reticulated pythons, boa constrictors, yellow anacondas, or carpet pythons contained PyT2RV sequences (Table 2). The absence of PyT2RV in these other species was confirmed by a second PCR specific for sequences in pol (primers Q-4 and Q-5). Amplifications with DNAs of all of these animals except the reticulated python were also done under reduced stringency conditions (5 and 10°C below the optimal annealing temperature). All products of a possibly interesting size were reamplified and sequenced but were unrelated to retroviral sequences. DNA of a reticulated python was PCR tested at 2°C below the optimal annealing temperature for pol and also for LTR sequences (primers U3-2 and G-4) but yielded no visible products within the expected size range.
TABLE 2.
Detection of PyT2RV gag sequences by PCR in PBMC from 33 boid snakes and of RT activity in plasmaa
| Species | No. of animals tested | No. of animals positive for PyT2RV gag in PBMC | No. of plasma samples tested | RT activity (μU/ml) in plasma (mean ± SD) |
|---|---|---|---|---|
| Boa constrictor | 3 | 0 | 7 | 4.2 ± 2.6 |
| Yellow anaconda | 1 | 0 | n.d. | n.d. |
| Carpet python | 1 | 0 | 1 | 10 |
| Blood python | 4 | 4 | 3 | 0.6 ± 0.4 |
| Burmese python | 18 | 18 | 11 | 6800 ± 5500 |
| Royal python | 5 | 0 | 6 | 1.3 ± 0.9 |
| Reticulated python | 1 | 0 | 1 | 0.3 |
n.d., not done.
(ii) PyT2RV is an endogenous retrovirus.
Among the PyT2RV-positive animals were also 11 Burmese pythons from a population in which BIBD had never occurred. These findings thus suggested a possible endogenous nature of the virus in Burmese python. This hypothesis was further supported by the demonstration that the number of proviral copies in PBMC samples from 11 Burmese pythons and in erythrocyte samples from four animals was, with more than 200 copies per haploid genome, very high and, among different animals, constant with respect to both gag and pol (Table 3). That the number of pol copies was considerably lower than that of gag was due to the 5-kb deletion (gag-pol; nt 1590 to 3996) mutant, which did not contain the region assessed in pol (see Fig. 2). The ratio of pol/gag sequence copies was, with 0.38 for PBMC and 0.34 for erythrocytes, also constant, thus providing further support for the endogenous nature of these sequences. When amplicons generated from PBMC or erythrocyte DNA by long-distance PCR with primers in R and U3 were analyzed on agarose gels, the same band patterns with similar relative intensities were found in both cell types and in all animals. In addition to the 7- and 5-kb sequences, there were another major band of approximately 4 kb, which corresponded to the 5-kb sequence having an additional deletion in env (5895 to 6835) and a number of weaker bands not investigated further. Inverse PCR (see Materials and Methods) yielded a short host cell DNA fragment of a viral integration site. PCR combining a primer (endo-1) specific for this fragment and primer U3r-1 with PBMC DNA from seven Burmese pythons from two different populations generated a product in four of the pythons, two from each of the two populations. In all four pythons, and only in these, an identical product was also obtained from erythrocyte DNA, reflecting a conserved proviral integration site.
TABLE 3.
Estimated PyT2RV proviral copy numbers per haploid genome in Burmese pythons
| Sequence | Tissue | No. of samples (n) | Copy no. (mean ± SD) |
|---|---|---|---|
| gag | PBMC | 11 | 218 ± 64 |
| Erythrocytes | 4 | 216 ± 68 | |
| pol | PBMC | 11 | 82 ± 16 |
| Erythrocytes | 4 | 74 ± 10 |
PCR for PyT2RV in four healthy blood pythons was positive in all. Further analysis showed that one haploid genome contained about 30 proviral copies. Population-based sequencing of one amplicon generated with primers U3-2 and G-4 in the LTR-5′ untranslated region showed 85% identity to the sequence in U3-RU5 (nt −60 to 132) and 97% identity to that between the primer binding site and the putative start codon of gag. Amplification of a 2,829-bp (nt 1415 to 4229) long stretch in gag-pr-pol by primers Q-1 and P-2 showed 95.4% sequence identity, namely, 94.4% within 336 bp of gag, 95.4% within 648 bp of pr, and 95.7% within 1,845 bp of pol. Compared to the Burmese python, 71% of the diverging nucleotide positions in pol represented conservative changes. The ORFs of the blood python showed the same relative positioning of the reading frames as those in the Burmese python and did not contain additional stop codons. No deletion mutants were found. In order to differentiate the blood python virus from PyT2RV, we chose the designation PyB1RV.
(iii) Expression of PyT2RV and PyB1RV in vivo and absence of a relationship with BIBD.
PyT2RV had initially been isolated from a healthy Burmese python kept together with animals suspected to suffer from BIBD. In order to investigate whether expression of PyT2RV was associated with BIBD, PBMC from animals kept in BIBD-free populations were also cultured for 1 month after initial stimulation with PHA. A maximal RT activity of 490 ± 250 μU/ml was observed 1 week after stimulation and decreased within 1 month to 8 ± 9 μU/ml in all of the five investigated animals (data not shown). RT activity in 11 Burmese python plasma samples amounted to an average of 6,800 ± 5,500 μU/ml, while it was only 0.6 ± 0.4 μU/ml in the four blood pythons (Table 2). Analysis of one of these Burmese python plasma samples by sucrose density gradient centrifugation resulted in the typical RT peak at 1.15 g/ml and copurification of both the RNA and DNA of PyT2RV exactly as shown in Fig. 1 (data not shown). Thus, PyT2RV appears to be strongly expressed and to exhibit particle-associated active RT in all Burmese pythons while the related PyB1RV of blood pythons is expressed considerably less.
Since the inclusion bodies of BIBD are typically found in the kidney and because retroviral particles were isolated from primary kidney cultures by others (19), paraffin-embedded kidney tissues from seven boa constrictors with a histologically verified diagnosis of BIBD were investigated for PyT2RV sequences with primers Q-1 and Q-2 from gag and Q-4 and Q-5 from pol. None of the samples was positive. When the same DNA samples were quantified for the single-copy gene c-mos with primers mos-7 and mos-8, which were specifically designed for the boa constrictor, a median of 1,768 copies/μl (range, 22 to 47,986) was found. Since the amplicons generated by the primers for PyT2RV were shorter than that for c-mos (and the PCR for PyT2RV was thus probably more sensitive), the negative results are strong evidence against the presence of PyT2RV in the diseased tissues. Attempts to transmit the virus in vitro to PHA-stimulated PBMC from two healthy royal pythons and one boa constrictor were unsuccessful, suggesting that PyT2RV is probably not infectious (data not shown).
Electron microscopy.
All attempts to visualize PyERV by transmission electron microscopy of nonadherent PBMC or by negative staining (2% phosphortungstic acid) of ultracentrifugated plasma pellets failed (2, 3).
DISCUSSION
In an attempt to identify the retroviral agent long suspected to cause BIBD, we have isolated a novel retrovirus provisionally named PyT2RV from a Burmese python. This virus is present as a highly expressed endogenous retrovirus in all Burmese pythons; a closely related but less expressed endogenous retrovirus designated PyB1RV was found in blood pythons, another Asian python species. All other investigated members of the Boidae family were negative for these sequences, but the presence of more distantly related viruses of the same lineage can only be formally excluded if Southern blot hybridizations are also negative. Based on the clearly established endogenous nature of PyT2RV and the sequence homology of PyB1RV, we propose the name python endogenous retrovirus (PyERV) for this novel group.
PyERV was initially isolated from a healthy Burmese python housed together with an animal of the same species which exhibited clinical symptoms suggestive of BIBD. Our investigations have yielded no evidence of a causal relationship between PyERV and BIBD. Cell-free supernatant of a primary kidney cell culture from a boa constrictor with BIBD was successfully used by others to transmit the disease to Burmese pythons (19). Provided that BIBD of the boa constrictor and of the Burmese python are indeed caused by the same agent, as is suggested by the successful transmission experiments described above, the absence of PyERV from histologically confirmed inclusion-body-positive kidney tissues from seven boa constrictors with a clinical diagnosis of BIBD is strong evidence against an etiological role of PyERV in BIBD. This conclusion is further supported by the fact that the virus could not be transmitted to PBMC from the boa constrictor or royal python. Our data do not completely rule out the possibility, however, that BIBD may be caused by a retrovirus related to PyERV but different enough to remain undetected by the PCR amplifications used here. It will be interesting to see whether the C-type particles reactive with the anti-BIBD virus rabbit antibody (13) or those seen in neoplasms of P. molurus bivittatus (7) are indeed different from PyERV. If this is the case, it should be possible to identify the true agent of BIBD by using the procedures described here, provided that the suspected agent is indeed an exogenous retrovirus.
PyERV is a novel retrovirus which cannot be classified into one of the seven hitherto defined retroviral genera based on sequence and genome information alone. Unfortunately, morphology as a further criterion was not available since all attempts to visualize the virus failed, most likely because the analyzed concentrations of virus particles (5 × 107 particles/ml, assuming there are three virus particles/nU of RT [18]), although well detectable by PCR for viral RNA or PERT assay, were too low. Based on the region compared and the algorithm employed, the most closely, but still only distantly, related viruses are among either the B- and D-type (intergrase and RT) or the mammalian C-type (TM) viruses. PyERV is too far away to be related to the endogenous MLV-related C-type sequences present in reptiles, birds, and mammals (15). Whether PyERV is related to the endogenous viper retrovirus shown to be immunologically related to D-type primate retroviruses (1) is unknown since the sequence of the viper virus has not been determined. Interestingly, based on the immunologic crossreactivity of the major internal (CA) proteins, the viper retrovirus has been reported to also be related to an endogenous guinea pig retrovirus, which in turn was related to the CA proteins of the B- and D-type retroviruses MMTV and simian sarcoma virus (10). It is thus conceivable that a group of retroviruses including PyERV, viper endogenous retrovirus, and the guinea pig virus might represent an intermediate group between the B- and D-types and the other retrovirus types. A possible alternative is that PyERV is a true recombinant retrovirus analogous to RD-114 which has recently been shown to be a recombinant of gag-pol from the feline endogenous type C retrovirus and env from the baboon endogenous virus (20). However, due to the extensive sequence divergence of PyERV from all other retroviruses, it is at present not possible to determine potential breakpoints in order to support this hypothesis. If the distribution of PyERV is indeed restricted to only a few species within the boid snake family, the virus must have become endogenous relatively late. This would be in agreement with the absence of premature stop codons, a high level of virus expression both in vitro and in vivo, and the fact that a specific integration site identified in four Burmese pythons was not present in three others. While it is clear that PyERV is an endogenous retrovirus, the possibility that one of the proviruses may be infectious cannot be ruled out despite the fact that our attempts to transmit the virus to PBMC from the royal python or boa constrictor were not successful.
Acknowledgments
We thank Lucia Bertodatto, Antonietta Baumgartner, and Celestine Wolfensberger for expert technical assistance, Andreas Pospischil for providing DNAs from kidney tissues of snakes with BIBD, and Thomas Bächi for electron microscopy.
This work was financed in part by the Kanton Zurich and by the Foundation for Scientific Research of the University of Zurich.
REFERENCES
- 1.Andersen, P. R., M. Barbacid, S. R. Tronick, H. F. Clark, and S. A. Aaronson. 1979. Evolutionary relatedness of viper and primate endogenous retroviruses. Science 204:318-321. [DOI] [PubMed] [Google Scholar]
- 2.Biel, S. S., and H. R. Gelderblom. 1999. Diagnostic electron microscopy is still a timely and rewarding method. J. Clin. Virol. 13:105-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biel, S. S., and H. R. Gelderblom. 1999. Electron microscopy of viruses, p. 111-147. In A. J. Cann (ed.), Cell culture: a practical approach. Oxford University Press, Oxford, England.
- 4.Bürgisser, P., P. Vernazza, M. Flepp, J. Böni, Z. Tomasik, U. Hummel, G. Pantaleo, J. Schüpbach, et al. 2000. Performance of five different assays for the quantification of viral load in subjects infected with various subtypes of HIV-1. J. Acquir. Immune Defic. Syndr. 23:138-144. [DOI] [PubMed] [Google Scholar]
- 5.Carlisle-Nowak, M. S., N. Sullivan, M. Carrigan, C. Knight, C. Ryan, and E. R. Jacobson. 1998. Inclusion body disease in two captive Australian pythons (Morelia spilota variegata and Morelia spilota spilota). Austral. Vet. J. 76:98-100. [DOI] [PubMed] [Google Scholar]
- 6.Carneiro, S. M., H. Tanaka, J. J. Kisielius, and A. Sesso. 1992. Occurrence of retrovirus-like particles in various cellular and intercellular compartments of the venom glands from Bothrops jararacussu. Res. Vet. Sci. 53:399-401. [DOI] [PubMed] [Google Scholar]
- 7.Chandra, A. M., E. R. Jacobson, and R. J. Munn. 2001. Retroviral particles in neoplasms of Burmese pythons (Python molurus bivittatus). Vet. Pathol. 38:561-564. [DOI] [PubMed] [Google Scholar]
- 8.Clark, H. F., and P. D. Lunger. 1981. Viruses, p. 135-164. In J. E. Cooper and O. F. Jackson (ed.), Diseases of the reptilia, vol. 1. Academic Press, New York, N.Y.
- 9.Conrad, B., Weissmahr, R. N., J. Boni, R. Arcari, J. Schupbach, and B. Mach. 1997. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell 90:303-313. [DOI] [PubMed] [Google Scholar]
- 10.Dahlberg, J. E., S. R. Tronick, and S. A. Aaronson. 1980. Immunological relationships of an endogenous guinea pig retrovirus with prototype mammalian type B and type D retroviruses. J. Virol. 33:522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donahue, P. R., E. A. Hoover, G. A. Beltz, N. Riedel, V. M. Hirsch, J. Overbaugh, and J. I. Mullins. 1988. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J. Virol. 62:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ippen, R., Z. Mladenov, and A. Konstantinov. 1978. Leukosis with viral presence proven by means of an electron microscope in 2 boa constrictors. Schweiz. Arch. Tierheilkd. 120:357-368. (In German.) [PubMed] [Google Scholar]
- 13.Jacobson, E. R., J. Oros, S. J. Tucker, D. P. Pollock, K. L. Kelley, R. J. Munn, B. A. Lock, A. Mergia, and J. K. Yamamoto. 2001. Partial characterization of retroviruses from boid snakes with inclusion body disease. Am. J. Vet. Res. 62:217-224. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson, E. R., J. C. Seely, and M. N. Novilla. 1980. Lymphosarcoma associated with virus-like intranuclear inclusions in a California king snake (Colubridae: Lampropeltis). J. Natl. Cancer Inst. (Bethesda) 65:577-583. [PubMed] [Google Scholar]
- 15.Martin, J., E. Herniou, J. Cook, R. W. O'Neill, and M. Tristem. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 73:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oros, J., S. Tucker, and E. R. Jacobson. 1998. Inclusion body disease in two captive boas in the Canary Islands. Vet. Rec. 143:283-285. [DOI] [PubMed] [Google Scholar]
- 17.Orr, H. C., L. E. Harris, A. V. Bader, R. L. Kirschstein, and P. G. Probst. 1972. Cultivation of cells from a fibroma in a rattlesnake, Crotalus horridus. J. Natl. Cancer Inst. 48:259-264. [PubMed] [Google Scholar]
- 18.Pyra, H., J. Boni, and J. Schupbach. 1994. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc. Natl. Acad. Sci. USA 91:1544-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher, J., E. R. Jacobson, B. L. Homer, and J. M. Gaskin. 1994. Inclusion body disease in boid snakes. J. Zoo Wildl. Med. 25:511-524. [Google Scholar]
- 20.van der Kuyl, A. C., J. T. Dekker, and J. Goudsmit. 1999. Discovery of a new endogenous type C retrovirus (FcEV) in cats: evidence for RD-114 being an FcEVGag-Pol/baboon endogenous virus BaEVEnv recombinant. J. Virol. 73:7994-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogt, V. M. 1997. Retroviral virions and genomes, p. 27-69. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 22.Weissmahr, R. N. 1995. Development and evaluation of a highly sensitive method for the identification of particle-associated retroviral sequences. Ph.D. thesis. Zurich University, Zurich, Switzerland.
- 23.Weissmahr, R. N., J. Schupbach, and J. Boni. 1997. Reverse transcriptase activity in chicken embryo fibroblast culture supernatants is associated with particles containing endogenous avian retrovirus EAV-0 RNA. J. Virol. 71:3005-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wozniak, E., J. McBride, D. DeNardo, R. Tarara, V. Wong, and B. Osburn. 2000. Isolation and characterization of an antigenically distinct 68-kd protein from nonviral intracytoplasmic inclusions in Boa constrictors chronically infected with the inclusion body disease virus (IBDV: Retroviridae). Vet. Pathol. 37:449-459. [DOI] [PubMed] [Google Scholar]
- 25.Zeigel, R. F., and H. F. Clark. 1970. Current studies on a new "C'-type virus of presumed reptilian origin. Bibl. Haematol. 36:643. [DOI] [PubMed] [Google Scholar]
- 26.Zeigel, R. F., and H. F. Clark. 1969. Electron microscopic observations on a “C”-type virus in cell cultures derived from a tumor-bearing viper. J. Natl. Cancer Inst. 43:1097-1102. [PubMed] [Google Scholar]
- 27.Zeigel, R. F., and H. F. Clark. 1971. Histologic and electron microscopic observations on a tumor-bearing viper: establishment of a “C”-type virus-producing cell line. J. Natl. Cancer Inst. 46:309-321. [PubMed] [Google Scholar]