Abstract
Assembly of the infectious herpes simplex virus type 1 virion is a complex, multistage process that begins with the production of a procapsid, which is formed by the condensation of capsid shell proteins around an internal scaffold fashioned from multiple copies of the scaffolding protein, pre-VP22a. The ability of pre-VP22a to interact with itself is an essential feature of this process. However, this self-interaction must subsequently be reversed to allow the scaffolding proteins to exit from the capsid to make room for the viral genome to be packaged. The nature of the process by which dissociation of the scaffold is accomplished is unknown. Therefore, to investigate this process, the properties of isolated scaffold particles were investigated. Electron microscopy and gradient sedimentation studies showed that the particles could be dissociated by low concentrations of chaotropic agents and by moderate reductions in pH (from 7.2 to 5.5). Fluorescence spectroscopy and circular dichroism analyses revealed that there was relatively little change in tertiary and secondary structures under these conditions, indicating that major structural transformations are not required for the dissociation process. We suggest the possibility that dissociation of the scaffold may be triggered by a reduction in pH brought about by the entry of the viral DNA into the capsid.
In mature herpes simplex virus type 1 (HSV-1) virions, the viral genome is enclosed within a T=16 icosahedral capsid made up of 12 pentons, 150 hexons, and 320 connecting densities called triplexes (52). The capsid shell is composed of multiple copies of four sequence-unrelated proteins, VP5 (149 kDa), VP19C (50 kDa), VP23 (34 kDa), and VP26 (12 kDa). VP5, which makes up the pentons and the bulk of the hexons (60, 68), represents 70% by mass of the capsid shell. The hexons also contain VP26, six copies of which are arranged in a ring on the top of each hexon (59, 67). The two remaining proteins, VP23 and VP19C, associate in a 2:1 ratio as a heterotrimer, called the triplex, that forms connections between neighboring hexons and between hexons and pentons (36, 47, 66). The initial step in capsid assembly involves a cocondensation of shell and scaffolding proteins to form a spherical particle designated the procapsid. In the procapsid, the shell surrounds the internal scaffold, which is formed predominantly by a single protein, pre-VP22a (32, 34). Pre-VP22a provides the framework around which a shell of the correct size and symmetry is constructed (7, 53, 55). In order to fulfill this role, pre-VP22a must interact both with the capsid shell proteins and with itself. The association with the shell has been extensively studied, and it is known that sequences at the C terminus of pre-V22a interact directly with VP5 (20). These sequences are essential for capsid assembly (21, 30, 54). The self-interaction of pre-VP22a is less well characterized (38, 42), but it is known that it can occur in the absence of the capsid shell proteins (31, 39, 53), when it results in the formation of particles resembling free scaffolds (referred to in this paper as scaffold particles).
The scaffolding proteins are encoded by the gene UL26 locus, which comprises two 3′-coterminal overlapping genes (UL26 and UL26.5) that encode proteins from a single open reading frame (26, 27, 43). UL26.5 maps to the 3′ portion of UL26 and encodes pre-VP22a. UL26 encodes a protease, which cleaves itself at two positions known as the R (release) and M (maturation) sites. Cleavage at the R site generates two products. One of these, pre-VP21, shares amino acid sequences with pre-VP22a and is a minor scaffolding component, while the other, VP24, contains the protease activity. The M site is present 25 amino acids upstream from the C termini of both pre-VP21 and pre-VP22a (8, 26, 43). These C-terminal 25 amino acids include the residues that interact with VP5 (20), and cleavage of pre-VP22a at the M site, to produce the shortened form VP22a, destroys this interaction. This cleavage also triggers the reconfiguration of the spherical procapsid shell into the polyhedral form found in mature capsids (2, 14, 46). Three types of polyhedral capsid are generated, designated A (empty), B (intermediate), and C (full) (16). In B capsids the scaffold is retained, while A capsids contain no internal material. In C capsids the internal space is occupied by the viral genome. To accommodate the genome, the scaffold must exit from the capsids. However, since the available channels through the procapsid and capsid shells are too small to allow the passage of large structures, the effective size of the scaffold must be reduced. Therefore, mechanisms presumably exist to reverse the self-association of the scaffolding proteins. Studies of viral mutants suggest that DNA packaging and scaffold release are coordinated events that take place before or during the conversion from procapsids to polyhedral capsids (2, 40, 46) and that loss of scaffold takes place only in circumstances where DNA packaging is actively occurring (4). Under conditions where DNA packaging is blocked, only B capsids are usually formed (49). Since loss of scaffold appears to be closely linked to the actual process of DNA packaging, it seems likely that entry of the DNA itself into the capsid could be an important factor in the process. We speculate that the introduction of a large amount of negatively charged DNA into the interior of the capsid, by altering the ionic environment, affects the interaction of the scaffolding proteins. To investigate this process, we examined the structural integrity of purified scaffold particles under mildly acidic conditions and in the presence of low concentrations of urea, which appears to bring about analogous changes in the protein.
MATERIALS AND METHODS
Cells and virus.
Spodoptera frugiperda (Sf21) cells were grown in TC100 medium (Invitrogen) supplemented with 5% fetal calf serum. The construction of the recombinant baculoviruses AcUL26.5 (53) and AcVP22a (21), expressing pre-VP22a and VP22a, respectively, have been described previously.
Purification of scaffold particles.
Three-hundred-milliliter cultures of Sf21 cells at 106 cells/ml were infected at 5 PFU/cell with either AcUL26.5 or AcVP22a. After 48 h of incubation at 28°C, cells were harvested by centrifugation and the pellet was resuspended in 1/60 volume of phosphate-buffered saline (PBS) (170 mM NaCl, 3.4 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.2). Aliquots containing approximately 108 cells were pelleted and stored at −20°C until required. The method of scaffold purification was similar to that described by Newcomb et al. (33). Frozen aliquots were resuspended in 500 μl of PBS and subjected to five cycles of freeze-thawing in dry ice-methanol before the cellular debris was removed by centrifugation at 13,000 rpm in an MSE Microcentaur centrifuge. The supernatant was transferred to a fresh tube, and 90% saturated ammonium sulfate was added to a final concentration of 10%. The solution was mixed at 4°C for 30 min, and the precipitated proteins were pelleted by centrifugation at 13,000 rpm. The supernatant was discarded, and the pellet was resuspended in 0.5 ml of PBS. The suspension was layered onto a 13-ml 10 to 40% sucrose (in PBS) gradient and centrifuged in a Sorvall TsT41 rotor at 40,000 rpm for 1 h at 4°C. A single band was observed towards the middle of the gradient. This was removed and dialyzed against PBS to remove the sucrose. The sample was concentrated by dialysis against 40% polyethylene glycol in PBS and examined by electron microscopy. At each stage, aliquots were removed for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis.
Polyacrylamide gel electrophoresis.
Samples were prepared for electrophoresis, and proteins were separated on SDS-10% polyacrylamide gels cross-linked with 2.6% (wt/wt) N,N′-methylene bisacrylamide (28). Proteins were either visualized after fixation by staining with 0.25% Coomassie brilliant blue or transferred to a Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech) for Western blot analysis. Antibody binding was carried out as described by the manufacturer for the membrane protocol. Briefly, the membrane was incubated for 1 h with the appropriate mouse monoclonal antibody. For pre-VP22a and VP22a, MCA406 (Serotec Ltd.) was used at a dilution of 1:200. For VP5, DM165 (raised against purified VP5) was used at a dilution of 1:100. The membrane was washed and incubated for a further 1 h with a 1:1,000 dilution of goat anti-mouse immunoglobulin G-peroxidase conjugate (Sigma Immunochemicals). Bound antibody was detected by enhanced chemiluminescence (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Electron microscopy.
For examination by negative staining, preparations of scaffold particles or capsids were diluted in PBS to a suitable concentration, applied to a Parlodion-coated microscope grid, and stained with 1% phosphotungstic acid (pH 7). Capsids to be examined in thin section were pelleted and then fixed and embedded as described previously (46). Sections were stained with saturated uranyl acetate in 50% ethanol and poststained with 1% lead citrate. All samples were examined in a JEOL 1200EX electron microscope.
Sucrose gradient analysis.
Analysis was carried out on 5-ml gradients run on a Sorvall AH650 rotor (scaffolds) or on 13-ml gradients run on a Sorvall Tst41 rotor (capsids). Ten to 40% (wt/vol) sucrose gradients prepared in PBS that had been adjusted to the appropriate pH were formed using a Biocomp Gradient Master according to the manufacturer's instructions. All gradients contained 1 mM dithiothreitol (DTT) and were precooled to 4°C before use. The duration and speed of centrifugation were as described in Results.
Fluorescence and ANS binding.
Fluorescence measurements were recorded on a Perkin-Elmer LS-50B spectrofluorimeter in a 1-ml semimicrocuvette with a 1-cm path length at 25°C. For protein fluorescence, the excitation wavelength was 290 nm, and the emission spectra were recorded at between 300 and 400 nm. For 8-anilino-1-naphthalenesulfonate (ANS) fluorescence, the excitation wavelength was 370 nm, and the emission spectra were recorded at between 440 and 540 nm. ANS was added to protein samples to a final concentration of 20 μM. The protein concentration in all experiments was 0.2 mg/ml.
CD.
Circular dichroism (CD) spectra were obtained on a Jasco-600 spectropolarimeter (Japan Spectroscopic Co., Tokyo, Japan). Near-UV CD spectra (320 to 260 nm) were collected using a cylindrical quartz cell with a path length of 1 cm. The protein concentration was 1 mg/ml. Far-UV CD spectra (260 to 200 nm) were obtained using a cylindrical quartz cell with a path length of 0.02 cm. Secondary structure was estimated using the SELCON procedure (50). The protein concentration was in the range of 0.2 to 0.5 mg/ml for different experiments, and all measurements were performed at 25°C.
RESULTS
Scaffold particle stability.
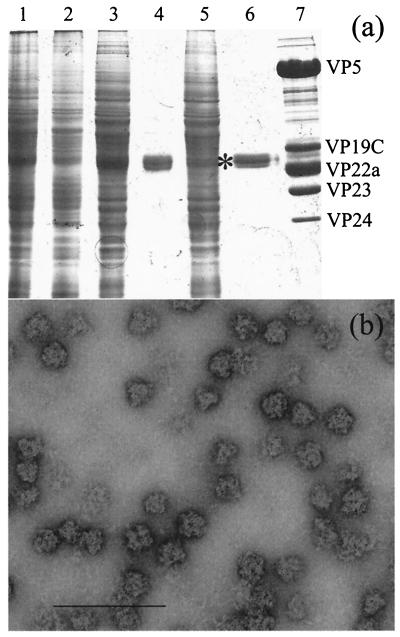
The preparation of scaffold particles purified from AcUL26.5-infected cells (see Materials and Methods) consisted almost entirely of pre-VP22a as determined by Coomassie brilliant blue staining (Fig. 1a, lane 6). When examined by electron microscopy, the particles appeared to be roughly spherical and approximately 60 nm diameter (Fig. 1b). Analysis by light scattering indicated a diameter of ∼70 nm with a molecular mass of 14,700 to 18,400 kDa (Brian Bowman, personal communication), corresponding to approximately 500 copies of pre-VP22a. This is considerably larger than the size of the scaffolds purified by Newcomb et al. (33), presumably reflecting the different properties of their human cytomegalovirus-HSV-1 fusion construct and the wild-type HSV-1 scaffolding protein used here.
FIG. 1.
Purification of scaffold particles. The scaffold particles were purified as described in Materials and Methods. (a) At each purification stage approximately 1/100 of the sample was removed for analysis on an SDS-10% polyacrylamide gel. Lane 1, infected cell extract; lane 2, pellet after freeze-thawing; lane 3, supernatant after freeze-thawing; lane 4, pellet from ammonium sulfate precipitation; lane 5, supernatant from ammonium sulfate precipitation; lane 6, purified pre-VP22a scaffolds after sucrose gradient banding, dialysis, and concentration; lane 7, purified HSV-1 B capsids. The names of the HSV-1 capsid proteins are indicated to the right of lane 7. The position of the pre-VP22a band in lane 6 is indicated by an asterisk. (b) Purified scaffold particles were absorbed onto a Parlodion-coated microscope grid, stained with 1% phosphotungstic acid, and examined by electron microscopy. Bar, 200 nm.
To investigate the stability of the scaffold particles, they were treated with a range of urea concentrations. Since the scaffolding protein undergoes extensive disulfide cross-linking when stored under nonreducing conditions (69), all experiments were carried out in the presence of 1 mM DTT. The presence of DTT did not alter the appearance of the scaffold particles when examined by negative staining (Fig. 2a). However, when exposed to urea at a concentration of 1 M or higher, no scaffold particles were visible (Fig. 2b), although on close inspection the grids were seen to be coated with a layer of protein that presumably arose from dissociated scaffold particles. This suggests that the presence of low concentrations of urea can cause the dissociation of the scaffold particles into much smaller units that do not form distinctive structures visible by electron microscopy.
FIG. 2.
Dissociation of scaffold particles by urea and pH reduction. Scaffold particles prepared as for Fig. 1 (a) were treated with 1 M urea (b) or dialyzed against PBS that had been adjusted to pH 6.0 (c) or pH 5.5 (d) as described in the text. The samples were absorbed onto Parlodion-coated microscope grids, stained with 1% phosphotungstic acid, and examined by electron microscopy. Bar, 500 nm.
The stability of scaffold particles under different pH conditions was tested by dialyzing them against PBS at a range of pHs between pH 5.0 and 6.5 at intervals of 0.5. At pH 6.5 (not shown) there was no obvious alteration in the numbers or appearance of the scaffold particles compared to the untreated control at pH 7.2 (Fig. 2a). At pH 6.0 (Fig. 2c) scaffold particles were still present in similar numbers, although they appeared to be smaller and less uniform, with a tendency to aggregate. However, at pH 5.0 (not shown) and pH 5.5 (Fig. 2d), no particles were visible and the grids were covered with a protein layer similar to that seen following treatment with 1 M urea.
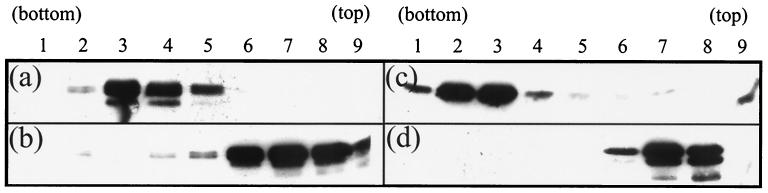
To confirm that lowering the pH was causing dissociation of the scaffold particles, their sizes were analyzed by sucrose gradient centrifugation. For these experiments both pre-VP22a and VP22a (the truncated form of pre-VP22a missing the C-terminal 25 amino acids [41]) scaffold particles were prepared. Purified scaffold particles at a concentration of 1 mg/ml in PBS (pH 7.2) were treated with 1 mM DTT for 1 h to ensure the removal of intermolecular disulfide cross-links before being layered on top of 10 to 40% sucrose gradients prepared at either pH 7.2 or 5.5 (see Materials and Methods). After centrifugation at 40,000 rpm (Sorvall AH650 rotor) for 105 min, the gradients were examined for the presence of visible refracting bands. For both pre-VP22a and VP22a, a single band was visible near the bottom of the pH 7.2 gradients, but in neither case could a band be seen on the pH 5.5 gradients (data not shown). To ascertain the distribution of the scaffolding proteins, each gradient was fractionated and analyzed by Western blotting. Figure 3 shows the profiles for both the full-length and truncated forms. At pH 7.2, the abundance of both pre-VP22a (Fig. 3a) and VP22a (Fig. 3c) was maximal in fraction 3, in agreement with the position of the visible scaffold particle band. In contrast, at pH 5.5 both pre-VP22a (Fig. 3b) and VP22a (Fig. 3d) were shifted towards the top of the gradient, suggesting that the scaffold particles were dissociating into smaller components.
FIG. 3.
Sucrose gradient analysis of scaffolds. Scaffold particles purified from AcUL26.5 (a and b)- or AcVP22a (c and d)-infected Sf21 cells were analyzed on 5-ml, 10 to 40% (wt/vol) sucrose gradients at pH 7.2 (a and c) or pH 5.5 (b and d). Centrifugation was at 40,000 rpm for 105 min in a Sorvall AH650 rotor. Nine fractions were collected from the bottom of the gradient, and 20-μl samples of each fraction were analyzed on SDS-10% polyacrylamide gels and transferred to nitrocellulose membranes. To reveal the distribution of the scaffolding proteins, the blots were probed with MCA406 antibody as described in Materials and Methods.
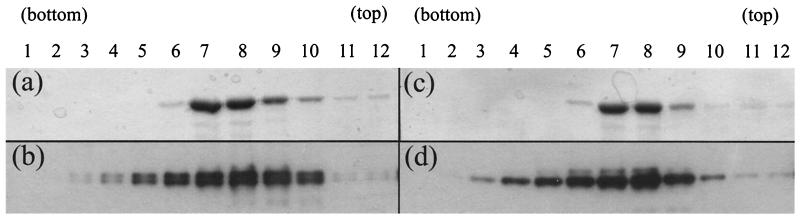
To determine the extent of dissociation, the sucrose gradient analysis at pH 5.5 was repeated as before, but the centrifugation was carried out at 40,000 rpm (Sorvall AH650 rotor) for 20 h. Bovine serum albumin (BSA) (250 μg/gradient; Sigma Aldrich) was added to each sample as a size marker. Following gel electrophoresis of the gradient fractions, the distribution of the BSA marker was determined by Coomassie brilliant blue staining (Fig. 4a and c), while the scaffolding proteins were detected by Western blotting (Fig. 4b and d). The gel profiles revealed that for both pre-VP22a (Fig. 4b) and VP22a (Fig. 4d) the peak fractions coincided with the BSA peak (Fig. 4a and c, respectively). Since BSA is a monomer with an Mr of 66,000, it is clear that under these conditions both pre-VP22a and VP22a, which have molecular masses of around 34 and 31 kDa, respectively, are sedimenting as small entities which are likely to be dimers or monomers.
FIG. 4.
Sizing of dissociated scaffolds. Scaffold particles purified from AcUL26.5 (a and b)- or AcVP22a (c and d)-infected Sf21 cells were mixed with BSA (250 μg/gradient; Sigma Aldrich) and layered onto 5-ml, 10 to 40% (wt/vol) sucrose gradients at pH 5.5. After centrifugation at 40,000 rpm for 20 h in a Sorvall AH650 rotor, 12 fractions were collected from the bottom of the gradient. Duplicate 20-μl samples of each fraction were analyzed on separate SDS-10% polyacrylamide gels. One gel from each pair was stained with Coomassie brilliant blue (a and c) to reveal the distribution of the BSA size marker, while the duplicate gel was blotted onto a nitrocellulose membrane (b and d) and probed with MCA406 antibody to reveal the distribution of the scaffolding proteins.
Effects of denaturants on the secondary and tertiary structures of the scaffolding protein.
The studies described above demonstrate that the association of scaffolding proteins into distinct scaffold particles is sensitive to experimental conditions and can be reversed by low concentrations of denaturants or by relatively small changes in pH. To determine whether these conditions were directly affecting the structure of the scaffolding proteins themselves, fluorescence spectroscopy and CD were used to assess the structural integrity of the scaffolding protein.
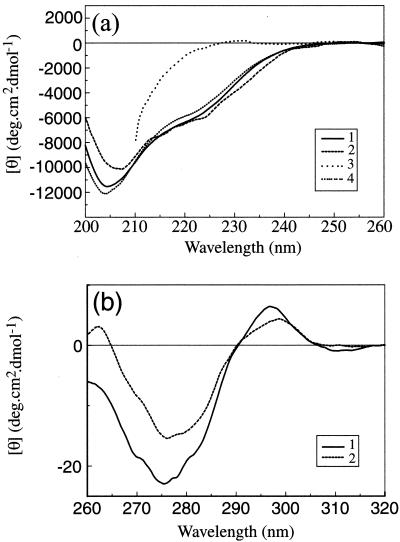
Figure 5 shows the CD spectra obtained from purified scaffold particles and the effects of pH changes and presence of urea on these spectra. The SELCON procedure (51) was used to estimate the secondary structure content of the sample from the far-UV CD spectrum (Fig. 5a). At pH 7.2, pre-VP22a was found to contain 19% α-helix, 24% antiparallel β-sheet, 2% parallel β-sheet, and 24% turn. The secondary structure prediction program PSIPRED gives a value of up to 21% α-helix but predicts less than 5% β-sheet. Lowering the pH from 7.2 to 5.5 or addition of 1 M urea led to small losses of secondary structure, with the changes in ellipticity at 225 nm of the order of 10% or less. Addition of 8 M urea led to a greater than 95% loss of ellipticity at 225 nm, indicating that the protein was completely unfolded under these conditions.
FIG. 5.
CD spectra of pre-VP22a. (a) Far-UV CD spectra. Purified pre-VP22a scaffold particles at a concentration in the range of 0.8 to 0.5 mg/ml in PBS (pH 7.2) containing 1 mM DTT (trace 1) were dialyzed against PBS that had been adjusted to pH 5.5 (trace 2) or were incubated for 15 min in the presence of 8 M (trace 3) or 1 M (trace 4) urea (diluted from a freshly prepared 10 M solution) and examined by far-UV CD as described in the text. Readings were taken at a final concentration in the range of 0.5 to 0.2 mg/ml. The presence of 8 M urea prevented the acquisition of reliable data below 210 nm. (b) Near-UV CD spectra. Purified pre-VP22a scaffold particles at 1.1 mg/ml in PBS (pH 7.2) containing 1 mM DTT were examined by near-UV CD as described in the text (trace 1). Freshly prepared 10 M urea was then added to a final concentration of 1 M, and the reading was repeated (trace 2). The spectra were corrected for the effects of buffer by subtracting the appropriate buffer controls and converted to molar ellipticity units to correct for concentration.
The near-UV CD spectrum of pre-VP22a at pH 7.2 (Fig. 5b) shows a small positive peak at 295 nm, characteristic of Trp side chains, and a larger negative peak at 275 nm, characteristic of Tyr side chains. Addition of 1 M urea led to a modest (less than 30%) decline in the amplitude of the near-UV CD spectrum, indicating that the tertiary structure of the protein had not been seriously affected. Despite several attempts, it proved impossible to obtain a satisfactory near-UV CD spectrum of the protein at pH 5.5. This was because at the concentrations (1 mg/ml or greater) required to record spectra, the sample had a tendency to precipitate while undergoing dialysis to lower its pH.
It should be noted that the amplitudes of the peaks in the near-UV CD spectrum of pre-VP22a are relatively small by comparison with those found in several well-characterized proteins such as insulin, lysozyme, and carbonic anhydrase (63). While there are several factors that influence the intensities of near-UV CD spectra, one explanation for the low intensities of the peaks in pre-VP22a is that the interactions stabilizing the tertiary structure of the protein may be relatively weak.
From the CD studies it can be concluded that the lowering of the pH from 7.2 to 5.5 or the addition of 1 M urea leads to the retention of most of the secondary and tertiary structural features of native pre-VP22a.
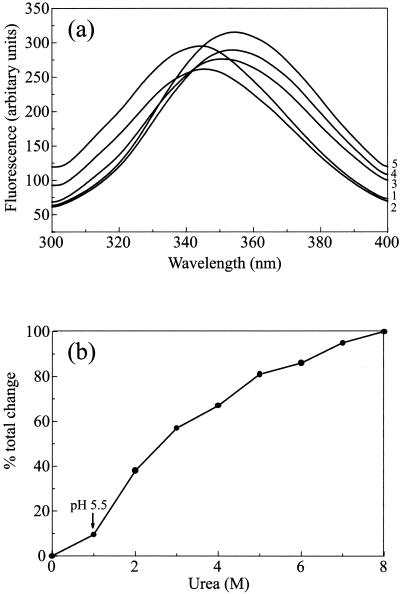
Fluorescence spectroscopy reports on the environment of the tryptophan side chains and therefore provides an additional measure of the integrity of the folded state of a protein. A completely unfolded protein would have a λmax (wavelength of maximum emission) of 356 nm, indicating that the Trp residues are fully exposed to the solvent. A shift to shorter wavelengths indicates that the Trp residues are at least partially shielded from the solvent (11). Changes in fluorescence intensity during unfolding can often show a more complex pattern because a variety of factors, such as efficiency of quenching by solvent and by energy transfer, are involved.
Pre-VP22a scaffold particles were incubated for 15 min with a range of urea concentrations of from 1 to 8 M, and their fluorescence spectra were recorded (Fig. 6a). The untreated control sample had a λmax of 344.5 nm. This is significantly shifted from the value of 356 nm observed for the model compound N-acetyltryptophanamide and is consistent with the scaffolding protein having a tertiary structure in which the Trp side chains are buried to a moderate extent. In the presence of 8 M urea, the protein had a λmax of 355 nm, indicating that the protein is essentially completely unfolded under these conditions. However, in the sample treated with 1 M urea (where the dissociation of scaffold particles still occurs), there was only a minor shift in λmax (to 345.5 nm), representing ∼10% of the total change seen between 0 and 8 M (Fig. 6b). When scaffold particles dialyzed against PBS at pH 5.5 were examined, they also gave a λmax of 345.5 nm. This suggests that reducing the pH has little effect on the average exposure of the Trp side chains and thus presumably the overall tertiary structural features of the protein.
FIG. 6.
Fluorescence spectra of pre-VP22a. (a) Purified pre-VP22a scaffold particles (in PBS [pH 7.2] with 1 mM DTT) at a final concentration of 0.2 mg/ml were incubated for 15 min in urea at concentrations of from 0 to 8 M, increasing in 1 M steps. Part of the sample was also dialyzed against PBS that had been adjusted to pH 5.5. The samples were examined by fluorescence spectroscopy as described in the text. All spectra were corrected for background by subtraction of a buffer control spectrum. To improve clarity, only the traces obtained with 0 M (trace 1), 1 M (trace 2), 3 M (trace 3), 6 M (trace 4), and 8 M (trace 5) urea are reproduced here. (b) The observed shift in λmax from the value with 0 M urea, expressed as a percentage of the total change in λmax induced by 8 M urea, is plotted against the urea concentration. The shift seen at pH 5.5 is indicated by an arrow.
These findings suggest that dissociation of scaffold particles is due to subtle changes affecting the interacting surfaces and does not involve extensive changes in the structure of pre-VP22a monomers themselves.
Pre-VP22a appears to have a weakly defined tertiary structure but does not behave like a typical molten globule.
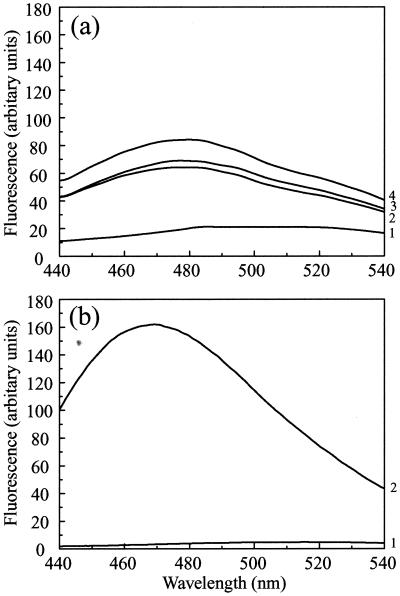
The molten globule is a relatively compact state, in which a protein retains most of its native secondary structure but the interactions between side chains stabilizing the tertiary structure are weak (1, 44). The fluorescent dye ANS binds preferentially to nonpolar regions of proteins. A characteristic large increase in fluorescence at 470 nm (with excitation at 370 nm) is generally considered to be a sensitive test for the molten globule state of proteins, where ANS has access to the hydrophobic interior (1, 44). Although ANS has been shown to bind to the native state of some proteins via clusters of nonpolar residues, binding to typical molten globule states (Fig. 7b) is usually enhanced by at least an order of magnitude compared with either the native or fully unfolded states (45, 48). Replicate samples of scaffold particles were tested for ANS binding ability at physiological pH (pH 7.2) or after dialysis against PBS adjusted to pH 6.5, 6.0, or 5.5.
FIG. 7.
ANS binding. (a) ANS (at a final concentration of 20 μM) was added to purified pre-VP22a scaffold particles (at 0.2 mg/ml) after they had been dialyzed against PBS containing 1 mM DTT that had been adjusted to pH 7.2 (trace 1), pH 6 (trace 2), or pH 5.5 (trace 3) as described in the text. The excitation wavelength was 370 nm, and fluorescence was recorded at between 440 and 540 nm. A control spectrum of ANS in PBS (pH 7.2) was measured (trace 4); there was no significant difference between the fluorescence of free ANS over the range from pH 5.5 to pH 7.2. (b) For comparative purposes, the ANS binding spectrum of the capsid shell protein VP23, which forms a molten globule, is reproduced from reference 23. Only the curves for ANS (20 μM) alone (trace 1) or mixed with VP23 (0.2 mg/ml) (trace 2) are shown here. The spectra in panel b were recorded using lower slit widths than for those in panel a, resulting in an approximately fourfold decrease in the intensities in panel b compared with panel a, as shown by comparing the spectra for 20 μM ANS in the two panels. When this factor is taken into account, it is clear that the enhancement of ANS fluorescence in the case of VP23 (b) is at least eightfold higher than that in the case of pre-VP22a at pH 5.5 (a).
Scaffold particles were found to bind ANS weakly at pH 7.2, as shown by a 3.5-fold increase in fluorescence at 470 nm compared with free ANS (Fig. 7a). At pH 6.5 and 6.0 the increases in fluorescence were of a similar order but, the increase at pH 5.5 was about 50% greater (fivefold greater fluorescence than free ANS), suggesting that some additional nonpolar sites on pre-VP22a for binding ANS had become available as a result of dissociation of the scaffold particles. However, the increase in ANS fluorescence under any of the conditions tested was only relatively modest, indicating that formation of a typical molten globule state was unlikely to have occurred in the case of pre-VP22a. This conclusion is supported by the data from the near-UV CD and fluorescence studies; namely, that although the interactions stabilizing the tertiary structure of pre-VP22a are weak, they persist under conditions where the scaffold is dissociated. In a typical molten globule state, the near-UV CD spectrum would be essentially abolished, reflecting the lack of stable tertiary interactions between side chains (1, 44).
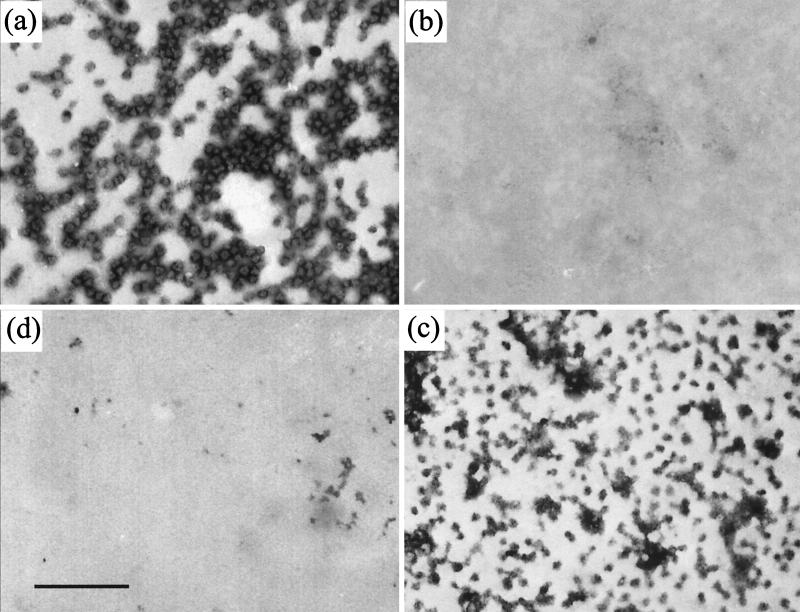
Effect of reduced pH on capsids.
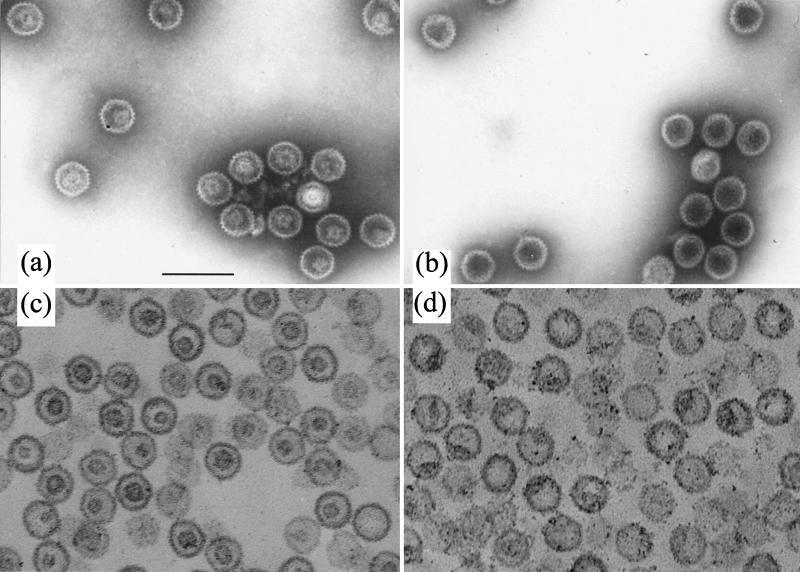
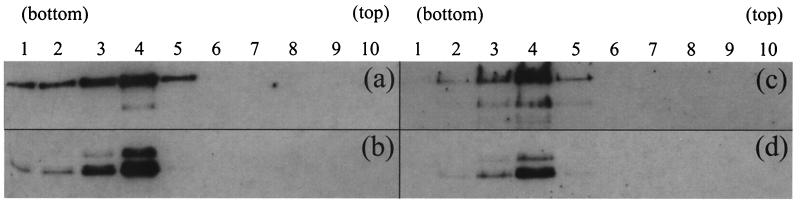
Since dissociation of the scaffold seems to be a necessary precursor to its exit from the capsid, the effect of reduced pH was examined by dialyzing purified B capsids against PBS at pH 5.5 for 16 h at 4°C in the presence of 1 mM DTT. Figure 8 shows electron micrographs of negatively stained B capsids treated at pH 7.2 (Fig. 8a) and pH 5.5 (Fig. 8b). While the scaffold density could clearly be seen inside the capsids at pH 7.2, it was not observed at pH 5.5, suggesting that the nature of the scaffold had altered. When capsids treated in a similar way were examined after thin sectioning, the characteristic ring-shaped, central scaffold cores were clearly defined at pH 7.2 (Fig. 8c) but were not seen at pH 5.5 (Fig. 8d). Instead the internal material had either congregated into one or more clumps on the inner surface of the capsid or was more evenly distributed around the inside of the capsid shell. To confirm that the scaffolding proteins had remained inside the capsids, B capsids that had been treated at pH 7.2 (Fig. 9a and b) or pH 5.5 (Fig. 9c and d) were layered onto sucrose gradients of the same pH. Western blot analysis showed that at both pHs, VP22a (Fig. 9b and d) cosedimented with the major capsid protein, VP5 (Fig. 9a and c), in the position occupied by the visible capsid band. A similar result was obtained when capsids were treated with 1 M urea (data not shown). No dissociated VP22a was detected at the top of the gradients, confirming that the scaffolding proteins had remained associated with the capsids.
FIG. 8.
Effects of lowered pH on scaffold-containing capsids. Purified B capsids were dialyzed against PBS at pH 7.2 (a and c) or pH 5.5 (b and d) for 16 h at 4°C in the presence of 1 mM DTT. The capsids were then either used for negative staining (a and b) or prepared for thin sectioning (c and d) and examined by electron microscopy. Bar, 200 nm.
FIG. 9.
Purified B capsids that had been treated at pH 7.2 (a and b) or pH 5.5 (c and d) as shown in Fig. 8 were loaded onto 13-ml, 10 to 40% (wt/vol) sucrose gradients of the same pH. After centrifugation at 40,000 rpm for 20 min in a Sorvall TsT41 rotor, 10 fractions were collected from the bottom of the gradient. Twenty-microliter samples of each fraction were analyzed on SDS-10% polyacrylamide gels and transferred to nitrocellulose membranes. To reveal the distribution of VP22a, the blots were probed with MCA406 antibody (b and d). The blots were then incubated in a solution containing 100 mM 2-mercaptoethanol, 2% (wt/vol) SDS, and 62.5 mM Tris-HCl (pH 6.7) at 50°C for 30 min to remove bound antibody and reprobed with DM185 antibody to reveal the distribution of the major capsid protein, VP5 (a and c).
DISCUSSION
It is well established that scaffolding proteins play a role in ensuring accurate and efficient assembly of capsid shells (9). Other functions have also been suggested, including the exclusion of material from the capsid interior during assembly (10) and the masking of DNA binding sites on the inner surface of the procapsid (22). However, the link between the procapsid-capsid transition and the breaking of the interaction between the scaffolding protein and VP5 in HSV suggests that they may have an additional role in stabilizing the procapsid structure.
When they first assemble, the proteins forming the HSV procapsids are apparently not in their most stable state, since they can rearrange to form polyhedral capsids in a process that does not require energy input (58). Studies on temperature-sensitive mutants have shown that the procapsid state can be maintained for several hours (46) but that once the conversion to polyhedral capsids is initiated, it progresses quite rapidly (2). From this it can be concluded that the procapsid shell is maintained in an energetically suboptimal conformation. This has been demonstrated for P22, where head expansion has been shown to be an exothermic process, implying that the mature phage head is in a lower energy state than the procapsid (13). The maintenance of the procapsid shell in this relatively unfavorable state may be explained if the shell and scaffold are considered as a single entity rather than separately. Thus, the scaffold-shell complex would have a minimum-energy state that did not correspond to that of either the shell or the scaffold on its own. This minimum-energy state is the procapsid and represents the natural end point of assembly. When the link between the scaffold and the shell is broken, the constraints on the capsid shell are released. The proteins can then reconfigure into the minimum-energy state for the shell, which corresponds to the polyhedral capsid conformation. The fate of the scaffold would depend on the conditions applying when the scaffold-shell link was broken. If cleavage is accompanied by DNA entry and packaging, the scaffold will dissociate and exit from the capsid to be replaced by the viral DNA. However, if there is no packaging, the scaffolding proteins can refold into their own minimum-energy state, represented by the small core of the B capsid. The ability of the scaffold to maintain the shell in the procapsid configuration may have an additional purpose, since this would ensure that it remains in a suitable condition for DNA packaging for a longer time, thereby enlarging the pool of procapsids available for packaging.
Status of scaffolding proteins during exit from capsids.
The form in which the scaffolding protein exits from the capsid is unknown, as is the route by which it leaves. In bacteriophage P22, the scaffolding protein is believed to be lost before the maturation from prohead to capsid is completed. The P22 prohead has a 30- to 45-Å-diameter channel at the center of each hexon (57). These channels are closed during the rearrangement of the shell proteins that accompanies DNA packaging, and the mature phage head forms a largely unbroken shell around the DNA (64). In HSV the mature capsid shell is not unbroken as in P22 but is perforated by channels through the pentons and hexons (60, 68), leaving open the question of when scaffold exit takes place. Indeed, it has been shown that moderate concentrations of chaotropic agents can remove VP22a from B capsids in the presence of 0.5 M NaCl, without apparently altering the structure of the capsid shell (36). However, by analogy with P22, it seems likely that the HSV scaffold is lost before or during the conversion from procapsid to capsid, as the HSV procapsid shell is more open than that of the mature capsid, with a poorly formed capsid floor and larger channels through the pentons and hexons (37, 58).
Treatment of B capsids at low pH showed that although the characteristic scaffold cores had disappeared, the scaffolding proteins remained inside the capsids. This suggests that the internal scaffold can be disrupted by low pH but either that suitable channels, through which the scaffolding protein can pass, are not present in the B capsid shell or that the individual protein molecules are not in an appropriate form to leave the capsid.
Monomeric P22 scaffolding protein has been suggested to form a molten globule (61). The adoption of a molten globule form would be expected to provide the P22 scaffolding protein with greater flexibility, which might aid its passage through the narrow channels in the prohead. One of the HSV-1 capsid shell proteins (VP23) has already been shown to have the properties of a molten globule (23). However, fluorescence, far-UV CD, and near-UV CD analyses showed that the conditions used to dissociate HSV-1 scaffold particles (1 M urea or pH 5.5) do not induce large alterations in the structure of pre-VP22a. Although there was an increase in ANS fluorescence at pH 5.5 compared to pH 7.2 (Fig. 7), it was not large enough to indicate that the protein had adopted a molten globule state. Therefore, there is no definite evidence that pre-VP22a adopts a classical molten globule structure as a means of exiting from the capsid. However, it should be pointed out that the relatively small magnitude of the near-UV CD spectrum indicates that the interactions involved in stabilizing the tertiary structure of the pre-VP22a protein may be relatively weak, and this degree of structural plasticity could be important for its role in forming a short-lived scaffold.
When purified scaffold particles, at relatively high concentrations (∼1 mg/ml), were maintained at pH 5.5, they tended to precipitate. This was observed, for example, when they were dialyzed to lower their pH for near-UV CD analysis. The propensity to aggregate may reflect the exposure of additional nonpolar sites on the protein as suggested by the increase in ANS binding (Fig. 7). However, when scaffold particles at 1 mg/ml were sedimented through gradients at pH 5.5, the subunits dissociated and aggregation did not take place (Fig. 3 and 4). The most probable explanation is that the subunits were able to separate quickly from each other when they migrated into, and were diluted by, the gradient, as aggregation was not a problem at the lower protein concentrations (∼0.2 mg/ml) used for far-UV CD and fluorescence. It seems likely that a similar process could occur during DNA packaging, as the exit of dissociated scaffolding proteins might be expected to be an efficient process that would remove the subunits before they had the opportunity to aggregate.
Role of DNA packaging in scaffold dissociation.
In order to exit from capsids, scaffolding proteins have to reverse both their self-interaction and their interaction with the capsid shell. In HSV, reversing the interaction with the capsid shell is achieved by proteolysis of pre-VP22a. Studies of mutants with defects in cleavage (2, 14, 40, 46) suggest that proteolysis of the scaffolding protein is normally needed to allow DNA packaging. However, Desai and Person (6) recently described a type of mutant that could still package DNA even though the cleavage site in pre-VP22a had been destroyed. These mutants contain secondary mutations in VP5 that weaken, but do not abolish, its interaction with pre-VP22a. The weakened interaction is still sufficient for capsid assembly while allowing the two intact proteins to separate subsequently to permit DNA packaging. Thus, it appears that it is the disruption of the connection between pre-VP22a and VP5 that is important for DNA packaging and scaffold loss rather than the cleavage of pre-VP22a itself. However, that the disruption of this connection is not sufficient to ensure exit of the scaffold is demonstrated by the existence of polyhedral B capsids containing the cleaved, VP22a, form of the scaffolding protein. Presumably, some additional step is required to ensure dissociation of the scaffold and convert VP22a into a form capable of passing through the shell.
It is well recognized that the nature of protein-protein interactions in virus particles can be significantly affected by changes in pH. For example, studies using the purified major capsid protein of rotavirus, VP6, have shown that its interactions are strongly influenced by pH (25). In this case, the interacting regions of the protein are negatively charged (29), and it is believed that the protonation of side chains at low pH counteracts the repulsive electrostatic forces, thereby allowing the proteins to interact. At pH values in the range of 3.5 to 5.5 VP6 forms spherical particles, while between pH 5.5 and 7 it forms long tubes. Interestingly, VP6 normally assembles on an inner shell of another protein, VP2, which is postulated to induce the formation of spherical capsids by supplying a suitably positively charged electrostatic environment (25). Low pH is also known to be an important factor in the dissociation of several types of virus that enter the cell by endocytosis. For example, during infection by influenza virus, separation of the M protein from the core is an important step in uncoating that is enhanced at the reduced pH found in the endosome (19, 65). In this case, the acidification is due to an influx of H+ ions through the ion channels in the virus envelope rather than to any effect brought about by the genomic RNA. The capsid of foot-and-mouth disease virus, a picornavirus, is also labile at acid pH. Interestingly, empty capsids are less acid labile than full, RNA-containing, capsids, requiring an extra 0.5-unit reduction in pH to cause their dissociation. Thus, the virion RNA appears to be modulating the response of the capsid to lowered pH (3). There are several other examples where the presence of nucleic acid has been proposed to exert a direct influence on protein conformations and interactions during the life cycles of various viruses. For example, Thuman-Commike et al. (56) suggested that the close proximity of significant amounts of negatively charged DNA could break the link between scaffold and shell in P22 procapsids, thereby acting as a trigger for head expansion. Also, Lata et al. (24) speculated that in the double-stranded DNA phage HK97, where head expansion can be induced in vitro by exposure of unexpanded proheads to acid pH, the acidification might mimic processes that occur during insertion of DNA into the phage prohead.
Pre-VP22a has a predicted isoelectric point (pI) of pH 6.62. Therefore, at neutral pH it should have a net negative charge, while at pH values lower than its pI, such as pH 5.5, it should have a net positive charge. From this it follows that relatively small changes in pH would have a large effect on the charge state of surface amino acids of the protein but would not necessarily have a dramatic effect on its overall structural properties. Studies on foot-and-mouth disease virus have demonstrated how pH-induced alterations in the protonation state of a small number of amino acids can overcome the ionic and hydrogen bonding forces holding the structural subunits together and lead to their disassociation (12). Self-interaction in pre-VP22a (5, 38, 42) and its human cytomegalovirus counterpart (62) has been shown to involve only limited portions of the protein, and it is easy to envisage how similar small-scale changes could cause the dissociation of scaffold particles without having any marked effect on the secondary or tertiary structures of the component proteins.
Based on the results described here, we propose that the disassembly of the scaffold core inside procapsids may result from a change in the local charge environment caused by the insertion of the viral genome. DNA is an acidic molecule with a substantial negative charge, which, within cells, is normally neutralized by being bound to basic proteins or to polyamines. Although spermine is found within the capsid in HSV-1 virions and is thought to be important for close packing of the genome, it is not known when it enters the capsid, and the amounts detected are sufficient to neutralize only about half the charges on the DNA molecule (15, 17). During packaging the viral DNA enters into a confined space by passing through a narrow channel, which may shield it from the cellular environment as has been suggested for the double-stranded RNA virus bluetongue virus (18). Recently it has been shown that the φ29 portal complex can exert a large force to counteract the combined resistance provided by the entropic, electrostatic, and bending energies of the packaged DNA (50). A similar portal complex is now known to be present in herpesvirus capsids (35). It is reasonable to assume, therefore, that the charges on the packaged DNA do not have to be fully neutralized. Indeed, it is likely that some residual repulsive force is important for initiating the release of the DNA from the capsid following infection (50). Nothing is known about the conditions within the capsid during DNA packaging, but again it seems reasonable to assume that localized reductions in pH would arise due to incomplete neutralization of the charges on the DNA. The experiments described here have clearly demonstrated that a relatively small reduction in pH induces the rapid dissociation of isolated scaffold particles into their constituent proteins. Equivalent local, and perhaps short-lived, alterations in pH within the capsid may provide a mechanism to drive the dissociation of the scaffold into a low-molecular-weight form capable of exiting through the available channels.
Acknowledgments
D. A. McClelland was supported by Human Frontier Science Program grant RG-537/96.
We thank the Biotechnology and Biological Sciences Research Council of the United Kingdom for provision of the CD facility.
REFERENCES
- 1.Christensen, H., and R. H. Pain. 1991. Molten globule intermediates and protein folding. Eur. Biophys. J. 19:221-229. [DOI] [PubMed] [Google Scholar]
- 2.Church, G. A., and D. W. Wilson. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J. Virol. 71:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curry, S., C. C. Abrams, E. Fry, J. C. Crowther, G. J. Belsham, D. I. Stuart, and A. M. Q. King. 1995. Viral RNA modulates the acid sensitivity of foot-and-mouth disease virus capsids. J. Virol. 69:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dasgupta, A., and D. W. Wilson. 1999. ATP depletion blocks herpes simplex virus DNA packaging and capsid maturation. J. Virol. 73:2006-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai, P., and S. Person. 1996. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology 220:516-521. [DOI] [PubMed] [Google Scholar]
- 6.Desai, P., and S. Person. 1999. Second site mutations in the N-terminus of the major capsid protein (VP5) overcome a block at the maturation cleavage site of the capsid scaffold proteins of herpes simplex virus type 1. Virology 261:357-366. [DOI] [PubMed] [Google Scholar]
- 7.Desai, P., S. C. Watkins, and S. Person. 1994. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J. Virol. 68:5365-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiIanni, C. L., D. A. Drier, I. C. Deckman, P. J. McCann III, F. Y. Liu, B. Roizman, R. J. Colonno, and M. G. Cordingley. 1993. Identification of the herpes simplex virus-1 protease cleavage sites by direct sequence analysis of autoproteolytic cleavage products. J. Biol. Chem. 268:2048-2051. [PubMed] [Google Scholar]
- 9.Dokland, T. 1999. Scaffolding proteins and their role in viral assembly. Cell. Mol. Life Sci. 56:580-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earnshaw, W. C., and S. R. Casjens. 1980. DNA packaging by the double-stranded DNA bacteriophages. Cell 21:312-331. [DOI] [PubMed] [Google Scholar]
- 11.Eftink, M. R., and C. A. Ghiron. 1976. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry 15:672-680. [DOI] [PubMed] [Google Scholar]
- 12.Ellard, F. M., J. Drew, W. E. Blakemore, D. I. Stuart, and A. M. Q. King. 1999. Evidence for the role of His-142 of protein 1C in the acid-induced disassembly of foot-and-mouth disease virus capsids. J. Gen. Virol. 80:1911-1918. [DOI] [PubMed] [Google Scholar]
- 13.Galisteo, M. L., and J. King. 1993. Conformational transformations in the protein lattice of phage P22 procapsids. Biophys. J. 65:227-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, M., L. Matusick-Kumar, W. Hurlburt, S. F. DiTusa, W. W. Newcomb, J. C. Brown, P. J. McCann III, I. Deckman, and R. J. Colonno. 1994. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 68:3703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, W., and B. Roizman. 1971. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA 68:2818-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson, W., and B. Roizman. 1973. The structural and metabolic involvement of polyamines with herpes simplex virus, p. 123-135. In D. H. Russell (ed.), Polyamines in normal and neoplastic growth. Raven Press, New York, N.Y.
- 18.Grimes, J. A., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. C. Mertens, and D. L. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 19.Helenius, A. 1992. Unpacking the incoming influenza virus. Cell 69:577-578. [DOI] [PubMed] [Google Scholar]
- 20.Hong, Z., M. Beaudet-Miller, J. Durkin, R. Zhang, and A. D. Kwong. 1996. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J. Virol. 70:533-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennard, J., F. J. Rixon, I. M. McDougall, J. D. Tatman, and V. G. Preston. 1995. The 25 amino acid residues at the carboxy terminus of the herpes simplex virus type 1 UL26.5 protein are required for the formation of the capsid shell around the scaffold. J. Gen. Virol. 76:1611-1621. [DOI] [PubMed] [Google Scholar]
- 22.King, J., and W. Chiu. 1997. The procapsid-to-capsid transition in double-stranded DNA bacteriophages, p. 288-311. In W. Chiu, R. M. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 23.Kirkitadze, M. D., P. N. Barlow, N. C. Price, S. M. Kelly, C. Boutell, F. J. Rixon, and D. A. McClelland. 1998. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J. Virol. 72:10066-10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lata, R., J. F. Conway, N. Q. Cheng, R. L. Duda, R. W. Hendrix, W. R. Wikoff, J. E. Johnson, H. Tsuruta, and A. C. Steven. 2000. Maturation dynamics of a viral capsid: visualization of transitional intermediate states. Cell 100:253-263. [DOI] [PubMed] [Google Scholar]
- 25.Lepault, J., I. Petitpas, I. Erk, J. Navaza, D. Bigot, M. Dona, P. Vachette, J. Cohen, and F. A. Rey. 2001. Structural polymorphism of the major capsid protein of rotavirus. EMBO J. 20:1498-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, F., and B. Roizman. 1991. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J. Virol. 65:5149-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, F., and B. Roizman. 1991. The promoter, transcriptional unit, and coding sequences of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J. Virol. 65:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsden, H. S., N. D. Stow, V. G. Preston, M. C. Timbury, and N. M. Wilkie. 1978. Physical mapping of herpes simplex virus-induced polypeptides. J. Virol. 28:624-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathieu, M., I. Petitpas, J. Navaza, J. Lepault, E. Kohli, P. Pothier, B. V. V. Prasad, J. Cohen, and F. A. Rey. 2001. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J. 20:1485-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matusick-Kumar, L., W. W. Newcomb, J. C. Brown, P. J. McCann III, W. Hurlburt, S. P. Weinheimer, and M. Gao. 1995. The C-terminal 25 amino acids of the protease and its substrate ICP35 of herpes simplex virus type 1 are involved in the formation of sealed capsids. J. Virol. 69:4347-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newcomb, W. W., and J. C. Brown. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 33.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Q. Cheng, A. Steven, F. Booy, and J. C. Brown. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newcomb, W. W., F. L. Homa, D. R. Thomsen, Z. Ye, and J. C. Brown. 1994. Cell-free assembly of the herpes simplex virus capsid. J. Virol. 68:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 37.Newcomb, W. W., B. L. Trus, N. Q. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelletier, A., F. Do, J. J. Brisebois, L. Lagace, and M. G. Cordingley. 1997. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J. Virol. 71:5197-5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston, V. G., M. F. Al-Kobaisi, I. M. McDougall, and F. J. Rixon. 1994. The herpes simplex virus gene UL26 proteinase in the presence of the UL26.5 gene product promotes the formation of scaffold-like structures. J. Gen. Virol. 75:2355-2366. [DOI] [PubMed] [Google Scholar]
- 40.Preston, V. G., J. A. V. Coates, and F. J. Rixon. 1983. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 45:1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preston, V. G., J. Kennard, F. J. Rixon, A. J. Logan, R. W. Mansfield, and I. M. McDougall. 1997. Efficient herpes simplex virus type 1 (HSV-1) capsid formation directed by the varicella-zoster virus scaffolding protein requires the carboxy-terminal sequences from the HSV-1 homologue. J. Gen. Virol. 78:1633-1646. [DOI] [PubMed] [Google Scholar]
- 42.Preston, V. G., and I. M. McDougall. 2002. Regions of the herpes simplex virus scaffolding protein that are important for intermolecular self-interaction. J. Virol. 76:673-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preston, V. G., F. J. Rixon, I. M. McDougall, M. McGregor, and M. F. Al-Kobaisi. 1992. Processing of the herpes simplex virus assembly protein ICP35 near its carboxy terminal end requires the product of the whole of the UL26 reading frame. Virology 186:87-98. [DOI] [PubMed] [Google Scholar]
- 44.Ptitsyn, O. B. 1995. Molten globule and protein-folding. Adv. Prot. Chem. 47:83-229. [DOI] [PubMed] [Google Scholar]
- 45.Ptitsyn, O. B., R. H. Pain, G. V. Semisotnov, E. Zerovnik, and O. I. Razgulyaev. 1990. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 262:20-24. [DOI] [PubMed] [Google Scholar]
- 46.Rixon, F. J., and D. McNab. 1999. Packaging competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saad, A., Z. H. Zhou, J. Jakana, W. Chiu, and F. J. Rixon. 1999. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol. 73:6821-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semisotnov, G. V., N. A. Rodionova, O. I. Razgulyaev, V. N. Uversky, A. F. Gripas, and R. I. Gilmanshin. 1991. Study of the molten globule intermediate state in protein folding by a hydrophobic fluorescent-probe. Biopolymers 31:119-128. [DOI] [PubMed] [Google Scholar]
- 49.Sherman, G., and S. L. Bachenheimer. 1988. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology 163:471-480. [DOI] [PubMed] [Google Scholar]
- 50.Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson, and C. Bustamante. 2001. The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature 413:748-752. [DOI] [PubMed] [Google Scholar]
- 51.Sreerama, N., and R. W. Woody. 1993. A self consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209:32-44. [DOI] [PubMed] [Google Scholar]
- 52.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 53.Tatman, J. D., V. G. Preston, P. Nicholson, R. M. Elliott, and F. J. Rixon. 1994. Assembly of herpes simplex virus type 1 capsids using a panel of recombinant baculoviruses. J. Gen. Virol. 75:1101-1113. [DOI] [PubMed] [Google Scholar]
- 54.Thomsen, D. R., W. W. Newcomb, J. C. Brown, and F. L. Homa. 1995. Assembly of the herpes simplex virus capsid: requirement for the carboxyl-terminal twenty-five amino acids of the proteins encoded by the UL26 and UL26.5 genes. J. Virol. 69:3690-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thuman-Commike, P. A., B. Greene, J. Jakana, A. McGough, P. E. Prevelige, and W. Chiu. 2000. Identification of additional coat-scaffolding interactions in a bacteriophage P22 mutant defective in maturation. J. Virol. 74:3871-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thuman-Commike, P. A., B. Greene, J. Jakana, B. V. V. Prasad, J. King, P. E. Prevelige, Jr., and W. Chiu. 1996. Three-dimensional structure of scaffolding-containing phage P22 procapsids by electron cryo-microscopy. J. Mol. Biol. 260:85-98. [DOI] [PubMed] [Google Scholar]
- 58.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 59.Trus, B. L., F. L. Homa, F. P. Booy, W. W. Newcomb, D. R. Thomsen, N. Cheng, J. C. Brown, and A. C. Steven. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 69:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. USA 89:11508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tuma, R., and G. J. Thomas. 1997. Mechanisms of virus assembly probed by Raman spectroscopy: the icosahedral bacteriophage P22. Biophys. Chem. 68:17-31. [DOI] [PubMed] [Google Scholar]
- 62.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J. Virol. 71:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woody, R. W., and A. K. Dunker. 1996. Aromatic and cystine side-chain CD in proteins, p. 109-157. In G. D. Fasman (ed.), Circular dichroism and the conformational analysis of biomolecules. Plenum Press, New York, N.Y.
- 64.Zhang, Z. X., B. Greene, P. A. Thuman-Commike, J. Jakana, P. E. Prevelige, J. King, and W. Chiu. 2000. Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J. Mol. Biol. 297:615-626. [DOI] [PubMed] [Google Scholar]
- 65.Zhirnov, O. P. 1990. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology 176:274-279. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, Z. H., J. He, J. Jakana, J. D. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026-1030. [DOI] [PubMed] [Google Scholar]
- 68.Zhou, Z. H., B. V. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]
- 69.Zweig, M., C. J. Heilman, and B. Hampar. 1979. Identification of disulfide-linked protein complexes in the nucleocapsids of herpes simplex virus type 2. Virology 94:442-450. [DOI] [PubMed] [Google Scholar]