Abstract
Reverse transcription in avian sarcoma virus (ASV) initiates from the 3′ end of a tRNATrp primer, which anneals near the 5′ end of the RNA genome. The region around the primer-binding site (PBS) forms an elaborate stem structure composed of the U5-inverted repeat (U5-IR) stem, the U5-leader stem, and the association of the tRNA primer with the PBS. There is evidence for an additional interaction between the viral U5 RNA and the TψC loop of the tRNATrp (U5-TψC). We now demonstrate that this U5-TψC interaction is necessary for efficient replication of ASV in culture. By randomizing specific biologically relevant regions of the viral RNA, thereby producing a library of mutant viruses, we are able to select, through multiple rounds of infection, those sequences imparting survival fitness to the virus. Randomizing the U5-TψC interaction region of the viral RNA results in selection of largely wild-type sequences after five rounds of infection. Also recovered are mutant viruses that maintain their ability to base pair with the TψC loop of the tRNATrp. To prove this interaction is specific to the tRNA primer, we constructed a second library, in which we altered the PBS to anneal to tRNAPro, while simultaneously randomizing the viral RNA U5-TψC region. After five rounds of infection, the consensus sequence 5′-GPuPuCPy-3′ emerged, which is complementary to the 5′-GGTTC-3′ sequence found in the TψC loop of tRNAPro. These observations confirm the importance of the U5-TψC interaction in vivo.
Shortly after entering a susceptible host cell, the RNA genome of avian sarcoma virus (ASV) is reverse transcribed into a double-stranded DNA, capable of undergoing integration and all subsequent steps of the viral life cycle. Reverse transcription utilizes a host tRNATrp annealed near the 5′ end of the viral genome as the primer. Regulation of this reverse transcription is largely at the level of initiation and is related to the presence of multiple stem structures in the viral RNA surrounding the primer-binding site (Fig. 1). These structural elements have been preserved in a wide variety of retroviruses and transposable genetic elements (4, 6, 8, 10, 12; for a review, see reference 11). In ASV, several of these structures have been extensively studied using viral infectivity assays and in vitro reverse transcription analysis (1, 2, 6, 7, 13). One of the less understood interactions is that of the viral U5 RNA with the TψC loop of the tRNATrp primer. It has been shown that base substitutions into this region of the U5 RNA that block interactions with the TψC loop of tRNATrp negatively impact virus replication in culture as well as initiation of reverse transcription reconstituted in vitro (1). These studies have shown that this region of the viral RNA is important for replication, but they examined only a small number of specific mutations and therefore have not been able to confirm the mechanism for these defects.
FIG. 1.
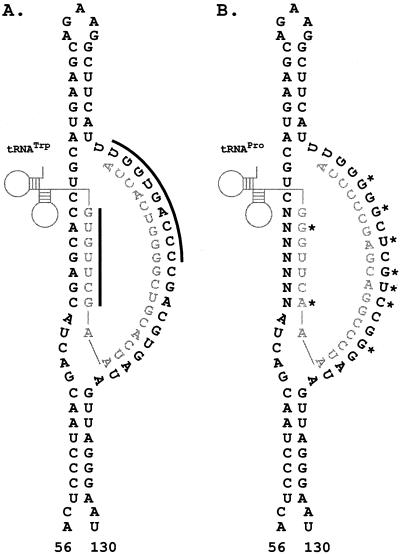
Secondary structure around the PBS of ASV RNA. (A) The nucleotide sequence of wild-type ASV, nucleotides 56 to 130 (boldface type), is shown annealed to the 3′ end of tRNATrp (in gray type). The solid bars indicate ASV RNA sequences in either the PBS or the TψC interaction region that were randomized. (B) The nucleotides of ASV RNA, altered to be complementary to tRNAPro (stars), annealed to the tRNAPro are shown. N, randomized position in viral RNA.
We have adopted a mutagenic technique that allows one to analyze in vivo, simultaneously, many possible mutations that can be made within a small biologically important region of the viral genome (5). This method involves placing randomized sequences into an infectious plasmid clone of ASV, transfecting the library into turkey embryo fibroblasts (TEFs), and allowing the virus to replicate. Recovered viruses are serially passaged, and the range of biologically acceptable sequences and/or structures is determined by direct sequencing methods. We have applied this method to examine the primer-binding site (PBS) and U5-TψC regions of the ASV genome and the potential relationship between the two. When randomizing the U5-TψC region of the viral RNA, sequences recovered after several rounds of infection included both wild-type as well as mutant sequences that maintain the ability to base-pair to the TψC loop of the tRNATrp primer. These results suggest that while the wild-type TψC-interacting sequence in the ASV genome is not absolutely required for virus replication, maintaining the ability to form this structure provides a growth advantage to the virus. Furthermore, when the PBS sequence was modified to accept tRNAPro as the primer, and the randomized U5-TψC region was subjected to selection, those bases selected from the pool were predicted to maintain base pairing to the TψC loop tRNAPro instead of that to the wild-type tRNATrp. This shows conclusively that the selection of this region of the viral RNA is not directly for sequence but is for maintaining the U5-TψC structure.
MATERIALS AND METHODS
Reagents and cells.
All enzymes were purchased from New England Biolabs. Deoxynucleotides were purchased from Boehringer Mannheim. Oligodeoxynucleotides were obtained from Genosys Biotechnologies, Inc., and from Integrated DNA Technologies, Inc. Escherichia coli DH10B bacteria and Lipofectamine transfection reagent were purchased from Gibco BRL. Primary TEFs, which do not contain endogenous retroviruses capable of recombining with ASV, were a generous gift from Rebecca Craven (Pennsylvania State Medical Center). The plasmids pDC101S and RCAS have been described previously (7, 9, 13). pDC101S.linker was created by removing a 206-bp SalI/SacI fragment encompassing nucleotides 52 to 259 of the ASV viral genome (including the PBS and surrounding stem structure) and replacing it with a small linker fragment which preserves the SalI and SacI sites and inserts a unique KpnI site to the vector. RCAS.Δ.3′PBS was derived from RCAS by removing an RsrII/EcoRV fragment immediately downstream of R in the 3′ long terminal repeat (LTR), thereby deleting a second PBS found in the original RCAS vector. RCAS.linkerΔ.3′PBS was created by removing a BsmI/SacI fragment, encompassing the 5′ LTR, from RCAS.Δ.3′PBS, and replacing it with a small linker fragment which preserves the BsmI and SacI sites and inserts a unique SpeI site to the vector.
Construction of randomized libraries.
Wild-type pDC101S was amplified with mutagenic primers to create two distinct randomized PCR products, one with a randomized PBS (rPBS) and one with a randomized U5-TψC interaction region (rU5-TψC). After amplification, the PCR products were purified by agarose gel electrophoresis using the QIAquick agarose gel extraction kit (Qiagen), digested with SacI and SalI, and treated with shrimp alkaline phosphatase to remove the 5′ phosphate groups. PDC101S.linker was digested with SalI and SacI, and the linker was removed using Microcon 50 spin columns (Amicon Bioseparations). The mutant inserts were ligated to the digested vector, using T4 DNA ligase and an insert/vector ratio of 4:1. After ligation, the reaction mixtures were heated to 65°C for 20 min to inactivate the T4 ligase, digested with KpnI to linearize any residual pDC101S.linker (without insert), and electroporated into E. coli DH10B. Each plasmid was prepared from bacterial cultures and sequenced to confirm the location of the randomization. RCAS.linkerΔ.3′PBS and the randomized pDC101S plasmids were digested with BsmI and SacI. The linker fragment was removed from the RCAS.linkerΔ.3′PBS digestion reaction mixture as described above. The BsmI/SacI inserts cleaved from the mutant pDC101S plasmids were gel purified, treated with shrimp alkaline phosphatase, and ligated to the digested RCAS vector as described above. After ligation, the plasmid DNA was digested with SpeI to linearize any residual RCAS.linker.Δ3′PBS. This product was then electroporated into E. coli DH10B. Plasmid DNA was prepared using the Qiagen EndoFree Plasmid Maxi kit.
Construction of rPBS and rU5-TψC randomized libraries in the context of tRNAPro as primer.
Mutagenic primers and the SacI/SalI sites of pDC101S were used to create pDC101S.PBS-Pro, a plasmid where the PBS has been modified to be complementary to tRNAPro. pDC101S.TψC-Pro, a plasmid in which the U5-TψC interaction region has been modified to be complementary to tRNAPro, was created in the same fashion. The mutant libraries with either a randomized PBS or U5-TψC interaction region in the context of a tRNAPro primer were created as described above except that pDC101S.PBS-Pro and pDC101S.TψC-Pro were used as the base plasmids from which the mutagenic inserts were amplified.
Transfection and infection of cells.
TEFs were transfected with the mutant RCAS.linker.Δ3′PBS plasmids using the Lipofectamine PLUS reagent according to manufacturer's instructions. Cells (2 × 106) in a 100-mm-diameter dish were transfected with 8 μg of DNA. One to two days postlipofection, the medium on the cells was changed in preparation for a 24- to 48-h virus collection period. Three days postlipofection, mutant virus was harvested from the medium by centrifugation through 20% sucrose gradients in the presence of STE (0.1 M NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) at 4°C for 90 min at 26,000 rpm in a Spinco SW27 rotor. Virus pellets were suspended in STE, and aliquots were assayed for reverse transcriptase (RT) activity as described previously (7). Mock-transfected control cells were treated in an identical fashion. Equal quantities of virus (as measured by RT activity) in 1 ml of serum-free Dulbecco's modified Eagle medium were used to infect Polybrene-treated (5 μg/ml) TEFs for 1 h (2 × 106 cells in 100-mm-diameter dishes). At three days postinfection, virus was harvested from the medium, as was done following lipofection. Serial passage into uninfected TEFs was performed after each harvest and continued through five rounds of infection, or until the library reverted to wild type.
Analysis of selected viral sequences.
Infected TEFs were trypsinized, washed with phosphate-buffered saline, suspended in 200 μl of phosphate-buffered saline, and frozen at −20°C. Cellular DNA was purified from these cell samples using the QIAamp tissue kit from Qiagen. The region of viral DNA surrounding the randomized region was PCR amplified from the cellular DNA. The PCR products, which represent the pool of sequences still present within the library, were purified from a 1% agarose gel using the QIAquick agarose gel extraction kit and sequenced using the Thermo Sequenase radiolabeled terminator cycle sequencing kit from U.S. Biochemical Corporation. Equivalent molar amounts of each PCR product were used in each sequencing reaction mixture so that direct comparisons could be made between samples. Additionally, the purified PCR products were digested with SacI and ligated into pUC19 linearized with SacI. Ligation products were electroporated into E. coli DH10B, and the transformed bacteria were plated onto ampicillin selection medium. Individual colonies were picked, suspended in 10 μl of distilled water, heated to 95°C for 5 min, and cooled on ice for 5 min, and debris was removed by centrifugation for 3 min. DNA from each of these colony preparations was individually sequenced.
RESULTS
Creating randomized libraries and maintaining a representative sample of each randomized pool.
To investigate the role of the PBS and the U5 RNA-TψC interaction region in virus replication, we introduced random sequences into these RNA elements and allowed the virus to select from the random libraries those sequences that supported replication. Random libraries were assembled by a PCR-based mutagenesis technique using PCR primers with specifically randomized sequences, a wild-type pDC101S template (7), and an acceptor pDC101S.linker plasmid with a unique KpnI site introduced into a SalI/SacI fragment (see Materials and Methods). After ligation of the randomized PCR product inserts into the pDC101S.linker acceptor plasmid, each sample of ligated DNA was digested with KpnI so that only circular ligation products with a randomized insert were propagated in bacteria. Because a pDC101S-based vector cannot produce infectious virus, the randomized sequences were shuttled into RCAS (a virus vector) (7, 9). We created an RCAS.linker vector in which we removed the second copy of the PBS (downstream of the 3′ LTR) as well as the U5 sequences of the 3′ LTR and inserted a unique SpeI site into a BsmI-SacI linker fragment (see Materials and Methods). Removal of these sequences is crucial, because we found that in the presence of the second PBS, recombination events that recover wild-type sequence from the 3′ LTR, rather than by selecting it from the library, are possible. A pool of BsmI-SacI fragment derived from the pDC101S library was treated with phosphatase and ligated to RCAS cut with BsmI and SacI. The ligation products were then digested with SpeI prior to transformation of bacteria again to ensure that only circular products with the randomized inserts were propagated. Even if recircularized linker plasmids somehow escape linearization with KpnI (pDC101S-based ligations) or SpeI (RCAS-based digestions) and make it into the final RCAS pools, they would be replication incompetent, because important regions of the 5′ LTR were missing. This cloning strategy ensures that when wild-type ASV sequences are recovered in viable viruses, they were derived from the library and not from a contaminant.
Our largest randomized region encompasses 9 nucleotides, which results in a library of over 106 possible sequence variations. Therefore, for the selection experiments to produce a complete data set, we must adequately sample well over 106 possibilities at every step in the experimental process. This includes during cloning, when each vector library is ligated and propagated in bacteria; during transfection of TEFs with the vector libraries; and during infection of TEFs with the virus libraries. With respect to cloning of the libraries, each ligation reaction mixture contained a significant excess of both vector and insert, so that at least 108 molecules of each was represented. After the ligation products were electroporated into bacteria, the number of individual colonies in each electroporated bacterial culture was estimated by plating a fraction of the culture and counting the colonies. The number of colonies in the vector-plus-insert culture was compared to the number of colonies produced from the culture of negative control vector alone (ligation reaction mixtures in which no insert was included). This method estimated the number of individual colonies, and presumably individual sequences, in each bacterial pool and verified that each possible sequence combination was represented in the vector library harvested from the bacteria.
To ensure adequate library sampling during the transfection step during which the RCAS-based vector libraries were introduced into TEFs using a highly efficient lipofection technique, an estimate of transfection efficiency was made using a β-galactosidase expression vector in which the ASV LTR drove β-galactosidase expression. In parallel with the RCAS-based vector libraries, TEFs were transfected with the β-galactosidase vector. The cells were then stained for β-galactosidase expression so that the transfection efficiency was estimated. This allowed us to verify that an adequate number of cells was transfected to have a representative sampling of each library.
Several days posttransfection or postinfection, the virus libraries were harvested and used to infect a fresh plate of previously uninfected TEFs. At the same time, wild-type virus was harvested from our virus producer QT6 quail cell lines and used as a positive control to monitor the course of infection and estimate the multiplicity of infection (MOI). Experiments using wild-type virus and TEFs provided an estimate of the ratio of RT activity to 50% tissue culture infective dose (a measure of infectious virus). Calculation of the MOI in this manner verified that adequate sampling of each library was achieved during the infection process. In addition, by using a low MOI (0.1) and choosing tissue culture lines (QT6 and TEF) that do not contain endogenous virus sequences related to ASV, we significantly reduce the possibility of recombination between the mutant viral libraries and exogenous or endogenous viral sequences.
Randomization of the PBS.
In ASV, the 18 nucleotides of the PBS are complementary to tRNATrp. If we randomized all 18 nucleotides of the PBS in a single library, there would be too many sequence possibilities to adequately screen the library. We therefore targeted the 5′ half of the PBS, which is complementary to the 3′ end of the primer (Fig. 1). As such, there should be a very strong selection for sequences in the viral RNA that maintain the proper positioning of the 3′ hydroxyl end of the primer used to initiate minus strand synthesis. In fact, the selection is strong enough that wild-type sequence dominated the library after only a single round of infection (Fig. 2). The presence of wild type was confirmed by sequencing several individual clones isolated from three replicates of selected sequences. Thus, there was no tolerance for mismatch base pairing between the 5′ half of the PBS and the 3′ end of the tRNATrp. The results from this library also demonstrated that this in vivo mutagenesis procedure is capable of rapidly returning wild-type sequences when they are required by the virus.
FIG. 2.
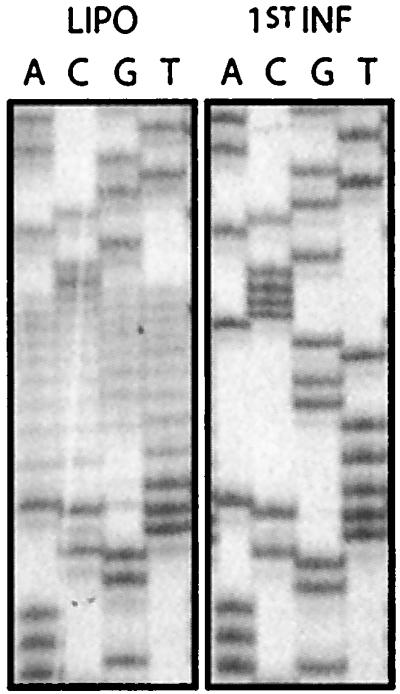
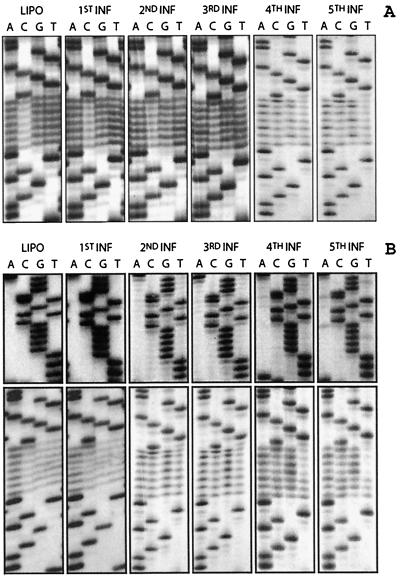
Sequences recovered from the pool of viruses derived from a library in which the 5′ half of the primer binding site was randomized. The PBS sequences randomized in the viral RNA are presented in Fig. 1A. “LIPO” refers to the randomized library transfected into cells. “1st INF” and refers to the pool recovered after the first round of infection derived from the transfected plate. Viral DNA was recovered from cells and sequencing as described in Materials and Methods.
Randomization of the U5-TψC interaction region.
We have previously shown that block mutations within the U5-TψC region of ASV RNA (nucleotides 71 to 77) results in a defect in growth and initiation of reverse transcription (1). In the present study, random sequences were introduced into the U5-TψC-interacting region (Fig. 3A) in order to examine the range of sequences capable of supporting virus growth in vivo. In contrast to the results observed with targeting the PBS, a significant amount of sequence variation is tolerated at these RNA nucleotide positions, even after five rounds of infection (Fig. 3A); however, analysis of the pool of sequences derived from the fifth round of replication, indicates a preference for wild-type bases at some positions (i.e., 72G, 73A, 74G, and 77C). Nineteen individual clones were isolated from the pool and sequenced (Table 1). Five of nineteen clones returned wild-type bases at all seven positions. Two additional clones were able to maintain the base pairing at all seven positions. When examining the selected mutant sequences more carefully, it was found that several additional clones could base pair to the TψC loop if the base pairing used nucleotides that were directly adjacent to and including bases in the U5 RNA TψC interaction region. The range of the base pairing on the viral RNA is indicated for each clone (Table 1). In the wild type, positions 71 to 77 are involved in this interaction, whereas the most extreme shifting resulted in the interaction with positions 69 to 75 or 73 to 79. Thus, in 80% of the clones that were sequenced, the potential for base pairing between the U5 RNA and the TψC arm of the tRNA is maintained. The fact that there are four clones with minimal potential to base pair argues that this interaction provides a growth advantage but is not absolutely required for initiation of reverse transcription.
FIG. 3.
Sequences recovered from the pool of viruses derived from a library in which the U5-TψC interaction region was randomized. The sequences of the ASV U5 TψC-interaction region that were randomized are as described in the legend to Fig. 1. All notations are as described in legend to Fig. 2. Designations such as 1st INF and 2nd INF refer to the pool recovered after the respective round of infection. (A) The U5-TψC randomization is done in a wild-type background. (B) The U5-TψC randomization is done in virus which has been altered to accept a tRNAPro as the priming tRNA. In the top panel the sequence of the PBS is shown for each round of infection, demonstrating that use of the alternate tRNA is maintained.
TABLE 1.
Sequences of individual clones from the randomized U5 RNA TΨC interaction region library
| AGCTTGTGCa | Shiftc | Predicted size of bp regiond | U5 RNA nucleotides |
|---|---|---|---|
| ACGAGCACCb | 0 | 7 | 71-77 |
| ACGAGCACC | 0 | 7 | 71-77 |
| ACGAGCACC | 0 | 7 | 71-77 |
| ACGAGCACC | 0 | 7 | 71-77 |
| ACGAGCACC | 0 | 7 | 71-77 |
| ATGAGCACC | 0 | 7 | 71-77 |
| ACGAACGCC | 0 | 7 | 71-77 |
| TCGGACTGC | +3 | 6 | 73-78 |
| CTAGACGCA | −2 | 6 of 7 | 69-75 |
| CTAGACACG | −2 | 6 of 7 | 69-75 |
| ACGGACTGC | +3 | 5 | 74-78 |
| TGCAGCGCT | +1 | 5 | 74-78 |
| TAGAGCCTG | +2 | 5 of 6 | 74-79 |
| TAGAGTCTC | −1 | 5 of 6 | 71-76 |
| GTAGACCTG | +2 | 5 of 7 | 73-79 |
| ATGTAGTTC | |||
| ACGCAAAAC | |||
| ACGCAAAAC | |||
| ATAATAGCC |
The sequence of the TΨC loop of the tRNATrp primer that can base pair with U5 RNA and which is presented as deoxynucleotides for sequence comparison.
Sequences obtained from selected clones. The underlined area indicates bases that were randomized in the original library. The bases in boldface type are those that can still base pair to the TΨC loop of the tRNATrp.
Direction and size of the shift required to maintain base pairing between the TΨC loop of the tRNATrp and U5 RNA from individual clones selected.
Predicted number of base pairs. “6 of 7” denotes the potential to form 6 bp within a 7-nucleotide sequence.
Randomization of the U5-TψC interaction region in virus clones that utilize tRNAPro.
Selection of non-wild-type nucleotides, which maintain the ability to base pair to the TψC arm of the tRNATrp, strongly suggests that there is a selective pressure to maintain this interaction. To prove that this is the case, we have engineered a library in which the PBS was mutated to accept tRNAPro as the primer (Fig. 1B), and we simultaneously randomized the U5-TψC region of the viral RNA. As seen in Fig. 3B, there is again a significant variation in the sequences recovered after five rounds of infection. The PBS sequences remain stable and complementary to tRNAPro throughout all five rounds of infections (Fig. 3B, top panel). By comparing the data in Fig. 3A and B, subtle differences can be seen between the pools recovered from the two libraries. For example, a preference towards A and G is seen at position 75 from the PBS-Pro rTψC library, whereas a stronger C preference is seen at that position in the rTψC library; however, the real differences between these libraries become obvious only after examining individual clones. The sequences recovered from the PBSPro rTψC library are shown in Table 2. These differ considerably from the sequences selected with the wild-type PBS shown in Table 1. Aligning the sequences reveals that a consensus sequence of G-Pu-Pu-C-Py was maintained in 20 of 24 sequenced clones. This consensus sequence has the potential to base pair to the G-G-G-U-U-C-A sequence found in the TψC loop of tRNAPro and not the G-U-G-U-U-C-G sequence found in tRNATrp. Again, as seen with the tRNATrp experiments, maintenance of base pairing between the viral five-base consensus sequence and tRNAPro required, in many cases, the shifting of the relative position of annealing up or down the viral RNA by a few nucleotides. These shifts are indicated in Table 2. The most extreme shifts utilize viral nucleotides 71 to 76 or nucleotides 76 to 79, as opposed to the wild-type region of nucleotides 71 to 77, so again these shifts are small and not expected to greatly distort the overall structure of the RNA complex. In all 24 clones, at least four nucleotides were able to base pair with the TψC loop.
TABLE 2.
Sequences of individual clones from the PBS-Pro rTΨC libraryb
| AACTTGGGCa | Shift | Predicted size of bp region | U5 RNA nucleotides |
|---|---|---|---|
| TAGGACCCGb | −1 | 7 | 71-77 |
| ACGAGCCTG | +2 | 7 | 74-80 |
| CCGGGCCTG | +2 | 7 | 74-80 |
| TAGAACCCA | −1 | 6 | 71-76 |
| TAGAACCCA | −1 | 6 | 71-76 |
| CGGAACCCT | +1 | 6 | 73-78 |
| AAGAACCCT | +1 | 6 | 73-78 |
| ACGGACCCT | +1 | 6 | 73-78 |
| CGGGACCCT | +1 | 6 | 73-78 |
| AAGAACTCC | 0 | 6 | 72-77 |
| CTGGACTGC | +3 | 6 | 74-79 |
| GAGCACCTG | +2 | 6 of 7 | 74-80 |
| ACGAGCTGC | +3 | 5 | 75-79 |
| ACGAGCTGC | +3 | 5 | 75-79 |
| CGGAGCTGC | +3 | 5 | 75-79 |
| ACGAGCTGC | +3 | 5 | 75-79 |
| CGGGACTGC | +3 | 5 | 75-79 |
| CGGGACTGC | +3 | 5 | 75-79 |
| CCGGACTGC | +3 | 5 | 75-79 |
| GCGGGCTGC | +3 | 5 | 75-79 |
| TCGGGCTGC | +3 | 5 | 74-78c |
| GATAACCCT | +1 | 5 | 74-78 |
| ACGAACACC | 0 | 5 of 6 | 72-77 |
| GACAGCTGC | +3 | 4 | 76-79 |
The sequence of the TΨC loop of the tRNAPro primer that can base pair with U5 RNA and which is presented as deoxynucleotides for sequence comparison. Nucleotides differing from the TΨC loop of the tRNATrp are shown in boldface type.
Notations are explained in the legend to Table 1.
Numbering 74-78 instead of 75-79 is because one of the upstream randomized bases was deleted in this clone.
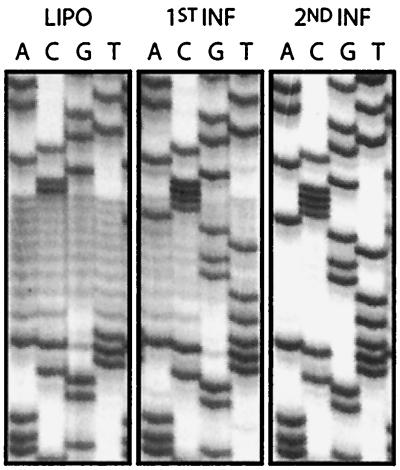
To complement the results described above, we have created an additional library, in which the viral RNA U5-TψC region was mutated to be complementary to the TψC loop of tRNAPro, and the 5′ end of the PBS was simultaneously randomized. In contrast to the results shown in Fig. 2, selection of exclusively the wild type was delayed to the second round of infection (Fig. 4). This indicates that even selection of the critical PBS sequences is affected by the nucleotides present within the U5-TψC region.
FIG. 4.
Sequences recovered from the pool of viruses derived from a library where the primer binding site was randomized in an RNA containing U5 sequences complementary to the TψC loop of tRNAPro. All notations are as indicated in the legend to Fig. 3.
DISCUSSION
The primary interaction between the tRNATrp and the viral RNA is the 18 bases of the primer binding site. Mutations in the primer-binding site that disrupted this interaction are lethal to virus growth. In the genetic analysis presented here, randomization of the 5′ half of the PBS, which is complementary to the 3′ end of the tRNA primer, resulted in immediate selection of wild-type clones from the virus library. This was expected since viral clones that are unable to initiate reverse transcription would be eliminated from the library before completion of even a single round of infection. This does not appear to be the case when the interaction between the primer tRNA TψC loop and the U5 viral RNA is disrupted. Previously, it had been shown that base substitutions that disrupt this base pairing caused a partial defect to viral replication in vivo (1). The virus grew but at lower rates. The defect could be reproduced using a reconstituted initiation of reverse transcription system (1). The lack of an absolute requirement for the maintenance of the U5-TψC interaction suggested that it is perhaps not part of the stable stem structure required at the initiation site. Instead, we theorized that this interaction could be directing the tRNATrp onto the correct primer binding site in the 5′ LTR, rather than several other regions within the viral RNA, which are similar enough in sequence to the PBS to base pair with tRNATrp. Since only properly annealed tRNAs can initiate reverse transcription, additional signals directing the tRNA to the proper site should increase the efficiency of assembly. An interaction such as the U5-TψC therefore may simply increase the efficiency of proper assembly, which would not be absolutely required for virus replication, but which would provide a growth advantage for viruses maintaining this interaction.
We have found that randomization of the U5-TψC interaction region allows for considerable sequence diversity even after five rounds of infection. Although wild-type sequence was selected in approximately 20% of the U5-TψC clones, they did not dominate the library as was seen when mutating the PBS. In order to understand this phenomenon, we examined the non-wild-type bases after five rounds of infection. In most of these mutant viruses, base pairing to the TψC loop of the primer was maintained, although in some cases this resulted from shifting the primer interaction a few bases up or down the U5 RNA. This suggests that small changes in the positioning of the TψC loop interaction with the U5 portion of the ASV RNA can be tolerated. Larger shifts in the TψC interaction would likely be prevented by the close proximity of the U5 leader stem and the U5-inverted repeat (U5-IR) stem.
To confirm that selection of the U5 sequences of the U5-TψC interaction is based on the requirement for base pairing to the TψC loop of the primer tRNA, we engineered a virus clone which could use tRNAPro to prime reverse transcription. tRNAPro was chosen for these experiments because it is present in an enriched concentration within the virus particle, thereby removing primer availability as a limiting factor for reverse transcription initiation with a non-wild-type tRNA. Within the context of the tRNAPro clone, we randomized the U5-TψC region. As in the TψC library, we found that a great deal of degeneracy was tolerated in the selected sequences after five rounds of infection. Again, we examined the mutant sequences and found that many of these clones could base pair to the TψC loop of tRNAPro with selection of interactions with as many as 6 or 7-base pairs. These clones, for the most part, lost the ability to base pair to the TψC loop of tRNATrp. Therefore, selection of the U5-TψC region was dependent on the sequence of the TψC loop, and the ability to base pair to this region of the tRNA was preserved regardless of which tRNA primer was used to initiate reverse transcription. In all of these clones, the mutant PBS was maintained through five rounds of infection (Fig. 3B, top panel). As an additional control, we infected TEFs with a mutant virus containing tRNAPro PBS with a wild-type TψC interaction region. These viruses also maintained the alternate tRNA usage through five rounds of infection (data not shown). This is consistent with the work of Whitcomb et al. (16), who have shown that tRNAPro can function for up to 15 rounds of infection in chicken embryo fibroblasts before reversion to use of the wild-type tRNA. It is likely with the added changes in the TψC interaction domain that usage of the alternate tRNA could persist for much longer. Also adding to the stability of the alternate tRNA usage in this study is the lack of ASV-related endogenous retroviral sequences in TEFs (see Materials and Methods).
This is the first study which clearly shows that in vivo, the role of specific ASV sequences upstream of the PBS is to base pair to the TψC loop of the primer. The fact that shifting of the interaction up or down the viral RNA does not cause a defect in initiation was somewhat unexpected, because previous studies showed that contorting the stem structure would cause a defect in viral growth (2). However, if the interaction was transient or weak, as suggested by previous nuclease mapping experiments (14), and only required for initial placement of the tRNA onto the correct PBS sequence, then shifting of the TψC interaction would not affect the overall stem structure. This type of biological role for the U5-TψC interaction region would explain previous data from in vitro reverse transcription initiation experiments, where the TψC interaction was found to be much less important when using very short RNA templates compared to longer templates. This was presumably because it was easier for the tRNA to find the correct priming location on a shorter RNA (unpublished observations). In addition, if the primary role of the U5-TψC interaction region was to increase the efficiency of proper tRNA placement on the viral RNA, then the lack of this interaction would not prevent virus replication, but simply decrease the efficiency of replication, as shown by previous in vivo data.
Another factor that may drive the selection of clones with a shifted interaction is that in the PBSPro-TψC library, many of the clones with a +3-shifted interaction use two nonmutated flanking nucleotides in the 5-bp interaction they maintain. This means that the virus needs only to select appropriate nucleotides at three positions (two of which need only to select a purine) so that 1 in 16 of the clones from the input library would meet the criteria for this interaction. The fact that these clones did not immediately dominate the library suggests that they are likely at least partially defective, but are still able to replicate well enough to persist through five rounds of infection.
The relative importances of the PBS and U5-TψC interaction in reverse transcription initiation and viral replication are reflected in how quickly certain sequences were selected in vivo after randomization of each region. The PBS is absolutely required for replication, a situation reflected in the fact that predominantly wild-type sequences were selected from the randomized PBS virus libraries after only one round of infection. In contrast, the U5-TψC interaction region, which we believe plays a role primarily during tRNA placement, and therefore is not absolutely required for replication, did not show definitive sequence selection until multiple rounds of selection were complete. This model of ASV reverse transcription initiation is supported by studies in HIV-1, which have found that secondary interactions between the tRNA primer and U5 sequences enhance proper placement of the tRNA and stabilize its interaction with the viral RNA (3). In the case of HIV-1, two interactions between the primer and the viral RNA are required for virus growth. There is a primary interaction between the 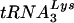 primer and the PBS and a secondary interaction between anti codon loop of the primer and the loop region of the U5-IR stem (7). When both the PBS and the U5-IR stem-loop are altered to be complementary to tRNAHis, the virus grows and stably maintains the use of the new tRNA during successive rounds of viral replication. If only the PBS is changed, the virus reverts to using the wild-type
primer and the PBS and a secondary interaction between anti codon loop of the primer and the loop region of the U5-IR stem (7). When both the PBS and the U5-IR stem-loop are altered to be complementary to tRNAHis, the virus grows and stably maintains the use of the new tRNA during successive rounds of viral replication. If only the PBS is changed, the virus reverts to using the wild-type 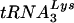 to replicate (15). Recently, it has been proposed that an additional interaction between the TψC loop of the primer RNA and a portion of the HIV-1 leader stem is involved in activation of reverse transcription (4). Both the primary and secondary interactions between the viral RNA and tRNA are similar in the avian and HIV model systems. In both cases the TψC interaction seems to be related to the very early stages of initiation, either by proper positioning of the tRNA or by signaling for initiation. While HIV maintains the additional interaction with the anticodon loop that is absent in the avian virus, the similarities observed with the TψC interaction demonstrate some level of conserved mechanism between different families of retroviruses.
to replicate (15). Recently, it has been proposed that an additional interaction between the TψC loop of the primer RNA and a portion of the HIV-1 leader stem is involved in activation of reverse transcription (4). Both the primary and secondary interactions between the viral RNA and tRNA are similar in the avian and HIV model systems. In both cases the TψC interaction seems to be related to the very early stages of initiation, either by proper positioning of the tRNA or by signaling for initiation. While HIV maintains the additional interaction with the anticodon loop that is absent in the avian virus, the similarities observed with the TψC interaction demonstrate some level of conserved mechanism between different families of retroviruses.
Acknowledgments
This work was supported in part by United States Public Health grants CA38046 and CA52047 (to J.L.) and CA43600 (to E.S.). S.M. is supported by grant GM07250 from the National Institutes of Health. M.J. is supported by Carcinogenesis Training Program grant CA09560 from the National Institutes of Health.
We thank Rebecca Craven for the generous gift of primary turkey embyro fibroblasts.
REFERENCES
- 1.Aiyar, A., D. Cobrinik, Z. Ge, H.-J. Kung, and J. Leis. 1992. Interaction between retroviral U5 RNA and the TψC loop of the tRNATrp primer is required for efficient initiation of reverse transcription. J. Virol. 66:2464-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar, A., Z. Ge, and J. Leis. 1994. A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J. Virol. 68:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beerens, N., B. Klaver, and B. Berkhout. 2000. A structured RNA motif is involved in correct placement of the tRNA3Lys primer onto the human immunodeficiency virus genome. J. Virol. 74:2227-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beerens, N., F. Groot, and B. Berkhout. 2001. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 276:31247-31256. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B., and B. Klaver. 1993. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res. 22:5020-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobrinik, D., L. Soskey, and J. Leis. 1988. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J. Virol. 62:3622-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobrinik, D., A. Aiyar, Z. Ge, M. Katzman, H. Huang, and J. Leis. 1991. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J. Virol. 65:3864-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freund, F., F. Boulme, S. Litvak, and L. Tarrago-Litvak. 2001. Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNA(Lys3) duplex. Nucleic Acids Res. 29:2757-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes, S. H., J. Greenhouse, J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeney, J. B., K. B. Chapman, V. Lauermann, D. F. Voytas, S. U. Astrom, U. von Pawel-Rammingen, A. Bystrom, and J. D. Boeke. 1995. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol. Cell. Biol. 15:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leis, J., A. Aiyar, and D. Cobrinik. 1993. Regulation of initiation of reverse transcription of retroviruses, p. 33-47. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Lin, J. H., and H. L. Levin. 1998. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem-loop of retroviruses. Mol. Cell. Biol. 18:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, J., Z. Ge, S. Morris, K. Das, and J. Leis. 1997. Multiple biological roles associated with the Rous sarcoma virus 5′ untranslated RNA U5-IR stem and loop. J. Virol. 71:7648-7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris, S., and J. Leis. 1999. Changes in Rous sarcoma virus RNA secondary structure near the primer binding site upon tRNATrp primer annealing. J. Virol. 73:6307-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakefield, J. K., S. M. Kang, and C. D. Morrow. 1996. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J. Virol. 70:966-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitcomb, J., B. Ortiz-Conde, and S. Hughes. 1995. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternative tRNAs as primers. J. Virol. 69:6228-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]