Abstract
Hepatitis C virus (HCV) replicates from a ribonucleoprotein (RNP) complex that is associated with the endoplasmic reticulum (ER) membrane. The replication activities of the HCV subgenomic replicon are shown here to induce ER stress. In response to this stress, cells expressing HCV replicons induce the unfolded protein response (UPR), an ER-to-nucleus intracellular signaling pathway. The UPR is initiated by the proteolytic cleavage of a transmembrane protein, ATF6. The resulting cytoplasmic protein fragment of ATF6 functions as a transcription factor in the nucleus and activates selective genes required for an ER stress response. ATF6 activation leads to increased transcriptional levels of GRP78, an ER luminal chaperone protein. However, the overall level of GRP78 protein is decreased. While ER stress is also known to affect translational attenuation, cells expressing HCV replicons have lower levels of phosphorylation of the α subunit of eukaryotic initiation factor 2. Interestingly, cap-independent internal ribosome entry site-mediated translation directed by the 5′ noncoding region of HCV and GRP78 is activated in cells expressing HCV replicons. These studies provide insight into the effects of HCV replication on intracellular events and the mechanisms underlying liver pathogenesis.
The hepatitis C virus (HCV) is a major worldwide health problem. HCV causes acute and chronic hepatitis, which can lead to chronic active hepatitis, liver cirrhosis, and hepatocellular carcinoma (9). The HCV viral genome consists of a positive-strand RNA, 9.6 kb in length, encoding a 3,000-amino-acid polyprotein (2, 33). The polyprotein is proteolytically cleaved into 10 distinct proteins (2, 33). The structural proteins include the core and highly glycosylated envelope proteins (E1 and E2); the nonstructural proteins (NS2 to NS5) encode enzymatic activities necessary for virus replication (20). The 5′ noncoding region (5′-NCR) and 3′-NCR of HCV contain sequences directly involved in RNA replication (10, 20). The HCV 5′-NCR also functions as an internal ribosome entry site (IRES) permitting cap-independent translation (35, 40, 41).
The absence of a reproducible and efficient cell culture system in HCV studies has hampered efforts to examine alterations in intracellular events by HCV replication. Recently, Lohmann et al. (22) developed selectable HCV subgenomic replicons in a human hepatoma cell line, Huh7, which supported RNA replication. These replicons were bicistronic constructs composed of the HCV IRES (nucleotides 1 to 377 of the 5′-NCR); the neomycin phosphotransferase (neo) gene; the encephalomyocarditis IRES, which mediates the translation of HCV nonstructural proteins NS3 through NS5; and the 3′-NCR (22) (Fig. 1). HCV replicons do not contain structural proteins and NS2 because they are probably not needed for replication of HCV RNA (22). A high level of subgenomic replicon replication was shown to result from adaptive mutations, which were found in several NS proteins (6, 23). HCV nonstructural proteins, including NS3, NS4, and NS5A/B, make up a ribonucleoprotein replication complex associated with the endoplasmic reticulum (ER) membrane (18, 27).
FIG. 1.
Structure of the HCV subgenomic replicon, I377/NS3-3′. The structure of the replicon is composed of the HCV 5′-NCR (solid line), the neo gene (open box), encephalomyocarditis virus IRES (solid line), coding region of NS3 to NS5B (open box), and the HCV 3′-NCR. The neomycin resistance marker allows the establishment of a transformed cell line that stably expresses replicons in RNA form.
The ER is sensitive to a variety of cellular stresses, including the alteration of calcium homeostasis and inhibition of protein glycosylation (19). These stresses inhibit protein folding in the ER, resulting in the accumulation of misfolded proteins in the organelle. Cells can respond to ER stress by activating two functionally separate signaling pathways, the unfolded protein response (UPR) and the ER overload response (EOR) (reviewed in reference 29). HCV proteins induce both of these signaling pathways. The HCV protein NS5A stimulates the EOR after disturbing intracellular calcium levels, leading to the activation of NF-κB and STAT-3 (13). The HCV structural protein E2 can accumulate in the ER as misfolded aggregates, inducing the UPR (21).
Intracellular events characteristic of the UPR include: transcriptional induction, translational attenuation, and protein degradation (reviewed in reference 28). In this study, we examine the role of HCV replication in activating the UPR. HCV replicons activate the ER stress responder, ATF6, by inducing the proteolytic cleavage of a transcriptionally active N-terminal domain from the ER membrane (46). This N-terminal domain of ATF6 is translocated to the nucleus, where it activates the transcription of selective genes containing conserved ER stress response elements (ERSE) (16, 46). Although a characteristic of the UPR is translational attenuation, surprisingly, both cap-dependent and cap-independent protein synthesis is enhanced in cells stably expressing HCV replicons. Both HCV and GRP78 cap-independent translation is increased in cells expressing HCV replicons. These results provide insight into the mechanisms by which HCV replication may alter intracellular events and contribute to liver pathogenesis.
MATERIALS AND METHODS
Plasmids.
GRP78 transcriptional activity was monitored with wild-type and mutant luciferase reporters with a 311-bp fragment of the human GRP78 promoter (−304 to +7 region; numbers indicate the nucleotide position relative to the transcription start site) (46). The wild-type GRP78 reporter contains three ERSE, and the same three ERSE sequences are eliminated in the mutant GRP78 reporter. Each GRP78 construct contains a simian virus 40 minimal promoter upstream of the GRP78 promoter and luciferase coding sequence. The plasmid containing GRP78 cDNA, p3C5, was supplied by A. Lee (University of Southern California, Los Angeles, Calif.). Cap-dependent and HCV IRES directed cap-independent translation were monitored with the luciferase reporter plasmid, pRL HCV 1b. This construct contains a T7 RNA polymerase promoter followed by a firefly luciferase gene, the HCV IRES, and a Renilla luciferase gene. mRNA was synthesized from this reporter plasmid in mammalian cells by T7 RNA polymerase. T7 RNA polymerase was produced in mammalian cells from a cytomegalovirus-directed protein expression vector supplied by D. Bentley (University of Colorado Health Sciences Center, Denver). GRP78 IRES-mediated translation was followed with a luciferase reporter directed by a simian virus 40 promoter upstream of the IRES element and luciferase gene. The GRP78 IRES reporter was a generous gift of P. Sarnow (Stanford University, Stanford, Calif.) (44). K. Shimotohno (Kyoto University, Kyoto, Japan) kindly provided the plasmids pCMV/729-3010 and pCMV/3010. pCMV/729-3010 carries the coding sequences for all the HCV nonstructural proteins (38), and pCMV/3010 expresses all proteins in the HCV genome (25). The HCV NS5A expression vector, pCNS5A, was generated from pCMV/729-3010 (13). The T7C1-341 plasmid DNA contains a T7 RNA polymerase promoter for in vitro transcription preceding the full-length 5′-NCR of HCV and the luciferase reporter gene (42). Plasmid DNA encoding wild-type HCV replicons (pHCVrep1b/BB7) and mutant HCV replicons (pHCVrep1b/BB7 pol−) were supplied by C. M. Rice (Apath, St. Louis, Mo.) (6). The RNA-dependent RNA polymerase-defective replicon (BB7 pol−) was constructed where the GDD sequence of NS5B was changed to AAG (6).
Cell culture.
The human hepatoma cell lines Huh7 and FCA4 were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin (100 U/ml), and streptomycin sulfate (100 μg/ml). FCA4 cells, a generous gift of C. Seeger (Fox Chase Cancer Center, Philadelphia, Pa.), were also grown in G418 (500 μg/ml; Geneticin; Gibco-BRL). FCA4 cells are a Huh7 cell line stably expressing a HCV subgenomic replicon with a single adaptive mutation, a deletion of serine residue 1176 (14). PKR0/0 mouse embryo fibroblasts, a gift from J. C. Bell (University of Ottawa, Ottawa, Ontario, Canada) and B. E. Magun (Oregon Health Sciences University, Portland, Oreg.), have been described previously (1). All cells were maintained in a 5% CO2 incubator at 37°C.
Luciferase assay.
Cells at ∼70% confluency in 60-mm-diameter dishes were transfected with the indicated plasmid(s) using Lipofectin reagent (Gibco-BRL). Huh7 and FCA4 cells were transfected with 0.5 μg of luciferase reporter plasmid or Huh7 cells were cotransfected with 0.5 μg of luciferase reporter along with 5 μg of pCMV-729-3010 or pCMV. The total amount of plasmid DNA was made constant in cotransfections by adding pCMV. Luciferase activity was determined by standard procedures. The transfection and plasmid transcription efficiency of Huh7 and FCA4 cells were normalized with a luciferase reporter plasmid.
RNase protection assay.
A radiolabeled cDNA probe specific for GRP78 was hybridized to Huh7 and FCA4 cellular RNA by standard methods (36). Unhybridized RNA and DNA were digested with mung bean nuclease. Samples were analyzed on a 4% denaturing polyacrylamide gel.
Immunoprecipitation of ATF6.
Endogenous ATF6 in Huh7 and FCA4 cells were labeled with Tran35S-label (ICN) for 5 h after cells were incubated for 30 min in methionine-free Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% fetal calf serum. After removal of the medium, cells were lysed by incubation for 20 min on ice with radioimmunoprecipitation (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM sodium orthovanadate, 1 mM sodium formate, 1 mM phenylmethylsulfonyl fluoride, aprotinin [10 μg/ml], and leupeptin [10 μg/ml]) or STE buffer (36) with 0.5% NP-40. Lysates in STE buffer with 0.5% NP-40 were centrifuged at 5,000 × g for 15 min, and the nuclear pellet was resuspended in RIPA buffer. All samples were immunoprecipitated with anti-ATF6 polyclonal antibody raised against the N-terminal region of ATF6 (16). After a 2-h incubation, immune complexes were bound to protein G-Sepharose and washed five times in RIPA buffer. Samples were fractionated on SDS-polyacrylamide gels.
Western blot analysis.
Huh7 and FCA4 cells were harvested, and cell extracts were prepared by incubating in lysis buffer (10 mM HEPES-KOH [pH 7.2], 142.5 mM KCl, 5 mM MgCl2, 1 mM EGTA, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride, aprotinin [3 μg/ml], and leupeptin [20 μg/ml]) for 20 min on ice. Samples were run on SDS-polyacrylamide gels that were transferred onto nitrocellulose membranes (Schleicher and Schuell) in a solution containing 25 mM Tris-HCl, 192 mM glycine, and 20% methanol. Membranes were blocked overnight in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.3% polyvinylpyrrolidone, and 0.5% Tween 20. The membranes were probed with antibody and developed with an enhanced chemiluminescence Western blotting detection system kit (Amersham Pharmacia Biotech). Mouse anti-GRP78 (clone 40) was obtained from Pharmingen (San Diego, Calif.), rabbit anti-eIF-2α (phosphoserine-51 specific) was obtained from StressGen Biotechnologies (Victoria, British Columbia, Canada), and rabbit anti-eIF-2α was a generous gift from J. W. Hershey (University of California, Davis).
RNA transcription and transfection.
Plasmid DNAs of T7C1-341 and pHCVrep1b/BB7 or pHCVrep1b/BB7 pol− were linearized for in vitro transcription with HpaI and ScaI, respectively. Linearized plasmids were transcribed using the Ampliscribe T7 transcription kit (Epicentre Technologies) as instructed by the manufacturer. RNA was transfected into Huh7 cells using Lipofectin reagent and methods described previously (41).
RESULTS
HCV replicons induce the transcription of ER chaperones.
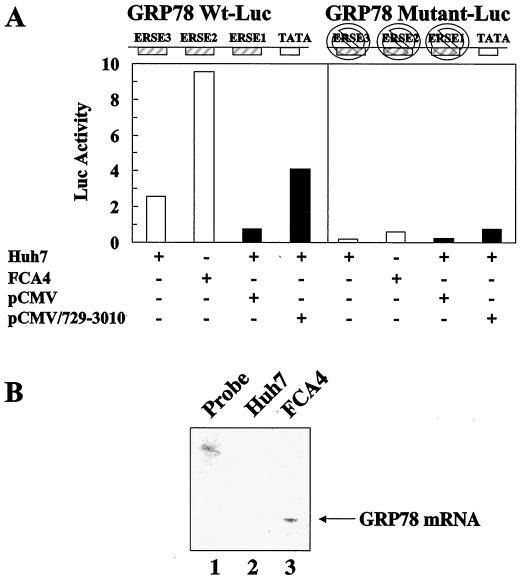
Mammalian ER chaperones contain conserved ERSE in their promoter region that are necessary for their transcriptional induction (46). ATF6 has been found to activate ER chaperone gene expression in an ERSE-dependent manner. Three ERSE motifs are found in the transcriptional control elements of the ER chaperone, GRP78. FCA4 cells expressing HCV subgenomic replicons stimulate the transcription of a GRP78 luciferase promoter about fivefold over that of Huh7 control cells (Fig. 2A, left panel). An RNase protection assay using a radiolabeled probe specific to cellular GRP78 mRNA also shows a similar increase in the transcription of GRP78 (Fig. 2B). However, removing all three ERSE sequences in the GRP78 promoter eliminates the ability of HCV replicon-expressing cells to stimulate GRP78 mRNA synthesis (Fig. 2A, right panel). These results imply that the transcriptional induction of ER chaperones with ERSE sequences is due to the HCV replicon-induced activation of ATF6.
FIG. 2.
GRP78 gene expression is stimulated by HCV replicons and HCV nonstructural proteins. (A) HCV replicons and HCV nonstructural proteins activate GRP78 transcription in an ERSE-dependent manner. Huh7 and FCA4 cells were transfected with GRP78 wild-type (left panel) or mutant (right panel) luciferase reporter plasmids (white). Huh7 cells were also cotransfected with pCMV/729-3010 or pCMV and wild-type (left panel) or mutant (right panel) GRP78-Luc (black). The GRP78 luciferase reporter has three ERSE sequences. The mutant GRP78-Luc has all three of these ERSE sequences eliminated. (B) RNase protection assay indicating an increase in GRP78 mRNA steady-state levels in FCA4 cells. RNA was extracted from Huh7 and FCA4 cells and analyzed using a probe specific to GRP78. Lane 1, probe alone; lane 2, Huh7 GRP78 mRNA analysis; lane 3, FCA4 GRP78 mRNA analysis.
FCA4 cells stably express all the nonstructural genes spanning from NS3 through NS5B and support both the replication and translation of HCV subgenomic replicons (Fig. 1), whereas the vector pCMV/729-3010, which contains the same HCV NS genes that are under the control of cytomegalovirus promoter and enhancer, allows for the expression of the NS proteins in mammalian cells. Huh7 cells transfected with this expression vector induce GRP78 transcriptional activity (Fig. 2A), although three times less than FCA4 cells, suggesting that the HCV replication makes a significant contribution in stimulating the ER stress response pathway.
ATF6 transactivating activity is stimulated by HCV replicons.
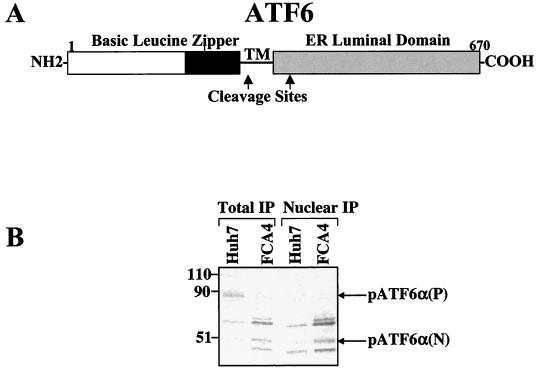
ATF6 is a transmembrane protein localized to the ER; the protein is regulated posttranslationally by ER stress (Fig. 3A) (16). ATF6 is synthesized as a 90-kDa protein that is cleaved in FCA4 cells expressing HCV subgenomic replicons (Fig 3B) but is intact in Huh7 control cells. Upon ER stress, ATF6 is cleaved at the cytoplasmic face of the ER membrane, and the resulting 50-kDa N-terminal domain, pATF6α(N), is translocated to the nucleus, where it transcriptionally activates ER chaperones whose promoters harbor several ATF6 binding sites (16, 46). The transcriptionally active form of ATF6, pATF6α(N), was successfully identified using anti-ATF6 raised against the N-terminal region of ATF6 (amino acids 6 to 307) (16). Immunoprecipitation of endogenous ATF6 in total cell lysates revealed a 50-kDa protein in FCA4 cells (Fig. 3B). This same 50-kDa protein was present in Huh7 cell lysates, but not to the extent found in FCA4 cells. The appearance of pATF6α(N) was also marked by the disappearance of the 90-kDa protein, pATF6α(P), in FCA4 cells lysates. After immunoprecipitating Huh7 and FCA4 cell nuclear extracts, pATF6α(N) was clearly observed again in FCA4 cells, but its presence was less obvious in Huh7 cell nuclear extracts (Fig. 3B).
FIG. 3.
HCV replicons activate ATF6. (A) Structure of ATF6. ATF6 is an ER transmembrane protein. Upon ER stress, the cytoplasmic N-terminal domain of ATF6 is cleaved at the cytoplasmic face of the ER membrane. The cleaved cytoplasmic domain is translocated to the nucleus where it activates the transcription of ER chaperone genes containing the ERSE. TM is the region of the protein that spans the ER membrane. The ATF6 cleavage sites are indicated by arrows (45). (B) HCV replicons induce the cleavage of endogenous p90ATF6 to p50ATF6. Total and nuclear Huh7 and FCA4 cell lysates were immunoprecipitated with anti-ATF6 antibody. Molecular masses (in kilodaltons) are indicated at left.
HCV replicons stimulate overall protein synthesis in ER-stressed cells.
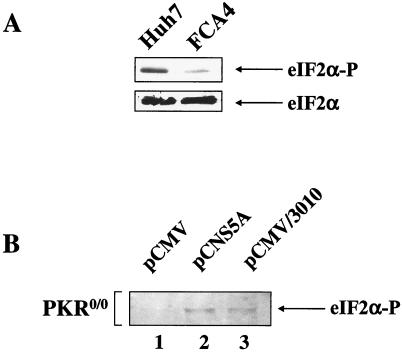
Cells adapt to ER stress by attenuating protein synthesis. However, Western blot analysis of lysates with antiserum that specifically detects the α subunit of eukaryotic initiation factor 2 (eIF-2α) phosphorylated on serine 51, revealed significantly lower levels of eIF-2α phosphorylation in FCA4 cells expressing HCV replicons (Fig. 4A). When eIF-2α is phosphorylated on serine residue 51, the initiation factor binds the guanine nucleotide exchange factor, eIF-2B, and inhibits the role of eIF-2B in forming translational preinitiation complexes. Therefore, lower levels of eIF-2α phosphorylation in FCA4 cells indicate that the overall level of protein synthesis is higher.
FIG. 4.
Effects of HCV replicons or HCV nonstructural proteins on eIF-2α phosphorylation levels in cells with or without PKR. (A) FCA4 cells with HCV replicons have lower levels of eIF-2α phosphorylation. Top panel, Huh7 and FCA4 cell lysates were analyzed by Western blot analysis using anti-eIF-2α serum specific to phosphoserine-51. Bottom panel, Western blot analysis of Huh7 and FCA4 cell lysates with antiserum reactive with all forms of eIF-2α. (B) NS5A stimulates eIF-2α phosphorylation in PKR knockout cells (PKR0/0). Western blot analysis of eIF-2α phosphoprotein from PKR0/0 transfected with pCMV (lane 1), pCNS5A (lane 2), and pCMV/3010 (lane 3) expression vectors.
Four different eIF-2α kinases have been identified in mammalian cells (3, 4, 8, 15), and two of these kinases, the double-stranded RNA-dependent kinase (PKR) and the PKR-like ER kinase (PERK), are activated by ER stress (15, 32, 37). The kinase activity of PKR has been shown to be inhibited by HCV NS5A protein (12). NS5A binds PKR and inhibits its kinase activity and dimerization (12). This interaction leads to the accumulation of unphosphorylated eIF-2α (12). To determine whether other eIF-2α kinases are stimulated or inhibited in response to HCV-induced ER stress, we monitored the levels of eIF-2α phosphorylation in PKR0/0 cells transfected with expression vectors that produce NS5A alone, pCNS5A, or all HCV proteins, pCMV/3010. Whether NS5A was expressed alone or in the context of all HCV proteins, NS5A stimulated eIF-2α phosphorylation in the absence of PKR (Fig. 4B). This result shows that NS5A is capable of activating other eIF-2α kinases, possibly in response to ER stress.
HCV replicons activate cap-independent and cap-dependent translation.
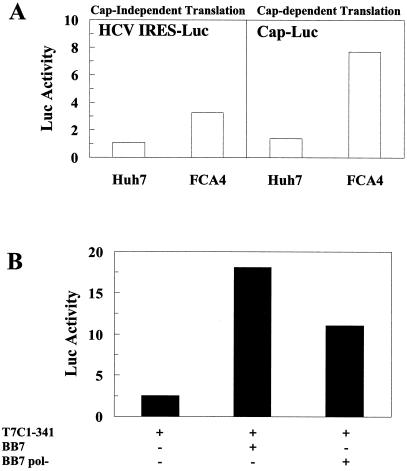
The functional relevance of lower levels of eIF-2α phosphorylation in the HCV replicon-expressing cells was examined further by monitoring cap-independent and cap-dependent translation in FCA4 cells. HCV translation is mediated through an IRES in a cap-independent manner (35, 40, 41). FCA4 cells transfected with an HCV IRES luciferase reporter stimulated HCV IRES-linked luciferase expression threefold (Fig. 5A; left panel). The combined replication and translation of HCV subgenomic replicons significantly enhances HCV IRES-mediated translation. This was confirmed when the HCV IRES activity in cells transiently transfected with HCV replicon RNA (BB7) was compared to the HCV IRES activity in cells transfected with RNA-dependent RNA polymerase-defective replicons (BB7 pol−) (6). Huh7 cells transfected with wild-type BB7 replicons stimulated HCV IRES-directed translation to levels 7.2 times higher than those in cells with an HCV IRES reporter alone, and 1.6 times higher than cells transfected with BB7 pol− (Fig. 5B). Cap-dependent translation was also enhanced 6-fold in FCA4 cells (Fig. 5A, right panel), suggesting that overall protein synthesis is increased in cells expressing HCV replicons.
FIG. 5.
HCV IRES-directed translation and cap-dependent translation increases in cells with HCV replicons. (A) HCV replicons induce HCV IRES translation and cap-dependent translation. Huh7 and FCA4 cells were transfected with a dual luciferase reporter plasmid containing the HCV IRES linked to Renilla luciferase (left panel). Cap-dependent translation activity was measured in Huh7 and FCA4 cells from firefly luciferase activity using the same dual luciferase reporter plasmid (right panel). (B) Wild-type HCV replicons enhance HCV IRES activity more than replication defective HCV replicons. Huh7 cells were transfected with an HCV IRES RNA luciferase reporter alone, T7C1-341, and a wild-type HCV replicon (BB7) or a replication-defective mutant HCV replicon (BB7 pol−) was transfected into Huh7 cells along with the luciferase reporter, T7C1-341.
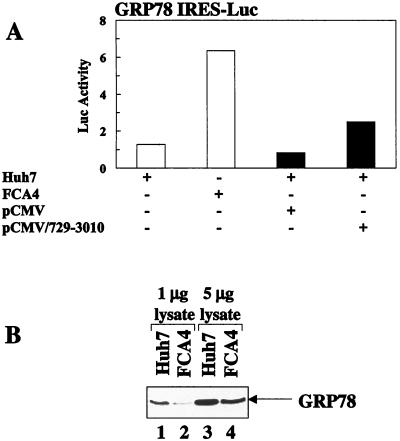
The ER chaperone, GRP78, is among a limited number of cellular proteins translated by a cap-independent mechanism (24). GRP78 IRES-mediated luciferase activity was enhanced fivefold in FCA4 cells expressing HCV replicons (Fig. 6A). Only a threefold increase in GRP78 IRES translation was observed in cells expressing all HCV nonstructural proteins synthesized from the pCMV/729-3010 expression vector with no replication (Fig. 6A). These results imply that the HCV replicon (or HCV nonstructural protein)-mediated stimulation of cap-independent translation is not specific to the HCV IRES and that cap-independent translation is generally enhanced.
FIG. 6.
HCV replicons induce GRP78 translation while lowering overall levels of GRP78 protein. (A) HCV replicons and HCV nonstructural proteins activate GRP78 IRES-mediated translation. Huh7 and FCA4 cells were transfected with an GRP78 IRES-linked luciferase reporter plasmid (white). Huh7 cells were also cotransfected with GRP78 IRES-Luc and pCMV/729-3019 or pCMV (black). (B) GRP78 protein levels in Huh7 and FCA4 cells. Shown is a Western blot analysis of Huh7 and FCA4 cell lysates with anti-GRP78 antibody. Lanes 1 and 2, 1 μg of Huh7 and FCA4 cell lysates, respectively; lanes 3 and 4, 5 μg of Huh7 and FCA4 cell lysates, respectively.
Although the level of GRP78 translation is increased in FCA4 cells, Western blot analysis of lysates revealed that the overall amount of GRP78 protein was lower in FCA4 cells in comparison to Huh7 control cells (Fig. 6B). The differences in the total GRP78 protein levels were more apparent when smaller amounts of cell lysates were analyzed. The reasons for these observations are not entirely clear.
DISCUSSION
The HCV nonstructural proteins are localized to the ER membrane in the reticular network of the perinuclear region (2, 18, 33). Here, they form a ribonucleoprotein (RNP) complex that engages in the replication of the viral genome, an event necessary for establishing the infectious process in hepatocytes. The association of the replication complex with the ER membrane induces ER stress. This stress elicits the activation of intracellular signaling pathways from the ER to the nucleus (reviewed in reference 29).
In the present study, we analyzed intracellular events following HCV replicon-induced ER stress, including activation of the unfolded protein-ER stress response. Cells expressing HCV replicons cleave ATF6 [pATF6α(P)] and convert it to a transcriptionally active, lower-molecular-mass form [50 kDa; pATF6α(N)] which leads to the induction of GRP78, a major target of the UPR (Fig. 2 and 3). Another characteristic of the UPR is translational inhibition. However, cells with HCV replicons have higher levels of protein synthesis. The phosphorylation level of eIF-2α is lower in HCV replicon-expressing cells, which is evidence of elevated translation initiation (Fig. 4A). In FCA4 cells, cap-independent translation directed by the HCV IRES and GRP78 IRES is stimulated (Fig. 5 and 6). Furthermore, cap-dependent translation is also enhanced in cells expressing HCV replicons (Fig. 5A, right panel).
Numerous viral proteins have been shown to induce the EOR, including influenza virus hemagglutinin (30), HBV MHBSt (26), adenovirus E3/19K (31), and HCV NS5A (13). However, a limited number of viral proteins activate the UPR in response to ER stress. The HBV large surface protein (43) and the HCV E2 envelope protein (21) both induce the UPR. The HCV E2 protein is a structural component of the viral envelope and is likely not required for viral replication (22).
HCV replicons express all of the HCV nonstructural proteins, including NS5A. NS5A is also capable of inducing ER stress, activating NF-κB by an ER overload stress response, which involves alteration of Ca2+ homeostasis and elevation of reactive oxygen species in the mitochondria (13). The ER overload response is functionally distinct from the unfolded protein ER stress response. Cells expressing HCV replicons and all the HCV nonstructural proteins activate ATF6, GRP78, and the UPR. However, when NS5A is expressed alone, the protein does not stimulate the UPR (data not shown), implying that UPR activation by HCV replicons and HCV nonstructural proteins in this study is distinct from the NS5A-induced ER overload response. This does not exclude the ability of NS5A to activate the UPR in the context of all HCV nonstructural proteins. It is unclear which HCV nonstructural protein(s) is responsible for inducing the UPR. The most likely explanation is that the combined translation and replication activities in the ER lead to this apparent ER stress.
Activation of ATF6 is required for the initiation of the UPR. ATF6 is activated by ER stress-induced proteolysis. pATF6α(P) is cleaved to a 50-kDa protein in FCA4 cells under ER stress (Fig. 3B). ATF6 initiates the transcriptional induction of ER chaperone and XBP1 genes in an ERSE-dependent manner (47). XBP1 mRNA is spliced by IRE1, an ER stress transducer (47). XBP1 is a transcription factor that activates the mammalian UPR through ERSE sequences, similar to ATF6 (47). Recent studies have identified XBP1 as a mammalian homolog of Saccharomyces cerevisiae Hac1p protein, a key component of the UPR (47). XBP1 knockout studies suggest an essential role of XBP1 in hepatocyte growth and differentiation (34). In light of recent studies, both ATF6 and XBP1 emerge as key transcription factors involved in activating UPR and contributing to liver pathogenesis associated with HCV infection.
Lower levels of eIF-2α phosphorylation observed in FCA4 cells are likely due to the NS5A-mediated inhibition of PKR (Fig. 4A) (12). In PKR0/0 cells, eIF-2α phosphorylation levels increase when NS5A is expressed (Fig. 4B). This suggests that NS5A may stimulate eIF-2α kinase activity. PKR and PERK are two of the eIF-2α kinases activated by ER stress. In the absence of PKR, NS5A may stimulate PERK activity, implying NS5A plays an important role in regulating eIF-2α phosphorylation. These results do not discount the ability of NS5A to stimulate eIF-2α kinases other than PERK. However, NS5A appears to physically interact with PERK in vitro (K. D. Tardif and A. Siddiqui, unpublished results).
FCA4 cells show increased cap-independent translation (Fig. 5A and 6A). Although GRP78 IRES-directed translation is enhanced in FCA4 cells, the overall level of GRP78 protein is decreased (Fig. 6B). GRP78 and ER-stressed cells undergo several changes that may explain this surprising result. First, posttranslational phosphorylation of GRP78 decreases with increased translation of GRP78 in ER stressed cells (17). Second, GRP78 dissociates from the ER luminal domain of ER stress transducers, PERK and IRE1, in the UPR (5). GRP78 changes in FCA4 cells may target the protein for ubiquitination and degradation by the 26S proteasome. Recent evidence suggests that the activation of the UPR is required for efficient degradation of proteins accumulated in the ER (7, 11, 39).
In this study, we show that the HCV subgenomic replicons induce ER stress. Cells expressing HCV replicons adapt to this stress by activating the UPR. HCV replication seems to alter the typical course of the UPR signaling pathway to prolong its survival in hepatocytes. The ability of HCV replicons to induce changes in intracellular events may provide insight into the mechanisms of HCV pathogenesis.
Acknowledgments
K.D.T. was supported by a postdoctoral training grant from the NIH (T3-DK07038).
We thank K. Shimotohno for the pCMV/729-3010 and pCMV/3010 plasmids, C. Seeger for the FCA4 cells, J. C. Bell and B. E. Magun for PKR0/0 cells, J. W. Hershey for eIF-2α antibody, A. Lee for GRP78 cDNA, C. M. Rice for plasmids encoding the BB7 and BB7 pol− selectable HCV replicons, P. Sarnow for the GRP78 IRES luciferase reporter, and D. Bentley for the T7 RNA polymerase mammalian expression vector.
REFERENCES
- 1.Abraham, V., D. F. Stojkl, P. I. Duncan, N. Methot, T. Ishii, M. Dube, B. C. Vanderhyden, H. L. Atkins, D. A. Gray, M. W. McBurney, A. E. Koromilas, E. G. Brown, N. Sonenberg, and J. C. Bell. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 274:5953-5962. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Berlanga, J. J., J. Santoyo, and C. De Haro. 1999. Characterization of a mammalian homologue of the GCN2 eukaryotic initiation factor 2 alpha kinase. Eur. J. Biochem. 265:754-762. [DOI] [PubMed] [Google Scholar]
- 4.Berry, M. J., G. S. Knutson, S. R. Lasky, S. M. Munemitsu, and C. E. Samuel. 1985. Mechanism of interferon from untreated and interferon-treated mouse fibroblasts. J. Biol. Chem. 270:11240-11247. [PubMed] [Google Scholar]
- 5.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Casagrande, R., P. Stern, M. Diehn, C. Shamu, M. Osario, M. Zuniga, P. O. Brown, and H. Ploegh. 2000. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5:729-735. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. J., and I. M. London. 1995. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem. Sci. 20:105-108. [DOI] [PubMed] [Google Scholar]
- 9.Di Biscegli, A. M. 1997. Hepatitis C and hepatocellular carcinoma. Hepatology 26:34.S-38S.9214449 [Google Scholar]
- 10.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedlander, R., E. Jarosch, J. Urban, C. Volkwein, and T. Sommer. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2:379-384. [DOI] [PubMed] [Google Scholar]
- 12.Gale, M. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural protein 5A. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 13.Gong, G., G. Waris, R. Tanveer, and A. Siddiqui. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proc. Natl. Acad. Sci. USA 98:9599-9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replication. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 398:271-273. [DOI] [PubMed] [Google Scholar]
- 16.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendershot, L. M., J. Ting, and A. S. Lee. 1988. Identity of the immunoglobulin heavy-chain-binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Mol. Cell. Biol. 8:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. USA 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman, R. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 20.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberman, E., Y.-L. Fong, M. J. Selby, Q.-L. Choo, L. Cousens, M. Houghton, and T. S. B. Yen. 1999. Activation of the grp78 and grp94 promoters by hepatitis C virus E2 envelope protein. J. Virol. 73:3718-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohmann, V., F. Korner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macajak, D. G., and P. Sarnow. 1991. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature 353:90-94. [DOI] [PubMed] [Google Scholar]
- 25.Marusawa, H., M. Hijikata, T. Chiba, and K. Shimotohno. 1999. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-κB activation. J. Virol. 73:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer, M., W. H. Caselmann, V. Schluter, R. Schreck, P. H. Hofschneider, and P. A. Baeuerle. 1992. Hepatitis B virus transactivator MHBst: activation of NF-kappa B, selective inhibition by antioxidants and integral membrane localization. EMBO J. 11:2991-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 28.Mori, K. 2000. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101:451-454. [DOI] [PubMed] [Google Scholar]
- 29.Pahl, H. 1999. Signal transduction from the endoplasmic reticulum to the cell nucleus. Physiol. Rev. 79:683-699. [DOI] [PubMed] [Google Scholar]
- 30.Pahl, H. L., and P. A. Baeuerle. 1995. Expression of influenza hemagglutinin activates transcription factor NF-κB. J. Virol. 69:1480-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pahl, H. L., M. Sester, H.-G. Burgert, and P. A. Baeuerle. 1996. Activation of transcription factor NF-κB by adenovirus E3/19K requires its ER retention. J. Cell Biol. 132:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prostko, C. R., J. N. Kholakia, M. A. Brostrom, and C. O. Brostrom. 1995. Activation of the double-stranded RNA-regulated protein kinase by depletion of endoplasmic reticular calcium stores. J. Biol. Chem. 270:6211-6215. [DOI] [PubMed] [Google Scholar]
- 33.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 34.Reimold, A. M., N. N. Iwakoshi, J. Manis, P. Vallabhajosyula, E. Szomolanyi-Tsuda, E. M. Gravallese, D. Friend, M. J. Grusby, F. Alt, and L. H. Glimcher. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412:300-307. [DOI] [PubMed] [Google Scholar]
- 35.Rijibrand, R., and S. Lemon. 1999. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 242:86-116. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Srivastava, S. P., M. V. Davies, and R. J. Kaufman. 1995. Calcium depletion from the endoplasmic reticulum activates the double-stranded RNA-dependent kinase (PKR) to inhibit protein synthesis. J. Biol. Chem. 270:16619-16624. [DOI] [PubMed] [Google Scholar]
- 38.Tanji, Y., M. Hijikata, Y. Hirowatari, and K. Shimotohno. 1994. Hepatitis C virus polyprotein processing: kinetics and mutagenic analysis of serine proteinase-dependent cleavage. J. Virol. 68:8418-8422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travers, K. J., C. K. Patil, L. Wodica, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 40.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, C., P. Sarnow, and A. Siddiqui. 1994. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 68:7301-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, Z., G. Jensen, and T. S. B. Yen. 1997. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J. Virol. 71:7387-7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, Q., and P. Sarnow. 1997. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 25:2800-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida, H., K. Haze, H. Yangi, T. Yura, and K. Mori. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. J. Biol. Chem. 273:33741-33749. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]