Abstract
Intracellular mature vaccinia virions are wrapped by cisternae, derived from virus-modified trans-Golgi or endosomal membranes, and then transported via microtubules to the cell periphery. Two viral proteins, encoded by the F13L and B5R open reading frames, are essential for the membrane-wrapping step. Previous transfection studies indicated that F13L induces the formation of post-Golgi vesicles that incorporate the B5R protein and that this activity depends on an intact F13L phospholipase motif. Here we show that the F13L protein has a general effect on the trafficking of integral membrane proteins from the Golgi apparatus, as both the vaccinia virus A36R protein and the vesicular stomatitis virus G protein also colocalized with the F13L protein in vesicles. In addition, increased expression of cellular phospholipase D, which has a similar phospholipase motif as, but little amino acid sequence identity with, F13L, induced post-Golgi vesicles that contained B5R and A36R proteins. Butanol-1, which prevents the formation of phosphatidic acid by phospholipase D and specifically inhibits phospholipase D-mediated vesicle formation, also inhibited F13L-induced vesicle formation, whereas secondary and tertiary alcohols had no effect. Moreover, inhibition of phospholipase activity by butanol-1 also reduced plaque size and decreased the formation of extracellular vaccinia virus without affecting the yield of intracellular mature virus. Phospholipase D, however, could not complement a vaccinia virus F13L deletion mutant, indicating that F13L has additional virus-specific properties. Taken together, these data support an important role for F13L in inducing the formation of vesicle precursors of the vaccinia virus membrane via phospholipase activity or activation.
Poxviruses are large, enveloped DNA viruses that replicate entirely within the cytoplasm of infected cells (30). The assembly of vaccinia virus, the most intensively studied member of the poxvirus family, can be divided into two phases. The first culminates in the formation of infectious intracellular mature virions (IMV) (13, 14, 21, 31, 43). The second involves the wrapping of IMV with cisternae derived from virus-modified trans-Golgi or endosomal membranes to form intracellular enveloped virions (IEV) (18, 39, 47) that are transported along microtubules to the periphery where the outer IEV membrane and the plasma membrane fuse (12, 20, 35, 49, 50). Most extracellular virus, called cell-associated enveloped virions (CEV), adhere to the outside of the plasma membrane and mediate direct cell-to-cell spread (4), which is facilitated by motile actin tails (7, 36, 38, 44, 52). The released virions, called extracellular enveloped virions (EEV), provide an additional mechanism for long-range spreading (32). The ratio of CEV to EEV varies with different vaccinia virus strains.
Of the seven known proteins associated with IEV- or EEV-specific membranes, the ones encoded by the F13L and B5R open reading frames (ORFs) are required for the wrapping of IMV to form IEV (3, 9, 51). The B5R product is a 42-kDa type I integral membrane component of the EEV (8, 23). The F13L ORF encodes a nonglycosylated, palmitylated protein that is the most abundant component of the EEV membrane (15, 16, 18, 19, 40). The F13L protein contains a variant of the HKD (His-Lys-Asp) motif that is conserved in a superfamily of phospholipases and phospholipid synthases (25, 33, 45) and has been reported previously to exhibit broad-specificity lipase activities in vitro (1). In addition, mutant vaccinia viruses with amino acid substitutions of either the conserved Lys or Asp of the phospholipase motif exhibited wrapping defects that inhibited IEV formation (37, 45).
To better understand the role of the F13L protein in membrane wrapping, we fused the F13L ORF to that of the enhanced green fluorescent protein (GFP) so that it could be visualized by microscopy. The GFP moiety had no deleterious effect, as the fusion protein complemented a mutant vaccinia virus with a deleted F13L gene and was localized in the IEV, CEV, and EEV membranes (22). When expressed by transfection in the absence of other viral gene products, F13L-GFP was localized in Golgi membranes and post-Golgi vesicles that contained early and late endosomal markers (22). Under similar transfection conditions, the B5R protein was targeted to juxtanuclear Golgi membranes (24, 29, 48). However, coexpression of F13L-GFP and B5R resulted in the colocalization of the two proteins in endosomal vesicles (22). Furthermore, colocalization was dependent on both an unmutated phospholipase motif and the palmitylation site in the F13L protein. These results were intriguing because phospholipase D (PLD) regulates the budding of vesicles from trans-Golgi membranes (2, 6, 10, 26, 41, 42). We therefore proposed a similar role for the F13L protein. To further examine this hypothesis, we posed the following questions. Is the colocalization of B5R with F13L specific, or will other Golgi transmembrane proteins colocalize with F13L? Will B5R or other viral proteins colocalize with PLD, instead of F13L, in post-Golgi vesicles? Can PLD complement an F13L deletion mutant? Will a PLD inhibitor prevent the localization of F13L in endosomes and inhibit extracellular vaccinia virus formation?
MATERIALS AND METHODS
Cells and viruses.
HeLa and BS-C-1 cells were grown and maintained in Dulbecco's modified Eagle's medium or Eagle's modified medium supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. The mutant vF13LΔ, originally referred to as vRB12 (3), has most of the F13L ORF replaced with the Escherichia coli xanthine guanine phosphoribosyltransferase ORF. Recombinant vF13L-GFP contains the GFP coding sequence at the C terminus of the F13L ORF (22). For virus titration and analysis of plaque size, infected BS-C-1 monolayers were fixed and stained with 0.1% crystal violet in 20% methanol.
Plasmid constructions.
Plasmids pF13L-GFP containing the GFP coding sequence at the C terminus of the F13L ORF and pB5R containing the B5R ORF were described previously (22). Plasmid pSVGL1, containing wild-type vesicular stomatitis virus (VSV) G protein, and VSVG-GFP, containing a GFP-tagged ts045 VSV G protein, were generously provided by J. Rose and J. Lippincott-Schwartz (34), respectively. Plasmids pCGN-hPLD1b and pCGN-hPLD1b (K898R) containing wild-type and mutated PLD ORFs (17), respectively, were kindly provided by M. Frohman. Additional plasmids were constructed by standard procedures. Briefly, the F13L-GFP fusion ORF in plasmid pGF13L (22) was replaced by PLD1-HA or PLD1-GFP fusion ORFs containing the hemagglutinin (HA) tag or GFP sequence at the N terminus. The HA tag or GFP ORF was joined with PLD1 coding sequence by two-stage PCR, and the final PCR products were cloned into plasmid pGF13L, yielding plasmids pGPLD-LR and pGPLD-LR2, respectively. These two plasmids contain PLD1-HA and PLD1-GFP fusion constructs flanked by ∼600 bp of left and right flanking sequences of the F13L gene. Similarly, the GFP coding sequence was appended at the N terminus of PLD1 in plasmid pCGN-hPLD1b. Relevant portions of all plasmids were verified by sequencing.
Recombinant virus construction.
Recombinant vaccinia virus expressing PLD1-GFP under the F13L late promoter was constructed according to the procedure described previously (22). Briefly, HeLa cells were infected with vF13LΔ and immediately transfected with plasmid pGPLD-LR2. After 2 days at 37°C, cells were harvested and the lysate was analyzed by plaque assay on BS-C-1 cells. Isolated green plaques were purified and amplified, and the recombinant virus called vPLD1-GFP was characterized as described earlier (22).
Antibodies.
A Golgi sampler kit containing monoclonal antibodies (MAbs) to marker proteins of cis- and trans-Golgi compartments and early endosomal antigen 1 (EEA1) were purchased from Transduction Laboratories. Rabbit anti-β-coat protein (anti-β-COP) polyclonal antibody was from Affinity Bioreagents; MAb HA.11, which recognizes an influenza virus HA epitope, was from Babco. Secondary antibody conjugates were purchased from Jackson ImmunoResearch Laboratories. Rabbit polyclonal antibody recognizing a peptide sequence at the C terminus of the A36R protein (53), 192C rat MAb against the B5R protein (39), and MAbI1 anti-VSV G (28) have been described previously. MAb to lysosome-associated membrane protein 2 (LAMP2) was a gift of Thomas August.
Transfections and infections.
For transfection, plasmids were prepared with the Qiagen plasmid Midi preparation kit. HeLa cells were grown on glass coverslips until they reached 80 to 90% confluence. Routinely, 2 to 10 μg of Lipofectamine 2000 (Invitrogen) and 0.5 to 2 μg DNA were diluted separately in Opti-MEM I medium (Invitrogen), mixed, incubated at room temperature for 20 min, and added to the cells for 4 to 5 h at 37°C. The Lipofectamine-DNA complex was replaced with Dulbecco's modified Eagle's medium supplemented with 10% FBS, and the incubation was continued for a total of 24 h.
For infections, virus stocks were diluted in growth medium supplemented with 2.5% FBS and added to cell monolayers in wells or on coverslips. After 2 h of incubation at 37°C, virus inocula were replaced with fresh medium containing 2.5% FBS and incubated for a further 17 to 18 h.
Confocal microscopy.
At 24 h after transfection, cells were fixed with cold 4% paraformaldehyde in phosphate-buffered saline (PBS) and then incubated at room temperature for 20 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. The permeabilized cells were incubated with primary antibodies diluted in 10% FBS in PBS for 1 h followed by secondary antibody diluted in 10% FBS in PBS for 30 min at room temperature. When two different antibodies were used, staining was sequential. The Golgi apparatus was visualized by staining with mouse anti-p115 MAb unless otherwise stated. Stained cells were washed extensively with PBS, and coverslips were mounted in 20% glycerol and sealed with rubber cement. Fluorescence was examined with a Leica TCS NT inverted confocal microscope, and images were overlaid by using Adobe Photoshop version 5.0.2.
Electron microscopy.
For immunoelectron microscopy, RK13 cells were grown in 60-mm-diameter dishes and infected with recombinant vaccinia virus expressing PLD1-GFP at a multiplicity of 10. After 24 h, the cells were prepared for freezing, and ultrathin sections were cut, collected, immunostained, and viewed as previously described (53). The rabbit anti-GFP polyclonal antibody (Clontech) was used at a dilution of 1:100.
Effects of alcohols on intracellular localization and virus formation.
Butanol-1, butanol-2, or propanol-2 (0.5 or 1%) was added to the medium overlaying cells at 4 h after transfection, and the cells were incubated for a total of 24 h at 37°C. Alternatively, BS-C-1 cells were infected with vaccinia virus for 2 h at 37°C and then the inoculum was replaced with fresh medium supplemented with butanol-1, butanol-2, or propanol-2 and incubated for 48 h at 37°C. Cells were stained with crystal violet to visualize the plaques. To measure the effect of butanol-1 on virus yields, HeLa cells were infected with the IHD-J strain of vaccinia virus and treated with alcohols as described above. Cells and medium were harvested separately, adjusted to equal volumes, and analyzed by plaque assay. As a control, medium of untreated, infected cells was harvested and treated with butanol-1 at 37°C for 24 h and then analyzed by plaque assay.
RESULTS
Colocalization of F13L and A36R membrane proteins.
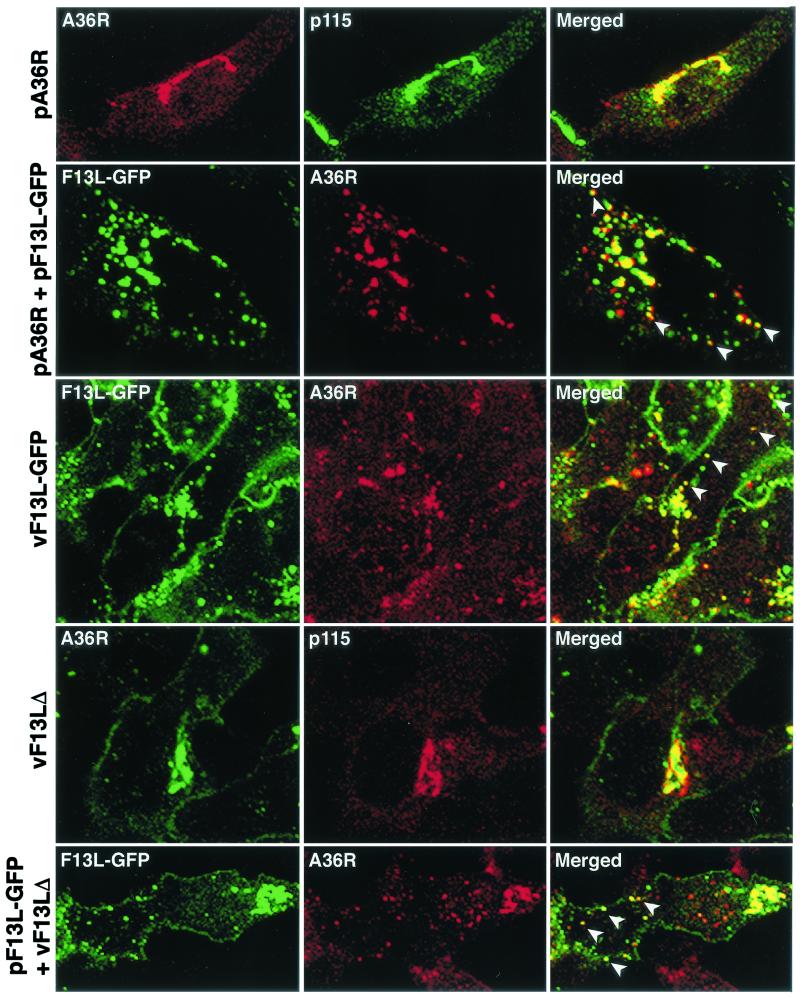
We reported previously that the F13L-GFP fusion protein colocalized with endosomal markers in transfected cells, whereas the B5R protein was found predominantly in juxtanuclear Golgi membrane under these conditions. However, when plasmids pF13L-GFP and pB5R were cotransfected, the B5R protein colocalized with F13L-GFP in endosomal vesicles. To determine whether this result was specific for the B5R protein, we analyzed the distribution of the A36R IEV protein. When expressed alone, the A36R protein was present in the juxtanuclear region and colocalized with Golgi markers (Fig. 1). However, when plasmids pF13L-GFP and pA36R were cotransfected, the distribution of the A36R protein was altered and greater than 60% appeared to colocalize with F13L (Fig. 1). In the latter experiments, only one or two viral proteins were synthesized. However, consistent results were obtained with recombinant viruses. Colocalization of A36R and F13L-GFP occurred in cells infected with vF13L-GFP, a virus expressing the fusion protein (Fig. 1). In contrast, A36R had a predominantly Golgi localization in cells infected with the mutant vaccinia virus vF13LΔ with a deleted F13L gene (Fig. 1). When cells transfected with pF13L-GFP were infected with vF13LΔ, about 45% of the A36R localized with F13L (Fig. 1). Thus, the F13L protein dramatically modulated the intracellular distribution of both the B5R protein as previously described (22) and the A36R protein as shown here.
FIG. 1.
Effect of F13L-GFP on the intracellular localization of A36R in transfected or infected cells. HeLa cells were transfected or infected for 24 or 18 h, respectively, with the name of the plasmid or virus indicated at the left of each row. First row, cells transfected with pA36R were stained with anti-p115 MAb followed by indodicarbocyanine (Cy5)-conjugated anti-mouse immunoglobulin antibody and then with anti-A36R polyclonal antibody followed by Texas red-conjugated anti-rabbit immunoglobulin antibody. Second row, cells were cotransfected with pA36R and pF13L-GFP and stained for A36R as in the first row. Third row, cells infected with vF13L-GFP (1 PFU/cell) were stained with anti-A36R polyclonal antibody followed by tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit immunoglobulin antibody. Fourth row, cells infected with vF13LΔ (1 PFU/cell) were stained with anti-p115 MAb followed by tetramethyl rhodamine isothiocyanate-conjugated anti-mouse immunoglobulin antibody and then stained with anti-A36R polyclonal antibody followed by Alexa 488-conjugated anti-rabbit immunoglobulin antibody. Fifth row, cells were transfected with plasmid pF13L-GFP; after 24 h the cells were infected with vF13LΔ as in the fourth row and stained with anti-A36R polyclonal antibody followed by tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit immunoglobulin antibody. Cells were analyzed by confocal microscopy. Green, GFP or Alexa 488; red, Texas red or tetramethyl rhodamine isothiocyanate; yellow, overlap of green and red. White arrowheads show the colocalization of F13L-GFP with A36R in vesicular structures.
Colocalization of the F13L and VSV G proteins.
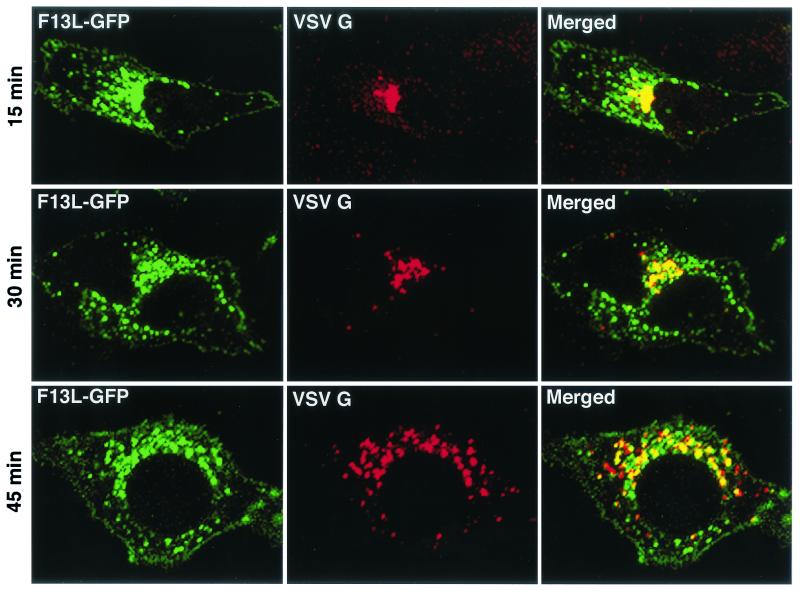
Since B5R and A36R are viral constituents of the IEV membrane, F13L might specifically interact with both, mediating the colocalization of these proteins. However, such interactions could not be confirmed by antibody coprecipitation experiments (M. Husain, unpublished data). This led us to consider the idea that the F13L protein has a general effect on the trafficking of integral membrane proteins from the Golgi apparatus. To evaluate this possibility, we analyzed the intracellular distribution of a well-characterized foreign protein, VSV G, in the presence of the F13L protein. Initial experiments indicated that VSV G extensively colocalized with F13L-GFP-containing vesicles when the two proteins were coexpressed. We then took advantage of the temperature sensitivity of VSV G-ts045 to synchronize the exit of this protein from the endoplasmic reticulum, where it is retained at 40°C (11, 34). Cells were cotransfected with pF13L-GFP and a plasmid encoding the VSV G-ts045 protein at 39°C. Although the mutated VSV G remained in the endoplasmic reticulum, the intracellular distribution of F13L-GFP was the same as that at 37°C. Within 15 min of shifting the temperature to 31°C, the mutated VSV G moved to juxtanuclear Golgi membranes, and by 45 min, more than 60% colocalized with F13L-GFP in vesicles (Fig. 2). Thus, the movement of VSV G into vesicles occurred after trafficking through the Golgi apparatus. Our finding that F13L influenced the intracellular distribution of VSV G as well as vaccinia virus membrane proteins strongly suggested that the mechanism did not involve sequence-specific protein-protein interactions.
FIG. 2.
Time course of VSV G colocalization with F13L-GFP. HeLa cells were cotransfected with plasmids pF13L-GFP and pVSVGts045 and incubated at 39°C. After 24 h, the cells were shifted to 31°C and chased for the time indicated on the left of each row. Cells were fixed, permeabilized, stained with anti-VSV G MAb followed by rhodamine red-conjugated anti-mouse immunoglobulin antibody, and analyzed by confocal microscopy. Green, GFP; red, rhodamine red; yellow, overlap of green and red.
Colocalization of PLD with F13L and B5R proteins.
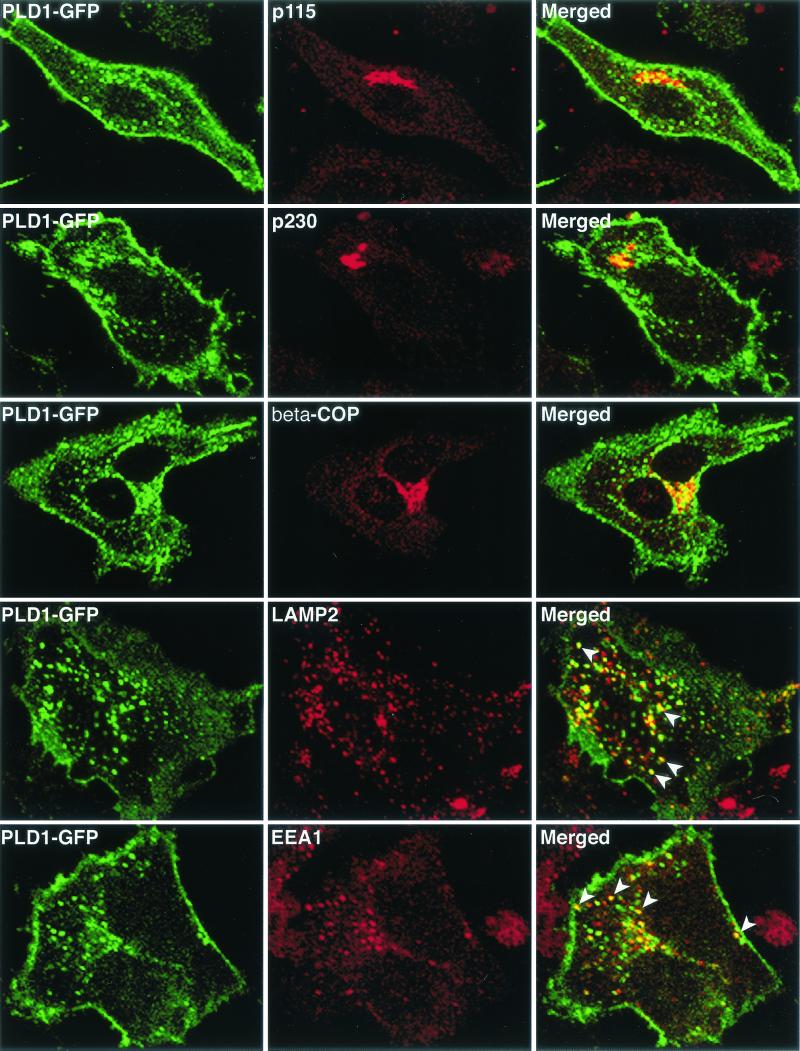
Based on the above results, and our previous finding that the phospholipase motif of F13L was required for localization of B5R in endosomal vesicles, we considered the possibility that increased expression of the tightly regulated cellular PLD could alter the intracellular trafficking of B5R. First, we expressed PLD1-GFP and analyzed its intracellular distribution. Juxtanuclear green fluorescence overlapped with Golgi membrane markers p115, p230, and β-COP, and approximately 50% of the punctate cytoplasmic fluorescence colocalized with late endosomal-lysosomal marker LAMP2 and 40% colocalized with the early endosomal marker EEA1 (Fig. 3). This pattern generally agrees with previous reports (5, 46) and was very similar to the intracellular distribution previously described for F13L-GFP (22), as was confirmed by cotransfection of F13L-GFP with PLD1-HA (Fig. 4). Nearly all of the PLD1-HA colocalized with F13L-GFP (Fig. 4).
FIG. 3.
Colocalization of PLD1-GFP with cellular markers. HeLa cells were transfected with plasmid pPLD1-GFP for 24 h and then fixed, permeabilized, and stained with the indicated antibodies. Transfected HeLa cells were stained with mouse MAbs to p115, p230, LAMP2, or EEA1 or rabbit polyclonal anti-β-COP antibodies followed by rhodamine red-conjugated anti-mouse immunoglobulin antibody or tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit immunoglobulin antibody, respectively. Cells were analyzed by confocal microscopy. Green, GFP; red, tetramethyl rhodamine isothiocyanate; yellow, overlap of green and red. White arrowheads indicate vesicles containing PLD1-GFP and endosomal marker LAMP2 or EEA1.
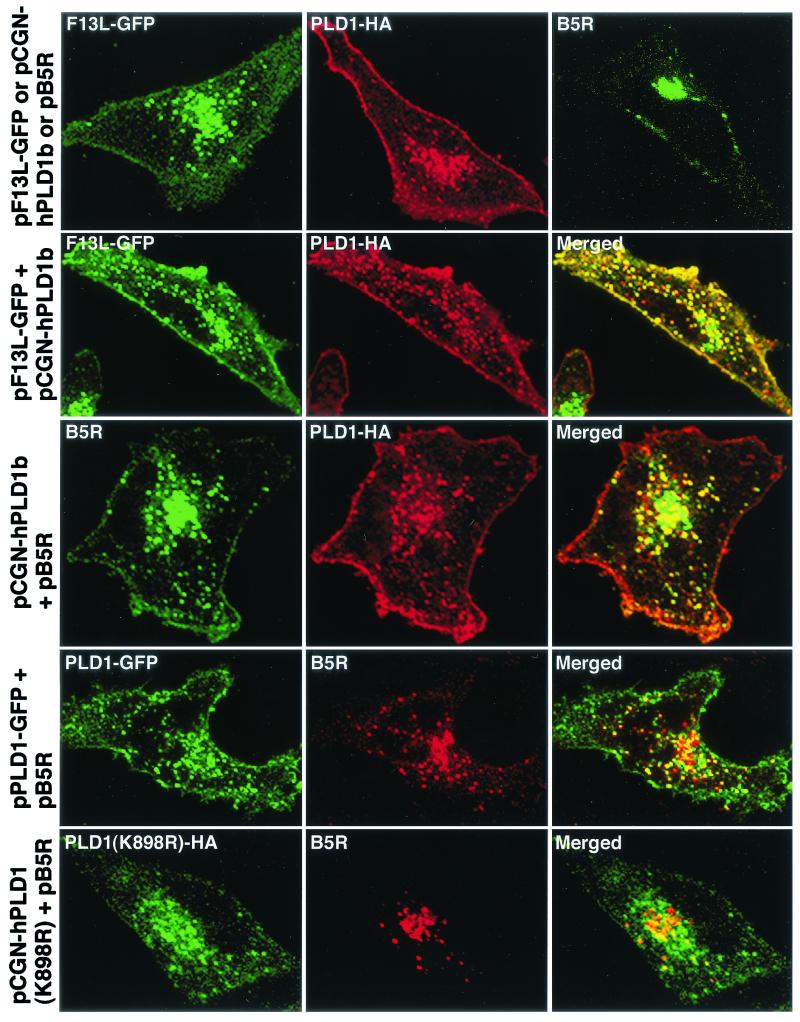
FIG.4.
Colocalization of PLD1 with F13L-GFP and B5R in transfected cells. HeLa cells were transfected with plasmids indicated at the left of each row and incubated for 24 h. First row, cells transfected with pF13L-GFP alone (left), pCGN-hPLD1b alone (middle), or pB5R alone (right) were unstained or stained with mouse anti-HA MAb or rat anti-B5R MAb followed by rhodamine red-conjugated anti-mouse immunoglobulin antibody or fluorescein isothiocyanate-conjugated anti-rat immunoglobulin antibody. Second row, cells cotransfected with plasmid pF13L-GFP and pCGN-hPLD1b were stained with mouse anti-HA followed by rhodamine red-conjugated immunoglobulin antibody. Third row, cells cotransfected with plasmids pCGN-hPLD1b and pB5R were stained for HA and then for B5R as in the first row. Fourth and fifth rows, cells cotransfected with pPLD1-GFP and pB5R or pPLD1(K898R)-HA and pB5R, respectively, were stained for B5R or HA and examined by confocal microscopy as in the third row. Green, GFP or fluorescein isothiocyanate; red, rhodamine red; yellow, overlap of green and red.
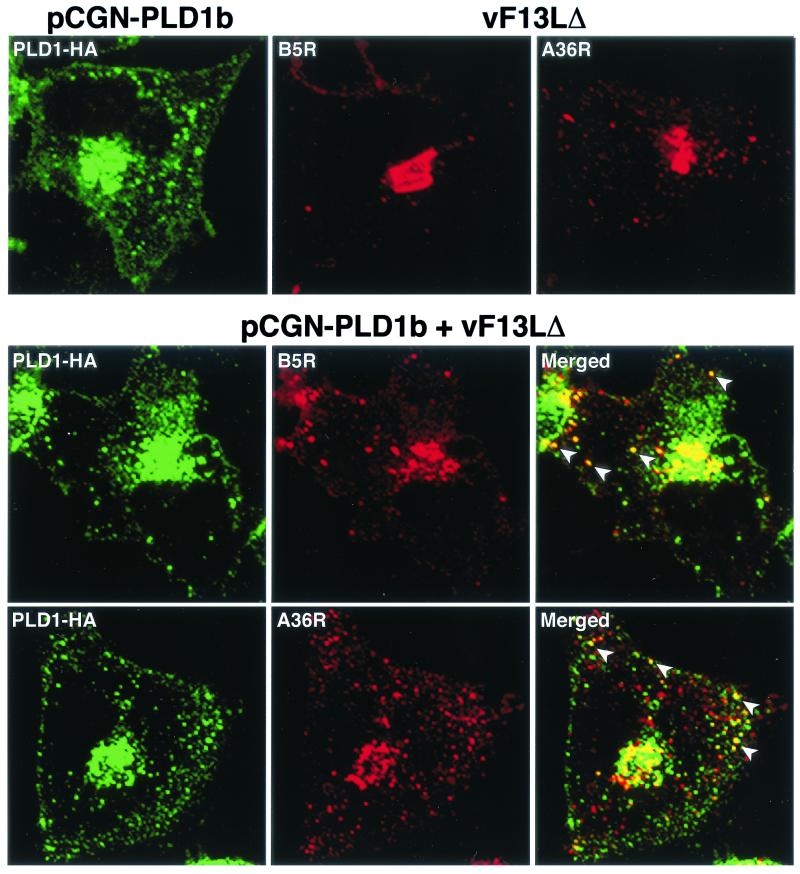
Next, we transfected a plasmid encoding B5R either by itself or with one that encoded either PLD1-HA or PLD1-GFP. Although the B5R protein was found predominantly in juxtanuclear Golgi membranes when expressed alone, more than 70% of the punctate fluorescence colocalized with PLD1-HA or PLD1-GFP after cotransfection (Fig. 4). In contrast, when a plasmid with PLD1 mutated at the active site (K898R) was transfected, the localization of the mutated protein was altered as described before (5) and importantly most of the B5R protein remained in the juxtanuclear Golgi region (Fig. 4). Additional experiments were carried out by transfecting cells with a plasmid expressing PLD1-HA and infecting them with vF13LΔ. Whereas B5R and A36R were predominantly juxtanuclear in cells infected with the mutant virus, colocalization with vesicles containing PLD1-HA was found in the transfection-infection experiment (Fig. 5).
FIG. 5.
Intracellular localization of B5R and A36R in vF13LΔ-infected cells expressing PLD1-HA. First row, HeLa cells were transfected with plasmid pCGN-PLD1b or infected with vF13LΔ. Second and third rows, transfected cells were infected with vF13LΔ and after 18 h were fixed, permeabilized, and stained with anti-HA MAb followed by Alexa 488-conjugated anti-mouse immunoglobulin antibody. Cells were then stained with either anti-B5R MAb or anti-A36R followed by tetramethyl rhodamine isothiocyanate-conjugated anti-rat or anti-rabbit immunoglobulin antibody, respectively. Stained cells were analyzed by confocal microscopy. Green, Alexa 488; red, tetramethyl rhodamine isothiocyanate; yellow, overlap of green and red. White arrowheads show the colocalization of PLD1 with B5R and A36R in vesicular structures.
Attempts to complement F13L with PLD1.
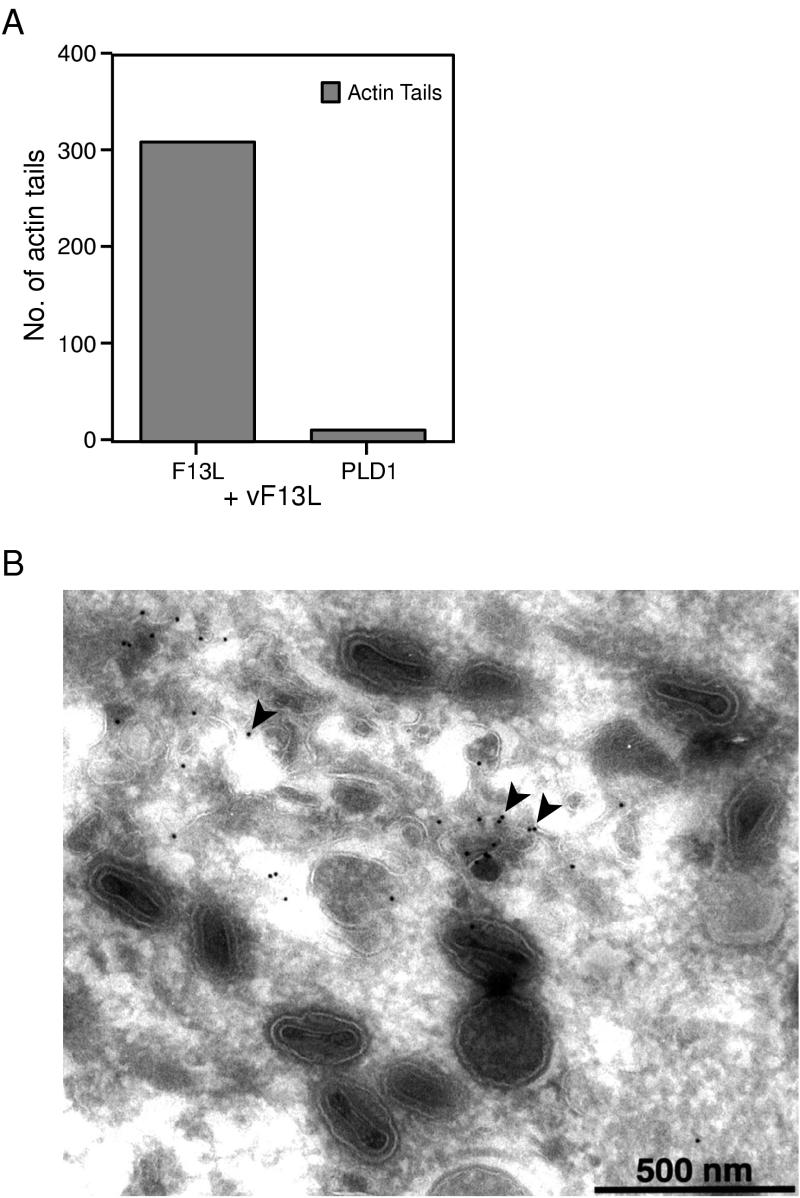
Further experiments were carried out to determine whether PLD1 could fully substitute for F13L or only for induced vesicles that contained B5R and A36R. A strong vaccinia virus late promoter was used to regulate expression of PLD1, and the plasmid was transfected into cells infected with vF13LΔ. As an initial measure of complementation, the cells were stained with phalloidin to discern IEV or CEV with actin tails. When cells were transfected with a plasmid expressing F13L, actin tails were present in the majority of cells whereas very few cells transfected with PLD1 had actin tails (Fig. 6A). In addition, we constructed a recombinant vaccinia virus in which the PLD1-GFP ORF replaced that of F13L. This recombinant virus (called vPLD1-GFP) expressed an expected ∼150-kDa fusion protein as demonstrated by immunoblotting and immunoprecipitation analyses (data not shown). Nevertheless, vPLD1-GFP formed small plaques, and IEV were not detected by electron microscopy. PLD1-GFP was associated with membranes, some of which were in the vicinity of IMV but not associated with them (Fig. 6B). We concluded that PLD1 could not fully substitute for F13L.
FIG. 6.
Failure of PLD1 to complement an F13L deletion mutant. (A) Quantitation of actin tail formation in vF13LΔ-infected cells transfected with either pGF13L or pGPLD1-LR. HeLa cells were infected with vF13LΔ for 2 h and then transfected with plasmid pGF13L or pGPLD1-LR. At 24 h after transfection, the cells were fixed and permeabilized. Cells transfected with plasmid pGPLD1-LR were stained with anti-HA MAb followed by Alexa 488-conjugated anti-mouse immunoglobulin antibody. Plasmid pGF13L has GFP at the C terminus of F13L. Data shown are the averages of three separate experiments. (B) vPLD1-GFP does not produce IEV particles. A confluent monolayer of RK13 cells was infected with vPLD1-GFP. After 24 h, cells were fixed, cryosectioned, and probed with rabbit anti-GFP polyclonal antibodies followed by protein A conjugated to 10-nm colloidal gold particles. Arrowheads point to PLD1-GFP fusion protein on membranes close to IMV particles.
Effects of PLD inhibitors on intracellular localization of F13L protein and formation of extracellular vaccinia virus.
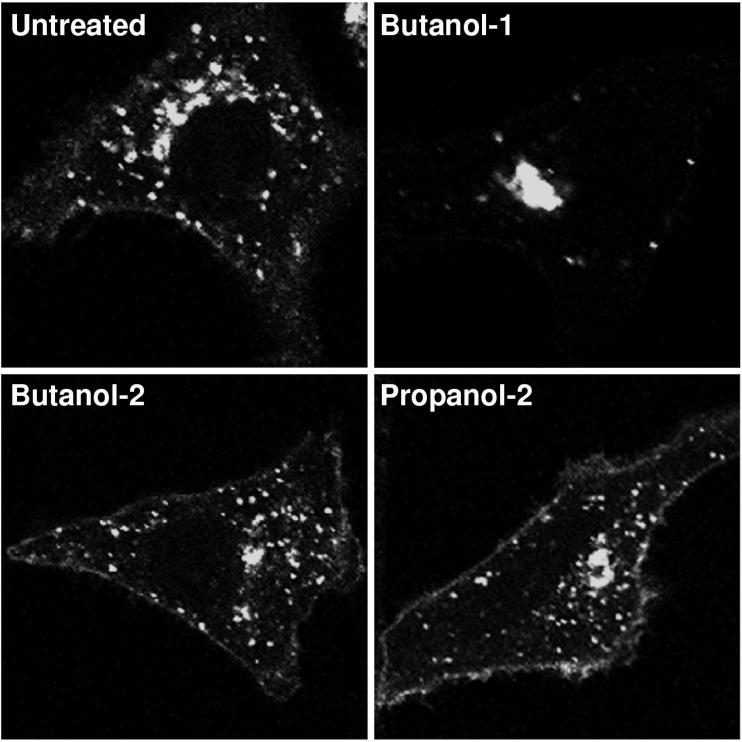
Primary alcohols such as butanol-1, but not secondary or tertiary alcohols, are specific inhibitors of PLD, as they can participate in PLD transphosphatidylation reactions, prevent synthesis of phosphatidic acid, and inhibit PLD-mediated vesicle budding (6, 26). When cells that were transfected with pF13L-GFP were incubated with 1% butanol-1, the fusion protein remained in the juxtanuclear Golgi region (Fig. 7). Neither butanol-2 nor propanol-2 had this effect, strongly suggesting that it was due to inhibition of phospholipase activity.
FIG. 7.
Effects of butanol-1, butanol-2, and propanol-2 on intracellular localization of F13L-GFP. HeLa cells were transfected with plasmid pF13L-GFP. After 4 h, 1% concentrations of the indicated alcohols were added to the media and the incubations were continued for a total of 24 h at 37°C. Cells were fixed and analyzed by confocal microscopy.
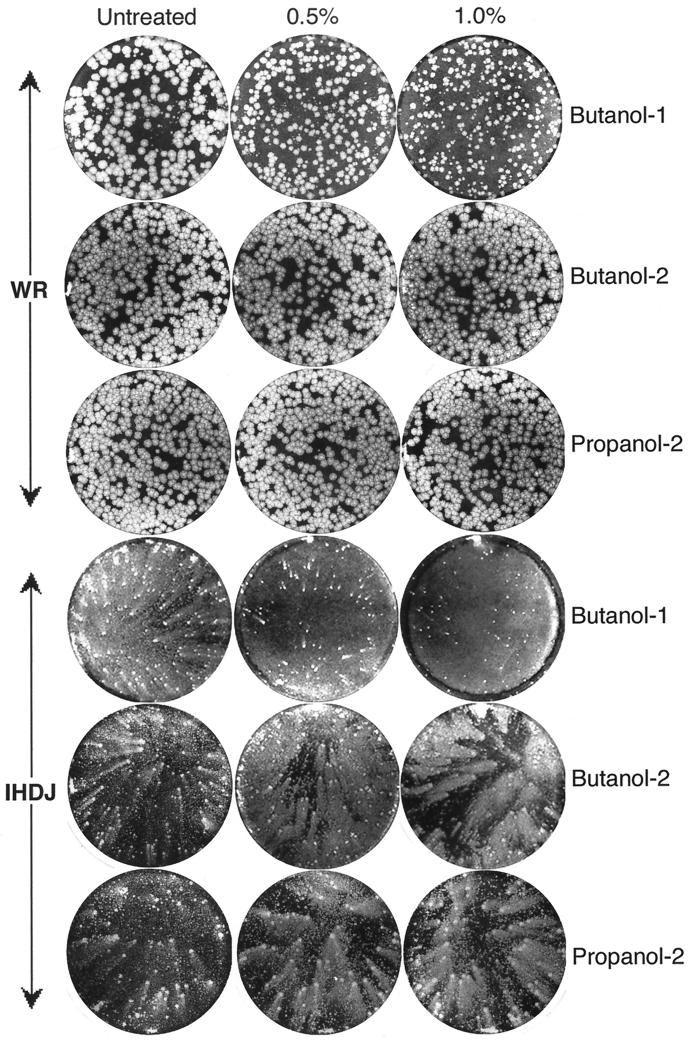
Additional experiments were carried out to ascertain the effects of butanol-1 on vaccinia virus infection. BS-C-1 cells were infected with either vaccinia virus strain WR or strain IHD-J. The WR strain produces large round plaques due to efficient direct cell-to-cell spread of the CEV and the release of only small amounts of EEV. In contrast, the IHD-J strain produces large amounts of EEV and consequently forms comet-shaped plaques in liquid overlay medium. Following infection, the monolayers were treated with 0, 0.5, or 1% butanol-1, butanol-2, or propanol-2. Butanol-1 had a profound effect, reducing the size of WR plaques and inhibiting IHD-J comet formation, whereas the secondary alcohols had little effect (Fig. 8).
FIG. 8.
Effects of butanol-1, butanol-2, and propanol-2 on plaque size of vaccinia virus. BS-C-1 cells, in six-well tissue culture plates, were infected with 50 to 100 PFU of vaccinia virus strain WR or IHD-J/well. After 2 h of adsorption of IHD-J, the virus inocula were replaced with liquid media supplemented with 0.5 or 1.0% concentrations of the indicated alcohol and the incubation was continued for 2 days. For vaccinia virus WR, methylcellulose was included in the overlay and the cells were incubated for 3 days. Plaques were visualized by staining with crystal violet.
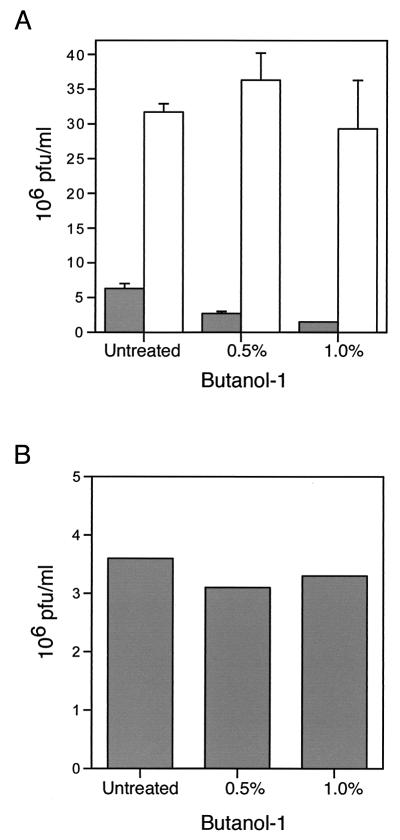
To ensure that the effects on plaque size and comet formation were not due to a nonspecific effect of butanol-1 on virus replication, we determined the yields of intracellular and extracellular IHD-J virus. As shown in Fig. 9A, butanol-1 had no effect on the formation of intracellular virus but reduced the yield of extracellular virus by fourfold. A control experiment, carried out by incubating harvested medium containing extracellular virus with 1% butanol-1 for 48 h at 37°C, showed that the reduction of extracellular virus was not due to the effect of butanol-1 on its infectivity (Fig. 9B).
FIG. 9.
Effect of butanol-1 on the production of IMV and EEV particles. (A) BS-C-1 cells were infected with vaccinia virus strain IHD-J. After 2 h, the inoculum was replaced with medium supplemented with 0.5 or 1.0% butanol-1. After 48 h at 37°C, the medium and cells were harvested separately in similar volumes and analyzed by plaque assay. Plaque numbers are presented as the averages of three separate counts from two different experiments with standard deviations. Filled and unfilled bars refer to extracellular and cell-associated virus, respectively. (B) BS-C-1 cells were infected with vaccinia virus strain IHD-J, and after adsorption, inoculum was replaced with normal medium. After 48 h, the medium was collected; cleared by low-speed centrifugation; and incubated with 0, 0.5, or 1.0% butanol-1 at 37°C for 48 h. Infectious virus was quantified by plaque assay.
DISCUSSION
We presented evidence that the vaccinia virus F13L protein, like PLD1, induces the formation of post-Golgi vesicles containing proteins that otherwise would localize in Golgi membranes. The post-Golgi vesicles contain endosomal markers, which may reflect increased trafficking induced by F13L and which appears to occur in vaccinia virus-infected cells as well (39, 47). Our finding that a specific PLD inhibitor prevented the induction of vesicles and also inhibited wrapping of IMV, cell-to-cell virus spread, and formation of extracellular virus suggests that F13L has or induces phospholipase activity that contributes to the formation of the virus-modified wrapping cisternae.
At the start of this study we posed several questions. The first was whether colocalization of B5R with F13L is specific or whether other Golgi transmembrane proteins colocalize with F13L. In a previous study (22), we demonstrated by cotransfection experiments that the B5R membrane protein colocalized with the F13L protein in endosomal vesicles. In the absence of F13L, B5R was predominantly associated with juxtanuclear Golgi membranes. Attempts to show interaction of the B5R and F13L proteins by coimmunoprecipitation were unsuccessful, however, leading us to suspect that the colocalization did not require sequence-specific interactions. Again using cotransfection methods, we found that another IEV protein, A36R, colocalized with F13L in endosomal vesicles. Moreover, the VSV G protein also colocalized with F13L. We took advantage of a VSV G protein harboring a ts mutation that prevented it from exiting the endoplasmic reticulum at the nonpermissive temperature. When the temperature was lowered, the G protein was first transported to the Golgi network and then into endosomal vesicles within 30 to 45 min. As sequence-specific interactions between vaccinia virus and VSV proteins seemed improbable, we concluded that F13L induced the formation of vesicles that incorporated Golgi membrane proteins relatively nonspecifically.
The second and third questions were whether B5R and A36R colocalize with PLD in place of F13L in post-Golgi vesicles and whether PLD can complement an F13L deletion mutant. Our previous study (22) demonstrated that mutations in the phospholipase motif of F13L abrogated its ability to induce colocalization of B5R. Since the present findings made sequence-specific interactions between F13L and colocalizing proteins unlikely, we considered that PLD1 might at least partially substitute for F13L. This prediction was borne out by cotransfection experiments. PLD1 and F13L colocalized with each other, and PLD1 induced the colocalization of B5R in endosomal vesicles. Moreover, B5R and A36R colocalized with PLD1 when cells expressing PLD1 were infected with an F13L deletion mutant virus. These results are consistent with reports that PLD stimulates release of nascent secretory vesicles from the trans-Golgi network (6, 26). PLD1, however, could not substitute for F13L in the formation of IEV, suggesting that PLD activity is insufficient for virus assembly.
The fourth question was whether a PLD inhibitor would prevent the localization of F13L in endosomes and inhibit extracellular vaccinia virus formation. We used a specific PLD inhibitor to correlate an enzymatic role of F13L in inducing vesicles with the wrapping process needed to form extracellular virus. In the presence of primary but not secondary alcohols, PLD produces phosphatidylalcohols at the expense of phosphatidic acid. Alcohol concentrations of 1 to 3% have been shown elsewhere to be inhibitory in vitro and in vivo (6, 26). We demonstrated that 1% butanol-1 prevented the localization of F13L in endosomal vesicles and severely reduced vaccinia virus plaque size and comet formation. No such effect was seen with the same concentrations of butanol-2 or propanol-2. The specificity of this effect was confirmed by showing that butanol-1 inhibited formation of extracellular virus without significantly reducing the yield of IMV.
The findings that F13L contains a region similar to the catalytic site of PLD (25, 33, 45); mutagenesis of the catalytic motif abrogates F13L function (22, 37, 45); F13L, like PLD, induces post-Golgi vesicle formation; and primary alcohols inhibit F13L and PLD function are all consistent with F13L having an essential PLD activity. Nevertheless, F13L lacks a conserved histidine of the PLD active site motif, and recombinant F13L fusion proteins were found previously only to have phospholipase A and C activities in vitro (1). Although this negative result for PLD activity could be due to a deficiency in the in vitro assay conditions or the absence of an activator, it is possible that the PLD-like effects of F13L seen in vivo are indirect and result from its other described phospholipase activities or other interactions. The inhibition of F13L-mediated vesicle formation by expression of amphiphysins (unpublished data), which specifically bind PLD (27), would be consistent with activation of PLD by F13L. PLD is a highly regulated protein and is activated by ADP-ribosylation factor 1, which indirectly induces the formation of coatomer-coated vesicles (17, 26).
In summary, our present and previous data support a model in which the F13L protein localizes in Golgi membranes and induces vesicle formation by direct or indirect means through a PLD-like activity. Viral membrane proteins present in the Golgi apparatus are included in the induced vesicles. Up to this stage, PLD1 can substitute for F13L. Further steps involving the membrane wrapping of IMV, however, depend on additional properties of the F13L protein that remain to be determined.
Acknowledgments
We thank M. Frohman for plasmids, A. Weisberg for electron microscopy, Norman Cooper for providing tissue culture cells, and Owen Schwartz for helping in confocal microscopy.
REFERENCES
- 1.Baek, S.-H., J.-Y. Kwak, S. H. Lee, T. Lee, S. H. Ryu, D. J. Uhlinger, and J. D. Lambeth. 1997. Lipase activities of p37, the major envelope protein of vaccinia virus. J. Biol. Chem. 272:32042-32049. [DOI] [PubMed] [Google Scholar]
- 2.Bednarek, S. Y., L. Orici, and R. Schekman. 1996. Traffic COPS and the formation of vesicle coats. Trends Cell Biol. 6:468-473. [DOI] [PubMed] [Google Scholar]
- 3.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, F. D., N. Thompson, K. M. Saqib, J. M. Clark, D. Powner, N. T. Thompson, R. Solari, and M. J. Wakelam. 1998. Phospholipase D1 localizes to secretory granules and lysosomes and is plasma-membrane translocated on cellular stimulation. Curr. Biol. 8:835-838. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y. G., A. Siddhanta, C. D. Austin, S. M. Hammond, T. C. Sung, M. A. Frohman, A. J. Morris, and D. Shields. 1997. Phospholipase D stimulates release of nascent secretory vesicles from the trans-Golgi network. J. Cell Biol. 138:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 8.Engelstad, M., S. T. Howard, and G. L. Smith. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 188:801-810. [DOI] [PubMed] [Google Scholar]
- 9.Engelstad, M., and G. L. Smith. 1993. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194:627-637. [DOI] [PubMed] [Google Scholar]
- 10.Frohman, M. A., and A. J. Morris. 1996. Rho is only ARF the story. Phospholipid signalling. Curr. Biol. 6:945-947. [DOI] [PubMed] [Google Scholar]
- 11.Gallione, C. J., and J. K. Rose. 1985. A single amino acid substitution in a hydrophobic domain causes temperature-sensitive cell-surface transport of a mutant viral glycoprotein. J. Virol. 54:374-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths, G., N. Roos, S. Schleich, and J. K. Locker. 2001. Structure and assembly of intracellular mature vaccinia virus: thin-section analyses. J. Virol. 75:11056-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimley, P. M., E. N. Rosenblum, S. J. Mims, and B. Moss. 1970. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J. Virol. 6:519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosenbach, D. W., and D. E. Hruby. 1998. Analysis of a vaccinia virus mutant expressing a nonpalmitylated form of p37, a mediator of virion envelopment. J. Virol. 72:5108-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosenbach, D. W., D. O. Ulaeto, and D. E. Hruby. 1997. Palmitylation of the vaccinia virus 37-kDa major envelope antigen. Identification of a conserved acceptor motif and biological relevance. J. Biol. Chem. 272:1956-1964. [DOI] [PubMed] [Google Scholar]
- 17.Hammond, S. M., Y. M. Altshuller, T. C. Sung, S. A. Rudge, K. Rose, J. Engebrecht, A. J. Morris, and M. A. Frohman. 1995. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 270:29640-29643. [DOI] [PubMed] [Google Scholar]
- 18.Hiller, G., and K. Weber. 1985. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirt, P., G. Hiller, and R. Wittek. 1986. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol. 58:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollinshead, M., A. Vanderplasschen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Husain, M., and B. Moss. 2001. Vaccinia virus F13L protein with a conserved phospholipase catalytic motif induces colocalization of the B5R envelope glycoprotein in post-Golgi vesicles. J. Virol. 75:7528-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isaacs, S. N., E. J. Wolffe, L. G. Payne, and B. Moss. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 66:7217-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz, E., E. J. Wolffe, and B. Moss. 1997. The cytoplasmic and transmembrane domains of the vaccinia virus B5R protein target a chimeric human immunodeficiency virus type 1 glycoprotein to the outer envelope of nascent vaccinia virions. J. Virol. 71:3178-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin, E. V. 1996. A duplicated catalytic motif in a new superfamily of phosphohydrolases and phospholipid synthases that includes poxvirus envelope proteins. Trends Biochem. Sci. 21:242-243. [PubMed] [Google Scholar]
- 26.Ktistakis, N. T., H. A. Brown, M. G. Waters, P. C. Sternweis, and M. G. Roth. 1996. Evidence that phospholipase D mediates ADP ribosylation factor-dependent formation of Golgi coated vesicles. J. Cell Biol. 134:295-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, C., S. R. Kim, J. K. Chung, M. A. Frohman, M. W. Kilimann, and S. G. Rhee. 2000. Inhibition of phospholipase D by amphiphysins. J. Biol. Chem. 275:18751-18758. [DOI] [PubMed] [Google Scholar]
- 28.Lefrancios, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology 121:157-167. [PubMed] [Google Scholar]
- 29.Lorenzo, M. M., I. Galindo, G. Griffiths, and R. Blasco. 2000. Intracellular localization of vaccinia virus extracellular enveloped virus envelope proteins individually expressed using a Semliki Forest virus replicon. J. Virol. 74:10535-10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Nagayama, A., B. G. T. Pogo, and S. Dales. 1970. Biogenesis of vaccinia: separation of early stages from maturation by means of rifampicin. Virology 40:1039-1051. [DOI] [PubMed] [Google Scholar]
- 32.Payne, L. G. 1980. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J. Gen. Virol. 50:89-100. [DOI] [PubMed] [Google Scholar]
- 33.Ponting, C. P., and I. D. Kerr. 1996. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 5:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Presley, J. F., N. B. Cole, T. A. Schrer, K. Hirschberg, K. J. M. Zaal, and J. Lippincott-Schwartz. 1997. ER-to-Golgi transport visualized in living cells. Nature 389:81-85. [DOI] [PubMed] [Google Scholar]
- 35.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmström, F. Frischknecht, M. Zettl, T. Zimmerman, and M. Way. 2001. Kinesin dependent movement on microtubules precedes actin based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 36.Roper, R., E. J. Wolffe, A. Weisberg, and B. Moss. 1998. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 72:4192-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roper, R. L., and B. Moss. 1999. Envelope formation is blocked by mutation of a sequence related to the HKD phospholipid metabolism motif in the vaccinia virus F13L protein. J. Virol. 73:1108-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanderson, C. M., F. Frischknecht, M. Way, M. Hollinshead, and G. L. Smith. 1998. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J. Gen. Virol. 79:1415-1425. [DOI] [PubMed] [Google Scholar]
- 39.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans-Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmutz, C., L. Rindisbacher, M. C. Galmiche, and R. Wittek. 1995. Biochemical analysis of the major vaccinia virus envelope antigen. Virology 213:19-27. [DOI] [PubMed] [Google Scholar]
- 41.Seaman, M. N. J. 1996. Phospholipase D in vesicle budding. Trends Cell Biol. 6:473.. [DOI] [PubMed] [Google Scholar]
- 42.Simon, J. P., I. E. Ivanov, M. Adesnik, and D. D. Sabatini. 1996. The production of post-Golgi vesicles requires a protein kinase C-like molecule, but not its phosphorylating activity. J. Cell Biol. 135:355-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stokes, G. V. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J. Virol. 18:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung, T.-C., R. L. Roper, Y. Zhang, S. A. Rudge, R. Temel, S. M. Hammond, A. J. Morris, B. Moss, J. Engebrecht, and M. A. Frohman. 1997. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J. 16:4519-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toda, K., M. Nogami, K. Murakami, Y. Kanaho, and K. Nakayama. 1999. Colocalization of phospholipase D1 and GTP-binding-defective mutant of ADP-ribosylation factor 6 to endosomes and lysosomes. FEBS Lett. 442:221-225. [DOI] [PubMed] [Google Scholar]
- 47.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 48.Ward, B. M., and B. Moss. 2000. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J. Virol. 74:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolffe, E. J., S. N. Isaacs, and B. Moss. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 67:4732-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolffe, E. J., E. Katz, A. Weisberg, and B. Moss. 1997. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J. Virol. 71:3904-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolffe, E. J., A. Weisberg, and B. Moss. 2001. The vaccinia virus A33R protein provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J. Virol. 75:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]