Abstract
The proliferation-specific Forkhead Box M1 (FoxM1 or FoxM1b) transcription factor is over expressed in a number of aggressive human carcinomas. Mouse hepatocytes deficient in FoxM1 fail to proliferate and are highly resistant to developing carcinogen induced liver tumors. We previously developed a transgenic (TG) mouse line in which the ubiquitous Rosa26 promoter was used to drive expression of the human FoxM1b cDNA transgene in all mouse cell-types. To investigate the role of FoxM1b in prostate cancer progression, we bred Rosa26-FoxM1b mice with both TRAMP and LADY TG mouse models of prostate cancer. We show that increased expression of FoxM1b accelerated development, proliferation and growth of prostatic tumors in both TRAMP and LADY double TG mice. Furthermore, development of prostate carcinomas in TRAMP/Rosa26-FoxM1b double TG mice required high levels of FoxM1 protein to overcome sustained expression of the ARF tumor suppressor, a potent inhibitor of FoxM1 transcriptional activity. Depletion of FoxM1 levels in prostate cancer cell lines PC-3, LNCaP or DU-145 by siRNA transfection caused significant reduction in proliferation and anchorage-independent growth on soft agar. This phenotype was associated with increased nuclear levels of the cyclin dependent kinase inhibitor protein p27Kip1 and diminished expression of S-phase promoting cyclin A2 and M-phase promoting cyclin B1 proteins. Finally, we show that elevated levels of FoxM1 protein correlate with high proliferation rates in human prostate adenocarcinomas. Our results suggest that the FoxM1 transcription factor regulates development and proliferation of prostate tumors, and that FoxM1 is a novel target for prostate cancer treatment.
Keywords: Forkhead Box M1 transcription factor, ARF Tumor Suppressor, growth on soft agar, prostate carcinoma, TRAMP and LADY prostate cancer mouse models, siRNA, cyclin B1, cyclin A2, cyclin dependent kinase Inhibitors, HFH-11B
Introduction
Development of cancer is a multi-step process involving gain of function mutations that activate signaling pathways, which stimulate cell cycle progression and cell survival (1, 2). Cancer progression also requires inactivation of tumor suppressor genes that function to arrest cell proliferation in response to oncogenic stimuli (3). Activation of the Ras-MAPK signaling pathway drives cell cycle progression by temporal expression of cyclin regulatory subunits, which activate their corresponding cyclin-dependent kinases (CDKs) through complex formation (3, 4). Progression into DNA replication (S-phase) requires phosphorylation of the Retinoblastoma (RB) protein by either the Cdk4/Cdk6 proteins in complex with cyclin D or cdk2 in complex with cyclin E or cyclin A2 (4). Hyper-phosphorylated RB dissociates from the E2F transcription factor and alleviates inhibition of E2F to allow transcriptional stimulation of Sphase promoting genes. The CDK inhibitor (CDKI) proteins p21Cip1 and p27Kip1 also negatively regulate CDK activity through protein complex formation, and their diminished nuclear expression is required for S-phase progression (4, 5). Activation of the cdk1-cyclin B complex is required to phosphorylate protein substrates essential for mitotic progression (4).
Expression of the Alternative Reading Frame (ARF) tumor suppressor is induced in response to oncogenic stimuli and prevents abnormal cell proliferation through increased stability of the p53 tumor suppressor by nucleolar targeting of Mdm2 (6–8). The ARF protein also targets both the E2F1 and c-Myc transcription factors to the nucleolus, thus preventing their transcriptional activation of their target genes involved in S-phase and/or transformation (9–12). Expression of the ARF protein is extinguished in a variety of tumors through DNA methylation and silencing of the ARF promoter region (3). Furthermore, ARF −/− mice are susceptible to developing spontaneous tumors (13, 14), underscoring the important role of the ARF tumor suppressor in preventing tumorigenesis.
Prostate cancer is one of the leading causes of cancer related death among men in the North America and Western Europe (15). The Transgenic (TG) Adenocarcinoma of the Mouse Prostate (TRAMP) recapitulates multiple stages of human prostate cancer by using the probasin promoter to drive prostate epithelial cell expression of the SV40 virus Large T and Small t Tumor Antigen (Tag) oncoprotein (16, 17), which function to inactivate the tumor suppressor proteins RB, p53 and PP2A Serine/Threonine-specific phosphatase (18). TRAMP TG mice develop prostate epithelial cell hyperplasia and prostatic intraepithelial neoplasia (PIN), which progresses to histological invasive prostatic carcinomas (16, 19). In LADY TG mice the rat probasin promoter drives prostate epithelial cell expression of only the SV40 virus large T antigen oncoprotein and they develop multifocal, low-grade PIN that progresses to high-grade PIN and early invasive prostate carcinomas with progressive neuroendocrine differentiation (20, 21). Both the reproducible and progressive nature of mouse prostate cancer development in the TRAMP and LADY TG mice has provided a greater understanding of the mechanisms involved in development and progression of prostate cancer (16).
The mammalian Forkhead Box (Fox) family of transcription factors consists of more than 50-mammalian proteins (22, 23) that share homology in the winged helix DNA binding domain (24). Expression of FoxM1 (or FoxM1b) is induced in all proliferating mammalian cells and tumor derived cell lines (25–29). Liver regeneration studies with mice in which the Albumin promoter-enhancer Cre Recombinase (Alb-Cre) mediated conditional deletion of the FoxM1 LoxP/LoxP (fl/fl) targeted allele in adult hepatocytes demonstrated that FoxM1 is required for hepatocyte DNA replication and mitosis (30, 31). FoxM1 was shown to be essential for diminishing nuclear accumulation of CDK inhibitor (CDKI) proteins p21Cip1 and p27Kip1 and for transcription of Cdc25B phosphatase, which is essential for stimulating activity of M-phase promoting Cdk1 (30, 32, 33). Using a well-established liver tumor initiation and promotion method, we showed that Alb-Cre FoxM1 −/− hepatocytes fail to proliferate and are highly resistant to chemical induced hepatocellular carcinomas (HCC) (32, 34). Furthermore, the FoxM1 transcription factor was identified as a novel inhibitory target of the mouse ARF tumor suppressor, which binds to FoxM1 and reduces FoxM1 transcriptional activity by targeting it to the nucleolus (32, 34). Moreover, we previously developed transgenic (TG) mice in which the Rosa26 promoter was used to drive ubiquitous expression of the human FoxM1b cDNA and increased FoxM1b levels stimulated proliferation of pulmonary cells in response to lung injury (35).
The FoxM1 transcription factor is highly expressed in human HCC (36), intrahepatic cholangiocarcinomas (37), basal cell carcinomas (38), infiltrating ductal breast carcinomas (39), anaplastic astrocytomas and glioblastomas (40) and in a number of other human tumors (41), suggesting that FoxM1 is involved in the proliferative expansion of many different human tumors. In this study we show that FoxM1 protein is expressed in highly proliferative human prostate cancer. To determine the potential role of FoxM1b in prostate cancer progression, we crossed Rosa26-FoxM1b TG mice with well established TRAMP (17) or LADY (20) TG mouse models of prostate cancer. We demonstrated that increased expression of FoxM1b accelerated development, proliferation and growth of prostate carcinomas in both TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice. Furthermore, inhibiting FoxM1 levels in human prostate cancer cell lines by siRNA transfection significantly diminished cell proliferation and anchorage independent growth on soft agar with altered expression of a number of cell cycle regulatory proteins. We also show that FoxM1 mediated increase in cyclin A2 expression correlates with enhanced proliferation and transformation of prostate cancer cells both in vitro and in vivo. Our results suggest that the FoxM1 transcription factor regulates development and proliferation of prostate tumors.
Materials and Methods
Transgenic mice.
Wild type (WT) C57BL/6 mice and TRAMP transgenic (TG) C57BL/6 mice expressing the SV40 Tag controlled by rat probasin regulatory elements were purchased from The Jackson Laboratory (Bar Harbor, ME). Female LADY (12T-10) TG mice were obtained from Dr. Robert J. Matusik (Vanderbilt Prostate Center, Nashville, TN). We previously described development of FVB/N TG mice in which the Rosa26 promoter drives ubiquitous expression of the human FoxM1b cDNA transgene in all cell types (35). Rosa26-FoxM1b TG mice were bred with TRAMP or LADY TG mice to generate TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice. Mice were sacrificed at approximately 90 or 165 days of age for TRAMP single or double TG mice and at approximately 120 or 150 days of age for LADY single and double TG mice. Prostate tissues were either used to prepare total RNA with RNA-STAT-60 (Tel-Test “B” Inc. Friendswood, TX) or fixed, paraffin embedded, and sectioned for morphological examination following histological staining with hematoxylin and eosin (H&E) or for immunohistochemical staining as described previously (35).
Immunohistochemical staining of prostate tissue sections.
Five μm sections of paraffin embedded mouse prostate tissue or human prostate cancer tissue microarrays (US BioMax, Inc., Ijamsville, MD; Catalog # PR801) were stained with H&E or used for immunohistochemical staining with mouse monoclonal Proliferation Cell Nuclear Antigen (PCNA) antibody (1:1000, clone PC-10, Roche Diagnostics) and mouse antibody conjugated with alkaline phosphatase (AP) followed by NBT/BCIP substrate (Vector Labs, Burlingame, CA) detection as described previously (35). PCNA immunostained prostate tissue sections were counterstained with Nuclear Fast Red as described previously (35). Prostate cancer tissue sections were also used for immunohistochemical staining with rabbit polyclonal FoxM1b antibody (1:200) (42) or mouse anti-Nucleophosmin (NPM/B23) monoclonal antibody (1:1000; Zymed, South San Francisco, CA; Catalog No. 32-5200) and secondary anti-rabbit antibody or anti-mouse antibody conjugated with biotin, avidin-horseradish peroxidase (HRP) complex, and DAB substrate (all from Vector Lab) for detection as described previously (29, 33).
Transfection of prostate cancer cell lines with siRNA duplexes
The human prostate cancer cell lines PC-3, LNCaP and DU-145 were obtained from American Type Culture Collection (ATCC, Manassas, VA) and were grown in culture as recommended by ATCC. In order to inhibit FoxM1 expression in prostate tumor cell lines, Dharmacon Research synthesized 21-nucleotide siRNA duplexes specific to either human FoxM1 (siFoxM1 #2; ggaccacuuucccuacuuu) or p27Kip1 (siP27; guacgaguggcaagagguguu) mRNA as described previously (42). These siRNA duplexes were transfected into PC-3, LNCaP and DU-145 cells using LipofectamineTM 2000 reagent (Invitrogen) in serum free tissue culture media as described previously (42). PC-3, LNCaP or DU-145 cells were harvested at 72 hours following siRNA transfection and used to prepare total RNA using RNA-STAT-60 (Tel-Test “B” Inc. Friendswood, TX).
Cell growth assay of prostate cancer cell lines depleted in mRNA levels of FoxM1.
To determine the growth rate of FoxM1 depleted cells, PC-3, LNCaP and DU-145 cells were transfected with siFoxM1 #2 or siP27 duplexes. Two days later FoxM1 or p27Kip1-depleted cells were trypsinized, replated and the cell growth rate was determined in triplicate at 3, 4 or 5 days after siRNA transfection. To determine the number of viable PC-3, LNCaP and DU-145 cells, we used Cell Titer-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) according to the manufacturer’s recommendations. Standard curves were made with increasing numbers of each of the prostate cancer cell lines to convert fluorescent readings into cell numbers. We plotted cell number ± SD versus days after siRNA transfection.
Flow cytometry and Soft agar assay of prostate cancer cell lines depleted in FoxM1 levels.
In order to examine the stage of the cell cycle affected by diminished FoxM1 levels, PC-3, LNCaP and DU-145 cells were transfected (in triplicate) with 100 nM of either siFoxM1 #2 or siP27 duplexes or left untransfected and then 72 hours after transfection the cells were subjected to flow cytometry to analyze their cell cycle profile as described previously (42). For soft agar assay, PC-3, LNCaP and DU-145 cells were transfected with siFoxM1 #2 or siRNA p27Kip1 duplexes (in triplicate) and the cells were trypsinized 24 hours after transfection and then plated on soft agar for three weeks to assay for anchorage-independent cell growth as described previously (32). Triplicate plates were used to count colonies and determine the mean number of colonies ± SD.
Quantitative Real-Time RT-PCR to determine mRNA expression levels.
To prepare total RNA for quantitative Real Time Reverse Transcriptase (QRT)-PCR, the RNA was first digested with DNaseI (RNase free) to remove contaminating genomic DNA and then it was affinity purified on the Qiagen RNeasy Micro Kit (Cat. # 74004). We used the BioRAD cDNA Synthesis Kit containing both oligo-dT and random hexamer primers to synthesize cDNA from 10 μg of total RNA. The following reaction mixture was used for all QRT-PCR samples: 1X of IQ SybrGreen Supermix (Biorad, Carlsbad, CA), 200 nM of each primer, and 2.5 μl of cDNA in a 25μl total volume. Reactions were amplified and analyzed in triplicate using a MyiQ Single Color Real-Time PCR Detection System (Biorad, Carlsbad, CA). The annealing temperature and human and mouse primers for FoxM1 and Cyclophilin and mouse ARF primers to measure mRNA levels by QRT-PCR were described previously (42). These QRT-PCR mRNA levels were normalized to human Cyclophilin mRNA levels ± SD.
Western blot analysis to determine protein expression levels.
Nuclear protein extracts were prepared from prostate cancer cell lines at 48 hours following siRNA transfection or mock transfection using Nuclear/Cytosol Fractionation Kit (K266-100, BioVision) following protocols provided by the manufacturer and Western blot analysis was performed as described previously (32, 42, 43). We also performed Western blot analysis with prostate tissue extracts from TRAMP or Lady TG mice and TRAMP/Rosa26-FoxM1b or Lady/Rosa26-FoxM1b double TG mice. The following commercially available antibodies and dilutions were used for Western Blotting: mouse anti–Kip1/p27 antibody (1:3000), mouse anti–Cip1/p21 antibody (1:3000; BD Transduction Laboratories), mouse anti-cyclin A2 antibody (H-432; 1:2000; Santa Cruz Biotechnology, Inc.), rabbit anti-Cdk2 (M2; 1:1500; Santa Cruz Biotechnology, Inc.) and mouse anti-human cyclin B1 antibody (GNS-11; 1:500; BD PharMingen). The signals from the primary antibody were amplified by HRP conjugated anti-mouse IgG (Bio-Rad, Hercules, CA), and detected with Enhanced Chemiluminescence Plus (ECL-plus, Amersham Pharmacia Biotech, Piscataway, NJ) followed by autoradiography.
Statistical analysis.
We used Microsoft Excel Program to calculate SD and statistically significant differences between samples using the Student T Test. The asterisks in each figure indicates statistically significant changes with P values calculated by Student T Test: *P <0.05, **P ≤ 0.01 and ***P ≤ 0.001. P values <0.05 were considered statistically significant.
Results
FoxM1 protein is expressed in highly proliferative human prostate adenocarcinomas.
We used tissue microarray of human prostate cancers containing 17 adenocarcinomas grade I-II, 26 adenocarcinomas grade III and 25 adenocarcinomas grade IV human prostate cancers, which also contained 7 adjacent normal prostate tissue. Immunohistochemical staining of this human prostate cancer tissue microarray with a C-terminal FoxM1 antibody revealed that expression of FoxM1 increased with the severity or grade of the human prostate adenocarcinomas. We found that 60% of grades I-II, 68% of grade III and 80% of grade IV human prostate adenocarcinomas stained positive for FoxM1 protein. Increased nuclear levels of FoxM1 protein were found in highly proliferative prostate adenocarcinomas as evidenced by abundant staining for Proliferation Cell Nuclear Antigen (PCNA; Fig. 1A-B, right panel and data not shown), whereas low background FoxM1 staining was detected in adjacent normal prostate tissue (Fig 1A; left panel). In contrast, 34% of human prostate adenocarcinomas failed to exhibit PCNA staining (Fig. 1C, right panel) and correlated with low background FoxM1 staining (Fig. 1C, left panel). We also quantitated the number of Foxm1 and PCNA positive nuclei in the human prostate tumor tissue microarray (Fig. 1D). This analysis revealed a correlation between increased Foxm1 staining and PCNA positive proliferating cells in human prostate adenocarcinomas. These results indicate that highly proliferative human prostate adenocarcinomas expressed elevated levels of FoxM1 protein.
Fig. 1. FoxM1 is expressed in highly proliferative human prostate adenocarcinomas.

Prostate cancer tissue arrays (US Biomax; Cat No. PR801) were immunostained with FoxM1 antibody as described in Materials and Methods. (A) Increased nuclear levels of FoxM1b protein in human prostate adenocarcinomas (right panel) compared to corresponding adjacent normal prostate tissue (left panel). (B) FoxM1 and PCNA are co-expressed in human prostate adenocarcinomas. Adjacent paraffin sections of the prostate cancer tissue array were stained with antibody specific to either FoxM1 (left panels) or PCNA (right panels) proteins. Prostate cancer tumor sections immunostained for PCNA expression were counterstained with nuclear fast red (PCNA staining regions indicated by arrows). (C) FoxM1-negative tumor (left panel) displays diminished PCNA levels (right panel). (D) Correlation between PCNA and FoxM1 staining in human prostate carcinomas. We counted the number of Foxm1 and PCNA positive nuclei in five fields of proliferating human prostate adenocarcinomas or normal prostate tissue. Graphic representation of the mean number of nuclei staining positive for either Foxm1 or PCNA ± SD in proliferating human adenocarcinomas versus normal prostate tissue (***P<0.001). Magnification: A, 200x; B and C, 50x.
TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double transgenic mice exhibit accelerated growth and development of prostate carcinomas.
In order to examine whether increased expression of FoxM1b accelerates development and proliferation of prostate carcinomas, we bred Rosa26-FoxM1b TG mice, which ubiquitously expressed FoxM1b (35), with the well established TRAMP (17) and LADY (20) TG mouse models of prostate cancer. We harvested and weighed prostates from male wild type (WT), Rosa26-FoxM1b, TRAMP and LADY single TG mice and TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice at the indicated two time points after birth (Fig 2A-B). Paraffin embedded mouse prostate sections were histologically stained with H&E to evaluate the types of prostatic tumors found in the single and double TG mice (Supplemental Fig. 1A) as described previously (44).
Fig. 2. TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double transgenic mice exhibit accelerated growth and development of prostate carcinomas.
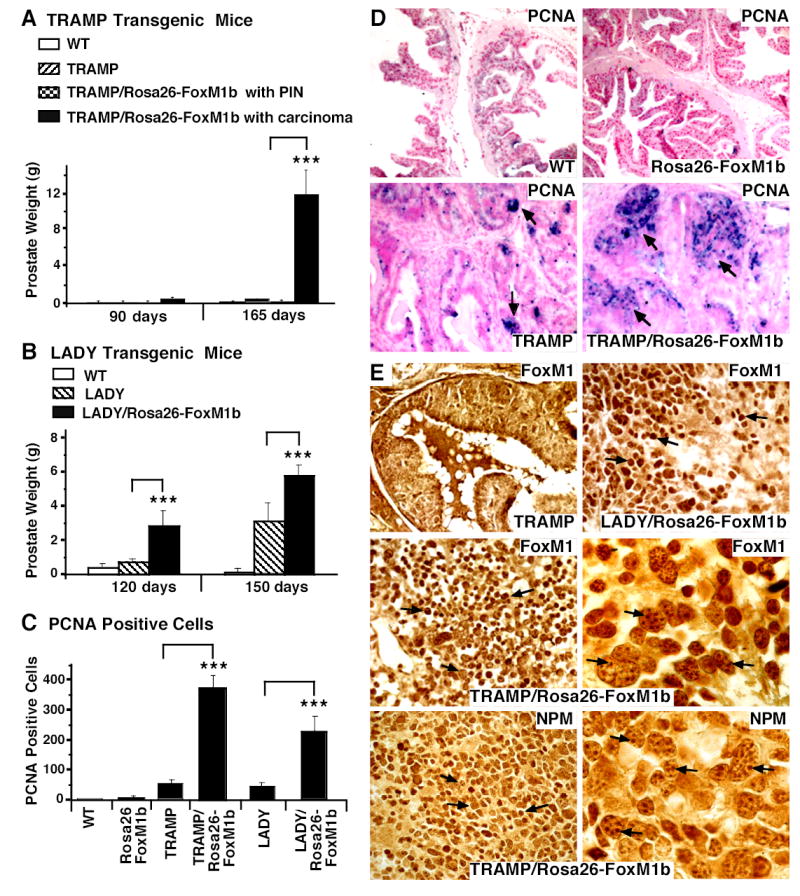
We sacrificed TRAMP and LADY single transgenic (TG) or TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice at the indicated time points. Prostate glands were collected and weighed from wild type (WT; n=4), Rosa26-FoxM1b (n=7), TRAMP (n=7) and LADY (n=7) single TG mice and TRAMP/Rosa26-FoxM1b (n=12) and LADY/Rosa26-FoxM1b (n=10) double TG mice. Prostate tissue sections were immunochemically stained with antibody specific to either the Proliferation Cell Nuclear Antigen (PCNA) or the FoxM1 protein. (A) Graphically shown are the weights of prostate glands containing carcinomas from TRAMP/Rosa26-FoxM1b double TG mice compared to those of TRAMP TG mice. We determined the mean weight of mouse prostate gland ± SE. Prostate weight is significantly increased in the subset of TRAMP/Rosa26-FoxM1b double TG mice that developed prostate carcinomas compared to dual or single TRAMP TG mice that developed PIN. (B) Statistically significant increase in weight of prostate glands from LADY/Rosa26-FoxM1b double TG mice compared to those of LADY single TG mice. We determined the mean weight of mouse prostate glands ± SE. (C) Elevated number of PCNA positive cells in TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice. Prostate tumor cells undergoing proliferation were detected with antibody specific to PCNA and prostate tumor sections were then counterstained with nuclear fast red (panel D). We counted the number of PCNA-positive cells in 5 random microscope fields from different mouse prostates to determine the mean number of PCNA positive cells ± SD. The asterisks in panels A, B and C indicate statistically significant increases with P values calculated by Student T Test: *P <0.05, **P ≤ 0.01 and ***P ≤ 0.002. (E) Nucleolar localization of FoxM1 protein in mouse prostate tumors from TRAMP/Rosa26-FoxM1b double TG mice. Prostate tissue sections from TRAMP, TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice were immunostained with antibody specific to either FoxM1 protein or the nucleolar Nucleophosmin (NPM) protein (49, 50). High magnification of FoxM1 staining in prostate carcinoma cells from TRAMP/Rosa26-FoxM1b double TG mice shows partial nucleolar staining of FoxM1 protein (right middle panel), which is similar to the nucleolar staining pattern of the NPM protein (right lower panel). The nucleolar staining pattern is indicated in these two panels by arrows. Magnification: D and E, 100x; right middle and right lower panels of panel E, 630X.
Analysis of prostate tissue from either TRAMP/Rosa26-FoxM1b or LADY/Rosa26-FoxM1b double TG mice established that the increased expression of the FoxM1b transcription factor significantly accelerated the development and progression of prostatic carcinomas in both groups of double TG mice (Table 1; Fig. 2A-B). As early as 90 days of age, 40% of the TRAMP/Rosa26-FoxM1b double TG mice exhibited prostate carcinomas, while the other 60% of the double TG mice developed prostate hyperplasia and prostatic intraepithelial neoplasia (PIN) (Table 1 and Fig. 1A). In contrast, all of the TRAMP TG mice developed only prostate hyperplasia and PINs at both time points examined (Table 1 and Supplemental Fig. 1A). By 165 days of age, weights of the prostate glands from the TRAMP/Rosa26-FoxM1b double TG mice containing carcinomas were between 64 to 87 times greater than those from TRAMP single TG mice or TRAMP/Rosa26-FoxM1b double TG mice displaying prostate hyperplasia and PINs (Fig. 2A and Table 1). All of the LADY/Rosa26-FoxM1b double TG mice developed prostate carcinomas at both time points examined, whereas only 50% of the Lady single TG mice exhibited prostate carcinomas (Table 1). At 120 days of age, weights of the prostate glands from the LADY/Rosa26-FoxM1b mice were approximately 3.5 times greater than those of the LADY single transgenic mice and these prostate tumors grew larger by 150 days of age (Fig. 2B and Table 1). No prostatic tumors, PINs or prostate hyperplasia were found in Rosa26-FoxM1b single TG mice or WT mice. These results indicate that elevated expression of the FoxM1b transcription factor accelerated the development and growth of prostate tumors in TRAMP and LADY double TG mice.
Table 1.
Histological analysis of prostate tumors and weight of mouse prostate
| †Diagnosis | TRAMP transgenic mice | TRAMP/Rosa26-FoxM1b double transgenic mice | LADY transgenic mice | LADY/Rosa26-FoxM1b double transgenic mice | ||||
|---|---|---|---|---|---|---|---|---|
| *90 days | 165 days | 90 days | 165 days | 120 days | 150 days | 120 days | 150 days | |
| Prostate Hyperplasia and PIN | ‡100% §(4/4) | 100% (3/3) | 60% (3/5) | 55.6% (5/9) | 50% (2/4) | 50% (3/6) | 0% (0/5) | 0% (0/5) |
| || Prostate Weight (g) | 0.05 ± 0.02 | 0.17 ± 0.1 | 0.03 ± 0.02 | 0.13 ± 0.03 | 0.4 ± 0.1 | 0.8 ± 0.1 | **NA | NA |
| Prostate carcinoma | 0% (0/4) | 0% (0/3) | 40% (2/5) | 44.4% (4/9) | 50% (2/4) | 50% (3/6) | 100% (5/5) | 100% (5/5) |
| Prostate Weight (g) | NA | NA | 0.44 ± 0.1 | 11.4 ± 2.5 | 0.9 ± 0.2 | 3.3 ± 0.4 | 2.9 ± 0.8 | 5.9 ± 0.5 |
Prostates from male mice were collected, weighed and sectioned at two time points: 90 or 165 days for TRAMP transgenic (TG) mice or TRAMP/Rosa26-FoxM1b double TG mice, and 120 or 150 days for LADY TG mice or LADY/Rosa26-FoxM1b double TG mice.
Histological sections of mouse prostates were stained with H&E, analyzed by a pathologist using guidelines described previously (44). The mouse prostatic tumors were separated into two groups: one group includes all the prostates with prostate epithelial cell hyperplasia and prostatic intraepithelial neoplasia (PIN), and the other group includes prostate carcinomas.
Percent of mice with prostate hyperplasia and PIN or with prostate carcinomas.
Below the percent is the number of prostates with diagnosis over the number of mouse prostate tissue examined.
Mean prostate weight in grams ± standard error.
NA-Not applicable.
TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b Double TG mice display increased proliferation of prostate carcinomas.
To correlate proliferation with FoxM1 expression in mouse prostate cancer, we used immunohistochemical staining of prostate sections with either PCNA or FoxM1 antibodies (42). This analysis revealed that prostate carcinomas from TRAMP/Rosa26-FoxM1b double TG mice displayed a 10-fold increase in PCNA positive cells compared to those in prostate hyperplasia and PINs from TRAMP single and double TG mice (Fig. 2C, D and data not shown). A 5-fold increase in PCNA staining was found in prostate carcinomas from LADY/Rosa26-FoxM1b double TG mice compared to those of LADY single TG mice (Fig. 2C). Abundant nuclear levels of FoxM1 protein were also found in highly proliferative prostate carcinoma cells from both double TG mice (Fig. 2E), whereas diminished nuclear levels of FoxM1 protein were found in prostate hyperplasia and PINs from both TRAMP and LADY single TG mice (Fig. 2E and data not shown). These results suggest that nuclear expression of FoxM1 correlates with proliferation of prostate carcinomas.
TRAMP/Rosa26-FoxM1b TG carcinomas display increased expression of FoxM1 mRNA and continue to express the ARF tumor suppressor.
To determine molecular mechanisms involved in development of carcinomas versus prostate hyperplasia and PINs in TRAMP/Rosa26-FoxM1b double TG mice, we used quantitative Real-Time Reverse Transcriptase (QRT)-PCR analysis of RNA isolated from prostate tissue with mouse primers specific to the FoxM1 gene (42). This QRT-PCR analysis showed that TRAMP/Rosa26-FoxM1b double TG mouse prostates containing carcinomas exhibited statistically significant 6-fold increase in mRNA levels of FoxM1 compared to those containing prostate hyperplasia and PINs (Fig. 3A; P≤ 0.001). Prostates from Rosa26-FoxM1b TG mice exhibited a two-fold increase in FoxM1 mRNA levels than those of either TRAMP or LADY TG mice (Fig. 3A). However, FoxM1 protein was cytoplasmic in non-proliferating prostate epithelial cells of Rosa26-FoxM1b TG mice (Supplemental Fig. 1B), a finding consistent with previous studies (29). We also used QRT-PCR analysis to determine that prostates containing carcinomas from LADY/Rosa26-FoxM1b double TG mice expressed 3-fold higher levels of FoxM1 mRNA than those of the Lady single TG mice (Fig. 3A). These studies showed that elevated FoxM1 levels correlated with increased development of prostatic carcinomas in both TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice.
Figure 3. Significant increase in FoxM1 mRNA levels and sustained ARF tumor suppressor mRNA expression in prostate carcinomas.

(A) A 6-fold increase in FoxM1 mRNA levels was found in prostate carcinomas compared to PIN in TRAMP/Rosa26-FoxM1b double TG mice. Total RNA was prepared from either prostate carcinomas (CaP) or prostatic intraepithelial neoplasia (PIN) in TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice and TRAMP or LADY single TG mice. We also prepared total RNA from normal prostate tissue from wild type (WT) and Rosa26-FoxM1b transgenic (TG) mice. To determine relative levels of FoxM1 mRNA, Quantitative Real-time reverse transcriptase (QRT)-PCR was performed with RNA from either prostate tissue, carcinoma or PIN and FoxM1 specific primers. (B) Prostate carcinomas in TRAMP/Rosa26-FoxM1b TG mice exhibit sustained expression of ARF tumor suppressor mRNA. Total RNA was prepared from either prostate carcinomas or PIN in TRAMP/Rosa26-FoxM1b double TG mice and analyzed for mRNA levels of the ARF tumor suppressor by QRT-PCR using primers specific to mouse ARF gene. Note that ARF mRNA levels in these prostate tumors were significantly less than those found in early passage FoxM1 − /− mouse embryonic fibroblasts (MEFs) (Fig. 3B), which undergo premature senescence and express high levels of nuclear ARF protein (42). Levels of Cyclophilin mRNA were used to normalize expression levels of FoxM1 and ARF mRNA as described in Materials and Methods. The asterisks indicate statistically significant increases with P values calculated by Student T Test: *P <0.05, **P ≤ 0.01 and ***P ≤ 0.001.
Higher magnification of FoxM1 staining in prostate carcinoma sections from TRAMP/Rosa26-FoxM1b double TG mice revealed nucleolar staining of the FoxM1 protein (Fig. 2E, right middle panel), which is similar to the nucleolar staining pattern of the Nucleophosmin (NPM) protein (Fig. 3E right lower panel). These results suggest that the ARF tumor suppressor targets endogenous FoxM1 protein to the nucleolus of prostate carcinoma cells (32). We therefore used QRT-PCR to measure mRNA levels of ARF in prostate glands from TRAMP/Rosa26-FoxM1b double TG mice. Consistent with the nucleolar localization of FoxM1 protein in prostate carcinoma cells (Fig. 2E), QRT-PCR analysis of ARF mRNA expression revealed that prostate glands containing carcinomas exhibited statistically significant increase in levels of ARF mRNA compared to those containing prostate hyperplasia and PINs (Fig. 3B). These results suggest that development of prostate carcinomas in TRAMP/Rosa26-FoxM1b double TG mice required high levels of FoxM1 protein to overcome sustained expression of the ARF tumor suppressor, a potent inhibitor of FoxM1 transcriptional activity (32).
FoxM1 is essential for growth, proliferation and transformation of prostate adenocarcinoma cell lines in vitro.
To examine the role of FoxM1 in proliferation of human prostate cancer cells, we reduced levels of FoxM1 protein in human androgen-dependent LNCaP or androgen-independent PC-3 and DU-145 prostate cancer cell lines by transfection with siFoxM1 #2 duplex, which was previously shown to significantly diminish endogenous FoxM1 expression in transfected osteosarcoma U2OS cells (42). Total RNA from these prostate cancer cell lines was prepared at 48 hours after transfection with siFoxM1 #2 duplex and QRT-PCR analysis of mRNA confirmed that all three transfected prostate cancer cell lines displayed a significant reduction in FoxM1 mRNA levels (Fig. 4A). As a control for siRNA silencing of gene expression, we transfected the prostate cancer cell lines with p27Kip1 siRNA (siP27) and this siRNA transfection did not influence expression of FoxM1 mRNA (Fig. 4A). To investigate the effect of depleting FoxM1 levels on growth of these prostate cancer cell lines, LNCaP, PC-3 and DU-145 cells were re-plated 48 hours after siRNA transfection and the growth curve in culture was determined by measuring cell numbers at 3, 4 or 5 days after transfection (Fig. 4B). All three prostate cancer cell lines exhibited a significant decrease in cell growth after siFoxM1 #2 transfection compared to untransfected cells or cells transfected with p27Kip1 siRNA (Fig. 4B). Silencing of FoxM1 expression by siRNA transfection caused the most dramatic reduction in growth of the LNCaP cells, followed by the PC-3 cells, and growth of DU-145 cells was the least severely affected by siFoxM1 transfection (Fig. 4B). These results suggest that depletion of FoxM1 resulted in reduced growth rate of prostate cancer cell lines.
Fig. 4. FoxM1 is essential for growth and proliferation of PC-3, LNCaP and DU-145 prostate cancer cell lines.

We transfected siRNA duplexes specific to either FoxM1 (siFoxM1 #2) or p27Kip1 (siP27) into PC-3, LNCaP and DU-145 prostate cancer cell lines and at 48 hours after transfection they were used for growth or flow cytometry analysis and at 72 hours after transfection they were used to prepare total RNA. (A) Transfection of FoxM1 siRNA inhibits expression of FoxM1 in PC-3, LNCaP and DU-145 cells. Total RNA was prepared from prostate cancer cell lines PC-3, LNCaP and DU-145 at 48 hours after transfection with either siRNA duplexes specific to FoxM1 (siFoxM1 #2) or p27Kip1 (siP27) or left untransfected and then was analyzed for expression levels of FoxM1 and Cyclophilin by quantitative Real-Time RT-PCR (QRT-PCR) as described in Material and Methods. FoxM1 mRNA levels in each individual sample were normalized to its corresponding Cyclophilin mRNA level. (B) Transfection of FoxM1 siRNAs into prostate cancer cell lines decreases their growth in culture. Prostate cancer cell lines PC-3, LNCaP and DU-145 were transfected with either siFoxM1 #2 or siP27 duplexes or left untransfected and were then re-plated 48 hours after siRNA transfection and cell numbers were counted at day 3, day 4 or day 5 after transfection. A statistically significant decrease in the growth of FoxM1 depleted prostate cancer cells compared to untransfected cells. (C) Flow cytometry analysis of FoxM1 depleted prostate cancer cell lines shows decreased S-phase progression. The indicated prostate cancer cell lines were transfected with siFoxM1 #2 or siP27 duplexes or left untransfected and then subjected to flow cytometry analysis at 72 hours post transfection after staining with propidium iodide. Graphically shown is the percentage of cells accumulating in G1, S, and G2/M (4N) in FoxM1 or p27Kip1 depleted prostate cancer cells compared to untransfected prostate cancer cells ± SD in triplicate. The asterisks indicate statistically significant increases with P values calculated by Student T Test: *P <0.05, **P ≤ 0.01 and ***P ≤ 0.001.
To determine cell cycle profiles, we performed flow cytometry analysis of FoxM1 depleted LNCaP, PC-3 and DU-145 prostate cancer cell lines or cells transfected with control siP27 duplex or left untransfected. Depletion of FoxM1 levels in LNCaP and PC-3 cells caused a significant accumulation of cells in the G1 phase and a 5-fold decrease in S-phase cells compared to untransfected or siP27 transfected cells (Fig. 4C). FoxM1 depleted DU-145 cells exhibited a less severe 50% decrease in S-phase progression (Fig. 4C), a finding consistent with less efficient down regulation of FoxM1 mRNA levels by siRNA transfection (Fig. 4A). Only slight changes in G2/M phase cells were found in the FoxM1 depleted prostate cancer cell lines (Fig. 4C). These studies demonstrated that siRNA mediated depletion of FoxM1 levels in prostate cancer cell lines decreased their growth and proliferation.
All of the prostate cancer cell lines with depleted FoxM1 levels showed significant decrease in anchorage-independent growth as evidenced by a reduction in the number of cell colonies that grew on soft agar (Fig. 5A and B) (32). In contrast, untransfected or siP27 transfected prostate cancer cell lines exhibited similar numbers of cell colonies using the soft agar assay (Fig. 5A-B). These results suggest that FoxM1 depleted prostate cancer cells displayed reduced transformation or anchorage-independent growth and that diminished growth of these FoxM1 depleted cells is contributing to this reduced transformation.
Figure 5. Depletion of FoxM1 reduces anchorage-independent growth of prostate cancer cell colonies on soft agar.
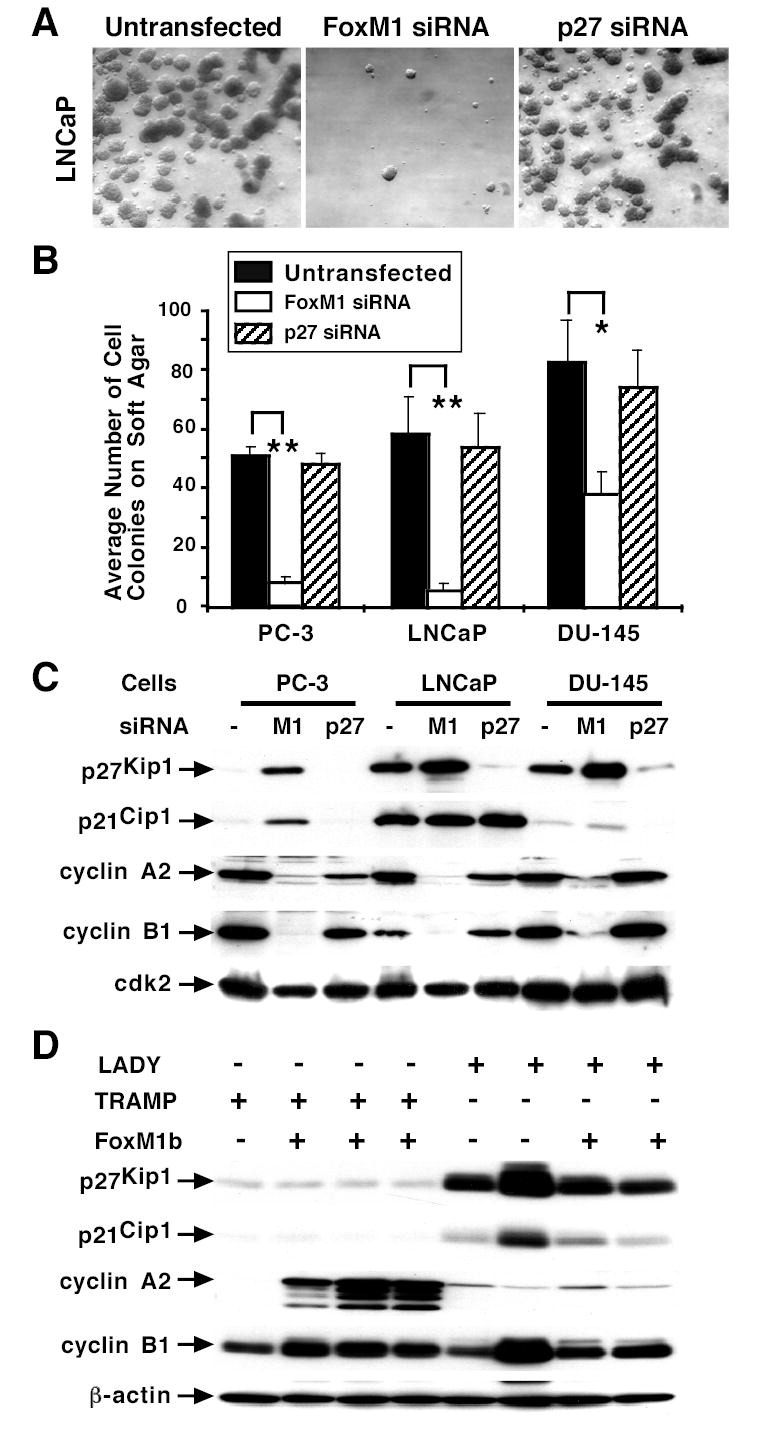
The prostate cancer cell lines PC-3, LNCaP and DU-145 were transfected with 100 nM of either siFoxM1 #2 or siP27 duplexes or left untransfected. One day after transfection, the cells were trypsinized and plated on soft agar for three weeks to analyze the anchorage-independent cell growth as described previously (32). (A) Representative micrographs of LNCaP prostate cancer cell colonies on soft agar after transfection with the indicated siRNA or left untransfected. (B) Graphic presentation of the number of prostate cancer cell colonies that grew on soft agar with or without depletion of FoxM1 or p27Kip1 protein levels. Triplicate plates were used to count the number of cell colonies and determine the mean number of colonies that grew on soft agar ± standard deviation (SD). FoxM1 depleted PC-3 (**P= 0.003), LNCaP (**P = 0.007), or DU-145 (*P= 0.03) prostate cancer cells displayed a statistically significant decrease in the number of cell colonies that grew on soft agar compared to untransfected or siP27 transfected controls. (C) FoxM1 depleted prostate cancer cell lines exhibit increased nuclear levels of CDKI proteins and diminished expression of cyclin A2 and cyclin B1 protein. PC-3, LNCaP, or DU-145 prostate cancer cells were transfected with siFoxM1 #2 or siP27 duplex or left untransfected (-) and nuclear extracts were prepared at 72 hours following transfection and analyzed for nuclear levels of p21Cip1, p27Kip1, cyclin A2 and cyclin B1 proteins by Western blot analysis. The protein levels of cdk2 were used as a loading control. (D) Increased proliferation in TRAMP/Rosa26-FoxM1b prostate carcinomas correlates with elevated expression of cyclin A2. Expression of p21Cip1, p27Kip1, cyclin A2 and cyclin B1 proteins was determined by Western blot analysis with prostate tissue extracts isolated at later time points from TRAMP or LADY single TG mice and TRAMP/Rosa26-FoxM1b or LADY/Rosa26-FoxM1b double TG mice.
Prostate cancer cells with depleted FoxM1 levels exhibited increased nuclear levels of the CDKI proteins and decreased expression of cyclin A2 and cyclin B1 proteins.
Liver regeneration studies demonstrated that FoxM1 deficient hepatocytes exhibited diminished S-phase progression through increase in nuclear levels of the CDKI proteins p21Cip1 and p27Kip1 (30, 32). FoxM1 depleted LNCaP, PC-3 or DU-145 prostate cancer cells were used to prepare nuclear extracts at 48 hours following transfection and analyzed for nuclear levels of p21Cip1 and p27Kip1 protein by Western blot analysis (Fig. 5C). Nuclear levels of CDKI inhibitor proteins were most dramatically increased in FoxM1 depleted PC-3 cells (20-fold) compared to controls, while a 2-fold increase in these CDKI inhibitor proteins was found in DU-145 cells with low levels of FoxM1. A 2-fold increase in nuclear levels of p27Kip1 protein was found in FoxM1 depleted LNCaP cells (Fig. 5C), whereas high levels of nuclear p21Cip1 protein were found in LNCaP cells with or without FoxM1 (Fig. 5C). These results suggest that increased nuclear expression of CDKI protein p27Kip1 contributes to reduced G1/S progression.
Western Blot analysis of FoxM1 depleted prostate cancer cell lines revealed a significant reduction in nuclear levels of cyclin B1 protein (Fig. 5C), which activates Cdk1 through complex formation and is critical for progression into mitosis (3, 4). Published studies have demonstrated that increased levels of the S-phase promoting cyclin A2 are responsible for the c-Jun transcription factor in stimulating anchorage independent growth of Rat1A cells on soft agar (45). Interestingly, FoxM1 depleted cancer cell lines exhibited reduced nuclear levels of cyclin A2 protein (Fig. 5C) and this correlated with their decreased colony formation on soft agar. We also performed Western blot analysis with prostate tissue protein extracts derived from either TRAMP or Lady single TG mice or TRAMP/Rosa26-FoxM1b or LADY/Rosa26-FoxM1b double TG mice harvested at the later time points (Fig. 5D). High proliferation rates in prostate carcinomas from TRAMP/Rosa26-FoxM1b double TG mice correlated with increased expression of cyclin A2 protein compared to prostatic tissue from other TG mice (Fig. 5D). Low expression levels of CDKI proteins p27Kip1 and p21Cip1 were found in prostate tissue from both TRAMP single and TRAMP/Rosa26-FoxM1b double TG mice compared to LADY TG mice (Fig. 5D). Moreover, a slight increase in levels of cyclin B1 protein were found in prostate tissue from TRAMP/Rosa26-FoxM1b double TG mice compared to that of TRAMP TG mice (Fig. 5D).
Discussion
The proliferation-specific Forkhead Box M1 (FoxM1) transcription factor is over expressed in a number of aggressive human carcinomas (36–41). Published studies showed that Alb-Cre FoxM1 −/− hepatocytes fail to proliferate and are highly resistant to development of hepatic tumors in response to chemical carcinogens and that the ARF tumor suppressor inhibits FoxM1 transcriptional activity by targeting it to the nucleolus (32, 34). To determine the role of FoxM1b in prostate cancer progression, we crossed Rosa26-FoxM1b transgenic (TG) mice, which ubiquitously express FoxM1b transgene (35), with the TRAMP (16, 17) or LADY (16, 20) TG mouse models of prostate cancer. We show that increased expression of FoxM1b significantly accelerated development, proliferation and growth of prostate carcinomas in TRAMP/Rosa26-FoxM1b and LADY/Rosa26-FoxM1b double TG mice. Furthermore, development of prostate carcinomas in TRAMP/Rosa26-FoxM1b double TG mice required high levels of FoxM1 protein to overcome sustained expression of the ARF tumor suppressor, a potent inhibitor of FoxM1 transcriptional activity (32). We also show that elevated expression of FoxM1 protein is found in highly proliferative human prostate carcinomas correlating with the aggressiveness or grade of the human prostatic tumors. These results indicate that high levels of FoxM1 protein are associated with greater proliferation in human prostate carcinomas and thus identify FoxM1 as a novel target for prostate cancer treatment.
We showed that depletion of FoxM1 levels in PC-3, LNCaP and DU-145 prostate cancer cells by siRNA transfection caused a significant decrease in cell growth and progression into DNA replication (S-phase) and reduced transformation or anchorage-independent growth on soft agar. Diminished G1/S progression and growth rate was associated with increased nuclear levels of the cyclin dependent kinase inhibitor (CDKI) protein p27Kip1, which negatively regulate CDK activity through protein complex formation (4, 5). Consistent with published studies (46), LNCaP cells already exhibits high nuclear levels of p21Cip1 protein and therefore the slight increase in nuclear levels of p27Kip1 protein in FoxM1 deficient LNCaP cells contributes to their diminished cell growth in culture. Moreover, prostate tissue from TRAMP TG mice displayed low expression levels of CDKI proteins p27Kip1 and p21Cip1 (Fig. 5D), a finding consistent with development of larger, highly proliferative prostate carcinomas in the TRAMP/Rosa26-FoxM1b double TG mice (Fig. 2).
FoxM1 depleted prostate cancer cells also displayed reduced nuclear levels of the FoxM1 target cyclin B1 (47, 48), which activates cdk1 through complex formation and the cdk1-cyclin B1 complex phosphorylates proteins that are critical for progression into mitosis (4). Diminished nuclear levels of cyclin B1 protein are therefore contributing to decreased cell proliferation of FoxM1 deficient cancer cell lines through reduced mitotic entry. Published studies have demonstrated that increased cyclin A2 levels are responsible for causing the c-Jun transcription factor to induce anchorage independent growth of Rat1A cells on soft agar (45). A significant decrease in nuclear levels of cyclin A2 was found in all of the FoxM1 depleted prostate cancer cell lines, which correlated with reduced cell proliferation and anchorage-independent growth on soft agar. Interestingly, we found that high proliferation rates in prostate carcinomas from Rosa26-FoxM1b/TRAMP double TG mice correlated with increased expression of cyclin A2 protein. These studies suggest that FoxM1 mediated increase in cyclin A2 expression correlates with enhanced proliferation and transformation of prostate cancer cells both in vitro and in vivo.
Loss of ARF function is a critical event for tumor promotion as evidenced by extinguished expression of the ARF protein in a variety of tumors through DNA methylation and silencing of the ARF promoter region (3). Expression of ARF tumor suppressor is induced in response to oncogenic stimuli and prevents abnormal cell proliferation through stabilization of p53 tumor suppressor (6), yet expression of Large T antigen, an inhibitor of both the p53 and RB tumor suppressor proteins (18), in both TRAMP and LADY models would therefore prevent this p53-dependent cell cycle arrest by ARF. However, ARF tumor suppressor also mediates p53-independent cell cycle arrest because the mouse ARF protein targets the FoxM1, E2F1 and c-Myc transcription factors to the nucleolus, thus preventing their transcriptional activation of cell cycle regulatory genes (9-12, 32, 34, 42).
We observed that 40–44% of the TRAMP/Rosa26-FoxM1b double transgenic mice developed prostate carcinomas, whereas the remaining double transgenic mice displayed only prostate hyperplasia and PINs. To our surprise, we found that levels of ARF mRNA were not decreased in prostate carcinomas of TRAMP/Rosa26-FoxM1b double transgenic mice and that the ARF protein was functional, as evidenced by nucleolar localization of FoxM1 protein in these prostate carcinoma cells. We propose that prostate carcinomas from TRAMP/Rosa26-FoxM1b double TG mice display a significant increase in FoxM1 expression to overcome FoxM1 inhibition by the ARF tumor suppressor protein. Because activation of the Ras-MAPK signaling pathway is required for high levels of FoxM1 in proliferating cells (25–29), we hypothesize that highly proliferative prostate carcinomas from these double TG mice exhibit mutations that activate the Ras-MAPK signaling pathway to stimulate FoxM1 expression.
In summary, we show that elevated levels of FoxM1 protein correlate with high proliferation rates in human prostate adenocarcinomas tumors. We demonstrate that increased expression of FoxM1b accelerated development, proliferation and growth of prostate carcinomas in TRAMP/Rosa26 FoxM1b or LADY/Rosa26 FoxM1b double TG mice. Furthermore, inhibiting FoxM1 levels in prostate cancer cell lines by siRNA transfection significantly diminished cell proliferation and anchorage independent growth using a soft agar assay. This phenotype was associated with increased nuclear levels of the CDKI protein p27Kip1 and diminished expression of cyclin A2 and cyclin B1 proteins. Our results demonstrate that the FoxM1 transcription factor regulates proliferation and growth of prostatic tumors, and thus identify FoxM1 as a new target for prostate cancer treatment.
Supplementary Material
Acknowledgments
We thank Dr. Gail Prins for critically reviewing our manuscript and Dr. Robert Matusik for providing the LADY (12T-10) TG mice. We also thank H. Yoder, B. Shin, W. N. Yu, A. Monson, M. Reyes, S. Carr, B. Jordan and I. Mankovskaia for excellent technical assistance.
References
- 1.McCormick F. Signalling networks that cause cancer. Trends Cell Biol. 1999;9(12):M53–M6. [PubMed] [Google Scholar]
- 2.Franke TF, Hornik CP, Segev L, Shostak GA, Sugimoto C. PI3K/Akt and apoptosis: size matters. Oncogene. 2003;22(56):8983–98. doi: 10.1038/sj.onc.1207115. [DOI] [PubMed] [Google Scholar]
- 3.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2(2):103–12. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 4.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432(7015):298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 6.Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13(1):77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 7.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 8.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12(19):2984–91. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 9.Datta A, Nag A, Raychaudhuri P. Differential regulation of E2F1, DP1, and the E2F1/DP1 complex by ARF. Mol Cell Biol. 2002;22(24):8398–408. doi: 10.1128/MCB.22.24.8398-8408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta A, Nag A, Pan W, et al. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J Biol Chem. 2004;279:36698–707. doi: 10.1074/jbc.M312305200. [DOI] [PubMed] [Google Scholar]
- 11.Martelli F, Hamilton T, Silver DP, et al. p19ARF targets certain E2F species for degradation. Proc Natl Acad Sci U S A. 2001;98(8):4455–60. doi: 10.1073/pnas.081061398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19(ARF) directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;431:712–7. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- 13.Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59(9):2217–22. [PubMed] [Google Scholar]
- 14.Kamijo T, Zindy F, Roussel MF, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91(5):649–59. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 15.Crawford ED. Epidemiology of prostate cancer. Urology. 2003;62(6 Suppl 1):3–12. doi: 10.1016/j.urology.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 16.Abate-Shen C, Shen MM. Mouse models of prostate carcinogenesis. Trends Genet. 2002;18(5):S1–S5. doi: 10.1016/s0168-9525(02)02683-5. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg NM, DeMayo F, Finegold MJ, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995;92(8):3439–43. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janssens V, Goris J, Van Hoof C. PP2A: the expected tumor suppressor. Curr Opin Genet Dev. 2005;15(1):34–41. doi: 10.1016/j.gde.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Gingrich JR, Barrios RJ, Morton RA, et al. Metastatic prostate cancer in a transgenic mouse. Cancer Res. 1996;56(18):4096–102. [PubMed] [Google Scholar]
- 20.Kasper S, Sheppard PC, Yan Y, et al. Development, progression, and androgen-dependence of prostate tumors in probasin-large T antigen transgenic mice: a model for prostate cancer. Lab Invest. 1998;78(3):319–33. [PubMed] [Google Scholar]
- 21.Masumori N, Thomas TZ, Chaurand P, et al. A probasin-large T antigen transgenic mouse line develops prostate adenocarcinoma and neuroendocrine carcinoma with metastatic potential. Cancer Res. 2001;61(5):2239–49. [PubMed] [Google Scholar]
- 22.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250(1):1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 23.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14(2):142–6. [PubMed] [Google Scholar]
- 24.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364(6436):412–20. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 25.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25(9):1715–9. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luscher-Firzlaff JM, Westendorf JM, Zwicker J, et al. Interaction of the fork head domain transcription factor MPP2 with the human papilloma virus 16 E7 protein: enhancement of transformation and transactivation. Oncogene. 1999;18(41):5620–30. doi: 10.1038/sj.onc.1202967. [DOI] [PubMed] [Google Scholar]
- 27.Yao KM, Sha M, Lu Z, Wong GG. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J Biol Chem. 1997;272(32):19827–36. doi: 10.1074/jbc.272.32.19827. [DOI] [PubMed] [Google Scholar]
- 28.Ye H, Kelly TF, Samadani U, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17(3):1626–41. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye H, Holterman A, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S-phase. Mol Cell Biol. 1999;19(12):8570–80. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A 2002 December 23. 2002;99(26):16881–6. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa RH, Kalinichenko VV, Holterman AX, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38(6):1331–47. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 32.Kalinichenko VV, Major M, Wang X, et al. Forkhead Box m1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes & Development. 2004;18:830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Krupczak-Hollis K, Tan Y, Dennewitz MB, Adami GR, Costa RH. Increased hepatic Forkhead Box M1B (FoxM1B) levels in old-aged mice stimulated liver regeneration through diminished p27Kip1 protein levels and increased Cdc25B expression. J Biol Chem. 2002;277:44310–6. doi: 10.1074/jbc.M207510200. [DOI] [PubMed] [Google Scholar]
- 34.Costa RH, Kalinichenko VV, Major ML, Raychaudhuri P. New and unexpected: forkhead meets ARF. Curr Opin Genet Dev. 2005;15:42–8. doi: 10.1016/j.gde.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 35.Kalinichenko VV, Gusarova GA, Tan Y, et al. Ubiquitous expression of the forkhead box M1B transgene accelerates proliferation of distinct pulmonary cell-types following lung injury. J Biol Chem. 2003;278:37888–94. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- 36.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–76. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 37.Obama K, Ura K, Li M, et al. Genome-wide analysis of gene expression in human intrahepatic cholangiocarcinoma. Hepatology. 2005;41(6):1339–48. doi: 10.1002/hep.20718. [DOI] [PubMed] [Google Scholar]
- 38.Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 Is a Downstream Target of Gli1 in Basal Cell Carcinomas. Cancer Res. 2002;62(16):4773–80. [PubMed] [Google Scholar]
- 39.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65(12):5181–9. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 40.van den Boom J, Wolter M, Kuick R, et al. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am J Pathol. 2003;163(3):1033–43. doi: 10.1016/S0002-9440(10)63463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pilarsky C, Wenzig M, Specht T, Saeger HD, Grutzmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia. 2004;6(6):744–50. doi: 10.1593/neo.04277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang I-C, Chen Y-J, Hughes DE, et al. Forkhead Box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) Ubiquitin Ligase. Mol Cell Biol 2005;In Press. [DOI] [PMC free article] [PubMed]
- 43.Major ML, Lepe R, Costa RH. Forkhead Box M1B (FoxM1B) Transcriptional Activity Requires Binding of Cdk/Cyclin Complexes for Phosphorylation-Dependent Recruitment of p300/CBP Co-activators. Mol Cell Biol. 2004;24(7):2649–61. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shappell SB, Thomas GV, Roberts RL, et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64(6):2270–305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- 45.Katabami M, Donninger H, Hommura F, et al. Cyclin A is a cJun target gene and is necessary for cJun-induced anchorage independent growth in Rat1a cells. J Biol Chem. 2005;280:16728–38. doi: 10.1074/jbc.M413892200. [DOI] [PubMed] [Google Scholar]
- 46.Wang LG, Ossowski L, Ferrari AC. Overexpressed androgen receptor linked to p21WAF1 silencing may be responsible for androgen independence and resistance to apoptosis of a prostate cancer cell line. Cancer Res. 2001;61(20):7544–51. [PubMed] [Google Scholar]
- 47.Leung TW, Lin SS, Tsang AC, et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507(1):59–66. doi: 10.1016/s0014-5793(01)02915-5. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Quail E, Hung N-J, Tan Y, Ye H, Costa RH. Increased levels of Forkhead Box M1B transcription factor in transgenic mouse hepatocytes prevents age-related proliferation defects in regenerating liver. Proc Natl Acad Sci U S A. 2001 Sept 25;98:11468–73. doi: 10.1073/pnas.201360898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24(3):985–96. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korgaonkar C, Hagen J, Tompkins V, et al. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25(4):1258–71. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


