Abstract
Purification of hepatitis C virus (HCV) from sera of infected patients has proven elusive, hampering efforts to perform structure-function analysis of the viral components. Recombinant forms of the viral glycoproteins have been used instead for functional studies, but uncertainty exists as to whether they closely mimic the virion proteins. Here, we used HCV virus-like particles (VLPs) generated in insect cells infected with a recombinant baculovirus expressing viral structural proteins. Electron microscopic analysis revealed a population of pleomorphic VLPs that were at least partially enveloped with bilayer membranes and had viral glycoprotein spikes protruding from the surface. Immunogold labeling using specific monoclonal antibodies (MAbs) demonstrated these protrusions to be the E1 and E2 glycoproteins. A panel of anti-E2 MAbs was used to probe the surface topology of E2 on the VLPs and to compare the antigenicity of the VLPs with that of truncated E2 (E2660) or the full-length (FL) E1E2 complex expressed in mammalian cells. While most MAbs bound to all forms of antigen, a number of others showed striking differences in their abilities to recognize the various E2 forms. All MAbs directed against hypervariable region 1 (HVR-1) recognized both native and denatured E2660 with comparable affinities, but most bound either weakly or not at all to the FL E1E2 complex or to VLPs. HVR-1 on VLPs was accessible to these MAbs only after denaturation. Importantly, a subset of MAbs specific for amino acids 464 to 475 and 524 to 535 recognized E2660 but not VLPs or FL E1E2 complex. The antigenic differences between E2660, FL E1E2, and VLPs strongly point to the existence of structural differences, which may have functional relevance. Trypsin treatment of VLPs removed the N-terminal part of E2, resulting in a 42-kDa fragment. In the presence of detergent, this was further reduced to a trypsin-resistant 25-kDa fragment, which could be useful for structural studies.
Hepatitis C virus (HCV), the major cause of non-A, non-B hepatitis, is an enveloped virus classified in the Flaviviridae family (31, 52). The positive-strand viral RNA genome encodes a single polyprotein of approximately 3,010 amino acids that is processed into functional proteins by host and viral proteases (52, 53). The putative HCV structural proteins comprising the core and the two envelope glycoproteins E1 and E2 are located within the N terminus of the polyprotein, while the nonstructural proteins reside within the C-terminal part. Analysis of the structural features of the HCV virion has been hampered by the inability to propagate the virus in vitro. In addition, it has been difficult to purify sufficient quantities of virus from infected patient plasma or tissue. By analogy with other flaviviruses, the core is presumed to be enveloped in a lipid bilayer containing the viral glycoproteins. Glycoproteins E1 and E2 are believed to be type I integral transmembrane proteins, with C-terminal hydrophobic anchor domains (reviewed in reference 40). In vitro expression studies have shown that both glycoproteins associate to form two types of complexes: (i) a heterodimer stabilized by noncovalent interactions and (ii) high-molecular-weight disulfide-linked aggregates representing misfolded proteins (9, 11, 13-15, 50). Both types of complex accumulate within the endoplasmic reticulum (ER), the proposed site for HCV assembly and budding. ER retention signals have been mapped to the C-terminal transmembrane domains (TMD) of the two glycoproteins, removal of which results in their secretion from expressing cells (10, 11, 16, 20, 34). A number of conformation-dependent monoclonal antibodies (MAbs) which specifically recognize non-disulfide-bridged E2 have been reported, allowing the study of glycoprotein complexes which may represent “native” prebudding forms of the HCV glycoprotein complex (11, 13, 34).
The mechanism of HCV attachment and entry into its target cell is not known. Several reports have implicated glycoprotein E2 in virus-cell attachment. Antibodies targeted against the N-terminal 27-amino-acid (aa) region of E2 (aa 384 to 411), which is the most variable region (known as hypervariable region 1 [HVR-1]) of the HCV polyprotein, inhibit the binding of glycoprotein E2 to cells and block HCV infectivity in vitro and in vivo (17, 23, 51, 55, 64). Due to the lack of availability of HCV virions in sufficient quantities, it has been difficult to examine the native structure of the viral glycoproteins and to study their precise role in cell attachment and entry. Truncated and soluble forms of E2 lacking its TMD have been used in functional assays to study interaction with its putative receptor, CD81 (6, 18, 21, 24, 33, 45, 48, 59). Recently, Yagnik et al. (61) generated a model for similar truncated E2 of HCV using fold recognition methods. The authors reported that the model was consistent in terms of the relative positions of exposed or buried epitopes with the published data on MAb recognition of different regions of E2660. However, it is not known whether the truncated and soluble forms of E2 glycoprotein fully mimic the corresponding E2 structures on the surfaces of HCV virions. It is important to consider whether antigenic differences exist between soluble and “native” full-length (FL) oligomeric forms of the glycoproteins and whether antibodies elicited by such recombinant antigens will be protective and able to bind HCV. At present, no biochemical information exists to support or refute computer-predicted structures for the E2 glycoprotein (61). Many glycosylated proteins do not yield readily to structural analysis via the classical approaches of crystallography or nuclear magnetic resonance spectroscopy. However, low-resolution models of protein surface topography can be predicted by studying the binding properties of peptide-reactive MAbs to native and denatured proteins (35, 36). Such a model for human immunodeficiency virus type 1 glycoprotein, gp120, was subsequently confirmed by crystal structures of truncated versions of the human immunodeficiency virus type 1 glycoprotein (27, 60). Indeed, such studies demonstrated that conserved regions readily available for MAb binding on the gp120 monomer were occluded on the oligomeric gp120-gp41 complex, probably as a result of their participation in intersubunit interactions. In addition, protease treatment and specific antibody probes can be used to examine the structures of viral glycoproteins. Such protease treatment allowed isolation of a soluble and crystallizable form of the flavivirus tick-borne encephalitis virus envelope glycoprotein E (25).
Baumert et al. (2, 3) have previously reported the assembly of HCV genotype 1b-derived virus-like particles (VLPs) in insect cell systems. We and others (42, 58) have recently shown that such particles can also be generated by using cDNA sequences derived from the infectious clone of HCV type 1a strain H77c (62). In this report, we compare the antigenic characteristics of mammalian cell-derived soluble truncated E2 (E2660) and FL E1E2 complexes with those present on insect cell-generated VLPs by using a panel of well-characterized anti-E1 and anti-E2 MAbs and polyclonal antisera. We demonstrate important differences in antigenicity, which suggest that the various forms of E2 adopt different structures. We further examine the E2 structure on VLPs by using protease treatment, and we show differences between the proteolytic profiles of E2 in the presence and absence of detergent.
MATERIALS AND METHODS
Cell culture.
COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO BRL) supplemented with 10% fetal calf serum (FCS), 5% nonessential amino acids, and penicillin-streptomycin (EFC10). Spodoptera frugiperda (Sf) insect cells were propagated at 28°C in TC100 medium (GIBCO BRL) supplemented with penicillin-streptomycin and 10% FCS.
Generation of plasmid constructs and recombinant viruses.
The cDNA sequences used in this study were derived from an infectious cDNA clone (pCV-H77c) of strain H77c (a kind gift from Jens Bukh [62]). The cDNA sequences from nucleotides 336 to 2850, encoding aa 1 to 836, representing the entire structural region (core, E1, and E2), the p7 protein, and the N-terminal 27 aa residues of NS2, were cloned into the baculovirus and vaccinia virus transfer vectors pAcCL29.1 and pMJ601, respectively (12, 28). Plasmid pAcCL29.1 carrying the HCV sequences was cotransfected with Bsu36I-cleaved genomic DNA of wild-type Autographa californica nuclear polyhedrosis virus strain AcPAK6, and recombinant baculovirus rbac-B45, expressing the HCV structural proteins, was generated as described by Bishop (5). Recombinant baculovirus rbac-DDX3, expressing human cellular protein DDX3 (43), was used as a control. The HCV sequences in pMJ601 were inserted into the thymidine kinase gene of vaccinia virus strain WR by homologous recombination, and a recombinant virus, v1-836, was isolated as described elsewhere (12). The recombinant vaccinia virus expressing E2660 of HCV strain H77c has been described previously (45).
Production, purification, and electron microscopy of HCV VLPs.
Approximately 2 × 108 Sf9 cells were infected with recombinant baculovirus rbac-B45 at a multiplicity of infection (MOI) of 3. At 96 h postinfection, cells were harvested, washed with phosphate-buffered saline (PBS), and lysed in lysis buffer, and the VLPs were partially purified by sucrose density gradient centrifugation essentially as described by Baumert et al. (2). The relative purity and quality of the VLP preparation were verified by negative staining with transmission electron microscopy as follows. Approximately 5 μl of the particle preparation was loaded onto a carbon-coated electron microscope (EM) grid (Agar Scientific, Stanstead, United Kingdom) for approximately 1 min. Grids were then washed in distilled water, stained with 2% sodium phosphotungstate (pH 7.0) for 30 s, drained, and examined under JEOL 100 S and 1200 EX EMs. For immunogold labeling, samples were loaded onto a collodion-coated EM grid for 3 min, drained, and incubated with an appropriately diluted primary antibody for 5 h at room temperature. Grids were then washed in distilled water twice for 10 min at room temperature and incubated for 1 h with appropriately diluted anti-mouse or anti-rabbit immunoglobulin G (IgG) conjugated with gold particles (Nanoprobe). Following two washes as above, the samples were stained with Nanovan (Nanoprobe) for 2 min, drained, and examined under the EM.
VLPs were prepared for electron cryomicroscopy by applying 3 μl of the particle suspension to a holey carbon-coated grid and plunge-freezing in liquid ethane. Vitrified specimens were imaged under low-electron-dose conditions at 120 kV under a JEOL 1200 EX EM fitted with a LaB6 filament and an Oxford Instruments cryotransfer stage. Images were recorded at a nominal magnification of ×30,000 on Kodak SO163 film at an approximate defocus of 1 to 2 μm. Particle radii were estimated from computed radial density profiles that were calculated as follows. Cryomicrographs were digitized on a Hi-Scan research drum scanner (Dunvegan S.A., Lausanne, Switzerland) by using a raster of 7 Å/pixel. Particles that were judged to be circular in projection were selected and cut out into boxes of 150 pixels2. Due to the polydisperse distribution of particle radii, particles could not be reliably centered by cross-correlation against models. Each particle was therefore centered by the rotational cross-correlation method (39). Projected radial density profiles were then calculated for each particle, and their radii were estimated by inspection of these plots and the raw images. All image processing was performed with the SPIDER image-processing package (22).
Preparation of protein samples.
COS-7 cells were infected with recombinant vaccinia viruses at an MOI of 10. Following incubation for 18 h at 37°C, cells were washed with PBS and lysed in lysis buffer (20 mM Tris-HCl [pH 7.4], 20 mM iodoacetamide, 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100), and the lysate was pelleted to remove nuclei. The supernatant was collected and analyzed by enzyme-linked immunosorbent assay (ELISA) as described below. To analyze the secreted proteins, the medium of recombinant vaccinia virus-infected cells was clarified to remove cell debris and adjusted to contain 20 mM Tris-HCl (pH 7.4), 20 mM iodoacetamide, 150 mM NaCl, 1 mM EDTA, and 0.5% Triton X-100 (final concentrations). Sf cells were infected with wild-type or recombinant baculoviruses at an MOI of 5 for 96 h at 28°C. Cell lysates were prepared and analyzed as described above. For antigen denaturation experiments, infected cells were lysed in PBS by freeze-thawing and sonication. Lysates were clarified by brief centrifugation and were either left untreated (native) or boiled (denatured) for 10 min after addition of sodium dodecyl sulfate (SDS) and dithiothreitol to final concentrations of 0.1% and 50 mM, respectively. The boiled lysates were allowed to cool, and NP-40 was added to a final concentration of 1% to both denatured and native samples. VLPs were denatured as described above except that NP-40 was excluded from both the native and the denatured preparations in order to ensure that the enveloped glycoproteins were not stripped from the native particles.
For SDS-polyacrylamide gel electrophoresis (PAGE) and Western immunoblot analysis, protein samples were mixed with SDS-PAGE denaturation buffer (200 mM Tris-HCl [pH 6.7], 0.5% SDS, 10% glycerol) and boiled for 3 min in the presence (reducing conditions) or absence (nonreducing conditions) of β-mercaptoethanol. Samples were subjected to SDS-PAGE (10% polyacrylamide), and fractionated proteins were transferred to Hybond-ECL membranes (Amersham, Little Chalfont, Buckinghamshire, United Kingdom) and probed with antibodies to HCV structural proteins followed by horseradish peroxidase-conjugated anti-mouse, rabbit, or rat IgG. Bound antibodies were detected by using enhanced chemiluminescence reagents (Amersham).
GNA capture ELISA.
Briefly, GNA (Galanthus nivalis) lectin (Boehringer Mannheim, Mannheim, Germany) was used to coat Immulon II ELISA plates (Dynal, Stone, United Kingdom) at 1 μg/ml overnight at 4°C. After a wash in PBS, the plates were blocked with PBS containing 4% milk powder (Cadburys, London, United Kingdom), and either E2660, FL E1E2 complex, or VLPs were allowed to bind for 2 h at room temperature. The wells were washed either with PBS containing 0.05% Tween 20 (for E2660 and FL E1E2) or with PBS alone (for VLPs). GNA-captured antigens were incubated with saturating concentrations of MAbs specific for E1 or E2 for 2 h at room temperature. Following a wash with PBS-Tween (for all three ligands), bound MAbs were detected by using horseradish peroxidase-conjugated anti-mouse or anti-rat IgG (Seralabs, Crawley, United Kingdom) and tetramethyl benzidine (TMB) substrate. Absorbance was determined at 450 nm. The detergent Tween 20 was excluded from the washing buffer and any diluent for VLPs up to the primary antibody incubation stage in order to prevent stripping of the envelope glycoproteins from the particles.
Trypsin treatment of HCV VLPs.
Approximately 1011 VLPs in PBS were digested with 25 μg of trypsin/ml (10,500 U/mg; Sigma) in the presence or absence of Triton X-100 (1%, wt/vol) in a total volume of 100 μl at 37°C for 30 min. Samples were then adjusted to a final volume of 200 μl by addition of SDS-PAGE denaturation buffer, boiled for 3 min, fractionated by SDS-PAGE, and analyzed by Western immunoblotting.
Antibodies.
To generate MAbs, BALB/c mice were immunized subcutaneously with 20 μg of a recombinant form of mammalian cell-expressed HCV strain Gla (45, 46) E1E2 emulsified in Freund's complete adjuvant (Sigma). After 4 weeks, the animals were boosted with E1E2 emulsified in Freund's incomplete adjuvant (Sigma). Mice were further boosted twice at 2-week intervals. Eight days following the last injection, the sera from tail bleeds were tested for E1E2-specific antibodies. Three weeks following the last boost, the animals were given a final intraperitoneal injection of 100 μg of purified recombinant E1E2. Spleen cells from immunized animals were removed 5 days postinjection and fused with Sp2/0-Ag 14 myeloma cells (ratio, 10:1) by using 50% polyethylene glycol 1500 (Koch Light, Haverhill, United Kingdom) in DMEM. Cells were then resuspended in HAT medium (EFC10 supplemented with histone acetyltransferase [Gibco Life BRL, Paisley, United Kingdom]) and 20% conditioned medium (i.e., clarified EFC10 that had previously been used to grow Sp2/0 cells), plated out into 96-well microtiter plates, placed in a CO2 humidified incubator at 37°C, and left for 12 to 14 days. Medium from wells containing colonies was removed and screened by ELISA for E1E2-specific MAbs. Cells from positive wells were removed and propagated. MAbs AP33, AP320, AP436, AP109, AP255, ALP42, AP266, ALP98, ALP11, ALP1, and AP274 were generated by this procedure. The epitopes recognized by these MAbs were mapped by using overlapping peptides (42). Mouse MAbs H53 and H60 (11), and rat MAbs 7/59, 6/82a, 6/16, 9/86a, 9/27, 3/11, 2/69a, 1/39, 6/41a, 2/64a, 9/75, and 6/53 (19) were kind gifts from Jean Dubuisson and Jane McKeating, respectively.
To generate polyclonal antisera, New Zealand rabbits were immunized with a secretory form of HCV strain Gla E1 (aa 197 to 334 of the HCV polyprotein) or strain H77c E2 (aa 364 to 660 [E2660]) (45) expressed in mammalian cells or with bacterially expressed aa 1 to 59 of the strain Gla core fused in-frame to glutathione S-transferase. The immunization protocol used has been described previously (44). Antisera R528, R646, and R526, specific for E1, E2, and core, respectively, were used in this study.
RESULTS
Characterization of HCV VLPs.
We previously generated a recombinant baculovirus, rbac-B45, carrying sequences encoding the core, E1, E2, and p7 proteins of the infectious clone strain H77c (42, 62). Sf cells infected with rbac-B45 produced VLPs, which were partially purified by sucrose gradient centrifugation essentially as described by Baumert et al. (2). The VLPs were imaged by using both negative staining and electron cryomicroscopy (Fig. 1a and b). These images revealed a population of highly pleomorphic particles with radii ranging from 20 to 44 nm. Figure 1c is a histogram of radii measured from 248 particles that were judged to be circular in projection. These data indicate a number of possible size classes, with many particles measuring 26 or 32 nm. It is not uncommon for VLPs to assemble in a range of sizes, and this may indicate a progression of icosahedral T-numbers (1, 26). Attempts to demonstrate unambiguous icosahedral symmetry have proved fruitless so far, although inspection of both negatively stained micrographs and cryomicrographs revealed a proportion of particles that were angular in projection (Fig. 1a, inset). Particle morphology, when imaged in the transmission electron microscope, was largely consistent with that predicted by analogy to related viruses, such as the pestiviruses bovine viral diarrhea virus (4, 30, 38, 57) and classical swine fever virus (57) and the flavivirus tick borne encephalitis virus (54). Particles are at least partially enveloped, as bilayers could be seen in both negatively stained micrographs and close-to-focus cryomicrographs (Fig. 1b). Large spikes protruded from the surfaces of the particles, which were clearly visible in both negatively stained micrographs and cryomicrographs, and in some particles these spikes appeared Y- or T-shaped in projection. Immunogold labeling suggested that these protrusions are composed of the E1 and E2 glycoproteins (see below). Internal features of the particles were poorly realized by negative staining. While many particles appeared to be penetrated by the stain, it still was not possible to discern the presence or absence of a nucleocapsid. Cryomicroscopy, however, revealed that these particles are dense, suggesting either the possibility of a nucleic acid-filled core or the possibility that these particles are formed by envelopment of large aggregates of core protein.
FIG. 1.
(a) Negatively stained electron micrograph of HCV VLPs. (Inset) Higher-magnification image of a VLP that is angular in appearance, with many spikes that appear Y-shaped. (b) Cryomicrograph of HCV VLPs, showing the presence of a lipid bilayer (arrowheads), with protruding spikes, surrounding the core structure. Bars, 100 nm. (c) Histogram of radii measured from 248 VLPs imaged by electron cryomicroscopy.
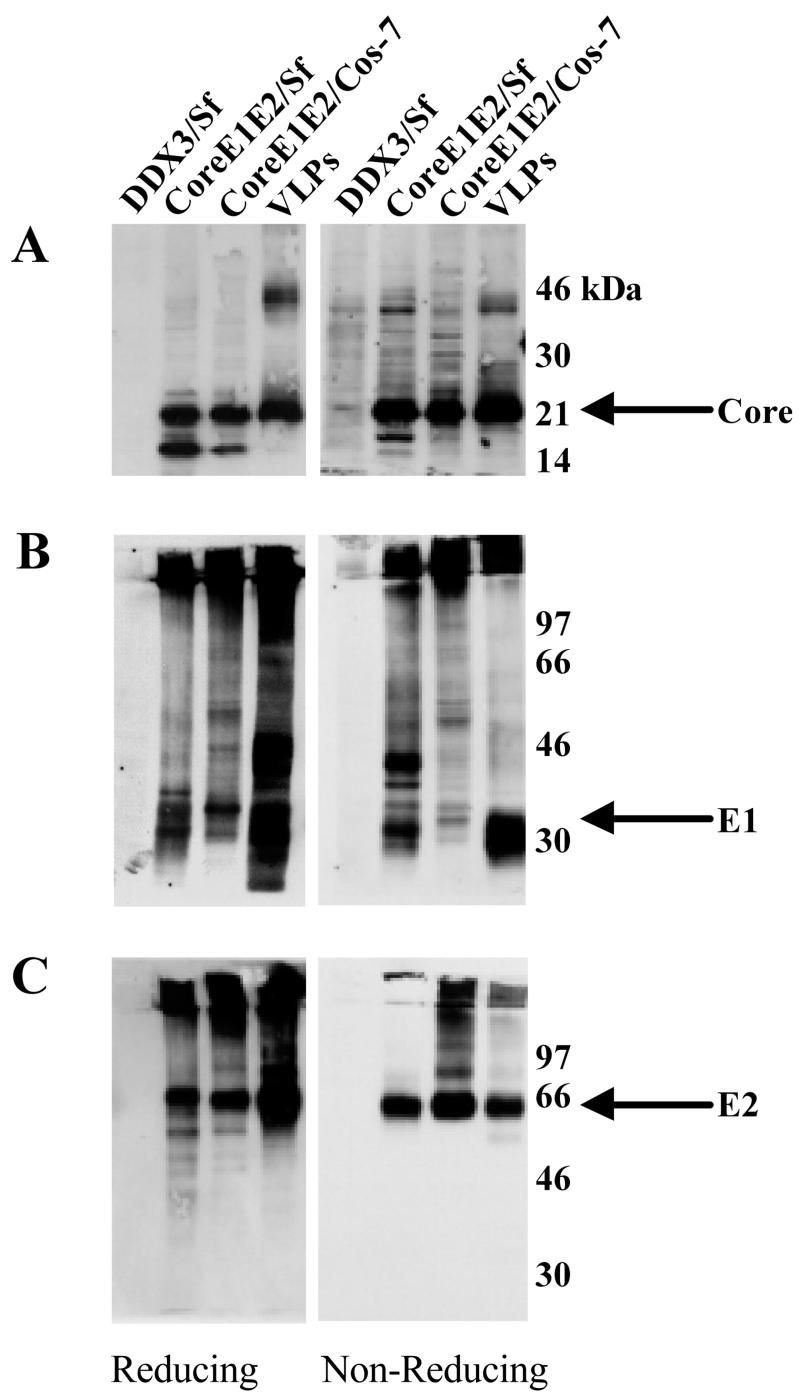
Direct analysis of the VLP preparation on SDS-PAGE followed by silver staining or Coomassie brilliant blue staining to detect recombinant HCV proteins revealed the presence of numerous contaminating Sf and baculovirus proteins, indicating that the method used does not yield purified VLPs (data not shown). Instead, the presence of HCV structural proteins in VLPs was confirmed and compared with the same proteins expressed in insect and mammalian cells as follows. Sf and COS-7 cells were infected with the appropriate recombinant baculovirus or vaccinia virus, respectively. Infected-cell lysates and VLPs were subjected to SDS-PAGE under reducing or nonreducing conditions followed by Western immunoblotting. As shown in Fig. 2, the VLPs contained all three HCV structural proteins. A mature core product of approximately 21 kDa was seen in VLPs and infected Sf or COS-7 cells under both reducing and nonreducing conditions (Fig. 2A). A smaller product, possibly representing an immature core protein, was present in infected Sf and COS-7 cells but not in the VLPs. Instead, the VLPs contained a ∼42-kDa protein, possibly representing a dimer of the mature core product (Fig. 2A). Immunoblotting, using an anti-E1 or anti-E2 MAb, of proteins fractionated under both reducing and nonreducing conditions demonstrated that the VLP preparation contained both a high-molecular-weight disulfide-linked aggregate and native forms of glycoproteins E1 and E2 (Fig. 2B and C, respectively). In agreement with the data of Baumert et al. (3), the E1 glycoproteins expressed in Sf and COS-7 cells migrated differently in the gel (Fig. 2B), possibly due to the differences in glycosylation of proteins between mammalian and insect cells (29).
FIG. 2.
Western immunoblot analysis of HCV structural proteins. Extracts of Sf cells infected with rbac-DDX3 or rbac-B45, extracts of COS-7 cells infected with recombinant vaccinia virus v1-836, and VLPs were subjected to SDS-10% PAGE under reducing and nonreducing conditions. Fractionated proteins were immunoblotted with anti-core antiserum R526 (A), anti-E1 antiserum R528 (B), and anti-E2 MAb AP33 (C). Positions of protein size markers are shown.
Given the current difficulties in generating pure VLPs, detection of HCV glycoproteins in VLP preparations by immunoblotting does not exclude the possibility that at least some of the glycoprotein molecules detected are of a nonparticulate nature, as they may comigrate with the particles on the sucrose gradient. To confirm the presence of HCV glycoproteins on VLPs, we carried out immunogold EM using E1- and E2-specific antibodies. The anti-E1 MAb AP21.010, anti-E2 MAbs AP33 and AP320 (which recognize E2 aa 412 to 423 and 464 to 471, respectively [see Table 1 and Fig. 4]), and the anti-E2 polyclonal serum R646 bound specifically to VLPs (Fig. 3a, b, c, and g, respectively). In addition, MAbs H53 and H60, both recognizing conformation-sensitive epitopes in E2 (11), also bound efficiently to the VLPs (Fig. 3e and f). Interestingly, the anti-E2 MAb AP436, which recognizes the E2 region comprising aa 464 to 475 (see Table 1) and shares this epitope with MAb AP320, did not recognize the VLPs (Fig. 3d). The presence of both E1 and E2 glycoproteins on the VLPs could be shown by double-labeling with the anti-E1 polyclonal serum R528 (10-nm gold particles) and the anti-E2 MAb AP33 (5-nm gold particles) (Fig. 3h). Similar results were obtained by a GNA-capture ELISA in which the lectin-captured VLPs were detected by using the antibodies described above (data not shown).
TABLE 1.
MAb recognition of various E2 glycoprotein forms
| MAba | Epitopeb (aa) | MAb recognition of the indicated H77c E2 antigenc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FLE1E2, intracellular
|
E2660 Intracellular
|
Secreted
|
VLP
|
I-EM
|
||||||
| N | D | N | D | N | D | N | D | |||
| 7/59 | 384-395 (HVR-1) | +++ | +++ | +++ | +++ | +++ | +++ | − | + | − |
| 6/82a | 384-395 (HVR-1) | + | + | +++ | +++ | ++ | − | − | + | − |
| 6/16 | 384-395 (HVR-1) | + | + | +++ | +++ | +++ | + | − | + | − |
| 9/86a | Conf. (HVR-1) | +++ | − | +++ | +++ | +++ | +++ | ++ | − | ++ |
| 9/27 | 396-407 (HVR-1) | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + |
| 3/11 | 412-423 | ++ | ++ | ++ | ++ | ++ | ++ | +++ | +++ | +++ |
| AP33 | 412-423 | ++ | ++ | ++ | ++ | ++ | ++ | +++ | +++ | +++ |
| 2/69a | 432-443 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ |
| 1/39 | 436-443 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 11/20c | 436-447 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| 7/16b | 436-447 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ |
| AP320 | 464-471 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| AP436 | 464-475 | − | − | ++ | ++ | ++ | +++ | − | − | − |
| AP109 | 464-475 | − | − | ++ | ++ | ++ | ++ | − | − | − |
| 6/41a | 480-493 | +++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | ++ |
| 2/64a | 524-531 | ++ | +++ | +++ | +++ | +++ | +++ | ++ | ++ | ++ |
| ALP42 | 524-535 | − | − | ++ | ++ | + | + | − | − | − |
| 9/75 | 528-535 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| 6/53 | 544-551 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| AP266 | 644-651 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | +++ |
| ALP98 | 644-651 | ++ | ++ | ++ | ++ | ++ | ++ | +++ | +++ | ++ |
| ALP11 | 644-651 | + | ++ | ++ | ++ | ++ | +/− | ++ | ++ | ++ |
| ALP1 | 648-659 | ++ | ++ | ++ | ++ | ++ | ++ | +++ | +++ | +++ |
| H53 | Conf. | +++ | − | +++ | − | +++ | − | +++ | − | +++ |
| H60 | Conf. | +++ | − | +++ | − | +++ | − | +++ | − | ++ |
MAbs of particular interest that are discussed in the text are boldfaced.
Conf., conformational.
The FL E1E2 complex and E2660 were expressed in COS-7 cells, whereas VLPs were generated in Sf cells infected with rbac-B45. All ligands were tested by GNA-capture ELISA as described in Materials and Methods. In addition, VLPs were also analyzed by immunogold-EM (I-EM). +, ++, and +++, optical density readings between 5 and 10%, 10 and 30%, and 30 and 60%, respectively, above background values. N, native. D, denatured.
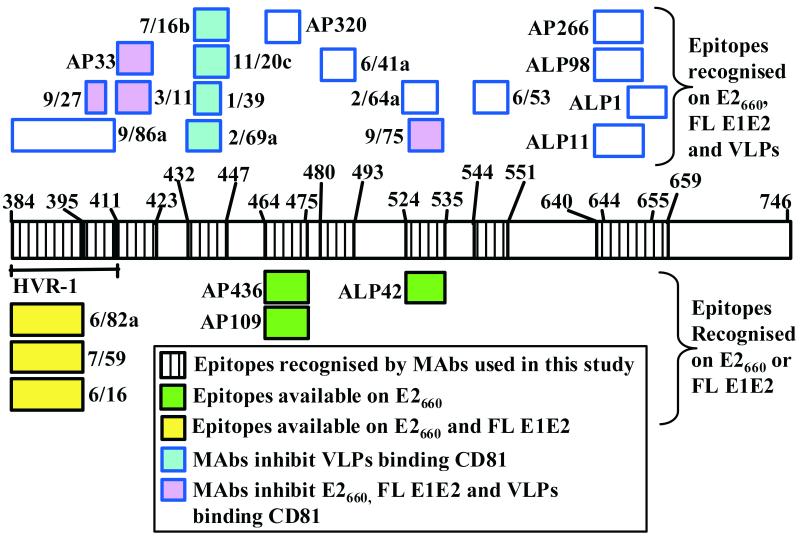
FIG. 4.
Surface topology of E2 epitopes exposed on VLPs. This map was constructed mainly on the basis of the data presented in Table 1, and also on the basis of recently published findings which identified some MAbs as inhibitors of E2-CD81 interaction (42).
FIG. 3.
Immunogold labeling of HCV VLPs. VLPs placed on EM grids were incubated either singly with MAbs specific for E1 (a) or E2 (b to f), with the polyclonal anti-E2 antiserum R646 (g), or doubly with the anti-E1 serum R528 and the anti-E2 MAb AP33 (h). Anti-mouse IgG and anti-rabbit IgG conjugated with 5- or 10-nm gold particles, respectively, were used as secondary reagents. Grids were washed, stained, and examined under the EM. Bar, 100 nm.
Antigenic characterization of various forms of E2 glycoprotein.
To date, it has been difficult to perform a proper structural and functional analysis of the HCV FL E1E2 glycoprotein heterodimer. In contrast with the insect cell system, HCV VLP assembly has not been detected in mammalian cells expressing viral structural proteins. The putative receptor for HCV, CD81, was identified by using a truncated soluble form of E2 (49). However, it is not certain whether the FL E1E2 complex synthesized in mammalian cells or the soluble derivative of E2 fully mimics the corresponding glycoprotein structures on HCV virions. To determine if antigenic differences exist between soluble E2 (E2660) or the FL E1E2 complex formed in mammalian cells and VLPs generated in insect cells, we tested a large panel of anti-E2 MAbs of known specificity and epitopes by GNA-capture ELISA. In addition, the VLPs were also tested for MAb recognition by immunogold-EM. All antigens were tested for MAb reactivity in their native forms or after denaturation with SDS and dithiothreitol (see Materials and Methods). As shown in Table 1, most MAbs bound to all forms of antigen while a number of others showed striking differences in efficiency of binding to the various forms of E2. Of the MAbs recognizing epitopes within HVR-1, 7/59 and 9/27 bound the native and denatured forms of all antigens expressed in mammalian cells with comparable efficiencies (Table 1). Similarly, MAb 9/86a recognized all antigens expressed in mammalian cells equally well, although it did not recognize the denatured FL E1E2 complex. In contrast, the reactivities of anti-HVR-1 MAbs 6/82a and 6/16 were dependent on whether the mammalian-cell-derived antigen was truncated or was obtained from intracellular or secreted sources. They bound the soluble E2660 glycoprotein more efficiently than the E2 present in the FL E1E2 complex. Interestingly, anti-HVR-1 MAbs 7/59, 6/82a, and 6/16 failed to recognize the native form of VLPs in a GNA-capture ELISA. When the VLPs were denatured, however, these MAbs were able to detect them, albeit weakly (Table 1). A similar effect was observed in immunogold-EM assays with these anti-HVR-1 MAbs, indicating that certain amino acid regions within HVR-1 of VLPs are not readily accessible to MAbs.
The differences in antigenicity between various forms of E2 were again highlighted by the observation that MAbs AP109 and AP436 (both specific for aa 464 to 475) and MAb ALP42 (specific for aa 524 to 535) recognized E2660 but not the E2 glycoprotein present in FL E1E2 complex or in VLPs (Table 1). In contrast to MAbs AP109 and AP436, MAb AP320, specific for aa 464 to 471, efficiently bound all forms of antigens with comparable affinities, indicating that amino acid residues present within these regions are not available for recognition by certain MAbs (Table 1; Fig. 3). As expected, the conformation-sensitive anti-E2 MAbs H53 and H60 both recognized only the native forms of all antigens tested (Table 1). The data presented in Table 1 were used to generate a surface topology map of E2 epitopes exposed on VLPs (Fig. 4). Taken together, these results indicate that the availability of certain epitopes on E2 for MAb recognition is dependent on the form of glycoprotein used and that important topological differences exist between the epitopes exposed on E2660, FL E1E2, and VLPs.
Trypsin treatment of VLPs.
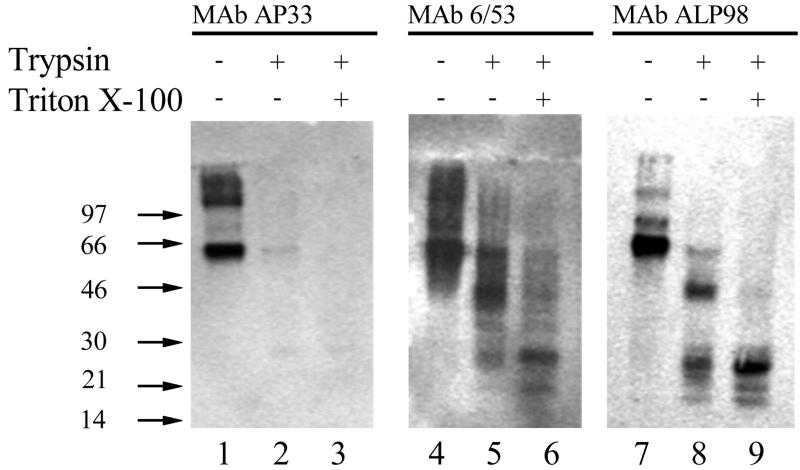
The topology of E2 glycoprotein on HCV VLPs was further investigated by using trypsin to identify domains accessible to proteolysis and/or those protected from digestion by the lipid bilayer. We chose MAbs AP33, 6/53, and ALP98, recognizing epitopes in the N-terminal, central, and C-terminal portions of the E2 ectodomain (Fig. 4), respectively, for identification of trypsin-digested products. HCV VLPs were either left untreated or treated with trypsin in the absence or presence of the detergent Triton X-100, and the digested products were analyzed by Western immunoblotting using the MAbs listed above. As expected, all three MAbs recognized the full-length E2 glycoprotein on untreated VLPs (Fig. 5, lanes 1, 4, and 7). However, MAb AP33 did not recognize E2 following digestion with trypsin in the absence or presence of Triton X-100 (Fig. 5, lanes 2 and 3), indicating removal of its epitope (aa 412 to 423). Interestingly, MAbs 6/53 (recognizing aa 544 to 551) and ALP98 (recognizing aa 644 to 651) recognized E2 fragments of lower molecular mass upon trypsinization of VLPs (Fig. 5, lanes 5 and 8, respectively). With both MAbs, a predominant E2 species of 40 kDa, a minor species of 25 kDa, and some smaller products were seen. This is presumably due to the removal of the N-terminal portion of E2 by trypsin. Furthermore, trypsinization in the presence of Triton X-100 resulted in the loss of the predominant 42-kDa species with a concomitant increase in the relative amount of the 25-kDa product (Fig. 5, lanes 6 and 9). This indicated that solubilization of the lipid membrane in the VLPs by the detergent rendered the N-terminal portion of the 42-kDa species (Fig. 5, lanes 5 and 8) susceptible to further proteolytic digestion (lanes 6 and 9). The 25-kDa species, representing the C-terminal portion of the E2 ectodomain, appeared to be completely protected from trypsin digestion.
FIG. 5.
Trypsin treatment of HCV VLPs. VLPs were either left untreated or treated with trypsin in the presence or absence of Triton X-100. Products were fractionated by SDS-PAGE, and proteins were detected by using the anti-E2 MAbs shown. Positions of molecular weight markers are indicated.
DISCUSSION
We are interested in performing a structure-function analysis of HCV glycoproteins. For that reason we sought to generate in insect cells HCV VLPs derived from the infectious clone genotype 1a strain H77c (62). Expression of H77c core, E1, E2, and p7 in insect cells yielded a population of VLPs that were pleomorphic in size. Particle morphology was consistent with that predicted by analogy to related flaviviruses and pestiviruses: partially enveloped particles with bilayer membranes and visible glycoprotein spikes protruding from the surface. Baumert et al. (2, 3) used a recombinant baculovirus carrying a portion of the 5′ untranslated region followed by sequences encoding the core, E1, and E2 of HCV genotype 1b to produce VLPs which could package short HCV RNA sequences. Interestingly, our expression construct lacks HCV 5′ untranslated region sequences, thus ruling out their possible requirement in VLP assembly. Our cryo-EM data reveal that VLPs contain a dense core, suggesting that the H77c VLPs used in this study may also have a nucleic acid-filled core, although additional experiments are required to confirm this.
From the data shown in Fig. 2A, we suggest that a 21-kDa (p21) mature form of the core is a component of VLPs. The core protein from patient sera has been shown to have a molecular weight identical to that of p21, leading to the suggestion that this form of the protein may be a component of the native virus particle (63). In addition to p21, the presence of a ∼42-kDa core species in VLP preparations (Fig. 2A) indicates that particles may also contain higher-order oligomers of this protein. The core protein is capable both of binding to viral RNA (56) and of homotypic interaction to form multimers (32, 37). Both of these properties are in keeping with its function of encapsidating the viral genome.
Immunogold labeling using E1- and E2-specific MAbs demonstrated the protrusions on the surfaces of VLPs to be the E1 and E2 glycoproteins. Furthermore, the presence of E1 and E2 in the VLP preparation was confirmed by Western immunoblotting. SDS-PAGE analysis under nonreducing conditions revealed the presence of both noncovalently associated E1 and E2 and their disulfide-linked aggregates in VLP preparations. The possibility that the virion envelope may contain a proportion of HCV glycoprotein in an aggregated form has not been ruled out (8). Current strategies of recombinant particle purification do not yield highly pure HCV VLP preparations. Hence, it is possible that the aggregated or nonparticulated form of E1E2 glycoproteins may comigrate with particles on sucrose gradients, thus contaminating the VLP preparation. Therefore, we are unable to unequivocally answer the question of the nature of the glycoprotein complexes present on VLPs. However, inspection of the VLP images in Fig. 1 reveals that the majority of the spikes appear ordered and regularly spaced, suggesting a predominantly nonaggregated form of E1E2. We have also shown that the conformation-sensitive anti-E2 MAbs H53 and H60 specifically recognize the VLPs. These antibodies bind predominantly non-disulfide-linked E2, either alone or complexed with E1 (13, 34). This confirms that at least a proportion of the E1E2 complexes on the VLP surface are in their native form. To further address this question, we are currently trying to develop methods to produce VLPs in a highly purified form.
Using mainly ELISA, Wellnitz et al. (58) recently analyzed the reactivities of a panel of anti-HCV envelope MAbs to HCV VLPs. However, the ELISA data would not necessarily distinguish between particulate and nonparticulate forms of HCV glycoproteins in the VLP preparation. In this study, we performed a more comprehensive analysis of the antigenicities of different forms of mammalian cell-expressed HCV glycoproteins and insect-cell produced VLPs using a wide range of well-characterized anti-E2 MAbs. In addition to ELISA, the reactivities of all MAbs to various antigenic forms (under denaturing and native conditions) of the viral glycoproteins were also analyzed by EM-immunogold assay (Table 1). Comparison of the antigenicity of the VLPs with that of mammalian cell-expressed truncated E2 (E2660) or FL E1E2 has yielded interesting results. In the assays described in this paper, MAb recognition of epitopes within HVR-1 (aa 384 to 411) show differential binding according to whether the antigen was truncated or was obtained from intracellular or secreted sources. Furthermore, the VLPs do not share the same HVR antigenicity as E2660 and FL E1E2 (Table 1; Fig. 4). While the anti-HVR-1 MAb 9/27 binds equivalently to E2660, FL E1E2, and VLPs (at least in an ELISA), HVR-1 epitopes (aa 384 to 395) recognized by MAbs 6/82a and 6/16 are preferentially exposed on E2660. Furthermore, MAbs 6/82a and 6/16, as well as another anti-HVR-1 MAb, 7/59, do not recognize the native form of VLPs in a GNA-capture ELISA or in an immunogold-EM assay. They are able to recognize HVR-1 on VLPs (albeit weakly) only upon denaturation, indicating that certain epitopes (particularly those within the region comprising aa 384 to 395 [Table 1]) within HVR-1 on particles may not be readily accessible by antibodies. Recently, a computer-aided model of E2661 proposed by Yagnik et al. (61) depicted the MAb 6/82a-binding region (aa 384 to 395) as highly exposed. Our data are in agreement with this, but only in the context of E2660 (Table 1; Fig. 4). Interestingly, MAb 9/86a exhibits conformational dependence within the context of FL E1E2 and VLPs, but not E2660. Taken together, our data suggest that different HVR-1 conformations can exist.
The E2 regions comprising aa 412 to 423, 432 to 447, 464 to 471, 480 to 493, 524 to 535, 544 to 551, and 644 to 659 are available for MAb recognition on all antigens tested. Intriguingly, a subset of MAbs specific for the regions comprising aa 464 to 475 (MAbs AP109 and AP436) and 524 to 535 (ALP42) recognized E2660 but not the FL E1E2 complex or VLPs (see Fig. 4 and Table 1). It seems that specific E2 amino acids in the 464-to-475 and 524-to-535 regions that are critical for recognition by MAbs AP109 and AP436 and MAb ALP42, respectively, are not available for recognition on the FL E1E2 complex or on VLPs. The reason for the disparity in recognition of E2660, FL E1E2, and VLPs is not known. It would be reasonable to rule out occlusion of the relevant amino acid residues due to conformational differences and/or heterodimer formation, since these MAbs do not recognize FL E1E2 and VLPs even when they are denatured. It is possible that differential posttranslational processing in E2660 and FL E1E2 complex may be responsible for this effect. In this context, it is interesting that immediately adjacent to E2 aa 464 to 475, there is a potential N-linked glycosylation site (the region comprising aa 476 to 478) which we have recently shown to be utilized at least in the truncated form of E2 (45). Moreover, the epitope recognized by MAb ALP42 (aa 524 to 535) overlaps a potential N-linked glycosylation site (aa 532 to 534). We are currently investigating these possibilities.
Based on certain known properties of E2660, Yagnik et al. (61) suggest that this protein represents the structural core of functional E2, while the C-terminal transmembrane region of E2 (aa 718 to 746) functions as a membrane anchor. However, we and others have recently shown that in addition to this, the TMD of E2 is also required for correct folding of E1 and for heterodimerization of E2 with E1 (41, 47). Therefore, caution should be used in interpretation of data derived by using truncated or “nonparticulated” ligands. We recently identified several MAbs that specifically inhibited the interaction of VLPs, but not E2660 or FL E1E2 complex, with a putative HCV receptor, CD81 (see Fig. 4), indicating that the existence of structural differences affects the functional characteristics of the various E2 forms (42).
The E2 structure on VLPs was further investigated by using protease treatment and specific antibody probes. As shown in Fig. 6, there are several potential trypsin recognition sites in E2. Our results clearly show that the E2 glycoprotein on VLPs is sensitive to trypsin. This is entirely consistent with the fact that the HCV glycoproteins are intraluminally disposed in the ER (40), the proposed budding site for the virus, and therefore, the glycoprotein regions are expected be exposed on the virion. The removal of the MAb AP33 epitope (aa 412 to 423) reduces E2 in size to a 42-kDa and a minor 25-kDa species recognized by MAbs 6/53 (aa 544 to 551) and ALP98 (aa 644 to 651). The loss of a large portion of the N terminus (containing the MAb AP33 epitope) of E2 occurs both in the presence and in the absence of Triton X-100, indicating that this region is well exposed on the surface of the VLP and is well hydrated. There are several trypsin sites within the region comprising aa 394 to 521, all or some of which could be utilized, resulting in degradation of this region into smaller species that are not retained in the gel. The difference between the trypsin profiles in the presence and absence of Triton X-100 of E2 detected by MAbs 6/53 and ALP98 is noteworthy. In the absence of the detergent, both MAbs recognize a major tryptic product of 42 kDa and a minor species of 25 kDa (Fig. 5). Interestingly, membrane solubilization of the VLPs by the detergent leads to almost-complete digestion of the 42-kDa product, with a concomitant increase in the relative concentration of the 25-kDa species, which retains the epitopes recognized by MAbs 6/53 and ALP98. It is difficult to predict which trypsin sites are utilized due to the presence of N-linked glycans scattered throughout E2 (Fig. 6). However, upon examination of the amino acid sequence of E2 (Fig. 6), it is apparent that there is a potential trypsin-sensitive site proximal to the ALP98 epitope (RDRSEL) at residues 656 to 661 and one at positions 713 to 717 (AIKWE), adjacent to the E2 TMD (aa 718 to 746). Our data suggest that the 25-kDa tryptic product recognized by MAbs 6/53 and ALP98 is released from the VLP by detergent solublization. It is conceivable that Triton X-100 exposes the potential trypsin site at aa 656 to 661 and/or that at aa 715 from a previously hydrophobic environment. Alternatively, the detergent treatment may remove steric hindrance due to the proximity of the hydrophobic environment of the nearby TMD, thus allowing cleavage and release of the partial ectodomain. Recently published data from computer-aided modeling have suggested that part of the E2 protein, distinct from the known transmembrane region, is adsorbed onto the membrane by an amphipathic alpha-helix corresponding to aa 700 to 715 (7). We suggest that detergent enables both sides of the amphipathic alpha-helix to be released from adsorption at the lipid bilayer, resulting in efficient spatial accessibility of trypsin to the sensitive site(s), thus generating the 25-kDa fragment. This fragment could be pursued as a candidate for crystallization in order to delineate the structure of at least a portion of the E2 protein.
FIG. 6.
Amino acid sequence of HCV E2 (aa 384 to 746 of the HCV polyprotein). Potential trypsin cleavage sites are boldfaced and underlined. Solid bars below amino acid residues, predicted N-linked glycosylation sites. Shaded boxes, epitopes recognized by MAbs AP33, 6/53, and ALP98. The sequence of the TMD (aa 718 to 746) is italicized.
Acknowledgments
We thank Jens Bukh, Jane McKeating, and Jean Dubuisson for kind gifts of reagents used in this study and Larry Loomis-Price for epitope mapping of MAbs.
REFERENCES
- 1.Al-Khayat, H. A., D. Bhella, J. M. Kenney, J. F. Roth, A. J. Kingsman, E. Martin-Rendon, and H. R. Saibil. 1999. Yeast Ty retrotransposons assemble into virus-like particles whose T-numbers depend on the C-terminal length of the capsid protein. J. Mol. Biol. 292:65-73. [DOI] [PubMed] [Google Scholar]
- 2.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumert, T. F., J. Vergalla, J. Satoi, M. Thomson, M. Lechmann, D. Herion, H. B. Greenberg, S. Ito, and T. J. Liang. 1999. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology 117:1397-1407. [DOI] [PubMed] [Google Scholar]
- 4.Bielefeldt Ohmann, H., and B. Bloch. 1982. Electron microscopic studies of bovine viral diarrhea virus in tissues of diseased calves and in cell cultures. Arch. Virol. 71:57-74. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, D. H. L. 1992. Baculovirus expression vectors. Semin. Virol. 3:253-264. [Google Scholar]
- 6.Chan-Fook, C., W. R. Jiang, B. E. Clarke, N. Zitzmann, C. Maidens, J. A. McKeating, and I. M. Jones. 2000. Hepatitis C virus glycoprotein E2 binding to CD81: the role of E1E2 cleavage and protein glycosylation in bioactivity. Virology 273:60-66. [DOI] [PubMed] [Google Scholar]
- 7.Charloteaux, B., L. Lins, H. Moereels, and R. Brasseur. 2002. Analysis of the C-terminal membrane anchor domains of hepatitis C virus glycoproteins E1 and E2: toward a topological model. J. Virol. 76:1944-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choukhi, A., A. Pillez, H. Drobecq, C. Sergheraert, C. Wychowski, and J. Dubuisson. 1999. Characterization of aggregates of hepatitis C virus glycoproteins. J. Gen. Virol. 80:3099-3107. [DOI] [PubMed] [Google Scholar]
- 9.Choukhi, A., S. Ung, C. Wychowski, and J. Dubuisson. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 72:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerel, L., S. Duvet, J. C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison, A. J., and B. Moss. 1990. New vaccinia virus recombination plasmids incorporating a synthetic late promoter for high level expression of foreign proteins. Nucleic Acids Res. 18:4285-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubuisson, J., H. H. Hsu, R. C. Cheung, H. B. Greenberg, D. G. Russell, and C. M. Rice. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubuisson, J., and C. M. Rice. 1996. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J. Virol. 70:778-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 2095. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32093. [DOI] [PubMed] [Google Scholar]
- 17.Farci, P., A. Shimoda, D. Wong, T. Cabezon, D. DeGioannis, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flint, M., and J. A. McKeating. 1999. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J. Gen. Virol. 80:1943-1947. [DOI] [PubMed] [Google Scholar]
- 21.Flint, M., J. M. Thomas, C. M. Maidens, C. Shotton, S. Levy, W. S. Barclay, and J. A. McKeating. 1999. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J. Virol. 73:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 23.Habersetzer, F., A. Fournillier, J. Dubuisson, D. Rosa, S. Abrignani, C. Wychowski, I. Nakano, C. Trepo, C. Desgranges, and G. Inchauspe. 1998. Characterization of human monoclonal antibodies specific to the hepatitis C virus glycoprotein E2 with in vitro binding neutralization properties. Virology 249:32-41. [DOI] [PubMed] [Google Scholar]
- 24.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinz, F. X., C. W. Mandl, H. Holzmann, C. Kunz, B. A. Harris, F. Rey, and S. C. Harrison. 1991. The flavivirus envelope protein E: isolation of a soluble form from tick-borne encephalitis virus and its crystallization. J. Virol. 65:5579-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenney, J. M., C. H. von Bonsdorff, M. Nassal, and S. D. Fuller. 1995. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure 3:1009-1019. [DOI] [PubMed] [Google Scholar]
- 27.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livingstone, C., and I. Jones. 1989. Baculovirus expression vectors with single strand capability. Nucleic Acids Res. 17:2366.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luckow, V. A., and M. D. Summers. 1988. Trends in the development of baculovirus expression vectors. Bio/Technology 6:47-55. [Google Scholar]
- 30.Magar, R., and J. Lecomte. 1987. Comparison of methods for concentration and purification of bovine viral diarrhea virus. J. Virol Methods 16:271-279. [DOI] [PubMed] [Google Scholar]
- 31.Major, M. E., B. Behermann, and S. M. Feinstone. 2001. Hepatitis C virus, p. 1127-1161. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 32.Matsumoto, M., S. B. Hwang, K. S. Jeng, N. L. Zhu, and M. M. C. Lai. 1996. Homotypic interaction and multimerization of hepatitis C virus core protein. Virology 218:43-51. [DOI] [PubMed] [Google Scholar]
- 33.Meola, A., A. Sbardellati, B. Bruni Ercole, M. Cerretani, M. Pezzanera, A. Ceccacci, A. Vitelli, S. Levy, A. Nicosia, C. Traboni, J. McKeating, and E. Scarselli. 2000. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol. 74:5933-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michalak, J. P., C. Wychowski, A. Choukhi, J. C. Meunier, S. Ung, C. M. Rice, and J. Dubuisson. 1997. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 78:2299-2306. [DOI] [PubMed] [Google Scholar]
- 35.Moore, J. P., Q. J. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolandt, O., V. Kern, H. Muller, E. Pfaff, L. Theilmann, R. Welker, and H. G. Krausslich. 1997. Analysis of hepatitis C virus core protein interaction domains. J. Gen. Virol. 78:1331-1340. [DOI] [PubMed] [Google Scholar]
- 38.Ohmann, H. B. 1990. Electron microscopy of bovine virus diarrhoea virus. Rev. Sci. Tech. 9:61-73. [DOI] [PubMed] [Google Scholar]
- 39.Olson, N. H., and T. S. Baker. 1989. Magnification calibration and the determination of spherical virus diameters using cryo-microscopy. Ultramicroscopy 30:281-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Op De Beeck, A., L. Cocquerel, and J. Dubuisson. 2001. Biogenesis of hepatitis C virus envelope glycoproteins. J. Gen. Virol. 82:2589-2595. [DOI] [PubMed] [Google Scholar]
- 41.Op De Beeck, A., R. Montserret, S. Duvet, L. Cocquerel, R. Cacan, B. Barberot, M. Le Maire, F. Penin, and J. Dubuisson. 2000. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 275:31428-31437. [DOI] [PubMed] [Google Scholar]
- 42.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 43.Owsianka, A. M., and A. H. Patel. 1999. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257:330-340. [DOI] [PubMed] [Google Scholar]
- 44.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 45.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 81:2873-2883. [DOI] [PubMed] [Google Scholar]
- 46.Patel, J., A. H. Patel, and J. McLauchlan. 1999. Covalent interactions are not required to permit or stabilize the non-covalent association of hepatitis C virus glycoproteins E1 and E2. J. Gen. Virol. 80:1681-1690. [DOI] [PubMed] [Google Scholar]
- 47.Patel, J., A. H. Patel, and J. McLauchlan. 2001. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology 279:58-68. [DOI] [PubMed] [Google Scholar]
- 48.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 50.Ralston, R., K. Thudium, K. Berger, C. Kuo, B. Gervase, J. Hall, M. Selby, G. Kuo, M. Houghton, and Q. L. Choo. 1993. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J. Virol. 67:6753-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. N. Lau, Q. L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg, S. 2001. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 313:451-464. [DOI] [PubMed] [Google Scholar]
- 53.Ryan, M. D., S. Monaghan, and M. Flint. 1998. Virus-encoded proteinases of the Flaviviridae. J. Gen. Virol. 79:947-959. [DOI] [PubMed] [Google Scholar]
- 54.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimizu, Y. K., H. Igarashi, T. Kiyohara, T. Cabezon, P. Farci, R. H. Purcell, and H. Yoshikura. 1996. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology 223:409-412. [DOI] [PubMed] [Google Scholar]
- 56.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiland, F., E. Weiland, G. Unger, A. Saalmuller, and H. J. Thiel. 1999. Localization of pestiviral envelope proteins E(rns) and E2 at the cell surface and on isolated particles. J. Gen. Virol. 80:1157-1165. [DOI] [PubMed] [Google Scholar]
- 58.Wellnitz, S., B. Klumpp, H. Barth, S. Ito, E. Depla, J. Dubuisson, H. E. Blum, and T. F. Baumert. 2002. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J. Virol. 76:1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 61.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355-366. [DOI] [PubMed] [Google Scholar]
- 62.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasui, K., T. Wakita, K. Tsukiyama Kohara, S. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zibert, A., E. Schreier, and M. Roggendorf. 1995. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology 208:653-663. [DOI] [PubMed] [Google Scholar]