Abstract
The mechanism(s) by which Sin Nombre (SN) hantavirus is maintained in deer mouse populations is unclear. Field studies indicate that transmission occurs primarily if not exclusively via a horizontal mechanism. Using an experimental deer mouse infection model in an outdoor laboratory, we tested whether infected rodents shed SN virus in urine, feces, and saliva, whether infected mice transmit infection to naïve cage mates, and whether infected dams are able to vertically transmit virus or antibody to offspring. Using pooled samples of urine, feces, and saliva collected from mice infected 8 to 120 days postinoculation (p.i.), we found that a subset of saliva samples, collected between 15 and 90 days p.i., contained viral RNA. Parallel studies conducted on wild-caught, naturally infected deer mice showed a similar pattern of intermittent positivity, also only in saliva samples. Attempts to isolate virus through inoculation of cells or naïve deer mice with the secreta or excreta of infected mice were uniformly negative. Of 54 attempts to transmit infection by cohousing infected deer mice with seronegative cage mates, we observed only a single case of transmission, which occurred between 29 and 42 days p.i. Dams passively transferred antibodies to neonatal pups via milk, and those antibodies persisted for at least 2 months after weaning, but none transmitted infection to their pups. Compared to other hantavirus models, SN virus is shed less efficiently and transmits inefficiently among cage mates. Transmission of SN virus among reservoir rodents may require factors that are not required for other hantaviruses.
Hantaviruses (Bunyaviridae family; Hantavirus genus) are rodent-borne viruses with a worldwide distribution. As with other members of the family Bunyaviridae, hantaviruses are enveloped and have a tripartite, negative-sense RNA genome. Two distinct human illnesses are associated with hantavirus infection: hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS) (36). HFRS occurs primarily in Europe and Asia, with a case/fatality ratio of 0.1 to 10% (36). Etiologic agents of HFRS include the prototypic Hantaan (HTN) virus, Seoul (SEO) virus, Puumala (PUU) virus, and Dobrava-Belgrade virus. HCPS occurs in North and South America (28). There are at least four etiologic agents of HCPS in North America: Sin Nombre (SN), Bayou, New York, and Black Creek Canal (BCC) viruses. Of these four viruses, the primary etiologic agent is SN virus, which is believed to be responsible for a large majority (>95%) of the more than 300 reported cases. The case/fatality ratio of HCPS is approximately 40%. The deer mouse (Peromyscus maniculatus) is the predominant carrier of SN virus (12, 32).
Natural transmission of hantaviruses among wild rodents is poorly understood, but both field and laboratory studies suggest that nearly all, if not all, transmission is via one or more horizontal routes and that male mice are differentially affected. In this study, we tested the hypothesis that deer mice experimentally infected with SN virus shed virus into urine, feces, and saliva. We also carried out experiments to determine (i) whether housing infected mice with naïve mice would result in horizontal transmission and (ii) whether infected dams would be able to vertically transmit virus and/or maternal antibody to offspring.
MATERIALS AND METHODS
Animal handling and viral inoculations.
We adhered strictly to the biosafety recommendations of the United States Centers for Disease Control and Prevention in all aspects of this work (8, 11, 30). Workers wearing respirators and personal protective gear carried out all inoculations and manipulations of infected animals in an outdoor quarantine facility as described previously (7). Animals were handled according to University of New Mexico animal research facility guidelines by using an approved protocol. Deer mice were obtained from an outbred colony and were used at the F1 to F3 generations (8, 9). For blood samples collected for nonterminal experiments, we anesthetized animals with methoxyflurane and collected blood from the retro-orbital sinus. For terminal experiments, we used tribromoethanol followed by exsanguination via cardiac puncture for euthanasia. We infected deer mice by intramuscular inoculation with SN virus strain SN77734, using five doses at which 50% of the inoculated animals became infected (ID50) for mice used as donors for shedding sample collection, donors of blood for viral isolation, index animals for intracage transmission studies, and dams in maternal transfer studies and using 500 animal ID50 for mice used in challenge studies (7). Intranasal (i.n.) inoculations were carried out by delivering either 25 or 40 animal ID50 of strain SN77734 in a volume of 50 to 80 μl of phosphate-buffered saline (PBS) while mice were under methoxyflurane anesthesia. To detect infection, we assayed for antibodies to viral nucleocapsid (N) antigen by strip immunoblot assay (SIA) on day 21 postinoculation (p.i.) or later (5, 7). With this virus under these conditions, 100% of deer mice that are inoculated intramuscularly become infected. These mice develop antibodies and are persistently infected, as judged by the consistent presence of N antigen observed in immunohistochemical (IHC) analysis and by the continued detection of viral genome by reverse transcription (RT)-PCR analysis of tissues obtained at necropsy (J. Botten et al., unpublished data) (7).
Collection of excreta and saliva.
For the viral shedding experiments, secretions and excretions were collected at the outdoor facility from a cohort of eight experimentally infected juvenile (4- to 6-week-old) deer mice between 16 May 2000 and 9 November 2000. Mice were provided food and water ad libitum. We collected urine, feces, and saliva from a subset of the eight animals, a minimum of two per time point, at days 8, 15, and 22 p.i. (May 2000), days 29, 43, and 51 p.i. (June 2000), days 60 and 78 p.i. (July 2000), day 90 p.i. (August 2000), and day 120 p.i. (November 2000). The monthly mean temperatures between May and September 2000 were lows of 14.4 to 18.3°C and highs of 29.1 to 33.1°C, while October and November registered lows of 7.3 and −1.9°C and highs of 18.6 and 9.6°C, respectively. The photoperiod (hours of light:hours of dark) during the study was ∼14:10 in May, June, and July, 13:9 in August, 12:12 in September, 11:13 in October, and 10:14 in November. The precipitation pattern was as follows: 0 mm in May, 15.5 mm in June, 36.3 mm in July, 37.2 mm in August, 6.8 mm in September, 49.6 mm in October, and 32 mm in November.
To collect urine, we placed infected animals over a sterile 100-mm-diameter petri dish and grasped them by the scruff of the neck, which induced micturation. Feces were also collected on petri dishes. Saliva was obtained by using a sterile oral swab (Harwood Products Company, Guilford, Maine). We attempted to avoid physically dislodging cells from the buccal mucosa during the collection of saliva. All samples were kept out of sunlight and placed directly onto ice following collection.
Urine samples from two to six animals per time point were pooled, diluted 10-fold with PBS containing 50 μg of gentamicin/ml, split into 110-μl aliquots, and frozen. Fecal samples were pooled from five to eight animals at each time point and diluted 10-fold in PBS containing 50 μg of gentamicin/ml. This suspension was then centrifuged at 900 × g for 20 min at 4°C. The supernatant was passed through a 0.45-μm filter and split into 110-μl aliquots and frozen. Swabs from at least four animals were pooled at each time point, and each swab was diluted with 325 μl of PBS containing 50 μg of gentamicin/ml. The contents of the swab were expressed into the supernatant, which was split into 110-μl aliquots and frozen. RNA extractions and in vitro viral isolation attempts from pooled shedding samples were carried out with 20 μl of urine, 20 mg of fecal supernatant, or two-thirds of the content of a single swab. For in vivo viral isolation attempts, we delivered a 10-fold dilution of the same material via intramuscular inoculation in the hind leg quadriceps of juvenile deer mice. Mice were screened for anti-N antibodies by SIA at day 35 p.i. to determine their infection status (5, 7). The quantities of urine, feces, and saliva we used for the RT-PCR and in vitro isolation attempts were chosen to assure that even very small amounts of viral RNA would be detected. These amounts were at least 20- to 250-fold more than was needed to consistently detect infectious virus in other hantavirus infection models (22, 25, 26).
Wild-caught shedding sample collection.
In a field collection conducted to obtain wild-caught deer mice for comparison to our experimentally infected subjects, we were able to capture a single 19-g male infected deer mouse by using methods described previously (8). This specimen came from the Manzano mountains of New Mexico (latitude, 37°37.37′; longitude, 106°24.78′; elevation, 2,621 m). After a positive SIA demonstrated that he was seropositive, we transported him to the outdoor quarantine laboratory (5). We collected serial samples of urine, feces, and saliva from this animal on days 13, 15, 20, 25, 35, 41, 42, 47, 59, 63, and 68 postcapture as described above (see Table 2).
TABLE 2.
Detection of viral RNA in samples of urine, feces, and saliva from a naturally infected deer mouse
| Day(s) postcapturea | Presence of viral RNA in indicated sample
|
||
|---|---|---|---|
| Urine | Feces | Saliva | |
| 13 and 15 | − | − | − |
| 20 and 25 | NDb | ND | − |
| 35, 41, and 42 | − | − | − |
| 47 | − | ND | − |
| 47 and 59 | ND | − | ND |
| 59 | − | ND | − |
| 63 and 68 | − | − | − |
For rows where more than 1 day is listed, samples of urine, feces, or saliva were pooled from the listed times postcapture.
ND, not done.
The five wild deer mice used as donors of saliva (see Table 3) were trapped by workers from the Museum of Southwestern Biology, near the Manzano mountains, as part of a routine protocol for an ongoing longitudinal trapping study. All of the mice were adult males and all were seropositive by SIA by the time the swabs were collected (5). The masses of the animals (identified by number) at the time when they were first shown to be seropositive were as follows: NK77724, 17 g; NK77731, 18 g; NK83610, 27.5 g; NK86291, 18 g; and NK85819, 21 g. Upon capture, oral swabs were taken and placed in 500 μl of minimal essential medium and frozen. RNA was extracted from two-thirds of the viral medium, and the entire RNA sample was used for nested RT-PCR analysis.
TABLE 3.
Detection of viral RNA in saliva samples obtained from naturally infected deer mice
| Animal | Presence of viral RNA at indicated date of sample collection
|
|||
|---|---|---|---|---|
| November 1998 | January 1999 | April 1999 | June 1999 | |
| NK77724 | − | − | NDa | ND |
| NK77731 | + | + | ND | ND |
| NK83610 | − | − | ND | ND |
| NK86291 | ND | ND | − | − |
| NK85819 | − | + | − | + |
ND, not done.
Intracage transmission.
To test for horizontal intracage transmission, 5- to 7-week-old naïve, colony-bred deer mice were exposed in the same cage to experimentally infected mice at the outdoor laboratory. For each exposure interval, approximately equal numbers of naïve male and female mice were exposed to infected mice of the same or opposite sex in approximately equal ratios. Ten to 13 naïve mice were exposed to groups of one or two infected animals over intervals of 14 days through the first 60 days p.i. In addition, one to three naïve mice were exposed to experimentally infected animals over the first 73, 81, 163, or 213 days of infection in index animals.
Naïve mice were quarantined for a period of 35 days following their exposure to infected cage mates and then screened for anti-N antibodies by SIA (5). Visible wounds and pregnancies were recorded. The single animal that became infected via intracage transmission was sacrificed 37 days after being placed in quarantine. We removed 15 tissues from this animal for IHC analysis for comparison to experimentally infected tissues as previously described (7).
Nested RT-PCR.
We extracted RNA from the pooled excreta, secretions, and cell culture supernatants by using a Qiamp Viral RNA Mini Kit (Qiagen, Valencia, Calif.), which was designed for extracting RNA from cell-free materials. Each shedding sample was eluted in a final volume of 30 μl of water, and we then used the entire volume of RNA for RT-PCR with nested primers for the small (S) genomic segment of SN virus as described previously (7). The coordinates of the outer primers were 167 and 423 on the SN virus S segment, with inner primers at 190 and 401. To reduce RNase activity during RT, we added 0.4 U of RNasin per reaction.
Quantitative TaqMan RT-PCR.
We used a PE Biosystems (Foster City, Calif.) model 5700 sequence detection system and the two-step TaqMan Gold RT-PCR protocol as described by the manufacturer. Each sample was tested in triplicate. For RT, 5 μl of template RNA was mixed either with random hexamers to determine total S-segment RNA copies or with our S-segment coordinate-167 sense AGC ACA TTA CAG AGC AGA CGG GC primer to determine total negative-strand S-segment RNA copies in a volume of 100 μl at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min (7). Five microliters of cDNA was removed for subsequent PCRs. The S-segment primers used for the PCR were coordinate-179 sense GCAGACGGGCAGCTGTG and coordinate-245 antisense AGATCAGCCAGTTCCCGCT. The fluorescent probe was a positive sense oligonucleotide at coordinate 198, TGCATTGGAGACCAAACTCGGAGAACTT. The probe was covalently labeled with the reporter dye FAM at the 5′ end and the quencher dye TAMRA at the 3′ end (PE Biosystems). All oligonucleotides were used at 200 nM.
During the reaction, the quencher is dissociated from the reporter by endonucleolytic cleavage of the probe, and a sample is judged positive when the FAM fluorescence exceeds 0.05 reporter units. In our experiments, all curves that exceeded that value progressed with the expected sigmoid amplification curve in later cycles. At the point that the absorption curve exceeds 0.05 units, the threshold cycle number (Ct) is determined, which is inversely related to the template copy number. PCR was conducted at 95°C for 10 min followed by 40 repetitions of 95°C for 10 s, 50°C for 10 s, and 72°C for 30 s. A standard curve containing dilutions ranging from 50 to 5 × 105 copies of template was used on each 96-well plate. All of our standard curves produced a −0.995 or better correlation coefficient between the quantity of template loaded and the log of the Ct value.
In vitro viral isolation.
Attempts to isolate SN virus in early passage Vero E6 cells were conducted in either T-25 flasks (shedding samples) or 48-well plates (blood samples). Our positive control for viral isolation was a 1% lung homogenate derived from an experimentally infected animal at day 13 p.i. Samples were incubated on cells (80 to 100% confluent) in a volume of 1 ml (T-25 flasks) or 200 μl (48-well plates) of supplemented minimal essential medium for 1 h at 37°C (4). We used 20 μl of urine, 20 mg of fecal supernatant, two-thirds of a swab, or serial dilutions (10−1 through 10−5) of 20 μl of blood for inoculation. After incubation, cells were washed with PBS and overlaid with medium (5 ml for T-25 flasks, 500 μl for 48-well plates). Supernatants were harvested at weekly intervals for the first 4 weeks after inoculation and frozen for RNA analysis by nested RT-PCR and viral titration.
SIA.
In separate lanes, we loaded 2 ml of a 2-μl/ml dilution of deer mouse serum in PBS for a 3+ intensity control, 2 ml of a 0.2-μl/ml dilution of serum for a 1+ intensity control, and 3 μg of recombinant, affinity-purified SN virus N antigen (5, 19). We then suctioned the antigens onto a wetted nitrocellulose membrane under vacuum. The membrane was cut lengthwise with a paper shredder into 1.6-mm strips, and each strip was placed into a well in a Western blot tray containing 1 ml of milk-PBS buffer and 5 μl of rodent periorbital sinus blood. The membranes were rocked gently overnight at room temperature. We applied a 1:1,000 dilution of alkaline phosphatase-conjugated goat anti-Peromyscus leucopus immunoglobulin G antibodies (Kirkegaard and Perry, Gaithersburg, Md.) and rocked gently for 1 h. Bound alkaline phosphatase was then detected with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate substrate (24, 37).
IHC.
At necropsy, 15 tissues (heart, lung, kidney, liver, spleen, pancreas, thymus, brain, salivary gland, brown fat, white fat, testis or ovary, urinary bladder, skeletal muscle, and large intestine) were placed in Z-fix formalin (Anatech Ltd., Battle Creek, Mich.) for at least 24 h before being embedded in paraffin. The paraffin-embedded tissues were cut into 4- to 6-μm sections. We mounted the sections on glass slides coated with poly-l-lysine, deparaffinized them, and then stained them with anti-N antiserum (1:10,000) on an automated processor following antigen retrieval as described previously (17). Immunodetection was performed with a hyperimmune polyclonal rabbit serum directed against the recombinant N antigen of SN virus strain 3H226 (3, 7, 20). The immune complexes were detected with a biotinylated anti-rabbit secondary antibody and then a horseradish peroxidase-avidin conjugate, followed by detection with an amino-ethyl carbazole chromogen. Specific stain consisted of punctate, cytoplasmic granules. After applying hematoxylin as a counterstain, we mounted the slides with aqueous mounting media. Preimmune rabbit serum was extensively used initially to verify the specificity of the test during the development of the IHC procedure, which verified that the observed staining was specific for the viral N antigen.
Spiking controls.
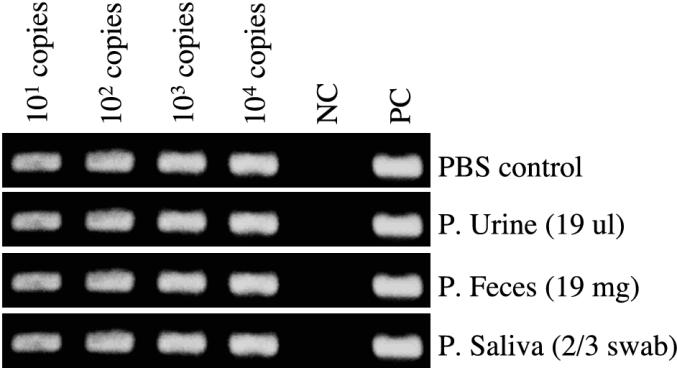
To ensure that our RNA extraction protocol was effectively extracting RNA from the samples, we spiked pooled urine, feces, and saliva samples with supernatants of infected Vero E6 cells containing 10, 100, 1,000, or 10,000 copies of total S-segment RNA. To prepare cell-free virus for experimental spiking of urine, feces, or saliva samples, we isolated strain SN77734 in Vero E6 cells essentially as described by Schmaljohn et al. (35) (R. Xiao, unpublished data). To determine the concentration of viral particles in the supernatants, we used TaqMan RT-PCR assays with two-tube RT and PCRs and random hexamer primers as described previously (7). Known amounts of viral RNA (in the form of virus) were then added into negative-control saliva, urine, or feces pools (with the same quantities of shedding samples described in “Collection of excreta and saliva”) before preparation of RNA to verify the efficiency of the extraction and nested RT-PCR processes.
RESULTS
Viral shedding.
All eight mice utilized as donors of shedding samples were positive for anti-N antibodies, N antigen in the heart, and S-segment RNA in heart, lung, and brown adipose tissue at the time of sacrifice (day 120 p.i., n = 5; day 180 p.i., n = 3) (data not shown). Using nested RT-PCR to screen our pooled shedding samples, we found no detectable viral RNA in the urine or fecal samples (Table 1). We found saliva samples positive for viral RNA on days 15, 43, 60, 78, and 90 p.i. The samples in each of the positive pools originated from three females and one male. We were able to screen individual swabs at the day 43 and 78 p.i. time points to determine the proportion of mice that had viral RNA present in saliva. These swabs were taken from mice that were not among those used for the pooled samples. Two of three mice at day 43 p.i. and one of four mice at day 78 p.i. were positive for viral RNA (Table 1). The donor mice at day 43 p.i. consisted of two females and one male, while at day 78 p.i. we screened saliva obtained from three females and one male. The positive samples at each time point were obtained from female mice. To verify that negative RT-PCR results were not due to inefficient extraction of RNA, we spiked pooled samples of urine, feces, and saliva with known quantities of cell-free virus particles (Fig. 1). With RT-PCR, our sensitivity threshold was no more than 10 copies of viral RNA from urine, fecal, and saliva samples.
TABLE 1.
Detection of viral RNA and infectious virus in samples of pooled urine, feces, and saliva from infected deer mice
| Day p.i. | Presence of viral RNA
|
Cell culture isolation
|
No. infected/no. inoculateda
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Urine | Feces | Saliva | Urine | Feces | Saliva | Urine | Feces | Saliva | |
| 8 | − | − | − | − | − | − | 0/3 | 0/3 | 0/3 |
| 15 | − | − | + | − | − | − | 0/3 | 0/3 | 0/3 |
| 22 | − | − | − | − | − | − | 0/3 | 0/3 | 0/2 |
| 29 | − | − | − | − | − | − | 0/3 | 0/3 | 0/3 |
| 43 | − | − | +b | − | − | − | 0/3 | 0/3 | 0/3 |
| 51 | − | − | − | − | − | − | 0/2 | 0/3 | 0/3 |
| 60 | − | − | + | − | − | − | 0/3 | 0/3 | 0/1 |
| 78 | − | − | +c | NDd | − | ND | 0/3 | 0/3 | 0/3 |
| 90 | − | − | + | − | − | − | 0/3 | 0/3 | 0/3 |
| 120 | − | − | − | − | − | − | 0/3 | 0/1 | 0/3 |
Indicates the total number of animals that seroconverted out of the total number of animals inoculated with a given pool.
Two of three animals were positive for viral RNA.
One of four animals was positive for viral RNA.
ND, not done.
FIG. 1.
Viral RNA spiking experiments. Viral supernatants of SN77734-infected cultures that contained known numbers of copies of SN virus S-segment RNA were added into either PBS or pooled samples of urine, feces, and saliva before extraction of viral RNA. These RNAs were then used in nested RT-PCRs. NC, negative control; PC, positive control.
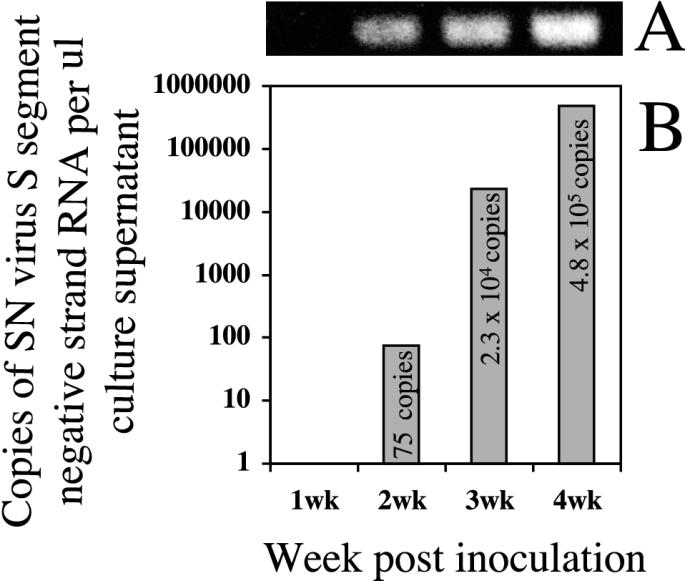
Pooled shedding samples were screened for infectious virus both in vitro and in vivo. Using our positive control (day 13 p.i. lung homogenate) for the in vitro isolation attempts, we were able to determine the kinetics of viral release into the cell culture supernatants after inoculation (Fig. 2). Viral RNA did not become detectable in the culture supernatants until 2 weeks p.i. (75 copies per μl of supernatant), and the quantity of viral RNA increased 306-fold between weeks 2 and 3 p.i. and another 20-fold between weeks 3 and 4 p.i. (Fig. 2). We were unable to detect infectious virus in any of the shedding samples, including the saliva pools positive for viral RNA, by either in vivo or in vitro inoculation (Table 1).
FIG. 2.
Kinetics of SN virus replication after primary isolation in Vero E6 cells from a positive control tissue (day 13 p.i. lung) homogenate. (A) Nested RT-PCR specific for viral S-segment RNA carried out on supernatant RNAs; (B) copies of negative-strand S-segment RNA per microliter of supernatant as determined by TaqMan quantitative RT-PCR.
To test whether blood might be serving as a vehicle for the dissemination of virus among mice, we attempted to isolate infectious virus from blood obtained from mice at day 13 p.i. (n = 3) and day 21 p.i. (n = 2). Based on the results of previous studies, we reasoned that blood samples obtained during acute infection might be positive for infectious virus, because viral RNA levels reach peak titers and neutralizing antibody titers are low at these time points after infection (7). We were unable to detect infectious virus from any of the samples tested.
In order to compare our experimental shedding results to natural infections, we trapped a naturally infected male deer mouse at the same location from which the founders of our deer mouse colony were obtained (9). This animal was moved to and maintained in our outdoor facility, where samples of urine, feces, and saliva were collected at several time points after capture (Table 2). Upon sacrifice (day 68 postcapture), IHC studies revealed N-antigen staining in the heart, lung, kidney, spleen, salivary gland, and brown fat of this animal. This pattern of antigen distribution was consistent with that observed with experimentally infected animals during persistent infection (data not shown). RNA was extracted from urine, feces, and saliva and screened for viral RNA by nested RT-PCR. All samples were negative for viral RNA. In order to increase our sample size, we tested oral swabs obtained from naturally infected deer mice (n = 5) that had been trapped on at least two occasions as part of an ongoing longitudinal trapping study (Table 3). Saliva from two of the five mice had viral RNA detectable by nested RT-PCR. Animal NK85819 displayed intermittent shedding of viral RNA at ≥1 and ≥6 months after seroconversion, and animal NK77731 had RNA in saliva at ≥8 and ≥10 months postseroconversion. The duration of infection we report is the length of time between the first positive antibody test and the collection of the sample in question and thus represents a minimum figure.
Because it remains unclear how natural infections are transmitted, we wished to test whether an alternative route of inoculation, such as i.n., would have an effect on the ability of infected mice to shed virus. Using i.n. inoculation, we delivered 25 and 40 animal ID50 of SN virus strain SN77734 to 6 and 10 mice, respectively. We were unable to demonstrate seroconversion in any of the mice, indicating that the titer of our SN virus stock administered by the i.n. route was much lower than that of the same stock administered intramuscularly.
Horizontal intracage transmission.
Naïve animals were introduced into and removed from cages to allow 14-day intervals of exposure to experimentally infected animals in the same cage. Of 46 naïve deer mice that were exposed to infected animals at various intervals p.i., only a single mouse, 2785, became infected (Table 4). This male animal had been exposed to a pair of infected males between days 29 and 42 p.i. of their infection. We were not able to document any wounding on this animal or its infected cage mates, nor could we detect any ectoparasites by routine brushing when it was sacrificed 35 days after the end of its exposure period. The tissue distribution of viral N antigen seen for animal 2785, as assessed by IHC, was indistinguishable from that seen for experimentally infected mice (7). We observed positive staining in the heart, lung, kidney, liver, spleen, thymus, pancreas, white fat, brown adipose tissue, and skeletal muscle. To determine whether exposure to index animals after day 60 p.i. would result in intracage transmission, other naïve animals were housed with infected animals for the first 73, 81, 163, or 213 days p.i., but again, no transmission was observed (Table 4).
TABLE 4.
Intracage transmission of SN virus and wounding events in deer mice
| Interval of exposure (days) after inoculation of index animal | No. of mice infected/no. of mice exposed | No. of mice with wounds |
|---|---|---|
| 0-14 | 0/13 | 0 |
| 15-28 | 0/12 | 2 |
| 29-42 | 1/11 | 0 |
| 46-60 | 0/10 | 1 |
| 0-73 | 0/1 | 0 |
| 0-81 | 0/2 | 0 |
| 0-163 | 0/3 | 0 |
| 0-213 | 0/2 | 0 |
All experimentally inoculated animals used in these experiments were shown to have seroconverted as expected (data not shown).
A limited number of wounding events were observed during the exposure intervals of days 15 to 28 p.i. (n = 2) and days 46 to 60 p.i. (n = 1). Two were tail lesions and one consisted of scabs about the ears. These events were not associated with transmission. Furthermore, three females became impregnated during the cohabitation studies. One infected male impregnated two naïve cage mates between days 46 and 60 p.i. Both females delivered litters during the quarantine period, and both they and their pups (n = 2 apiece) remained uninfected after quarantine. In the second instance, a naïve male impregnated an infected female, which we back-calculated to be at day 170 p.i. This mouse did not become infected. Thus, sexual transmission was not observed.
Vertical transmission and maternal antibody transfer and protection.
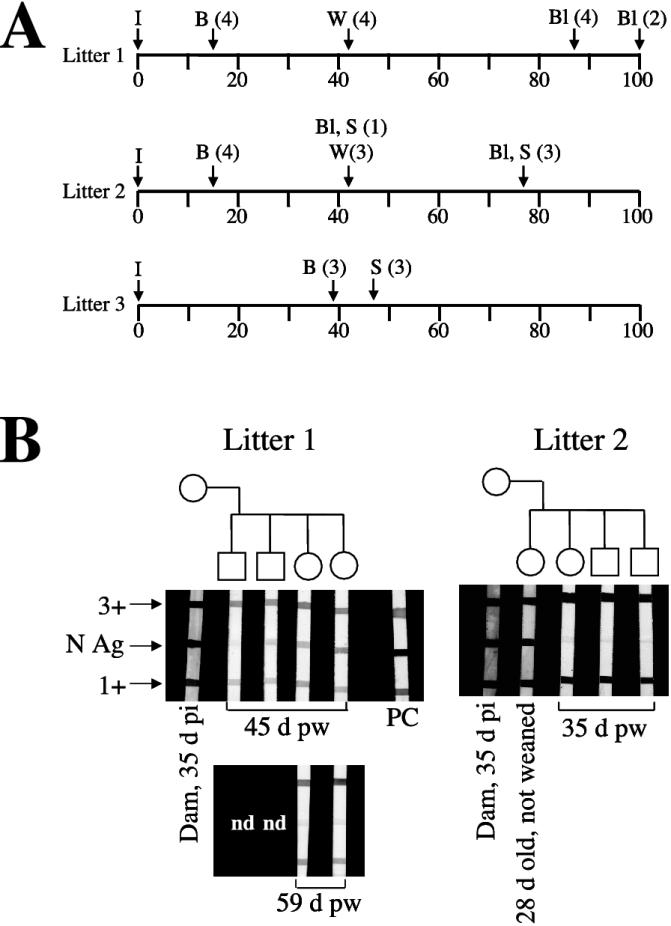
To address whether infected dams are able to vertically transmit virus and/or passively transfer anti-SN virus antibodies to offspring, we allowed three experimentally infected females to become pregnant and deliver litters (Fig. 3A). Two dams gave birth to four pups each (litters 1 and 2) on day 15 p.i., while the third dam delivered a litter of three (litter 3) on day 39 p.i. We sacrificed the three pups in litter 3 at day 4 after birth, one pup in litter 2 at day 28 after birth, and the remaining three pups in litter 2 at day 63 after birth (35 days postweaning) and carried out IHC analysis in an attempt to demonstrate viral N antigen (17). No tissue was positive in any of the pups. We also took blood from each of the four mice in litter 2 at the time of sacrifice for SIA. The pup (from litter 2) taken at 28 days old was positive for antibodies to N antigen (SIA titer, 1:500), and faint seroreactivity (titer, 1:125) was evident in the remaining three pups, which were 63 days old and 35 days postweaning at the time of sampling (Fig. 3B). The pups from litter 1 were bled at two time points: day 45 (n = 4) and day 59 (n = 2) postweaning. SIA titers were 1:2,000 at day 45 and 1:500 at day 59 (Fig. 3B; titration data not shown) (5).
FIG. 3.
(A) Timeline (in days) for manipulations performed on litters born to infected dams. I, dams inoculated; B, pups born; W, weaning; Bl, phlebotomy performed; S, sacrifice. (B) Sera from pups (litters 1 and 2) born to infected dams were examined at a dilution of 1:200 for reactivity to the SN virus N antigen by SIA (central band). In addition to the viral band, high-intensity (3+) and low-intensity (1+) bands of deer mouse serum were loaded, as described previously (5). Although each lot of SIA strips was independently tested for reactivity to the viral N antigen with a standard positive control serum, a wider degree of lot-to-lot variation was allowed for the conjugate reactivity bands (3+ and 1+). For that reason, some variation in the intensities of those control bands was observed in this study. PC, positive control; pw, postweaning. All sera tested were reactive at the 1:125 dilution.
The absence of vertical transmission led us to attempt to challenge a pup born to an infected dam and determine whether the passive transfer of maternal antibody can protect against challenge with live virus. A single pup was born to the dam from litter 1 when this female was at day 192 p.i. We inoculated the pup on day 43 postweaning with 500 ID50 of SN virus strain SN77734. The pup had an anti-N antibody titer of ≥1:8,000 on the day of inoculation (data not shown). Upon sacrifice at day 38 p.i. (day 81 postweaning), we found no evidence of N-antigen expression in tissues and the anti-N antibody titer had declined to 1:125 (data not shown).
DISCUSSION
Epidemiological studies indicate that SN virus and other hantaviruses are transmitted to humans primarily through inhalation of aerosols generated from urine, feces, and/or saliva shed by infected rodents. In the case of SN virus, the overwhelming majority of infected individuals are exposed in indoor settings (18). The mechanisms which maintain SN virus in nature are unclear, but males are differentially affected (1, 29). It is possible that the transmission of hantaviruses among mice occurs by mechanisms similar to those that result in transmission to humans.
To address hantavirus transmission mechanisms, animal models have been used to investigate the ability of infected host rodents to both shed and transmit hantaviruses in the laboratory (Table 5). All models have shown that infected rodents shed infectious virus and/or viral RNA in urine, feces, and/or saliva (2, 22, 23, 25, 26, 38). Furthermore, housing infected and naïve animals together results in horizontal, intracage transmission in each model (2, 15, 16, 22, 23, 25-27, 38). In the HTN virus and PUU virus animal models, intercage transmission can occur between cages and over 1.5 m of airspace (16, 26). Typically, infected rodents in these models shed and transmit virus readily during the acute phase of infection, but intermittent shedding and transmission may occur during persistent infection as well (2, 27, 38).
TABLE 5.
Summary of findings for hantavirus shedding and transmission in animal models
| Virus (host) | Presence of viral RNA and/or infectious virus
|
Occurrence of transmission
|
Reference(s) | ||||
|---|---|---|---|---|---|---|---|
| Urine | Feces | Saliva | Intracage | Intercage | Vertical | ||
| HTN (Apodemus agrarius and Fischer rats) | +b | +b | +b | + | + | − | 26, 27 |
| SEO (Wistar, Fischer, and Norway rats) | +a,b | +a | +a | + | − | 14, 25, 31 | |
| PUU (Clethrionomys glareolus) | +b | +b | +b | + | + | − | 2, 16, 38 |
| BCC (Sigmodon hispidus) | +b | +b | + | Possible | 22, 23 | ||
| SN (Peromyscus maniculatus) | −a,b | −a,b | +a, −b | + (rare) | − | This study | |
Viral RNA.
Infectious virus.
Laboratory studies generally confirm that vertical transmission is not a mode of transmission among rodents. Maternally acquired antihantavirus antibodies are protective against vertical transmission and subsequent challenge (13, 14, 31, 39). Furthermore, infected dams do not transmit PUU virus or HTN virus to pups (2, 16, 26). The BCC model appears to be atypical. In that model, not only do infected animals transmit virus horizontally to 100% of naïve cage mates, regardless of sex, age, or time of inoculation (p.i.), but dams readily transmit virus to offspring (23).
Field studies of wild deer mouse populations in the United States show that (i) the majority of infected animals are adults, (ii) significantly more adult males are infected than females, (iii) in juvenile mice, seroprevalence is highest in the youngest animals and tends to wane as they reach sexual maturity, and (iv) infections are persistent, presumably lasting the life of the rodent (1, 29). Because seroprevalence wanes as juvenile mice mature, it is believed that antiviral antibodies of juvenile mice are passively acquired from the dam and are not caused by infection. Using mark-release methods, Borucki et al. showed that wild mice that were seropositive as juveniles later became seronegative. However, the authors could not completely exclude the possibility that the juvenile mice were indeed infected but were able to clear the infection by adulthood (6). To date, the only direct testing for vertical transmission of SN virus involves a single example, wherein a wild-caught dam seroconverted and delivered four pups during a 5-week quarantine. The pups were seropositive but lacked detectable viral RNA in the their tissues (10). Thus, the preponderance of data suggests that transmission of SN virus among deer mice occurs through horizontal mechanisms. The exact mechanisms of transmission are still unclear. Potential mechanisms include aggressive behavior (especially among older males), sexual contact, cross-grooming, and inhalation of infectious aerosols (21).
SN virus appears to behave differently from other pathogenic hantaviruses, including the old-world viruses HTN, SEO, and PUU as well as the North American BCC virus (Table 5). All of these viruses are shed in urine, feces, and/or saliva from the host rodent, with viral RNA and/or infectious virus readily detectable in excretions and secretions. In addition, all of these viruses are efficiently transmitted from infected hosts to naïve cage mates during the acute phase of infection (2, 15, 16, 22, 23, 25-27, 38).
Although we were able to detect viral RNA shed into the saliva at days 15, 43, 60, 78, and 90 p.i., we were unable to isolate infectious virus from any of the pooled excreta or secretions. Nor were we able to detect infectious virus in the blood during acute infection. Similar studies to screen urine, feces, and saliva for SN virus RNA in both naturally and experimentally infected deer mice have reported negative results (K. Hutchinson, L. H. Elliott, S. Zaki, P. E. Rollin, T. G. Ksiazek, and S. T. Nichol, Abstr. 14th Annu. Meet. Amer. Soc. Virol., abstr. W19-4, 1995) (33). These results were surprising to us, considering that viral RNA and antigen are detectable in the salivary gland and kidney during the first 35 days of experimental infection (7). In addition, we demonstrated that SN virus is not readily transmitted from experimentally infected deer mice to naïve cage mates when the animals are housed together through as many as 213 days of infection. Only a single animal out of 54 that were exposed became infected through sharing a cage with an experimentally infected animal. The lack of infectiousness of excreta and secretions combined with the low rate of transmission seen in our studies indicates that inhalation and/or ingestion of excreta from an experimentally infected rodent may not be a highly efficient means of contracting infection. Indeed, the fact that attempts to infect deer mice via i.n. inoculation of materials of known infectiousness were unsuccessful further supports this claim. These findings are also supported by less formal observations of naturally infected deer mice that were detected incidentally in longstanding deer mouse colonies (J. Nestler, personal communication) (33). Wounding (n = 3) and sexual interactions that led to impregnation (n = 3) of virus-discordant pairs of animals during the cohabitation experiments did not result in infection.
In an earlier study, Botten et al. were unable to find any evidence of viral antigen expression in the reproductive organs (uterus, testis, and ovary) of experimentally infected deer mice (7). The provocative experiments described herein support previous observational studies that suggest that vertical transmission is very rare (1, 6, 10, 29). These studies indicate that seropositive juvenile deer mice passively acquire maternal antibodies that eventually wane. Our experiments lend additional support to the hypothesis that maternally transferred antibodies, rather than transient infection, are the likely cause of most of the antibody reactivity that is observed among young pups in field studies (6, 29). Furthermore, pups born to infected dams in our study did not become infected. These pups were seropositive, but no viral antigen could be detected via IHC in the multiple tissues examined. In addition, passively acquired maternal antibody was protective against challenge (n = 1) with a high dose of virus. The experience with SEO virus also suggests that maternally acquired antibodies are in fact protective (13, 14). Because our sample size (three litters, when the data of Camaioni et al. are added) was rather small, we cannot exclude completely the possibility that there are rare circumstances in which a recently infected deer mouse can transmit the virus to her pups, for instance, if she becomes viremic immediately after delivery (in the absence of high titers of neutralizing antibody) (10). In human infection, viral RNA can be found in the particulate fraction of breast milk, but neutralizing antibodies are also present (34).
Our experimental results do not enable us to further define how humans become infected with SN virus, which is almost certainly through the inhalation of aerosolized excreta from infected rodents (18, 28, 36). We would expect that infectious virus must first be excreted or secreted by a deer mouse for a person to become infected by the inhalation route. We believe that the most likely explanation for the apparent discrepancy is that natural shedding of infectious virus does occur in nature, but with very poor efficiency. This explanation would help explain why many-hundred-fold more people report exposures to rodent excreta than actually acquire the infection, even in areas where the virus is highly endemic (B. Hjelle, unpublished data). By contrast, human infection is much more common in Asia and Europe, where the predominant reservoir rodents appear to excrete infectious virus more readily (16, 26).
Another possible explanation may be that our experimental infections do not completely replicate naturally occurring infections. We used deer mice that had been lab reared for less than 2 years and fewer than 4 generations from the wild. We were careful to use a virus that was low passage, at low dosages, and that had never been passed in tissue culture, but subtle adaptations involving either virus or rodent cannot be completely excluded. We also addressed viral salivary shedding and viral tissue distribution in naturally infected deer mice obtained from the same population from which we founded our breeding colony and our virus, and the results were similar. Because we used nucleic acid detection technologies and spiking experiments, direct, sensitive technologies for detection of infectiousness, and intracage transmission experiments, we believe that many but perhaps not all trivial errors can be excluded as an explanation for our results. It is possible that in the wild, deer mice may undergo periods of immunosuppression or that stresses or seasonal signals may induce a specific host factor(s) required for SN virus to replicate to high enough titers to be effectively shed from the host (15). We are currently investigating the role of seasonal influences and immunosuppression in SN virus replication to determine whether environmental signals may lead to viral reactivation and secondary transmission. Our negative data may also suggest the need to reconsider a role of arthropod vectors in the transmission cycle of certain hantaviruses, because all other genera of viruses in the family Bunyaviridae use such vectors in their maintenance.
Acknowledgments
We thank M. Buchmeier, R. Lyons, D. Goade, J. Nestler, R. Nofchissey, R. Ricci, D. Theele, and R. Xiao for helpful comments, unpublished data, and/or technical assistance. We thank T. Yates, J. Dunnum, J. Dragoo, B. Frank, P. Polechla, R. Schwarz, and D. Tinnin from the Museum of Southwestern Biology and Cheryl Parmenter from the Division of Genomic Resources at the University of New Mexico for providing oral swab specimens from naturally infected deer mice. We also thank the United States Fish and Wildlife Service (Sevilleta National Wildlife Refuge) for their cooperation in this study.
This work was supported by U.S. Public Health Service grants RO1 A1 41692 and PO1 AI39780 and Defense Advanced Research Projects Agency grant MDA972-97-1-0013. J.B. was an Infectious Diseases and Inflammation Training Grant Fellow (Public Health Service grant T32 A107538) during the course of this work.
REFERENCES
- 1.Bennett, S. G., J. P. Webb, Jr., M. B. Madon, J. E. Childs, T. G. Ksiazek, N. Torrez-Martinez, and B. Hjelle. 1999. Hantavirus (Bunyaviridae) infections in rodents from Orange and San Diego counties, California. Am. J. Trop. Med. Hyg. 60:75-84. [DOI] [PubMed] [Google Scholar]
- 2.Bernshtein, A. D., N. S. Apekina, T. V. Mikhailova, Y. A. Myasnikov, L. A. Khlyap, Y. S. Korotkov, and I. N. Gavrilovskaya. 1999. Dynamics of Puumala hantavirus infection in naturally infected bank voles (Clethrinomys glareolus). Arch. Virol. 144:2415-2428. [DOI] [PubMed] [Google Scholar]
- 3.Bharadwaj, M., J. Botten, N. Torrez-Martinez, and B. Hjelle. 1997. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am. J. Trop. Med. Hyg. 57:368-374. [DOI] [PubMed] [Google Scholar]
- 4.Bharadwaj, M., C. R. Lyons, I. A. Wortman, and B. Hjelle. 1999. Intramuscular inoculation of Sin Nombre hantavirus cDNAs induces cellular and humoral immune responses in BALB/c mice. Vaccine 17:2836-2843. [DOI] [PubMed] [Google Scholar]
- 5.Bharadwaj, M., R. Nofchissey, D. Goade, F. Koster, and B. Hjelle. 2000. Humoral immune responses in the hantavirus cardiopulmonary syndrome. J. Infect. Dis. 182:43-48. [DOI] [PubMed] [Google Scholar]
- 6.Borucki, M. K., J. D. Boone, J. E. Rowe, M. C. Bohlman, E. A. Kuhn, R. DeBaca, and S. C. St. Jeor. 2000. Role of maternal antibody in natural infection of Peromyscus maniculatus with Sin Nombre virus. J. Virol. 74:2426-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botten, J., K. Mirowsky, D. Kusewitt, M. Bharadwaj, J. Yee, R. Ricci, R. M. Feddersen, and B. Hjelle. 2000. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus). Proc. Natl. Acad. Sci. USA 97:10578-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botten, J., R. Nofchissey, H. Ahern, P. Rodriguez-Moran, I. A. Wortman, D. Goade, T. Yates, and B. Hjelle. 2000. Outdoor facility for quarantine of wild rodents infected with hantavirus. J. Mammal. 81:250-259. [Google Scholar]
- 9.Botten, J., R. Ricci, and B. Hjelle. 2001. Establishment of a deer mouse (Peromyscus maniculatus rufinus) breeding colony from wild-caught founders. Comp. Med. 51:291-295. [PubMed] [Google Scholar]
- 10.Camaioni, M., J. Botten, B. Hjelle, and S. S. Loew. 2001. Hantavirus seroconversion of wild-caught Peromyscus during quarantine. Emerg. Infect. Dis. 7:14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 1994. Laboratory management of agents associated with hantavirus pulmonary syndrome: interim biosafety guidelines. Morb. Mortal. Wkly. Rep. 43:1-7. [PubMed] [Google Scholar]
- 12.Childs, J. E., T. G. Ksiazek, C. F. Spiropoulou, J. W. Krebs, S. Morzunov, G. O. Maupin, K. L. Gage, P. E. Rollin, J. Sarisky, and R. E. Enscore. 1994. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J. Infect. Dis. 169:1271-1280. [DOI] [PubMed] [Google Scholar]
- 13.Dohmae, K., U. Koshimizu, and Y. Nishimune. 1993. In utero and mammary transfer of hantavirus antibody from dams to infant rats. Lab. Anim. Sci. 43:557-561. [PubMed] [Google Scholar]
- 14.Dohmae, K., and Y. Nishimune. 1995. Protection against hantavirus infection by dam's immunity transferred vertically to neonates. Arch. Virol. 140:165-172. [DOI] [PubMed] [Google Scholar]
- 15.Dohmae, K., M. Okabe, and Y. Nishimune. 1994. Experimental transmission of hantavirus infection in laboratory rats. J. Infect. Dis. 170:1589-1592. [DOI] [PubMed] [Google Scholar]
- 16.Gavrilovskaya, I. N., N. S. Apekina, A. Bernshtein, V. T. Demina, N. Okulova, Y. A. Myasnikov, and M. P. Chumakov. 1990. Pathogenesis of hemorrhagic fever with renal syndrome virus infection and mode of horizontal transmission of hantavirus in bank voles. Arch. Virol. 1(Suppl. 1):57-62. [Google Scholar]
- 17.Green, W., R. Feddersen, O. Yousef, M. Behr, K. Smith, J. Nestler, S. Jenison, T. Yamada, and B. Hjelle. 1998. Tissue distribution of hantavirus antigen in naturally infected humans and deer mice. J. Infect. Dis. 177:1696-1700. [DOI] [PubMed] [Google Scholar]
- 18.Hjelle, B., and G. E. Glass. 2000. Outbreak of hantavirus infection in the Four Corners region of the United States in the wake of the 1997-1998 El Nino-southern oscillation. J. Infect. Dis. 181:1569-1573. [DOI] [PubMed] [Google Scholar]
- 19.Hjelle, B., S. Jenison, N. Torrez-Martinez, B. Herring, S. Quan, A. Polito, S. Pichuantes, T. Yamada, C. Morris, F. Elgh, H. W. Lee, H. Artsob, and R. Dinello. 1997. Rapid and specific detection of Sin Nombre virus antibodies in patients with hantavirus pulmonary syndrome by a strip immunoblot assay suitable for field diagnosis. J. Clin. Microbiol. 35:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hjelle, B., S. Jenison, N. Torrez-Martinez, T. Yamada, K. Nolte, R. Zumwalt, K. MacInnes, and G. Myers. 1994. A novel hantavirus associated with an outbreak of fatal respiratory disease in the southwestern United States: evolutionary relationships to known hantaviruses. J. Virol. 68:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hjelle, B., and T. Yates. 2001. Modeling hantavirus maintenance and transmission in rodent communities, p. 77-90. In C. S. Schmaljohn and S. T. Nichol (ed.), Hantaviruses. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 22.Hutchinson, K. L., P. E. Rollin, and C. J. Peters. 1998. Pathogenesis of a North American hantavirus, Black Creek Canal virus, in experimentally infected Sigmodon hispidus. Am. J. Trop. Med. Hyg. 59:58-65. [DOI] [PubMed] [Google Scholar]
- 23.Hutchinson, K. L., P. E. Rollin, W. J. Shieh, S. Zaki, P. W. Greer, and C. J. Peters. 2000. Transmission of Black Creek Canal virus between cotton rats. J. Med. Virol. 60:70-76. [DOI] [PubMed] [Google Scholar]
- 24.Jenison, S., T. Yamada, C. Morris, B. Anderson, N. Torrez-Martinez, N. Keller, and B. Hjelle. 1994. Characterization of human antibody responses to Four Corners hantavirus infections among patients with hantavirus pulmonary syndrome. J. Virol. 68:3000-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kariwa, H., M. Fujiki, K. Yoshimatsu, J. Arikawa, I. Takashima, and N. Hashimoto. 1998. Urine-associated horizontal transmission of Seoul virus among rats. Arch. Virol. 143:365-374. [DOI] [PubMed] [Google Scholar]
- 26.Lee, H. W., P. W. Lee, L. J. Baek, C. K. Song, and I. W. Seong. 1981. Intraspecific transmission of Hantaan virus, etiologic agent of Korean hemorrhagic fever, in the rodent Apodemus agrarius. Am. J. Trop. Med. Hyg. 30:1106-1112. [DOI] [PubMed] [Google Scholar]
- 27.Lee, P. W., R. Yanagihara, C. J. Gibbs, Jr., and D. C. Gajdusek. 1986. Pathogenesis of experimental Hantaan virus infection in laboratory rats. Arch. Virol. 88:57-66. [DOI] [PubMed] [Google Scholar]
- 28.Mertz, G. J., B. L. Hjelle, and R. T. Bryan. 1997. Hantavirus infection. Adv. Intern. Med. 42:369-421. [PubMed] [Google Scholar]
- 29.Mills, J. N., T. G. Ksiazek, B. A. Ellis, P. E. Rollin, S. T. Nichol, T. L. Yates, W. L. Gannon, C. E. Levy, D. M. Engelthaler, T. Davis, D. T. Tanda, J. W. Frampton, C. R. Nichols, C. J. Peters, and J. E. Childs. 1997. Patterns of association with host and habitat: antibody reactive with Sin Nombre virus in small mammals in the major biotic communities of the southwestern United States. Am. J. Trop. Med. Hyg. 56:273-284. [DOI] [PubMed] [Google Scholar]
- 30.Mills, J. N., T. L. Yates, J. E. Childs, R. R. Parmenter, T. G. Ksiazek, P. E. Rollin, and C. J. Peters. 1995. Guidelines for working with rodents potentially infected with hantavirus. J. Mammal. 76:716-722. [Google Scholar]
- 31.Morita, C., S. Morikawa, K. Sugiyama, T. Komatsu, H. Ueno, and T. Kitamura. 1993. Inability of a strain of Seoul virus to transmit itself vertically in rats. Jpn. J. Med. Sci. Biol. 46:215-219. [DOI] [PubMed] [Google Scholar]
- 32.Nerurkar, V. R., J. W. Song, K. J. Song, J. W. Nagle, B. Hjelle, S. Jenison, and R. Yanagihara. 1994. Genetic evidence for a hantavirus enzootic in deer mice (Peromyscus maniculatus) captured a decade before the recognition of hantavirus pulmonary syndrome. Virology 204:563-568. [DOI] [PubMed] [Google Scholar]
- 33.Otteson, E. W., J. Riolo, J. E. Rowe, S. T. Nichol, T. G. Ksiazek, P. E. Rollin, and S. C. St. Jeor. 1996. Occurrence of hantavirus within the rodent population of northeastern California and Nevada. Am. J. Trop. Med. Hyg. 54:127-133. [DOI] [PubMed] [Google Scholar]
- 34.Pai, R. K., M. Bharadwaj, H. Levy, G. Overturf, D. Goade, I. A. Wortman, R. Nofchissey, and B. Hjelle. 1999. Absence of infection in a neonate after possible exposure to Sin Nombre hantavirus in breast milk. Clin. Infect. Dis. 29:1577-1579. [DOI] [PubMed] [Google Scholar]
- 35.Schmaljohn, A. L., D. Li, D. L. Negley, D. S. Bressler, M. J. Turell, G. W. Korch, M. S. Ascher, and C. S. Schmaljohn. 1995. Isolation and initial characterization of a newfound hantavirus from California. Virology 206:963-972. [DOI] [PubMed] [Google Scholar]
- 36.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada, T., B. Hjelle, R. Lanzi, C. Morris, B. Anderson, and S. Jenison. 1995. Antibody responses to Four Corners hantavirus infections in the deer mouse (Peromyscus maniculatus): identification of an immunodominant region of the viral nucleocapsid protein. J. Virol. 69:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanagihara, R., H. L. Amyx, and D. C. Gajdusek. 1985. Experimental infection with Puumala virus, the etiologic agent of nephropathia epidemica, in bank voles (Clethrionomys glareolus). J. Virol. 55:34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X. K., I. Takashima, and N. Hashimoto. 1988. Role of maternal antibody in protection from hemorrhagic fever with renal syndrome virus infection in rats. Arch. Virol. 103:253-265. [DOI] [PubMed] [Google Scholar]