Abstract
We present TFAM, an automated, statistical method to classify the identity of tRNAs. TFAM, currently optimized for bacteria, classifies initiator tRNAs and predicts the charging identity of both typical and atypical tRNAs such as suppressors with high confidence. We show statistical evidence for extensive variation in tRNA identity determinants among bacterial genomes due to variation in overall tDNA base content. With TFAM we have detected the first case of eukaryotic-like tRNA identity rules in bacteria. An α-proteobacterial clade encompassing Rhizobiales, Caulobacter crescentus and Silicibacter pomeroyi, unlike a sister clade containing the Rickettsiales, Zymomonas mobilis and Gluconobacter oxydans, uses the eukaryotic identity element A73 instead of the highly conserved prokaryotic element C73. We confirm divergence of bacterial histidylation rules by demonstrating perfect covariation of α-proteobacterial tRNAHis acceptor stems and residues in the motif IIb tRNA-binding pocket of their histidyl-tRNA synthetases (HisRS). Phylogenomic analysis supports lateral transfer of a eukaryotic-like HisRS into the α-proteobacteria followed by in situ adaptation of the bacterial tDNAHis and identity rule divergence. Our results demonstrate that TFAM is an effective tool for the bioinformatics, comparative genomics and evolutionary study of tRNA identity.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRS) esterify or ‘charge’ specific amino acids to specific sets of tRNAs, called ‘isoacceptors’ (1). AaRS must distinguish highly similar tRNA substrates to maintain translational fidelity. This specificity is mediated by so-called ‘identity elements,’ encompassing features in tRNAs that either promote recognition or catalytic activity (‘determinants’) or inhibit non-specific interactions (‘antideterminants’), as well as aaRS features with comparable roles (2). Here, we use the term ‘identity rules’ to mean a complete set of identity determinants in a clade over all amino acid aminoacylation systems.
AaRS have played a central role in the construction of the modern view of the tree of life [encompassing the three domains of eukarya, bacteria and archaea (3)], such as its rooting with bacteria (4). Yet the majority of aaRS gene phylogenies are inconsistent with monophyly of the three domains (5–7). In the ‘complexity hypothesis’, this is explained by the relative modularity of aaRS function (7,8). Because aaRS interact primarily with only one other kind of genetically encoded substrate, tRNAs, they are more likely to function both correctly and without interference in novel cellular milieu. Relative to genes with more complex gene interactions, purifying selection should be weaker against substitution of aaRS genes acquired by lateral gene transfer (LGT) (7,8).
However, eukaryotes and prokaryotes quite often use incompatible tRNA identity rules, which can cause a barrier to cross-species charging of foreign tRNAs by aaRS of another domain (9–16). Surprisingly, aaRS gene trees have been obtained that suggest, with high confidence, that LGT has also occurred across such interdomain charging barriers (13,17). Despite this, in all such cases, there was no evidence that LGT of an incompatible aaRS had altered tRNA identity rules in the recipient lineage. Rather, if LGT was involved, the aaRS seems to have adaptively converged to function with the tRNAs in the recipient lineage, leaving its identity rules unperturbed.
LGT of an aaRS across an identity rule barrier could occur because of a compensating positive advantage, such as antibiotic resistance over the resident gene, as suggested to explain the eukaryotically derived, but functionally bacterial IleRS in Mycobacterium tuberculosis (17). Alternatively, lineage-specific gene losses can give the appearance of LGT (18), when ancient paralogs are independently lost in multiple lineages and treated in analysis as orthologs. We propose a third hypothesis to help explain aaRS evolutionary patterns: that low levels of ambiguity in tRNA identity rules may be tolerable. This would not only relax barriers to LGT but also facilitate divergence of resident aaRS and tRNA genes. tRNAs and aaRS could coevolve new identity rules while maintaining function through the compensation of mildly deleterious mutations. Also, tRNAs and aaRS might switch identities by evolving through transitionally ambiguous identity states.
To test these and other hypotheses we need an automated, statistical and systematic approach to analyzing tRNA identity rules over many species. The first bioinformatic approaches to tRNA identity rules were implemented for Escherichia coli, Salmonella typhimurium (19,20) and yeast (21), but were not fully probabilistic and limited by the available data. More recently, Marck and Grosjean (22) comprehensively compared tDNA sequences from sequenced genomes. But because they did not produce a statistical model, their results cannot be used to classify new tRNAs. Most tDNA data from genome projects, found by tRNAscan-SE (23) or other methods (24,25), are classified by their anticodons. The tool of choice for tRNA gene-finding, tRNAscan-SE, introduced identity-specific tRNA models, in particular for selenocysteine tRNAs, as well as evolutionary domain specific tRNA models (23). However, its anticodon-based approach to tRNA identity prediction will fail with suppressors, pseudo-tRNAs and tRNAs with unalignable or post-transcriptionally modified anticodons, assumes a given genetic code, is vulnerable to sequencing error, and cannot predict initiator tRNAs.
On the other hand, purely experimental approaches to tRNA identity are taxonomically limited but provide a wealth of mechanistic information. The year 2000 release of the database of Sprinzl et al. (26) (http://www.staff.uni-bayreuth.de/~btc914/search/index.html) contains manually aligned tRNA sequences where the majority of annotated identity classifications are experimentally obtained. However, interfaces to the database are designed for descriptive use by humans, rather than for automated classification.
In this study, we used the Sprinzl database to make profile-based models of tRNA identity rule families that we call ‘tFAMs,’ in analogy to ‘PFAMs’ used to annotate and classify protein domains (27). The tFAM models use a ‘profile contrast’ approach that, for a given tRNA identity class, facilitates the scoring of a test tRNA sequence in terms of how well it matches a ‘positive’ sequence profile (28) of isoaccepting tRNAs versus a ‘negative’ sequence profile of alloaccepting tRNAs. We used these models to make an automated statistical classifier of tRNA identity called TFAM. Using TFAM we found evidence of significant diversity of identity rules within bacteria. We discovered significantly aberrantly scoring tRNAHis in a clade of α-proteobacteria that on inspection were found to carry a eukaryotic identity element. Subsequent analysis showed perfect covariation between HisRS phylogenetic groups and tRNAHis identity elements in the α-proteobacteria. Until this work, to our knowledge, diversity in identity rules within a taxonomic domain has not been reported. We suggest that identity rules changed in an ancestor to this clade by a process of RNA-protein co-evolution, likely initiated by LGT of a foreign HisRS.
MATERIALS AND METHODS
Modified Sprinzl training tRNA database
All 820 eubacterial tRNA/tDNAs were downloaded from the 2000 edition of the Sprinzl database (26) (the last version of the database before a massive amount of post-genomic data was added) and converted to a DNA alphabet. Redundant entries were removed leaving 663 tDNAs. These were split into 423 ‘pre-genomic’ tDNAs and 240 (36%) ‘post-genomic’ tDNAs derived from the published genomes of Haemophilus influenzae, Mycoplasma genitalium, Synechocystis, Borrelia burgdorferi, Helicobacter pylori and Treponema palladum. In the pre-genomic training set, some identity annotations were corrected after verification and analysis using tRNAscan-SE v.1.12 (23) and BLASTN (29) and 5 selenocysteine and 3 alanine tRNAs were removed. The identity of the post-genomic tDNAs was checked using tRNAscan-SE, a preliminary version of TFAM only with the corrected pre-genomic data (to classify initiators), and BLASTN for verification, with tRNAscan-SE identity annotation taking priority, and checked manually (for details see Supplementary Data). After this screening the pre- and post-genomic datasets were combined for a total of 655 training tDNAs.
Eubacterial genomic test tDNA datasets
tRNAscan-SE was run on the 58 bacterial genomes available at GenBank on March 6, 2002, plus the Bartonella henselae and Bartonella quintana genomes (30). A later test dataset of 213 bacterial genomes (a superset of the preceding test dataset) was analyzed in the same way, downloaded on July 7th, 2005. In addition, 21 archaeal genomes downloaded on July 7th, 2005 were analyzed with tRNAscan-SE using the archaeal search mode. tDNA sequences were extracted from genome data by coordinate.
Construction and analysis of tFAMs
The TFAM program first aligns, using both primary and secondary structural information, all of the test tDNAs and all of the training tDNAs and then generates a collection of sequence profiles from the training tDNAs. For each tRNA identity class, the training data is partitioned into a ‘positive’ set of tDNAs that belong to that class and a ‘negative’ set-complement. The alignment calculation is sped up by pre-aligning the training data using the COVEMF package (available separately from D. H. Ardell at http://www.lcb.uu.se/~dave/COVEMF) modified from the COVE package v. 2.4.2 (31) to the TRNA2-prok.cm model (23). Alternative training datasets and models can be provided to the TFAM program. TFAM is available for download under the terms of the academic free license at http://www.lcb.uu.se/~dave/TFAM.
A ‘tFAM matrix’ or ‘tFAM’ is a 5× L matrix of five rows (one for each possible nucleotide symbol and a gap) and L columns, with L equal to the length of the alignment of test and training tDNAs. The i,jth entry of the tFAM matrix for identity class A is the log-odds of the ith symbol (in order of ‘A’,‘C’,‘G’,‘T’ and ‘–’) at the jth position in the positive training set versus the negative set for class A. LaPlace ‘add-one’ pseudocounts are used to estimate the probability of unobserved states (32). The score of a test tDNA against the tFAM for class A is the sum of log-odds of its sequence against that tFAM. The TFAM annotation of identity of a test tDNA is the class against which it has maximum score. TFAM excludes columns in which the entire training portion of the alignment contains only the gap state. This prevents long insertions in query tDNAs from distorting tFAM scores. TFAM scores each query tDNA against each tFAM using BioPerl (33) and the Perl Data Language (PDL) (http://pdl.perl.org).
We defined 21 tFAMs, one for initiator tRNAs and 20 types of elongator tRNAs. In a consistency check, where 663 unique Sprinzl tDNAs are used as both training and query datasets, 24 were classified by TFAM as incompatible with their Sprinzl annotations. We detected four additional incompatibilities with tRNAscan-SE. We believe that at least twenty of 820 entries in the Sprinzl database may be erroneously annotated as detected by tFAMs and other methods, and confirmed by BLASTN (29) (by criterion of top hit to training set, see Supplementary Data). Additionally, some initiator tRNAs from genome projects are annotated as elongators (DM1151, DM1231, DM2000 and DM2001). Annotations of all these incompatible entries were changed to make the final modified Sprinzl training Database (MSDB) used to obtain our results.
Analysis of tRNA acceptor stem regions and HisRS tRNA-binding domains
Upstream flanking regions of genomic tDNAs were extracted and aligned by their 3′ ends. RNase P cleavage sites were predicted by inspection (L. A. Kirsebom, personal communication), and tRNAs were folded using ARAGORN (24). Consensus mitochondrial tRNAHis sequences were computed from data in the Sprinzl database. Ancestral sequence reconstruction was computed by Fitch parsimony, using E.coli tDNAHis as an outgroup, in DNAPARS from PHYLIP (34) (http://evolution.genetics.washington.edu/phylip.html) and a phylogeny of α-proteobacteria from (35). Sequence logos were made from the motif IIb tRNA-binding domains (16) of a genomic set of inferred HisRS sequences translated from annotated genes using the Weblogo Server (36) based on Delila software (37). RefSeq data for the 213 bacterial genomes were downloaded from NCBI. Protein annotation (‘ptt’) files were parsed for matches to COG0124 or COG3705 (the Cluster of Orthologous Groups (38) for HisRS or HisZ) or the Perl regular expression (case-insensitive) ‘histid.*tRNA.*(lig|synth(et)?)ase|his[SZ]’. Up to four candidate HisRS genes were obtained for archaeal and bacterial genomes. Putative HisZ paralogs imputed in histidine biosynthesis (39) were identified by phylogenomic analysis (40) (neighbor-joining tree available as supplementary data) and removed. The remainder were aligned with CLUSTALW (41) (default settings), the motif IIB loop region (16) was extracted and logos made as before.
Phylogenetic analysis of HisRS sequences
Initially the GenBank™ nr database was queried with Agrobacterium tumefaciens HisRS using PSI-BLAST (42) for three iterations. The search yielded 251 protein sequence homologs; this data was combined with the genomic data previously obtained and HisZ sequences annotated from TrEMBL™. These were aligned with CLUSTALW using default settings. A pairwise maximum likelihood distance matrix was generated with the VT + F + Γ model (43) using Tree-Puzzle v.5.1 (44) and parameters estimated from data. A likelihood topology was then made for these 251 sequences using NJDIST (45) and PROTML (46) (http://www.lcb.uu.se/~dave/molphy-2.3b3.zip). A final alignment and tree of a smaller number of sequences was built progressively. A core alignment of 31 HisRS most closely related to the RC-clade (mostly BacII, see below for definition) was optimized with MULTICLUSTAL (47). Sequences were added successively in CLUSTALX profile mode in the following number and order: 15 archaeal, 13 eukaryotic, 11 BacI and 29 HisZ. In the final two analyses, 8 and 23 sequences, respectively, were taken away after alignment for clarity of presentation. Some additional α-proteobacterial HisRS sequences were added to this alignment in CLUSTAL profile mode after they appeared. Inspection showed that the Zymomonas mobilis (48) HisRS as annotated was truncated. We used the Silicibacter pomeroyi (49) ortholog to find a matching region of the genome with TBLASTX (50) and took an enclosing region from the genome between positions 1 530 000 and 1 533 000, translated in all three reading frames and aligned to the other data with CLUSTALX, followed by trimming of unaligned regions from the alignment. This alignment is available as supplementary data. The likelihood of various protein models with the data were tested with several alignments using PUZZLE-MODELTEST (D. H. Ardell, unpublished data), which favored the WAG model (51). Site-rate variation was included after likelihood ratio testing (52). In each alignment, gap-majority sites not correlated with major phylogenetic groups were removed by hand. The final bootstrapped (100 iterations) consensus likelihood tree was calculated using the WAG + F + Γ model in PHYML (53). The final boostrapped (1000 iterations) consensus BIONJ (54) tree was generated by CONSENSE (34) (http://evolution.genetics.washington.edu/phylip.html) from BIONJ trees calculated with WAG + F + Γ pairwise distance matrices calculated in turn with Tree-Puzzle v. 5.2 (44).
RESULTS
To study the co-evolution of tRNAs and their aminoacylation enzymes, we developed a system called TFAM to classify tDNAs from whole genome data. TFAM aligns a query set of genomic tDNAs to a training set (MSDB) derived from the Sprinzl database, generates, for each tRNA family, a log-odds ‘profile contrast’ model from the aligned positive and negative identity profiles for each tRNA identity family, and then scores each query against the model. The main distinction of this approach is that the positive and negative profiles are aligned previously to each other and with the data before scoring, which allows position-specific scores to be generated. Our approach assumes that tRNA secondary structure is highly conserved and that identity rules are homogeneous in the taxa sampled in the MSDB. We did not make a tFAM for tRNASel-Cys because of their unique structural differences.
Performance of TFAM
In a resubstitution analysis examining the ability of TFAM to recover the identity classifications of its own training data, TFAM sensitivity and specificity were both >98% on average with the MSDB, and 96% when anticodons were excluded from the analysis (Table 1). Sensitivities and specificities of thirteen tFAMs were perfect even excluding the anticodon. Thus, TFAM uses information outside the anticodon for tDNA classification. Anticodons affected recognition of Ala-, Met-, Asn-, Val- and Trp-class tDNAs and discrimination of Asn-, Arg-, Thr- and Val-class tDNAs. The low specificity of the Ala tFAM is almost entirely due to tDNAVal that match neither Ala nor Val tFAMs well but match the Ala tFAM better. Even if this is due to misannotation, it suggests that tRNAVal are diverse and sometimes share identity elements with tRNAAla. TFAM did not reliably distinguish tDNAIle with lysidine-modified anticodons from tDNAMet.
Table 1.
Resubstitution analysis of TFAM performance with the MSDB
| Model | Na | Snb | Spc | Seq1d | Seq2e |
|---|---|---|---|---|---|
| Af | 52 | 100 | 91 | A: 33 [48,5] | V: −3 [21,−26] |
| A w/o acg | 52 | 98 | 91 | A: 29 [44,3] | V: −1 [22,−24] |
| C | 13 | 100 | 92 | C: 24 [37,0] | S: −12 [5,−34] |
| C w/o ac | 13 | 100 | 92 | C: 21 [33,−3] | S: −12 [8,−35] |
| D | 21 | 100 | 100 | D: 20 [37,4] | E: −3 [7,−11] |
| D w/o ac | 21 | 100 | 100 | D: 17 [34,1] | E: −4 [6,−12] |
| E | 23 | 100 | 100 | E: 38 [54,15] | D: −11 [3,−37] |
| E w/o ac | 23 | 100 | 100 | E: 34 [51,11] | D: −11 [3,−37] |
| F | 22 | 100 | 100 | F: 23 [30,11] | C: −16 [−1,−33] |
| F w/o ac | 22 | 100 | 100 | F: 19 [26,7] | K: −11 [−1,−23] |
| G | 45 | 100 | 100 | G: 26 [46,6] | W: −14 [−2,−30] |
| G w/o ac | 45 | 100 | 100 | G: 22 [42,2] | H: −11 [0,−19] |
| H | 15 | 100 | 100 | H: 23 [36,12] | Q: −14 [−7,−22] |
| H w/o ac | 15 | 100 | 100 | H: 19 [32,8] | Q: −15 [−8,−23] |
| I | 61 | 96 | 95 | I: 27 [38,−25] | K: −10 [0,−35] |
| I w/o ac | 61 | 95 | 95 | I: 24 [35,−27] | R: −6 [8,−37] |
| K | 22 | 100 | 95 | K: 16 [28,7] | N: −10 [−1,−22] |
| K w/o ac | 22 | 100 | 95 | K: 13 [25,4] | F: −7 [0,−20] |
| L | 63 | 100 | 100 | L: 61 [79,30] | Y: −13 [4,−54] |
| L w/o ac | 63 | 100 | 100 | L: 59 [77,28] | Y: −10 [7,−51] |
| M | 14 | 92 | 100 | M: 13 [18,−7] | X: −10 [−4,−14] |
| M w/o ac | 14 | 78 | 100 | M: 10 [15,−10] | X: −13 [−7,−17] |
| N | 21 | 100 | 100 | N: 20 [32,1] | K: −12 [−4,−23] |
| N w/o ac | 21 | 95 | 95 | N: 16 [29,−2] | K: −12 [−5,−23] |
| P | 28 | 100 | 100 | P: 29 [36,6] | X: −12 [−4,−23] |
| P w/o ac | 28 | 100 | 100 | P: 25 [34,4] | X: −7 [0,−17] |
| Q | 19 | 100 | 100 | Q: 30 [39,0] | H: −16 [−5,−33] |
| Q w/o ac | 19 | 100 | 100 | Q: 27 [36,−2] | C: −15 [−6,−29] |
| R | 48 | 100 | 100 | R: 20 [32,5] | N: −8 [−2,−29] |
| R w/o ac | 48 | 100 | 96 | R: 18 [25,3] | N: −4 [1,−25] |
| S | 53 | 100 | 100 | S: 77 [104,44] | Y: 0 [18,−23] |
| S w/o ac | 53 | 100 | 100 | S: 74 [101,44] | Y: 0 [19,−22] |
| T | 43 | 100 | 100 | T: 17 [24,6] | N: −8 [4,−14] |
| T w/o ac | 43 | 100 | 93 | T: 14 [21,4] | N: −7 [5,−13] |
| V | 26 | 73 | 100 | V: 14 [25,0] | A: 4 [14,−13] |
| V w/o ac | 26 | 69 | 94 | V: 11 [22,−1] | A: 4 [14,−13] |
| W | 18 | 100 | 100 | W: 23 [30,1] | G: −17 [−2,−33] |
| W w/o ac | 18 | 94 | 100 | W: 20 [26,−2] | G: −17 [−1,−30] |
| Xh | 28 | 100 | 100 | X: 46 [51,29] | M:−28 [−8,−48] |
| X w/o ac | 28 | 100 | 100 | X: 42 [47,25] | P:−23 [−9,−54] |
| Y | 20 | 100 | 100 | Y: 45 [64,31] | S:7 [33,−11] |
| Y w/o ac | 20 | 100 | 100 | Y: 41 [60,28] | S:9 [35,−8] |
| Average | 655 | 98.1 | 98.7 | ||
| Average w/o ac | 655 | 96.6 | 97.7 |
aTraining set size.
bModel percent sensitivity.
cModel percent specificity.
dSequence class with highest median score against the model and its distribution: median and range.
eSequence class with second-highest median score against the model and its distribution: median and range.
fSequences are scored against the model including the anticodon positions.
gw/o ac = without anticodon. Sequences are scored against the model excluding the anticodon positions.
htRNAiMet.
We did a formal cross-validation comparison of accuracy of TFAM on the training data against comparable use of HMMer in both global and local search modes, COVE and its INFERNAL update (vers. 0.55, see Supplementary Data for details). All methods performed very similarly except INFERNAL, which performed slightly worse. Accuracies were in decreasing order: HMMer Local (638/655 = 97.4%), HMMer Global (637/655 = 97.3%), TFAM (636/655 = 97.1%), COVE (636/655 = 97.1%) and INFERNAL (623/655 = 95.1%). Misannotations were different for different methods. Thus, TFAM performs comparably to HMMer and COVE, which simultaneously align and classify data. There is no evidence that incorporating secondary structural information helps average discrimination with this training set. It is possible that COVE performance suffers from the greater number of parameters to be estimated. An advantage of TFAM over the HMMer approach despite its slight performance advantage is that TFAM outputs a single common alignment of all data and models, which facilitates studying the contribution of specific positions (such as the anticodon) and research into position-specific contributions to tRNA identity rules (55).
Initiator and non-standard tRNAs
Initiator tRNAs can be discriminated with very high confidence by tFAMs (Table 1). The tRNAs annotated as initiators all shared known initiator tRNA identity elements such as the C1 and A72 mismatch (22,56). We classified five of the nine tDNAMet in Vibrio cholerae as initiators, the maximum in any genome we examined in the 2002 dataset. This could be related to the record-high growth rates recorded among Vibrio species (57). Initiator tRNA concentration increases more with growth rate in E.coli than other tRNAs (58) and tRNA concentration at high growth rate is correlated with gene dosage in unrelated microbes (58,59), consistent with translational initiation being rate-limiting for growth.
Mollicutes, including Mycoplasmas and Ureaplasma, have an altered genetic code, coding the canonical termination codon UGA as Trp (60–62). These non-standard tDNATrp are identified as tDNASel-Cys by tRNAscan-SE, but by inspection they do not share structural features with known tDNASel-Cys. TFAM correctly classifies them as tDNATrp.
Taxonomic variation in genomic tDNAs
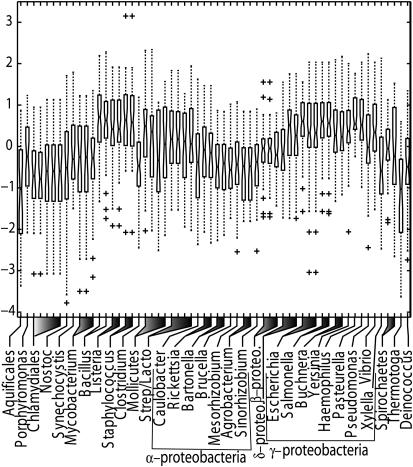
We applied tFAMs to a reduced set of 2309 tDNAs from the 2002 dataset of 60 sequenced bacterial genomes. To be conservative in assessing taxonomic diversity we excluded tDNAs with Sel-Cys, Ala, Met, Ile, Val, His or unalignable anticodons or pseudo-tDNAs, as well as tDNAs whose TFAM and anticodon classifications did not match. The remaining tDNA top-tFAM scores were statistically standardized by tFAM class to have a mean of zero and standard deviation one, and subjected to one-way ANOVA by genome of origin. Box-and-whisker plots showing class-standardized tFAM score distributions are shown in Figure 1. The analysis shows large variation in the distribution of standardized tFAM scores across taxa, and the presence of many outliers, even among tDNAs whose TFAM classifications match their anticodons. ANOVA confirmed the significance of this taxonomic variation in mean tFAM scores (F = 11.5 with 59 and 2249 degrees of freedom). The boxplots show a bias towards γ-proteobacteria and gram-positives, which are the over-represented taxa in the MSDB. The biological significance of this result comes from the fact that, because TFAM scores are built from contrasts of profiles the taxonomic variation (Figure 1) should derive mostly from features that are important in tRNA identity, at least in the γ-proteobacteria and gram-positives. The implication is that there is surprisingly extensive taxonomic diversity in tRNA identity elements in bacteria.
Figure 1.
Box-and-whisker plots of tFAM scores by eubacterial taxon, after standardization of TFAM scores by their tDNA identity class. Boxes show the interquartile range and median. Whiskers extend to 1.5 times the interquartile range. Plusses (+) show outliers.
It is likely that much of this diversity owes to base content variation in stable RNA, which is affected by ecological factors such as hyperthermophily (63). The median scores of the two hyperthermophiles Thermotoga maritima and Aquifex aeolicus were a full standard deviation below that expected by chance. This may be partly due to increased GC-content of stable RNA in hyperthermophiles in association with optimal growth temperature (63). We therefore explored the effect of tDNA base composition on mean standardized top-tFAM score by taxon (‘mean score’). We calculated the log-likelihood goodness-of-fit (‘base composition fit’) of a taxon's tDNA base composition to that of the MSDB. We excluded the two hyperthermophiles in this analysis as outliers. There was a significant regression of the mean score against base composition fit (slope = −0.018, P < 0.01, adj. R = 0.12, F = 8.706 and df = 1,56). Repeating the ANOVA without the hyperthermophiles but with the residuals of the regression, the variation by taxon was no longer significant (F = 1.23, P = 0.12 and df = 57, 2185). That is, the mean taxonomic variation was entirely explicable by differences in base content of genomic tDNAs from that of the tFAM training set. Excluding sampling effects, in a profile contrast model we do not expect base content variation at sites that do not contribute functional information to affect tFAM scores. Thus, these results suggest that evolutionary forces acting on tDNA base content, for instance increasing GC-content in the stable RNA of hyperthermophilic bacteria, have altered tRNA identity rules in those bacteria.
Screening for tDNA outliers (Figure 1) reveals a surprisingly large number of outlying tDNAs across the entire taxonomic range of bacteria surveyed. Many of these values are less than two standard deviations below respective taxonomic means. Both the proportion and occurrence of mismatches between TFAM and anticodon-based classifications were independent of base composition fit (proportions: ANOVA, arcsine-transformed, F = 2.286, P = 0.14 and df = 1,58; occurrences: logistic regression, z = 1.376 and P = 0.17).
Sequence diversity of α-proteobacterial tRNAHis
Applying TFAM to the complete 2002 genomic dataset of 3230 tDNAs, the TFAM classifications of 336 tDNAs (>10%) did not match tRNAscan-SE anticodon-based classifications. tDNAMet by anticodon were 227 and 108 of those were predicted to be initiators. Among the remaining mismatching tDNAs, a group of tDNAHis had highly unusual tFAM scores, from a group of genomes within a clade of the α-proteobacteria encompassing members of the Rhizobiales as well as Caulobacter cresentus (the RC-clade). This is a monophyletic group that excludes the Rickettsia species within α-proteobacteria. However, tDNAHis in all other bacteria in this dataset scored normally with top-ranking matches to the His tFAM. Table 2 shows that tDNAHis of the RC group scored significantly low against the His tFAM (P < 0.001; Wilcoxon rank sum tests with continuity correction). However, other tDNAs from the RC group scored only slightly lower against their respective tFAMs in comparison to other bacteria (P = 0.04). Thus, the very low scores of RC tDNAHis against the His tFAM are not explained by a generally bad fit of tFAMs to the RC-clade.
Table 2.
Scores of eubacterial tDNAs (60 genomes) with MSDB TFAMs
| Taxa | tRNA | N | His-tFAMa | Other tFAMb |
|---|---|---|---|---|
| RCc | His | 7 | −10 [−8, −21] | −4 [−2, −12] |
| Other | His | 62 | 15 [28, −11] | −10 [−1, −18] |
| ***d | NS | |||
| RC | Othere | 340 | −34 [−11, −58] | 22 [90, −8] |
| Other | Other | 2786 | −34 [−3, −55] | 24 [110, −16] |
| NS | * |
aScore distribution of tRNAs against His-TFAM (median and range).
bMedian and range of maximum scores against all other tFAMs but the His-tFAM.
cRhizobiales + Caulobacter.
dMann–Whitney test, ***P < 0.001, *P < 0.05, NS, not significant.
eAll tDNAs except His, Sel-Cys, Pseudo and Undet (according to tRNAscan-SE).
RC tDNAHis do not match other tFAMs very well either; their scores were not significantly different from those of other bacterial tDNAHis against other non-His tFAMs (P = 0.17) and are not different from the scores of tDNAs of other classes in other bacteria against the His tFAM (P = 0.24). Indeed, all seven aberrant tDNAHis that were analyzed scored negatively against all tFAM models. Furthermore, no other tDNAs in the RC genomes were identified with a His anticodon that could serve as a substitute for the tDNAHis. There was no difference between the RC-clade and other bacteria in the scores of other tDNAs to the His tFAM model (P = 0.22). This confirms the absence of other tDNAs in the RC genomes that ‘look like’ tDNAHis. The conclusion is that the RC-clade appears to lack any tDNAs that have tRNAHis prokaryotic identity elements. The results further show the advantage of TFAM in facilitating the statistical study of tRNA sequence variation putatively affecting tRNA identity.
Co-evolution of RC-clade tRNAHis and HisRS identity elements
To examine the nature of the deviant RC-clade tRNAHis, we looked at α-proteobacterial tDNAHis and their upstream sequences in the species analyzed in TFAM as well as more recently published α-proteobacteria: Wolbachia pipientis, Anaplasma marginale, Erlichia ruminatium, Zymomonas mobilis, Gluconobacter oxydans and Silicibacter pomeroyi. In a recent analysis (35), it was found that S.pomeroyi forms a clade with the RC group (hereafter called the RCS-clade). The other taxa above form a sister clade containing the Rickettsiales (a large group that contains W.pipientis, A.marginale and E.ruminatium), Z.mobilis and G.oxydans (hereafter called the RZG-clade). In an analysis of upstream sequences we confirmed that practically all bacterial tDNAHis present a G at the −1 position, i.e. they genetically encode the highly conserved −1G His identity element (22 and data not shown). This pattern shifts the RNase P cleavage site leaving bacterial and archaeal tRNAHis with an extra 5′ base and a mature 8 bp acceptor stem (64). Thus, tRNAscan-SE misspecifies the 5′ boundaries of prokaryotic tDNAHis.
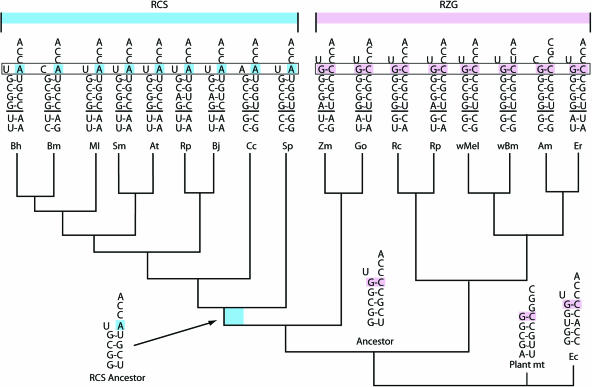
However, among the α-proteobacterial tRNAHis, as shown in Figure 2, the RCS-clade genetically encodes neither −1G nor the C73 discriminator base that is unique to bacterial tRNAHis and critical for His identity in E.coli (65,66). Instead, like eukaryotes, RCS-clade tRNAHis contain A73, which in yeast is a less essential but important His identity element (67,68). This suggests the possibility that LGT of a eukaryotic tRNA perturbed identity rules in the RCS ancestor. Yet parsimony-based sequence reconstruction of α-proteobacterial tRNAHis (Figure 2) suggests that the RCS-clade tRNAHis can be explained by only a small number of changes from an α-proteobacterial ancestor: three substitutions at bases 1, 72 and 73. Other changes in RCS tRNAHis not shown are apparent relaxation of constraint at bases 38 in the anticodon loop and four other bases in the T arm, and apparent novel substitutions of A44 in the variable loop and G26 just 3′ of the D-stem. Further inspection revealed that the variable loops and D-arms of RCS-clade and RZG-clade tRNAHis share fixed differences that distinguish them from eukaryotic tRNAHis (data not shown). Taken together, RCS-clade tRNAHis are best explained by in situ divergence rather than LGT of the tRNA from eukaryotes.
Figure 2.
tRNAHis acceptor stems (with adjacent 5′ leader bases) showing the different identity elements in the α-proteobacteria. The phylogeny is taken from (35,72). Also shown is a parsimony-based reconstruction of ancestral stems and leaders. The most obvious functionally significant differences are outlined in blue for the RCS-clade and in pink for the RZG-clade, to correspond to the covariation of HisRS shown in Figures 3 and 4. Abbreviations are as follows: Bh, Bartonella henselae; Bm, Brucella melitensis; Ml, Mesorhizobium loti; Sm, Sinorhizobium meliloti; At, Agrobacterium tumefaciens; Rp, Rhodopseudomonas palustris; Bj, Bradyrhizobium japonicum; Cc, Caulobacter crescentus; Sp, Silicibacter pomeroyi; Zm, Zymomonas mobilis; Go, Gluconobacter oxydans; Rc, Rickettsia conorii; Rp, Rickettsia prowazekii; wMel, Wolbachia strain wMel; wBm, Wolbachia strain wBm; Am, Anaplasma marginale; Er, Ehrlichia ruminantium.
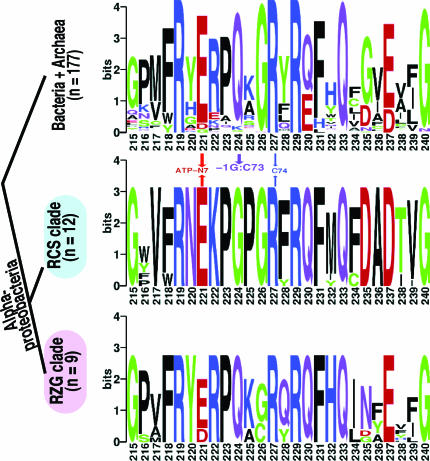
Next we examined the motif IIb loop tRNA-binding domain in bacterial HisRS. The HisRS residue that recognizes the tRNAHis C73 identity element (16,69) covaries perfectly with the presence of C73 in α-proteobacterial tRNAHis (Figure 3). Thus, Q224, predicted to recognize C73 (Q118 in E.coli), is highly conserved among bacteria but differs in the RCS-clade. In contrast, residues predicted (16) to be critical in adenylation [E221 (Rickettsia D221)] and aminoacylation (R227) are present in all bacterial HisRS. Interestingly, two other residues predicted (69) to recognize −1G, R229 and R219, are present in the HisRS of the RCS-clade. The α-proteobacterial RZG-clade with normal bacterial-type tRNAHis differs from the RCS-clade, but is like other bacteria, in that their HisRS sequences contain the highly conserved Q224.
Figure 3.
Sequence logos of the motif IIb loop in bacterial HisRS. The cladogram at left indicates source and number of sequences. Within α-proteobacteria, Q224 covaries perfectly with tRNAHis C73 in Figure 2.
HisRS phylogeny
The A73 discriminator base so unusual in RCS-clade tRNAHis is highly conserved among eukaryotic nuclear tRNAHis (2) where it serves as an important eukaryotic histidylation determinant (67). Furthermore, yeast hisRS histidylates E.coli tRNAHis, although with a 10-fold loss of catalytic efficiency in vitro that can be improved to a 4-fold loss upon mutation of C73 to A73 (68). This raises the possibility that the RCS-clade His identity rules may have been altered by substitution in the native tDNAHis to A73 after insertion by LGT of a eukaryotic-like HisRS gene. Suggestively, bacterial HisRS are paraphyletic, with one group more closely related to eukaryotic sequences (7,70), although it was not earlier suspected that any of those bacteria had a eukaryotic His-tRNA identity rule. To test this hypothesis we used likelihood methods to examine the evolutionary relationship of the HisRS sequences.
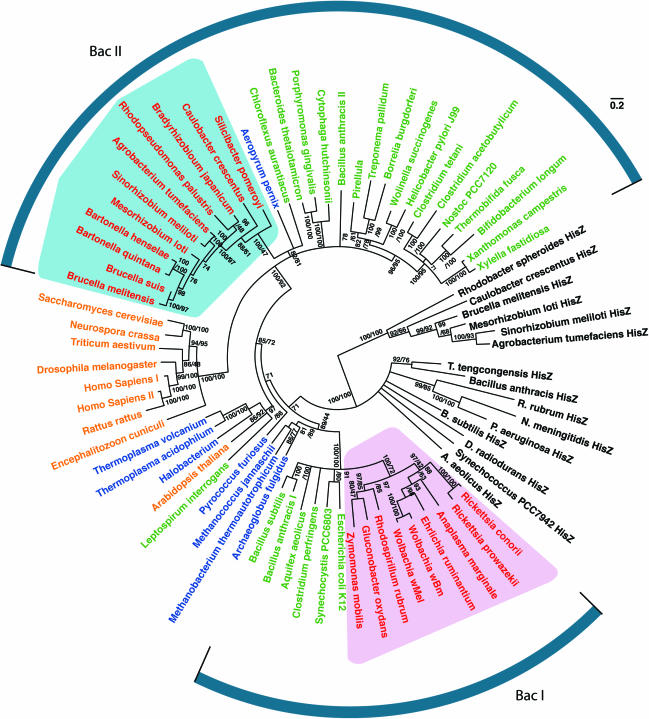
Our results (Figure 4 and Supplementary Data) confirmed the paraphyly of bacterial HisRS, with a predominant ‘BacI’ group (reduced for presentation in Figure 4) and a smaller ‘BacII’ group that treed with eukaryotes [terminology from (70)]. The paralogous HisZ proteins were monophyletic (70). RCS HisRS treed together with eukaryotic and BacII HisRS (bootstrap support 100%). However, with the known exception of an Arabidopsis paralog that trees with archaea (71), eukaryotic HisRS were monophyletic. Furthermore, we found moderate support (82–81%) for a prokaryotic origin of the RCS-clade HisRS. We confirmed that the BacII or archaeal HisRS most closely related to RCS HisRS have the canonical motif IIb Q224 and that the tRNAHis encoded by these genomes contained C73. Thus, although eukaryotes are undersampled, there is no direct evidence that LGT of a eukaryotic HisRS directly caused identity rule divergence in the RCS-clade.
Figure 4.
An unrooted protein likelihood HisRS/HisZ phylogram. Sequences are colored by taxonomic source: α-proteobacteria in red, other bacteria in green, eukaryotes in gold and archaea in blue. HisZ sequences are in black. The RCS-clade is shaded in blue and the RZG-clade is shaded in pink, to correspond to the covariation with tRNAHis acceptor stems shown in Figure 2. The tree topology shows the consensus of 100 bootstrapped PHYML likelihood trees, where edges of less than 70% bootstrap support with likelihood were collapsed. Splits with two support values show likelihood and BIONJ percent bootstrap values in that order. Single split support values indicate support by likelihood only. Units of branch length are substitutions per site.
Nonetheless, our phylogenetic results confirm the exceptional nature of RCS HisRS. RZG-clade α-proteobacterial species having the conventional tRNAHis with the −1G:C73 identity element contains HisRS of the ‘BacI’ group (bootstrap support = 100%, see also Supplementary Data), together with HisRS from other α-proteobacteria, including Rhodospirillum (Figure 4), Novosvingobium and Magnetospirillum (Supplementary Data). In contrast, the RCS-clade species with tRNAHis that have A73 contain HisRS sequences that are of the ‘BacII’ group (or eukaryotic) with perfect support. Also, the divergence of RCS HisRS follows the expected species divergence (72). Taken together, the data support vertical descent of RCS-clade HisRS with a distinct history from those of other α-proteobacteria.
DISCUSSION
We have an evolutionary result of perfect covariation between tRNA histidylation identity determinants and HisRS-types in a set of 20 α-proteobacterial species. Members of the RZG-clade use the canonical bacterial histidylation identity determinants (−1G:C73) and have HisRS genes of the bacterial BacI-type. In contrast, the RCS-clade bacteria use the major eukaryotic tRNA histidylation identity determinant (A73) and their HisRS genes, which are of the BacII-type, cluster with those of eukaryotes and other unrelated prokaryotes. Three hypothetical mechanisms may explain these two patterns: (i) lineage-specific loss of ancient paralogs, (ii) neutral or positively selected LGTs of both tRNAs and synthetases, or (iii) in situ co-evolution of identity determinants following (or followed by) LGT of either gene.
We find the simplest explanations involving paralogy insufficient to completely explain all our results. For instance, we do not see evidence of long-term retentions of ancient paralogs: BacI and BacII HisRS do not occur simultaneously in any genome except in Bacillus spp. Because HisRS of Bacillus subtilis and all other low-GC gram positive are BacI, like the majority of bacterial HisRS, these exceptions are more likely due to LGT than gene paralogy. Furthermore, the basal placement and the long branch of the Bacillus anthracis and Bacillus cereus BacII HisRS in the tree suggest that these genes may be evolving rapidly.
In contrast, LGT is evidently important in the evolution of HisRS genes. Along with the α-proteobacteria, the cyanobacteria and Clostridia also have paralogous HisRS—i.e. both BacI and BacII-type HisRS within the same clade, although not within the same genome. This is consistent with possible LGT of one or the other HisRS-types. We emphasize that the vast majority of bacterial HisRS are BacI and that BacI HisRS phylogeny is more consistent with expected bacterial phylogeny than the BacII phylogeny (Supplementary Data). This confirms, with much more taxonomic diversity, what was indicated in previous work and suggests a model of rare replacements of BacI HisRS by BacII HisRS by LGT (7,70).
The clear separation of the HisRS and histidylation identity determinants in the RCS-clade from all other α-proteobacteria and their subsequent divergence according to the expected phylogenetic pattern (72) may be consistent with their origin in a single LGT event. Yet our phylogenetic results indicate that LGT cannot completely explain the evolution of the eukaryotic-like identity rule in the RCS-clade. We find no evidence for an alien origin of RCS-clade tRNA genes, and fair support for a prokaryotic rather than eukaryotic donor of the HisRS gene to the RC-clade, from a lineage with a canonical prokaryotic histidylation identity rule.
Thus, tentatively, we suggest that LGT of a BacII HisRS gene (with a prokaryotic identity rule) into the RCS ancestor may have been followed by loss of the ancestral BacI HisRS and convergence of the HisRS and the ancestral tDNAHis in situ to the eukaryotic identity rule through a process of RNA-protein co-evolution. Perhaps BacII sequences, which are phylogenetically related to eukaryotic HisRS, are pre-adapted to specifically recognize eukaryotic tRNAHis better than BacI HisRS, and in the RCS-clade this resulted in a nearly neutral drift of histidylation identity determinants towards those of eukaryotes. Alternatively, a deleterious change of C73 to A73 in the RCS ancestral tDNAHis could have been rescued by coincidental LGT of a HisRS with improved histidylation efficiency for this mutant tRNA. Although E.coli HisRS tolerates A73 quite well and better than other discriminator base mutants in vitro (65), E.coli HisRS is completely out-competed in charging A73 mutants by other aaRS in vivo (66). So, perhaps this scenario of rescue by LGT is less likely than perturbation by LGT.
We should caution that the phylogenetic placement of the RCS-clade HisRSs could change upon further sampling of HisRS sequences from eukaryotes and from α-proteobacteria branching off ancestrally to S.pomeroyi in the lineage leading to the RCS-clade. Yet if the RCS-clade HisRS originated by LGT, regardless of the donor, subsequent conversion in the RCS-clade to the eukaryotic identity rule must have involved some in situ co-evolution of tRNAs and/or synthetases through potentially ambiguous or inefficient identity states. In the convergence hypothesis we propose that this happened in a nearly neutral sequence of slightly deleterious and compensatory substitutions driven largely by genetic drift. In defense of the plausibility of sustainable ambiguity, we point out that a single nuclear HisRS gene in yeast codes for both cytoplasmic and mitochondrial isoforms (73), even though yeast mitochondrial tRNAs, unlike in the cytoplasm, use prokaryotic histidylation identity determinants (data not shown).
The co-evolution of tRNA identity elements and the synthetase domains that recognize them so as to give new identity rules and genetic code expansions has been discussed in relation to the evolution of the GlnRS system from the GluRS system (74), the divergence of the mitochondrial and cytoplasmic tRNAAla identity elements in animals (75,76), and in other systems [see for instance (77,78)]. We suggest our findings and interpretation may be distinguished in that, and marshall circumstantial evidence for, the idea that in a nearly neutral process, spontaneous mutations that give rise to slightly deleterious charging ambiguities or inefficiencies may drive co-evolution of new identity rules in tRNAs and synthetases, and has done so particularly in the Histidine system. Alternatively, others have suggested that selection to preserve translational fidelity, which may be threatened by the presence of redundant, and therefore rapidly evolving, synthetases which can act in the same cellular compartment (generated by gene duplication or LGT) may drive the in situ divergence of identity rules, and may have done so particularly in the Alanine system of animal mitochondria (75,76).
We do not see the applicability of this selective hypothesis to the yeast histidylation system, where only one nuclear gene codes for both cytoplasmic and mitochondrial isoforms of HisRS. These isoforms are probably identical after cleavage of the mitochondrial targeting presequence (79), which does not affect the motif IIb tRNA-binding loop. Yet the acceptor stem of the yeast mitochondrial tRNAHis contains C73, while the cytoplasmic variant contains A73. Thus, the same enzyme must be able to recognize both substrates efficiently, consistent with the reduced importance of the discriminator base for identity in the yeast enzyme (68). The plant mitochondrial consensus tRNAHis also carries C73, but perhaps most plants are like Arabidopsis in using a prokaryotic HisRS paralog in organelles (71).
The absence of −1G and C73 in Sinorhizobium meliloti and of −1G in Methanobacterium thermoautotrophicum and Methanopyrus kandleri was reported previously (22), where they were regarded as exceptions to a general rule, and no analysis of HisRS was done or suggestion made that these bacteria have divergent His identity rules. In a later survey, we found another bacterium, Bacteroides thetaiotamicron and another archaeon Pyrobaculum aerophilum in which tDNAHis also lack a genetically encoded −1G. Perhaps tRNAHis biogenesis in prokaryotes that lack −1G is like in eukaryotes, where −1G is added post-transcriptionally by an enzyme called Histidine tRNA Guanynyltransferase or Thg1 (80,81). The gene encoding yeast Thg1 was recently identified, but it has no homologs in the RCS-clade nor in any other known bacterial genomes (82). Yet, consistent with the tRNA data, Thg1 homologs are found in M.thermoautotrophicum, M.kandleri and other Euryarchaeota, while a distant homolog is found in P.aerophilum, the only Crenarchaeote in which a Thg1 homolog was identified (82). Despite the absence of a homolog to the yeast Thg1 in the RCS-clade, data from the HisRS tRNA-binding site would seem to suggest that another analogous gene may be involved in maturation of RCS-clade tRNAHis to contain −1G.
It is interesting that, at least in yeast if not other eukaryotes, mitochondrial tRNAHis have not adapted to the eukaryotic HisRS that charge them. It is also ironic that the RCS-clade, and not the RZG-clade, evolved a eukaryotic identity rule, since symbiosis with eukaryotes probably occurred in the latter lineage [see also (83)], as also supported by their similar identity elements with those of mitochondria. Nonetheless, from current knowledge, it seems that only few substitutions are required to traverse between eukaryotic and prokaryotic His identity rules (16,67). Perhaps ambiguity between eukaryotic and prokaryotic His identity rules may be sustainable and, in the RCS-clade, has acted like an evolutionary transition state.
Our ‘ambiguous identity’ hypothesis extends the ‘ambiguous intermediate’ hypotheses for the evolution of alternative genetic codes (84), and is supported by several lines of evidence. First, the number of sequence changes required to alter the identity of tRNAs can be quite small. For example, only 1 nt change was required to alter both the reading and charging identities of a tRNA (85) or of species-specificity of charging in an acceptor stem model minihelix (12). Furthermore, identity rules in different aminoacylation systems quite often have overlapping determinants. For instance, determinants in two independently derived bacterial LysRS are substantially overlapping (86). These factors suggest that perhaps generally a small number of compensatory mutations may be necessary to diverge by co-evolution through mildly deleterious ambiguous identity states to new identity rules. With such added evolutionary flexibility, tRNA and aaRS genes might not only have lower barriers to LGT but also be more likely to fluidly evolve new identity rules in situ. Experimental and theoretical assessments of evolutionary dynamics would be very helpful in assessing these various hypotheses for the evolution of tRNA identity rules. The automated statistical discovery of altered tRNA identity rules in bacteria along with our other results such as the likely statistical variation of identity rules in hyperthermophilic bacteria highlight the potential utility of TFAM in future bioinformatic analysis of tRNA identity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation under a grant awarded in 2000 to D.H.A., by awards from the Swedish Research Council, the Swedish Foundation for Strategic Research and the Wallenberg Foundation to S.G.E.A., and the Wallenberg Consortium North under a grant awarded to Leif A. Kirsebom. We thank Leif Kirsebom for advice and discussion, and Eva Freyhult, Todd Lowe and anonymous reviewers for comments on the work and/or manuscript. Some computations were carried out on a high-performance computer cluster at UPPMAX with consultation from Ann-Charlotte Berglund Sonnhammer. Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council to S.G.E.A.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ibba M., Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Giege R., Sissler M., Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J.R., Doolittle W.F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl Acad. Sci. USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz-Lazcoz Y., Aude J.C., Nitschke P., Chiapello H., Landes-Devauchelle C., Risler J.L. Evolution of genes, evolution of species: the case of aminoacyl-tRNA synthetases. Mol. Biol. Evol. 1998;15:1548–1561. doi: 10.1093/oxfordjournals.molbev.a025882. [DOI] [PubMed] [Google Scholar]
- 6.Wolf Y.I., Aravind L., Grishin N.V., Koonin E.V. Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 7.Woese C.R., Olsen G.J., Ibba M., Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Micro. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain R., Rivera M.C., Lake J.A. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampson J.R., DiRenzo A.B., Behlen L.S., Uhlenbeck O.C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989;243:1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- 10.Lee C.P., RajBhandary U.L. Mutants of Escherichia coli initiator tRNA that suppress amber codons in Saccharomyces cerevisiae and are aminoacylated with tyrosine by yeast extracts. Proc. Natl Acad. Sci. USA. 1991;88:11378–11382. doi: 10.1073/pnas.88.24.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiba K., Schimmel P., Motegi H., Noda T. Human glycyl-tRNA synthetase. Wide divergence of primary structure from bacterial counterpart and species-specific aminoacylation. J. Biol. Chem. 1994;269:30049–30055. [PubMed] [Google Scholar]
- 12.Hipps D., Shiba K., Henderson B., Schimmel P. Operational RNA code for amino acids: species-specific aminoacylation of minihelices switched by a single nucleotide. Proc. Natl Acad. Sci. USA. 1995;92:5550–5552. doi: 10.1073/pnas.92.12.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiba K., Motegi H., Schimmel P. Maintaining genetic code through adaptations of tRNA synthetases to taxonomic domains. Trends Biochem. Sci. 1997;22:453–457. doi: 10.1016/s0968-0004(97)01135-3. [DOI] [PubMed] [Google Scholar]
- 14.Stehlin C., Burke B., Yang F., Liu H., Shiba K., Musier-Forsyth K. Species-specific differences in the operational RNA code for aminoacylation of tRNAPro. Biochemistry. 1998;37:8605–8613. doi: 10.1021/bi980364s. [DOI] [PubMed] [Google Scholar]
- 15.Xu F., Chen X., Xin L., Chen L., Jin Y., Wang D. Species-specific differences in the operational RNA code for aminoacylation of tRNA(Trp) Nucleic Acids Res. 2001;29:4125–4133. doi: 10.1093/nar/29.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawko S.A., Francklyn C.S. Covariation of a specificity-determining structural motif in an aminoacyl-tRNA synthetase and a tRNA identity element. Biochemistry. 2001;40:1930–1936. doi: 10.1021/bi0025316. [DOI] [PubMed] [Google Scholar]
- 17.Sassanfar M., Kranz J.E., Gallant P., Schimmel P., Shiba K. A eubacterial Mycobacterium tuberculosis tRNA synthetase is eukaryote-like and resistant to a eubacterial-specific antisynthetase drug. Biochemistry. 1996;35:9995–10003. doi: 10.1021/bi9603027. [DOI] [PubMed] [Google Scholar]
- 18.Salzberg S.L., White O., Peterson J., Eisen J.A. Microbial genes in the human genome: lateral transfer or gene loss? Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- 19.McClain W.H., Nicholas H.B., Jr Differences between transfer RNA molecules. J. Mol. Biol. 1987;194:635–642. doi: 10.1016/0022-2836(87)90240-3. [DOI] [PubMed] [Google Scholar]
- 20.Steinberg S.V., Kisselev L.L. Mosaic tile model for tRNA-enzyme recognition. Nucleic Acids Res. 1993;21:1941–1947. doi: 10.1093/nar/21.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholas H.B., Jr, McClain W.H. An algorithm for discriminating sequences and its application to yeast transfer RNA. Comput. Appl. Biosci. 1987;3:177–181. doi: 10.1093/bioinformatics/3.3.177. [DOI] [PubMed] [Google Scholar]
- 22.Marck C., Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laslett D., Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsui V., Macke T., Case D.A. A novel method for finding tRNA genes. RNA. 2003;9:507–517. doi: 10.1261/rna.2193703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sprinzl M., Vassilenko K.S., Emmerich J., Bauer F. tRNA compilation 2000. 1999 [Google Scholar]
- 27.Sonnhammer E.L., Eddy S.R., Birney E., Bateman A., Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gribskov M., McLachlan A.D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Alsmark C.M., Frank A.C., Karlberg E.O., Legault B.A., Ardell D.H., Canback B., Eriksson A.S., Naslund A.K., Handley S.A., Huvet M., et al. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl Acad. Sci. USA. 2004;101:9716–9721. doi: 10.1073/pnas.0305659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy S.R., Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durbin R., Eddy S., Krogh A., Mitchison G. Biological Sequence Analysis. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 33.Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., et al. The bioperl toolkit: perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felsenstein J. PHYLIP (Phylogeny Inference Package) 2004 [Google Scholar]
- 35.Sallstrom B., Andersson S.G. Genome reduction in the alpha-Proteobacteria. Curr. Opin. Microbiol. 2005;8:579–585. doi: 10.1016/j.mib.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sissler M., Delorme C., Bond J., Ehrlich S.D., Renault P., Francklyn C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl Acad. Sci. USA. 1999;96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisen J.A., Fraser C.M. Phylogenomics: intersection of evolution and genomics. Science. 2003;300:1706–1707. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- 41.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller T., Vingron M. Modeling amino acid replacement. J. Comput. Biol. 2000;7:761–776. doi: 10.1089/10665270050514918. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 45.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Adachi J., Hasegawa M. Molphy version 2.3b3, Programs for molecular phylogenetics based on maximum likelihood. 1996 [Google Scholar]
- 47.Yuan J., Amend A., Borkowski J., DeMarco R., Bailey W., Liu Y., Xie G., Blevins R. MULTICLUSTAL: a systematic method for surveying Clustal W alignment parameters. Bioinformatics. 1999;15:862–863. doi: 10.1093/bioinformatics/15.10.862. [DOI] [PubMed] [Google Scholar]
- 48.Seo J.S., Chong H., Park H.S., Yoon K.O., Jung C., Kim J.J., Hong J.H., Kim H., Kim J.H., Kil J.I., et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 2005;23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran M.A., Buchan A., Gonzalez J.M., Heidelberg J.F., Whitman W.B., Kiene R.P., Henriksen J.R., King G.M., Belas R., Fuqua C., et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 50.Gish W., States D.J. Identification of protein coding regions by database similarity search. Nature Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 51.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 52.Goldman N., Whelan S. Statistical tests of gamma-distributed rate heterogeneity in models of sequence evolution in phylogenetics. Mol. Biol. Evol. 2000;17:975–978. doi: 10.1093/oxfordjournals.molbev.a026378. [DOI] [PubMed] [Google Scholar]
- 53.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 54.Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 55.Freyhult E., Moulton V., Ardell D.H. Visualizing Bacterial tRNA Identity Determinants and Antideterminants Using Function Logos and Inverse Function Logos. Nucleic Acids Res. 2006. In press. [DOI] [PMC free article] [PubMed]
- 56.Mayer C., Stortchevoi A., Kohrer C., Varshney U., RajBhandary U.L. Initiator tRNA and its role in initiation of protein synthesis. Cold Spring Harb. Symp. Quant. Biol. 2001;66:195–206. doi: 10.1101/sqb.2001.66.195. [DOI] [PubMed] [Google Scholar]
- 57.Aiyar S.E., Gaal T., Gourse R.L. rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J. Bacteriol. 2002;184:1349–1358. doi: 10.1128/JB.184.5.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong H.J., Nilsson L., Kurland C.G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 59.Percudani R., Pavesi A., Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- 60.Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc. Natl Acad. Sci. USA. 1985;82:2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanchard A. Ureaplasma urealyticum urease genes; use of a UGA tryptophan codon. Mol. Microbiol. 1990;4:669–676. doi: 10.1111/j.1365-2958.1990.tb00636.x. [DOI] [PubMed] [Google Scholar]
- 62.Inamine J.M., Ho K.C., Loechel S., Hu P.C. Evidence that UGA is read as a tryptophan codon rather than as a stop codon by Mycoplasma pneumoniae, Mycoplasma genitalium, and Mycoplasma gallisepticum. J. Bacteriol. 1990;172:504–506. doi: 10.1128/jb.172.1.504-506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Galtier N., Lobry J.R. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 1997;44:632–636. doi: 10.1007/pl00006186. [DOI] [PubMed] [Google Scholar]
- 64.Kirsebom L.A., Vioque A. RNase P from bacteria. Substrate recognition and function of the protein subunit. Mol. Biol. Rep. 1995;22:99–109. doi: 10.1007/BF00988713. [DOI] [PubMed] [Google Scholar]
- 65.Himeno H., Hasegawa T., Ueda T., Watanabe K., Miura K., Shimizu M. Role of the extra G-C pair at the end of the acceptor stem of tRNA(His) in aminoacylation. Nucleic Acids Res. 1989;17:7855–7863. doi: 10.1093/nar/17.19.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan W., Francklyn C. Cytosine 73 is a discriminator nucleotide in vivo for histidyl-tRNA in Escherichia coli. J. Biol. Chem. 1994;269:10022–10027. [PubMed] [Google Scholar]
- 67.Rudinger J., Florentz C., Giege R. Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues −1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 1994;22:5031–5037. doi: 10.1093/nar/22.23.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nameki N., Asahara H., Shimizu M., Okada N., Himeno H. Identity elements of Saccharomyces cerevisiae tRNA(His) Nucleic Acids Res. 1995;23:389–394. doi: 10.1093/nar/23.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Connolly S.A., Rosen A.E., Musier-Forsyth K., Francklyn C.S. G-1:C73 recognition by an arginine cluster in the active site of Escherichia coli histidyl-tRNA synthetase. Biochemistry. 2004;43:962–969. doi: 10.1021/bi035708f. [DOI] [PubMed] [Google Scholar]
- 70.Bond J.P., Francklyn C. Proteobacterial histidine-biosynthetic pathways are paraphyletic. J. Mol. Evol. 2000;50:339–347. doi: 10.1007/s002399910037. [DOI] [PubMed] [Google Scholar]
- 71.Akashi K., Grandjean O., Small I. Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett. 1998;431:39–44. doi: 10.1016/s0014-5793(98)00717-0. [DOI] [PubMed] [Google Scholar]
- 72.Boussau B., Karlberg E.O., Frank A.C., Legault B.A., Andersson S.G. Computational inference of scenarios for α-proteobacterial genome evolution. Proc. Natl Acad. Sci. USA. 2004;101:9722–9726. doi: 10.1073/pnas.0400975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Natsoulis G., Hilger F., Fink G.R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986;46:235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- 74.Skouloubris S., Ribas de Pouplana L., De Reuse H., Hendrickson T.L. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc. Natl Acad. Sci. USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ribas de Pouplana L., Schimmel P. Operational RNA code for amino acids in relation to genetic code in evolution. J. Biol. Chem. 2001;276:6881–6884. doi: 10.1074/jbc.R000032200. [DOI] [PubMed] [Google Scholar]
- 76.Lovato M.A., Swairjo M.A., Schimmel P. Positional recognition of a tRNA determinant dependent on a peptide insertion. Mol. Cell. 2004;13:843–851. doi: 10.1016/s1097-2765(04)00125-x. [DOI] [PubMed] [Google Scholar]
- 77.Francklyn C., Perona J.J., Puetz J., Hou Y.M. Aminoacyl-tRNA synthetases: versatile players in the changing theater of translation. RNA. 2002;8:1363–1372. doi: 10.1017/s1355838202021180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ibba M., Soll D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev. 2004;18:731–738. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 79.Chiu M.I., Mason T.L., Fink G.R. HTS1 encodes both the cytoplasmic and mitochondrial histidyl-tRNA synthetase of Saccharomyces cerevisiae: mutations alter the specificity of compartmentation. Genetics. 1992;132:987–1001. doi: 10.1093/genetics/132.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cooley L., Appel B., Soll D. Post-transcriptional nucleotide addition is responsible for the formation of the 5′ terminus of histidine tRNA. Proc. Natl Acad. Sci. USA. 1982;79:6475–6479. doi: 10.1073/pnas.79.21.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pande S., Jahn D., Söll D. Histidine tRNA guanylyltransferase from Saccharomyces cerevisiae. I. Purification and physical properties. J. Biol. Chem. 1991;266:22826–22831. [PubMed] [Google Scholar]
- 82.Gu W., Jackman J.E., Lohan A.J., Gray M.W., Phizicky E.M. tRNA-His maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNA-His. Genes Dev. 2003;17:2889–2901. doi: 10.1101/gad.1148603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu M., Sun L.V., Vamathevan J., Riegler M., Deboy R., Brownlie J.C., McGraw E.A., Martin W., Esser C., Ahmadinejad N., et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schultz D.W., Yarus M. Transfer-RNA mutation and the malleability of the genetic-code. J. Mol. Biol. 1994;235:1377–1380. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 85.Saks M.E., Sampson J.R., Abelson J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–1670. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]
- 86.Ambrogelly A., Korencic D., Ibba M. Functional annotation of class I lysyl-tRNA synthetase phylogeny indicates a limited role for gene transfer. J. Bacteriol. 2002;184:4594–4600. doi: 10.1128/JB.184.16.4594-4600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.