Abstract
p63 is a developmentally regulated transcription factor related to p53, which activates and represses specific genes. The human AEC (Ankyloblepharon–Ectodermal dysplasia-Clefting) and EEC (Ectrodactyly–Ectodermal dysplasia–Cleft lip/palate) syndromes are caused by missense mutations of p63, within the DNA-binding domain (EEC) or in the C-terminal sterile alpha motif domain (AEC). We show here that p63 represses transcription of cell-cycle G2/M genes by binding to multiple CCAAT core promoters in immortalized and primary keratinocytes. The CCAAT-activator NF-Y and ΔNp63α are associated in vivo and a conserved α-helix of the NF-YC histone fold is required. p63 AEC mutants, but not an EEC mutant, are incapable to bind NF-Y. ΔNp63α, but not the AEC mutants repress CCAAT-dependent transcription of G2/M genes. Chromatin immunoprecipitation recruitment assays establish that the AEC mutants are not recruited to G2/M promoters, while normally present on 14-3-3σ, which contains a sequence-specific binding site. Surprisingly, the EEC C306R mutant activates transcription. Upon keratinocytes differentiation, NF-Y and p63 remain bound to G2/M promoters, while HDACs are recruited, histones deacetylated, Pol II displaced and transcription repressed. Our data indicate that NF-Y is a molecular target of p63 and that inhibition of growth activating genes upon differentiation is compromised by AEC missense mutations.
INTRODUCTION
p63 is a transcription factor homologous to p53 and p73 (1,2). It binds to DNA in a sequence-specific way in the promoter and enhancers, activating genes that block cell-cycle progression and promote apoptosis. Unlike p53, p63 and p73 are not ubiquitously expressed, and are involved in separate developmental processes. Three protein motifs are shared by the homologues: a transactivation domain—TA—at the N-terminal, a central DNA-binding domain and a tetramerization domain. There are two different transcription initiation sites generating proteins containing, TA, or lacking, ΔN, an activation domain. Furthermore, the 3′ end of the gene is involved in alternative splicing, which gives rise to three isotypes, alpha, beta and gamma; hence, six p63 isoforms are potentially present in cells, at various levels of relative expression. The C-terminal of p63 and p73 contains the sterile alpha motif (SAM) domain, found in >40 proteins involved in developmental regulation (3). It consists of five helices packed into a compact globular domain representing a protein–protein interaction module (4).
The association of ectodermal dysplasia with cleft lip/palate is found in several clinical entities often associated with dominant transmission (5,6). The Hay–Wells (Ankyloblepharon–Ectodermal dysplasia-Clefting) AEC, the Ectrodactyly–Ectodermal dysplasia–Cleft lip/palate (EEC) and the Split Hand/split Foot Malformation syndromes show clinical variability, with sparse hair, dry skin, philosebaceous gland dysplasia and oligodontia. Patients with the AEC syndrome do not show ectrodactyly or other limb defects, but have ankyloblepharon, fused eyelids, and severe scalp dermatitis, unlike EEC patients, who have widespread defects in ectodermal development, but less severe skin alterations. These syndromes are caused by mutations in the p63 gene (5).
The vast majority of the mutations found in EEC syndromes are missense mutations generating amino acid substitutions in the residues predicted to contact DNA [(5,7) and references therein]. All isoforms of p63 are affected by these alterations. On the other hand, mutations causative of the AEC syndrome are all missense mutations falling within exon 13, coding for most of the SAM domain; thus, only the p63α isoforms are affected. Interestingly, skin biopsies documented p63 staining in the differentiating cells of the suprabasal layer, where p63 is normally absent (6). Genetic experiments in mice confirmed the specificities of the p63 gene function and are well in agreement with the phenotypes in humans; mice lacking p63 die soon after birth with severe defects in limb, craniofacial and skin development (8,9). Additional clues to the function of p63 came from zebrafish, where the dominant isoform, corresponding to ΔNα, is required for epithelial development (10,11). The ΔNα is apparently the most abundant isoform found in keratinocytes, and indeed p63 has been shown to be a marker of epithelial stem cells of the skin and of the ocular limbus (12,13). Altogether, these data establish p63 as a master regulatory gene of skin development [reviewed in (14,15)].
In addition to being an activator, p63 can also repress transcription (16–19). p53 and p73 negatively regulate the expression of G2/M regulators such as CDC25B, CDC25C, Cyclin B1, Cyclin B2, Cdc2, Check 2, securin and Topoisomerase IIα upon DNA-damage (20–29). In these studies, the negative activity was shown to be exerted indirectly through the multiple conserved CCAAT boxes; in other reports SP1, or direct p53 binding were implicated in Cyclin B1 and CDC25C repression (27,30). As for p63, it was shown to have opposite effects on cdc2 and HSP70, in both cases acting on CCAAT boxes (22,31).
The CCAAT box is bound and regulated by the trimeric factor NF-Y, composed of three subunits, NF-YA, NF-YB and NF-YC, all necessary for DNA-binding (32). NF-YB and NF-YC contain histone fold motifs (HFMs) common to all core histones; dimerization is essential for NF-YA association and sequence-specific DNA binding (33). A recent bioinformatic analysis of cell-cycle promoters showed a remarkable and specific abundance of CCAAT boxes in promoters regulated during the G2/M phase (34). Chromatin immunoprecipitation (ChIP) experiments determined that NF-Y is dynamically bound in the different phases of the cell cycle (35). Finally, association of NF-Y to p63 has been detected (31,36).
To shed light on p63 repressive function in the CCAAT promoters circuitry, we investigated the relationship between p63 and NF-Y in human keratinocytes with protein–protein interaction, transfection and ChIP experiments, using wt p63 and AEC/EEC-derived mutants.
MATERIALS AND METHODS
Immunoprecipitation analysis
The AEC mutants were described in Ref. (6). NF-YC αC mutants were produced in the backbone of the His-YC5 mutant as detailed in Ref. (37). Immunoprecipitations were performed as in Ref. (20); 50–100 ng of recombinant proteins were incubated in 100 µl of NDB 100 [100 mM KCl, 20 mM HEPES, pH 7.9, 0.1% NP40, 0.5 mM EDTA and 1 mM phenylmethlysulfonyl fluoride (PMSF)] rotated for 2 h at 4°C and then added to 10 µl of ProteinG–Sepharose to which 5 µg of the anti-NF-YC antibodies had been bound previously. Incubation was pursued for 2 h at 4°C, unbound material recovered after centrifugation and the beads washed with NDB100. SDS buffer was added, the samples boiled at 90°C for 5′ and loaded on SDS gels. Western blots were performed according to standard procedures with the indicated primary antibody (anti-YB, anti-YC and anti-p63 4A4). For in vivo interactions, SaoS2 cells were transfected in 24-well plates with 100 ng of the NF-Y and p63 eukaryotic expression vectors. The cells were harvested after 24 h in lysis buffer (50 mM Tris–HCl, pH 8, 120 mM NaCl, 2 mM DTT, 10 µg/ml leupeptin and 10 µg/ml aprotinin, 1 mM PMSF, 0.5% NP40). Immunoprecipitations were performed by incubating whole-cell extracts with 2 µg of the relevant antibodies, with ProteinA–Agarose (Sigma) at 4°C overnight. The beads were washed twice with NDB100 plus protease inhibitors, eluted in SDS buffer and loaded on 12% SDS–polyacrylamide gels.
Chromatin immunoprecipitations
ChIP assays were performed as described (20,35). HaCaT cells (0.5/1 × 108) were washed in phosphate-buffered saline (PBS) and incubated for 10 min with 1% formaldehyde; after quenching the reaction with Glycine 0.125 M, cells were sonicated and chromatin fragments of an average length of 1 kb recovered by centrifugation. Immunoprecipitations were performed with ProtG–Sepharose (KPL) and 3–5 µg of the indicated antibodies: NF-YB purified rabbit polyclonal; p63 (4A4), anti-acetyl-H3, anti-acetyl-H4 (Upstate), anti-Pol II (Santa Cruz) and anti-HDAC1 (Sigma). Anti-HDAC4 (Active Motif), control anti-Flag (Sigma). ProtG–Sepharose was blocked twice at 4°C with 1 µg/µl salmon sperm DNA sheared at 500 bp length and 1 µg/µl BSA, for 2 h and overnight. Chromatin was precleared by adding ProtG–Sepharose for 2 h at 4°C, aliquoted and incubated with the antibodies overnight at 4°C. Semi-quantitative PCRs were performed with the following primers: Topoisomerase II α 5′-CCTGCACACTTTTGCCTCAG-3′, 5′-GACCAGCCAATCCCTGACTC-3′; CDC25C 5′-GCTGAGGGAACGAGGAAAAC-3′; 5′-CGCCAGCCCAGTAACCTATC-3′; Cyclin B1 5′-TGTCACCTTCCAAAGGCCACTA-3′, 5′-AGAAGAGCCAGCCTAGCCTCAG-3′; Cyclin B2 5′-AGAGGCGTCCTACGTCTGCTTT-3′, 5′-ATTCAAATACCGCGTCGCTTG-3′; OGG1 5′-CCGAGTGCAGACAATCCCGG-3′; 5′-CTCCTTGCGACTTATCTTCTCC-3′; PLK 5′-GAAAGGGAGAAACCCCGAAG-3′, 5′-GCTCCTCCCCGAATTCAAAC-3′; Luciferase 5′-TTGCTCTCCAGCGGTTCCAT-3′. HSP70 forward 5′-GGCGAAACCCCTGGAATATTCCCGA-3′ and reverse 5′-AGCCTTGGGACAACGGGAG-14-3-3σ 5′-CTCACTACCTCAAGATACCC-3′; 5′-CACAGGCCTGTGTCTCCC-3′.
Transient transfections
SaoS2 cells (2.5 × 104) were transfected with Lipofectamine (Gibco-BRL) using 0.1 µg of Cyclin B2-Luciferase (23), cdc2-CAT (gift of G. Piaggio, IRE, Rome, Italy), Topo II α Luciferase (Gift of M. Broggini, Negri Institute, Milan, Italy) or Calreticulin Luciferase vectors, 100 ng of the p63 expressing plasmids (Obtained from H. Van Bokhoven, Leyden, The Netherlands), 50 ng of NβGAL and carrier plasmid to keep the total DNA concentration constant at 1 µg. Cells were recovered 36 h after transfection, resuspended in TEN (150 mM NaCl and 40 mM Tris–HCl, pH 7.4) for measurements of CAT activity, or Lysis buffer (Triton-X 1%, Glycil-glycine 25 mM, MgSO4 15 mM and EGTA 4 mM) for luciferase. β-galactosidase was assayed to control for transfection efficiencies. Three to four independent transfections in duplicate were performed. SDs were calculated as <20% of the relative levels.
The ChIP analysis performed in Figure 6 was carried out in SaoS2 (15 cm plates, 6 × 106 cells) transfected with the p63 plasmids (10 µg) and the Cyclin B2 Luciferase vector (10 µg). After 36 h, cells were fixed with formaldehyde and ChIP analysis was performed as described above.
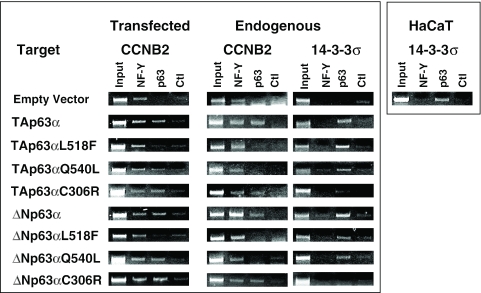
Figure 6.
In vivo recruitment of p63 AEC and EEC mutants to Cyclin B2. ChIP analysis of NF-Y and p63 in SaoS2 cells transfected with ΔN and TA isoforms of p63-wt, L518F, Q540L and C306R- and the control empty vector. Together with the p63 expressing vector, we cotransfected the Cyclin B2 Luciferase construct. The PCRs on the transfected templates are performed with a 5′ primer for Cyclin B2 and a 3′ primer specific for the Luciferase reporter (left panels). The PCRs on the endogenous Cyclin B2 (middle panels) and 14-3-3σ genomic loci (right panels) were performed with the specific oligonucleotides indicated in Materials and Methods on the same immunoprecipitated DNAs. In the right panel, we performed ChIPs with HaCaT chromatin as in Figure 2, to ascertain the positivity of the 14-3-3σ promoter for p63.
Immunofluorescence
Cells were washed twice in PBS at room temperature, fixed in cold methanol/acetone for 2′ and permeabilized with 0.05% TritonX 100 in PBS for 5′. Samples were preincubated with 1% BSA in PBS for 15 min and then incubated overnight at 4°C with the following primary antibodies diluted 1:100 in PBS plus BSA 1%: anti-CK8-18 (Santa Cruz sc-8020); anti-CK1 (Covance PRB-149P); anti-p63 (Dako 4A4). Cells were then washed twice with PBS and incubated for 15 min at room temperature in PBS plus BSA before a further incubation, 60 min at room temperature in the dark, with Hoechst and the relative secondary antibodies: FITC anti-rabbit 1:200 (F7512; Sigma), TRITC anti-mouse 1:150 (T5393; Sigma). Slides were taken with a Zeiss-Axioskop microscope.
RT–PCR analysis
RNA was extracted using an RNA-Easy kit (Quiagen, Germany), according to the manufacturer's protocol, from HaCaT cells. For cDNA syntesis, 2 µg of RNA were used with M-MLV-RT kit (Invitrogen). Semi-quantitative PCR analysis was performed with primers specific for the different p63 isoforms. TA and ΔNp63α reverse, 5′-ACTTGCCAGATCATCCATGG-3′; TA and ΔNp63β reverse, 5′-TCAGACTTGCCAGATCCTG-3′; TA and ΔNp63γ reverse, 5′AAGCTCATTCCTGAAGCAGG-3′; ΔNp63 forward, 5′-CCAGACTCAATTTAGTGAGC-3′; TAp63 forward, 5′-TTAGCATGGACTGTATCCGC-3′. CCNB1 forward, 5′-CACTTCCTTCGGAGAGCATC; reverse, 5′-CAGGTGCTGCATAACTGGAA-3′. CCNB2 forward, 5′-CAGTTCCCAAATCCGAGAAA-3′; reverse, 5′-TCTGAGACAAGCAGGAAGCA-3′. CDC25C forward, 5′-AAAGGCGGCTACAGAGACTTC-3′; reverse, 5′-AGCCCAGAGAGAAAGAGTTGG-3′. NF-YB forward, 5′-AGGTGCCATCAAGAGAAACG-3′; reverse, 5′-TGTTGTTGACCGTCTGTGGT-3′; Topo II α forward, 5′-GCCCTCCTGCTACACATTTC-3′; reverse, 5′-AACACTTGGGCTTTACTTCACTT-3′; HSP70 forward, 5′-TTTCGAGAGTGACTCCCGTT-3′; reverse, 5′-AAAGGCCAGTGCTTCATGTC-3′.
RESULTS
ΔNp63α is associated to G2/M promoters in vivo
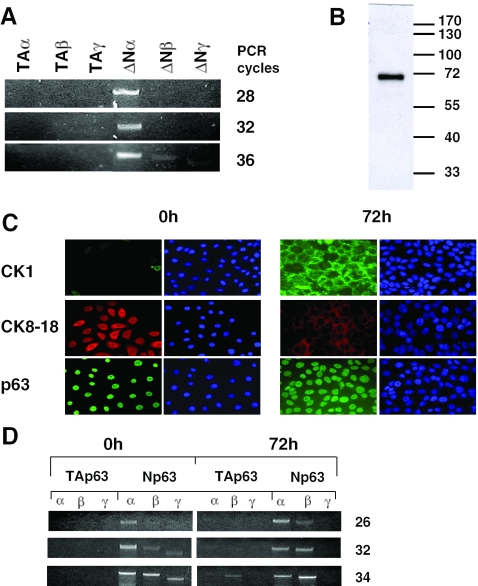
It is generally assumed that the major, if not the sole isoform present in HaCaT keratinocytes is ΔNp63α (2,38). To confirm this point in our system, we performed RT–PCR analysis, using isoform-specific combinations of oligonucleotides. PCR amplifications show that while ΔNp63α was promptly visible at a low number of cycles, the p63β or p63γ isoforms were far less abundant and the TAp63 isoforms essentially absent (Figure 1A). At the protein level, western blot analysis confirmed this finding, since a single polypeptide with a MW consistent with that of ΔNp63α was visualized (Figure 1B). HaCaT cells can be induced to differentiate into more mature keratinocytes by adding calcium and eliminating serum from the medium (39,40). We checked this with positive and negative markers by immunofluorescence analysis. In accordance with expectations, CK1 increases, while CK8-18 decreases after 72 h in differentiation medium, whereas staining with p63 was positive before and after (Figure 1C). RT–PCR analysis indicated that the ΔNβ isoform increases after differentiation. Note that growing cells under calcium-free medium, which is required for optimal differentiation, increases the levels of the ΔN β and γ isoforms in undifferentiated cells. These differences notwithstanding, the ΔNα is still very abundant after differentiation and the predominant isoform in all conditions tested. The TA isoforms are essentially absent in any conditions.
Figure 1.
HaCaT cells express ΔNp63α. (A) RT–PCR analysis of HaCaT cells exponentially growing in DMEM medium. Specific oligonucleotides for each of the six major isoforms were used with increasing PCR cycles. For details see Materials and Methods. (B) Western blot analysis of HaCaT nuclear extracts with the 4A4 antibody, which recognizes all p63 isoforms; a major band corresponding to the ΔNα isoform is predominant. (C) Immunofluorescence analysis of differentiation markers. HaCaT cells were grown in CaCl2-free medium containing 10% foetal calf serum (FCS) (0 h, left panels) and in 1.2 mM CaCl2, 0.1% FCS for three days (72 h, right panels). After fixation, cells were stained with anti-Cytokeratin 1, anti-Cytokeratin 8–18 differentiation markers, as well as anti-p63 antibodies. (D) RT–PCR analysis of p63 isoforms in HaCaT keratinocytes. mRNA was extracted from cells as in (C). Note that under these conditions, HaCaT cells tend to have higher levels of ΔNβ and γ isoforms. Nevertheless, the ΔNα isoform is still predominant.
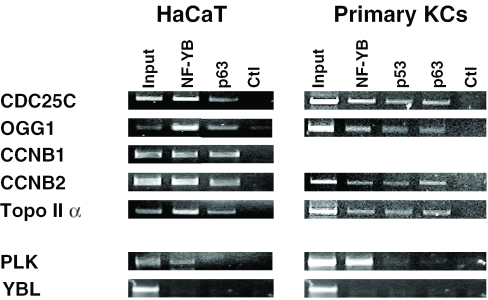
Recent ChIP experiments performed in our laboratory established that p53 binds to G2/M promoters in mouse NIH3T3 fibroblasts in the absence of DNA-damage and of p53 binding sites (20). Given the similarities, it is possible that p63 behaves similarly in the appropriate cellular context, such as keratinocytes. We prepared HaCaT chromatin and performed ChIPs with the 4A4 anti-p63 antibody. As a positive control, we used an anti-YB antibody. As shown in Figure 2, all G2/M promoters—Cyclin B1, Cyclin B2, CDC25C and Topoisomerase II α—are strongly positive for p63. On the other hand, PLK, a promoter with a single CCAAT, was negative for p63, in line with recent p53 ChIP experiments (20). Two additional p63 antibodies yielded the same results (data not shown). NF-Y was positive, an expected result, since PLK is known to be activated by NF-Y (41). We controlled a CCAAT-less cell-cycle regulated promoter, YBL1/Pole3; none of the antibodies tested significantly immunoprecipitated this target (lower panel), confirming the specificity of our ChIP procedure. We then switched to a normal system, using primary human keratinocytes for preparation of chromatin. In this setting, we added an anti-p53 antibody to our ChIPs; both p63 and p53 were positive on G2/M promoters, while negative on CCAAT-less, or single CCAAT promoters (Figure 2, right panels). Therefore, in immortalized or primary keratinocytes, p63 is associated in vivo to promoters of crucial cell-cycle regulatory genes containing double CCAAT boxes, in the absence of a p53-binding sequence.
Figure 2.
In vivo binding of ΔNp63α to promoters with multiple CCAAT boxes. ChIP analysis of growing HaCaT keratinocytes (left panels) and human primary keratinocytes (right panels) using the indicated anti-NF-YB, anti-p63 (4A4) and anti-53 antibodies.
Effects of p63 on CCAAT-dependent transcription
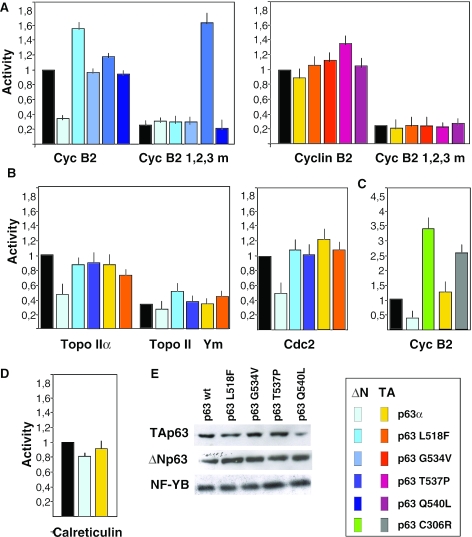
Having shown a negative role of p53 on multiple CCAAT-dependent transcription (20,25), we wished to ascertain whether p63 would also have a transcriptional effect. We transfected the Cyclin B2, which contains three functionally important CCAAT-boxes (23,25), with the TA and ΔN isoforms in the p53 negative SaoS2 cell line, in which low levels of p63 and p73 are expressed (16,42). Figure 3A shows that ΔNp63α represses the promoter. Parallel transfections with four AEC mutants had no significant effect on transcription. In parallel, we transfected a mutant Cyclin B2 construct, in which all CCAAT boxes were substituted with random pentanucleotides; this construct shows a considerably lower level of activity, but it is still quite active when compared with a promoter-less Luciferase construct (20). ΔNp63α had no effect on the lower activity of the promoter, indicating that repression is operative as long as the three CCAAT are intact. A surprising exception was ΔNp63α T537P, which somewhat activated the mutant promoter. It should be noted that this mutant was the only one behaving dissimilarly in repression assays on the HSP70 promoter (16). As this seems specific for Cyclin B2 (see below), the effect was not clarified further. Similar coexpression experiments were conducted with the TAp63α versions; none of the vectors showed any effect on either the wt, or mutant Cyclin B2.
Figure 3.
ΔNp63α represses Cyclin B2, cdc2 and Topoisomerase α promoters. (A) Functional analysis of the Cyclin B2 promoters [wt and mutated in the three CCAAT boxes, see Ref. (25)] in SaoS2 cells, cotransfected with the ΔNp63α (left panel) and TAp63α (right panel) isoforms. The different AEC SAM domain mutants were cotransfected in parallel, both in the ΔN and TA configurations. (B) In the middle panels, the indicated wt and AEC ΔN and TA p63 constructs (wt, L518F and T537P) were transfected with TopoII α, wt and CCAAT-mutated promoters (left panel), or a Cdc2 promoter Luciferase plasmid (middle panel). (C) The activity of the Cyclin B2 promoter was tested with the p63 EEC C306R mutant, both in the ΔN and TA configurations. (D) The CCAAT-dependent Calreticulin promoter was cotransfected with ΔN and TA p63 expressing vectors. (E) Western blot analysis detailing the relative levels of the wt and p63 mutant proteins overexpressed, together with endogenous NF-Y to normalize protein levels.
Next, we performed similar experiments with other target promoters, known for their NF-Y-dependence and repression by p53: cdc2 and TopoII α (21,22). The former contains two CCAAT boxes, the latter five, although in the version we used here, only the three most proximal CCAAT were present. In both cases, we observed a clear repression by the ΔNp63α, but not by the TA isoform, nor by L518F or T537P, representative of the AEC mutants used above (Figure 3B). In the case of Topo II α, we also checked the transcription of a CCAAT-mutated construct; if anything, p63 showed modest positive effects, and no change was scored with the T537P. We then transfected Cyclin B2 with wt and the C306R EEC mutant (Figure 3C); surprisingly, the mutant showed a clear activation potential, both in the ΔN and TA configurations. The difference with wt was particularly striking for ΔN, switching a repressor into an activator.
To control for the specificity of these data, calreticulin, a single CCAAT promoter, was also tested; the p63 vectors had no effect on the levels of transcription (Figure 3D). Finally, we checked the expression levels of the transiently transfected p63 isoforms; as shown in Figure 3E, equal amounts of the wt and mutants were produced; NF-YB, used as control, was similarly unaffected. From these data, we conclude that the specific negative role of ΔNp63α on cell-cycle regulated promoters is dependent on (i) intact multiple CCAAT-boxes, and (ii) an intact p63 SAM domain.
NF-Y–p63 association
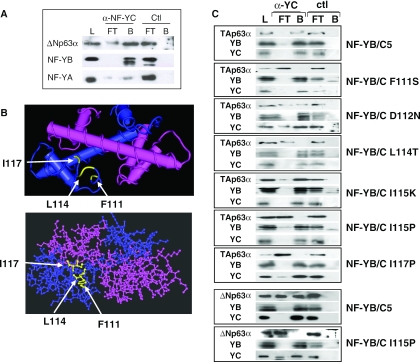
It was found recently that p53, and overexpressed p73 and p63 bind to NF-Y in vivo (20,31,36). To confirm these data in a physiological setting, we immunoprecipitated endogenous NF-Y and p63 from human primary keratinocytes with anti-NF-YC and control (anti-Flag) antibodies. The major isoform is, as in HaCaT, ΔNp63α. Western blot analysis with anti-p63, anti-YB and anti-YA antibodies revealed association in the NF-YC, but not control IPs (Figure 4A). We note that a consistently higher amount of NF-YB was immunoprecipitated, as compared with NF-YA, some of which remained in the unbound fraction. We then used NF-Y mutants to pinpoint the region of interaction on the NF-Y side. Extracts of cells in which TA (upper panels) or ΔN (lower panels) p63 was overproduced were incubated with recombinant NF-Y mutants. The HFM dimer was sufficient for p63 binding (data not shown); in particular, the NF-YC αC helix has been shown to be a docking site for NF-YA, MYC and, most relevantly here, p53/p73 (20,33,36). We therefore used single amino acid mutants in this NF-YC domain. We performed immunoprecipitations with anti-YC antibodies and western blot analysis with anti-YB, anti-YC and anti-p63. Figure 4C shows that all mutants are positive for anti-YC, as well as anti-YB, as expected, confirming that alteration of the αC has no effect on HFM dimer formation (37). However, several mutants were negative when probed with anti-p63: F111S, L114T and I117P. At the 115 position, an Isoleucine to Proline, but not to Lysine, mutation abolished binding. No effect on interactions was evident with a D112N mutation. No binding was scored when immunoprecipitating with a control antibody. Similar results were obtained with the ΔNp63α with NF-YB/YC and a representative I115P mutant (lower panels). These data indicate that the integrity of the NF-YC αC is crucial for p63 association, since Proline substitutions at positions 115 and 117 severely affect binding, and that hydrophobic residues are important, as both Phenylalanine 111 and Leucine 114 are required.
Figure 4.
In vivo NF-Y–p63 interactions. (A) Endogenous NF-Y and p63 were immunoprecipitated from human primary keratinocytes with anti-NF-YC and control (anti-Flag) antbodies. Load, flow through (FT) and bound (B) materials were loaded on SDS gels and assayed in western blot analysis with the indicated antibodies. (B) Structure of the NF-YC αC structure (37). The amino acids mutated in the analysis are indicated. (C) The indicated p63-TA or ΔN- were overproduced in transfections, and the NF-YC mutants indicated were produced in Escherichia coli, together with recombinant NF-YB, and assembled. Interactions were assayed in immunoprecipitations with in vivo produced wt TAp63α (upper panels with different NF-YC mutants) or ΔNp63α (lower two panels, with NF-YB/NF-YC and the NF-YC I115P mutant).
AEC mutants, but not an EEC mutant, fail to associate NF-Y
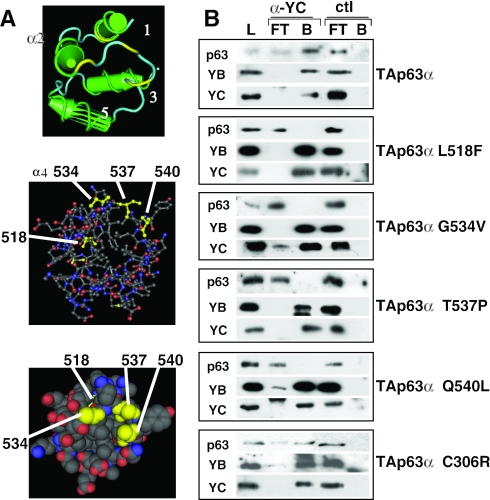
To map the protein–protein interactions on the p63 side, we coexpressed TAp63α and NF-Y in vivo and performed immunoprecipitations with an anti-NF-YC antibody. A robust interaction was evident (Figure 5B, upper panel), confirming the experiments shown above. We then coexpressed various p63 single amino acid mutants derived from AEC patients in the SAM domain (L518F, G534V, T537P and Q540L) and assayed them in immunoprecipitation: essentially none was capable to associate NF-Y. Interestingly, parallel coexpression of an EEC-derived mutant in the DNA-binding domain, C306R, was normal in NF-Y association (Figure 5B, lower panel). The expression of all these mutants was checked in western blot and all were equally abundant (data not shown and Figure 3). These data are in agreement with repression assays and we conclude that all SAM domain mutants, but not an EEC mutant, loose the capacity to interact with NF-Y in coexpression assays in vivo.
Figure 5.
NF-Y–p63 interactions: analysis of AEC p63 mutants. (A) Structure of the p73 SAM domain: the residues corresponding to AEC mutations in p63 are indicated. (B) Immunoprecipitation analysis of the different p63 (wt and mutants) transfected in SaoS2 cells with full-length NF-YB and NF-YC. Extracts were incubated with anti-YC and control anti-thioredoxin antibodies. Western blot analysis was performed with anti-p63 (upper panel; Santa Cruz H137), anti-YB and anti-YC (lower panels) antibodies. L, load; FT, flow through; B, bound material.
Differential recruitment of p63 mutants in vivo
The loss of repression capacity of the ΔNp63α AEC mutants lead us to verify whether the mutants were capable of interacting with the repressed promoters in vivo. We performed ChIPs by cotransfecting a wt Cyclin B2 Luciferase reporter and p63 expressing vectors in SaoS2 cells: both the wt and Q540L p63 were capable to associate to the reporter when compared with control ChIPs, in which an empty vector was cotransfected (Figure 6); the ΔN and TAp63α behaved similarly. Interestingly, the EEC C306R showed promoter association. On the other hand, neither the TA nor the ΔNp63α L518F associated with the transfected Cyclin B2 promoter. Although we only cotransfected p63, and not NF-Y vectors, binding of the latter was evident on the transfected Cyclin B2 Luciferase reporter, indicating the efficient association of the endogenous NF-Y (20). Thus, a negative effect on NF-Y binding as a possible cause of repression, was ruled out. In parallel, we also checked two endogenous promoters in the same ChIPs, Cyclin B2 and a bona fide p63 target, 14-3-3σ (18). That the latter is indeed a p63 target was confirmed with ChIPs performed in HaCaT cells (Figure 6, right panel). Interestingly, none of the AEC mutants was able to associate to the endogenous Cyclin B2 promoter, unlike the TA, ΔN and the EEC C306R mutants, in both configurations (Figure 6, middle panels). Conversely, all the AEC mutants were bound to 14-3-3σ, but not the C306R. In conclusion, the EEC C306R mutant is indeed defective in sequence-specific DNA-binding in vivo, as predicted by comparison with the corresponding p53 mutants isolated in human tumours, and shown with in vitro electrophoretic mobility shift assays (EMSAs) for the TAp63γ isoforms on p53 oligonucleotides (7). p63 AEC mutants normally bind DNA in a sequence-specific manner, but they are incapable to associate to CCAAT promoters. Under high, non-physiological levels of DNA targets, they behave somewhat dissimilarly with respect to promoter association: one, L518F, is totally incapable to promoter association, the other, Q540L, retaining this capacity only on transfected targets.
Repression of G2/M promoters following HaCaT differentiation
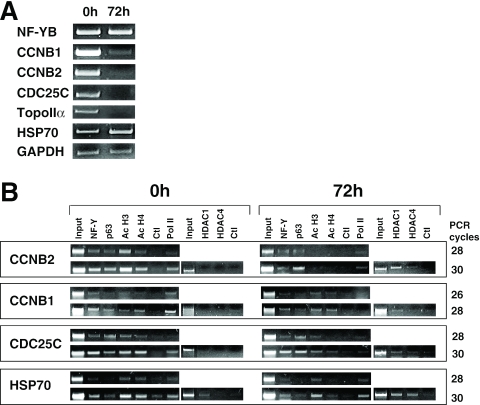
We showed recently that p53 and NF-Y are necessary for repression of G2/M genes following DNA damage. We used the HaCaT system to understand the role of p63 in differentiation. RT–PCR analysis indicates that Cyclin B1/B2, Topo II α and CDC25C mRNA levels dramatically decrease after 72 h in differentiation medium (Figure 7A), whereas the NF-YB mRNA was largely unchanged, as expected from experiments with other cell types (32). HSP70 and the internal control GAPDH also did not change (Figure 7A, lower panels). ChIP analysis were performed in growing and differentiated HaCaT with different antibodies; NF-Y and p63 were present before and after, whereas the levels of acetylation of histones H3 and H4 tails remarkably dropped in Cyclin B2 and CDC25C, less so in Cyclin B1 (Figure 7B). Consistent with this, HDAC1 was recruited at 72 h, more prominently on Cyclin B2 and CDC25; HDAC4 was also, albeit weakly, detectable (Figure 7B). On the heat inducible HSP70, a gene shown to be regulated by p63 (31), HDAC1 was observed in growing and differentiated cells. In keeping with promoter repression, RNA Pol II was completely absent on Cyclin B1 and decreased in CDC25C and Cyclin B2 (Figure 7B, right lanes). On HSP70, RNA Pol II, as well as NF-Y and p63, were present continuously. We notice a decrease in H4, but not H3, acetylation, which was not investigated further.
Figure 7.
Repression of G2/M genes upon HaCaT differentiation. (A) RT–PCR analysis of G2/M genes in growing (0 h) and differentiated HaCaT cells (72 h). The constitutive NF-YB and GAPDH transcripts served as internal controls. (B) ChIP analysis of Cyclin B2 promoter in undifferentiated and differentiated HaCaT cells with the indicated anti-YB, anti-p63 (4A4), anti-Acetyl-H3, anti-Acetyl-H4, anti-HDAC1, anti-HDAC4 and anti-Pol II antibodies.
Thus, consistent with a role in repression of growth regulating genes, the shut-off of promoter activity after differentiation is concomitant with p63, and NF-Y, remaining stably bound to the G2/M promoters, while repressive changes are installed.
DISCUSSION
Genotype–phenotype analysis of human syndromes, knockout experiments in mice, and genetic data in zebrafish unambiguously pointed to p63 as a master regulator of ectoderm and, more specifically, of multi-layered epithelia. Indeed, the ΔN isoform is a marker of keratinocytes stem cells. Extrapolating from our observations on p53, we find that p63 associates with NF-Y, residing on CCAAT-dependent G2/M promoters in vivo in growing and differentiated cells. Most importantly, it inhibits transcription, unlike its AEC missense derivatives. Loss of repression parallels the incapacity to contact NF-Y and to be recruited on Cyclin B2. Surprisingly, an opposite phenotype emerged for the DNA-binding mutant EEC, in terms of NF-Y-binding, promoter recruitment and transcriptional activation.
NF-Y–p53 versus NF-Y–p63 interactions
The requirement of NF-Y for repression of p53 family members is inferred from several findings. (i) The elimination of the CCAAT boxes leads to the abrogation of the negative effect of DNA-damaging agents, or p53/p73/p63 overexpression, on promoter function. (ii) In vivo ChIP experiments from our lab established that p53 is associated to G2/M promoters in NIH3T3 mouse fibroblasts, and it is activated through acetylation upon DNA-damage. Moreover, several control experiments strictly linked the presence of p53 to NF-Y bound promoters [(20) and references therein]. This picture is mirrored for p63 in the cells used here, HaCaT and primary keratinocytes, as representative of a tissue (skin) affected by p63 mutations in AEC patients and in knock-out mice. HaCaT cells harbour two DNA-binding mutant alleles of p53. In these cells, the ΔNp63α is by far the most abundant, if not the sole isoform present [(31) and Figure 1). Our ChIP data identify ΔNp63α associated to cell-cycle regulatory genes and confirm that p53 is also bound; importantly, this is observed both in immortalized and primary keratinocytes, indicating that this is not an epiphenomenon of immortalization. The PLK promoter, harbouring only one CCAAT, is devoid of ΔNp63 (and p53). Thus, CCAAT promoters are targeted not only by p53, but also by p63; the presence of multiple CCAAT boxes is important for p63 association. It is possible that p73, in the proper cellular context, behaves similarly, as evidenced by ChIP studies on the PDGF-β-R (43). In none of these promoters are there recognizable p53/p63-binding sequences, and the most obvious hypothesis is that recruitment is obtained through direct protein–protein interactions with NF-Y. Indeed, endogenous NF-Y and p63 do interact (Figure 4A). On the NF-Y side, the αC of the H2A-like NF-YC is required, a stretch clearly different from H2A, centered on the crucial, highly conserved Phenylalanine 111; a mutation of this residue abolishes binding to p63, as well as to NF-YA (37). The model resulting from the NF-YB-NF-YC crystal structure (37) suggests that multiple NF-Ys might predispose a peculiar, highly bent DNA structure for p63 association. This could reflect the tetrameric status of p63, needing multiple NF-Y docking sites to associate, therefore being limited to promoters, and cellular conditions, in which multiple CCAAT boxes are bound.
A simple explanation for promoter repression by p63 was that it directly inhibits binding of NF-Y to CCAAT. Jung et al. (22) observed a drastic reduction in NF-Y-binding capacity, as measured in vitro with EMSAs on the cdc2 promoter, in cells infected with adeno-p63. This is certainly not the case in ChIPs for p53 (20) and p73 (36); here, we confirm that even vast overexpressions of various p63 isoforms do not prevent endogenous NF-Y to associate to promoters in vivo (Figure 6). Indeed, endogenous NF-Y is readily capable to associate to a cotransfected promoter construct. Moreover, differentiated HaCaT have NF-Y bound to repressed promoters (Figure 7). Finally, we find that the effect of p63 overexpression on NF-Y binding is quite minute in in vitro EMSAs (B. Testoni and R. Mantovani, unpublished data). Therefore, we consider that inhibition of NF-Y DNA binding is not physiologically relevant to explain promoter repression. Rather, more complex alterations related to post-translational modifications, of p63 and NF-Y, and recruitment of corepressors are likely to be crucial.
p63 on growth regulating promoters: implications for the pathogenesis of the AEC syndromes
NF-Y is, to the best of our knowledge, the first transcription factor whose association is altered in p63 AEC missense mutants. Given its pivotal role for activation of growth-regulating genes, the lack of binding and promoter association of these mutants is relevant and could contribute to explain the pathogenesis of the skin alterations. The key to skin development is the α isoforms, ΔN (15) and TA (14). The ΔN isoform is a repressor of p53 activation. In both configurations, SAM domain mutations behave as dominant negatives on p53-activated transcription of artificial, as well as natural promoters (16,18). The p21 and 14-3-3σ promoters are repressed by ΔNp63α, and repression is only partially lost in AEC mutants (18). Here, we also find that the ΔN, but not TAp63α is capable to repress Cyclin B2, cdc2 and Topo II α promoters. The ΔN versus TA difference in repression could be explained by the findings of Serber et al. (44) that intramolecular interactions mask, in the TA isoforms, a repressive domain at the C-terminal of the protein present in the α, but not in the β or γ isoforms. In line with this, we find that the ΔN and TA isoforms are equally capable to bind NF-Y in IP analysis and to be recruited on Cyclin B2 in ChIPs (Figures 4 and 6); therefore the lack of repression by TA must be due to intrinsic properties of the extra N-terminal amino acids and not to lack of promoter recruitment.
The ChIP recruitment assays yielded several intriguing findings. First, the lack of association of the EEC C306R mutant to the 14-3-3σ promoter, a bona fide p63 target containing p53 target sequences, confirms recent data obtained with the TAp63γ in in vitro EMSAs (7) and it is a proof that indeed this is a DNA-binding defective mutant in vivo. In fact, it involves a residue that is predicted to be required for DNA binding and corresponds to one of the hotspots of mutations in DNA-binding defective, tumour derived p53 analogues (5). Second, the normal recruitment of C306R on Cyclin B2 is a further indication that direct DNA binding to p53/p63 target sequences is not required for Cyclin B2 association. Third, the two AEC mutants tested behave slightly differently in promoter recruitment assays. Note that both the L518F and Q540L are incapable to associate to NF-Y in solution (Figure 5), indicating a decrease in affinity for NF-Y. Q540L involves a residue predicted to be on the surface of the SAM domain (4,6); it is recruited in vivo, but only on cotransfected high-copy plasmids. Under the same conditions, the L518F mutant is negative; this latter mutation is predicted to deeply affect the overall structure of the SAM domain (6). In general, the lack of NF-Y binding of AEC mutants is in accordance with the lack of recruitment and the lack of repression (Figures 3 and 5). Fourth, AEC mutants have a normal DNA-binding domain and they are perfectly capable to associate 14-3-3σ; hence, the negative data in Cyclin B2 ChIP assays reinstate that binding is not elicited through a specific DNA element within the promoter. Finally, an unexpected finding emerging from the repression assays was that the EEC C306R not only lost this capacity (Figure 3), but even activated transcription, behaving as a gain of function mutant. This establishes that AEC and EEC have opposite transcriptional effects in terms of CCAAT-containing promoters. Interestingly, this is also the case for p53 tumour-derived mutants in the DNA-binding domain (S. d'Agostino, G. Blandino and G. Piaggio, personal communication).
The fundamental question is what is the role of p63 association to G2/M promoters? A hint might come from the p53 experiments. DNA-damage rapidly ‘activates’ p53 on repressed promoters, as checked by p300 and PCAF-mediated acetylation, triggering a set of events leading to HDACs recruitment, histones deacetylation, coactivators release and promoter shut-off (20). Similarly, p73-mediated recruitment of HDACs was reported on PDGF-β-R (43). The p53 pathway is physiologically linked to genotoxic stress, in as much as the involvement of p63 and p73 in response to DNA-damage has been detailed (45–47), genetic data point to p63 as an ectodermal regulator and a master gene of stem cells of multilayered epithelia, at the crossroad between self-renewal and differentiation. The RT–PCR and ChIP results of differentiated HaCaT, in which repressed promoters are associated by p63 and by the NF-Y activator (Figure 7), strongly suggest that p63 is permissive for transcription in growing cells, but behaves as a repressor of key cell-cycle controlling genes in differentiated cells. In selected cellular contexts, it would act as a sensor and transducer of developmental and environmental signals of differentiation versus growth. If the p53 paradigm is followed, we should expect to find a plethora of yet unknown post-translational modifications in p63, and possibly in NF-Y, regulating the access of coactivators and corepressors to different classes of promoters in different growth conditions. The mechanistic details of this will only be understood when such modifications, and their effects on the protein structure, will be available. One intriguing possibility is that the subversion of the repressive, on cell-cycle genes, or activating, on growth arrest genes, behaviours by AEC mutations favour unrestricted growth of keratinocytes stem cells, resulting in an early wasting of their proliferative potential. Indeed, colony formation assays support the idea that AEC mutants lose the capacity to suppress growth (16). Finally, it is important to reinstate that ΔNp63α activates specific set of genes through additional activation domains [(16,19) and B. Testoni and R. Mantovani, manuscript in preparation]. The panel of validated bona fide p63 targets in keratinocytes, as observed not only in overexpression assays, but most importantly, in in vivo ChIPs experiments, is still extremely skinny (48–51). A full understanding of the molecular mechanisms underlying the AEC mutant phenotype will have to await a considerable lengthening of this list, and the functional analysis of AEC-derived p63 on their expression.
Acknowledgments
We wish to thank M. Broggini and G. Piaggio for gift of reagents, H. Van Bokhoven for the p63 plasmids. We thank L. Guerrini for many helpful discussions. This work is supported by grants from Telethon, Fondazione Cariplo and FIRST to R.M. Funding to pay the Open Access publication charges for this article was provided by Fondazione Cariplo.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blandino G., Dobbelstein M. p73 and p63: why do we still need them? Cell Cycle. 2004;2:886–894. [PubMed] [Google Scholar]
- 2.Westfall M.D., Pietenpol J.A. p63: Molecular complexity in development and cancer. Carcinogenesis. 2004;6:857–864. doi: 10.1093/carcin/bgh148. [DOI] [PubMed] [Google Scholar]
- 3.Schultz J., Ponting C.P., Hofmann K., Bork P. SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 1997;6:249–253. doi: 10.1002/pro.5560060128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chi S-W., Ayed A., Arrowsmith C.A. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 1999;18:4438–4445. doi: 10.1093/emboj/18.16.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunner H.G., Hamel B.C., van Bokhoven H.H. p63 gene mutations and human developmental syndromes. Am. J. Med. Genet. 2002;112:284–290. doi: 10.1002/ajmg.10778. [DOI] [PubMed] [Google Scholar]
- 6.McGrath J.A., Duijf P.H., Doetsch V., Irvine A.D., de Waal R., Vanmolkot K.R., Wessagowit V., Kelly A., Atherton D.J., Griffiths W.A., et al. Hay–Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum. Mol. Genet. 2001;10:1–9. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- 7.Ying H., Chang D.L., Zheng H., McKeon F., Xiao Z.X. DNA-binding and transactivation activities are essential for TAp63 protein degradation. Mol. Cell. Biol. 2005;25:6154–6164. doi: 10.1128/MCB.25.14.6154-6164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills A.A., Zheng B., Wang X-J., Vogel H., Roop D.R., Bradley A. p63 is a homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 9.Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C., et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 10.Bakkers J., Hild M., Kramer C., Furutani-Seiki M., Hammerschmidt M. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev. Cell. 2002;2:617–627. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- 11.Lee H., Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev. Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- 12.Pellegrini P., Dell'Ambra E., Molisano O., Martinelli E., Fantozzi I., Bondanza S., Ponzin D., McKeon F., De Luca M. p63 identifies keratinocyte stem cells. Proc. Natl Acad. Sci. USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Iorio E., Barbaro V., Ruzza A., Ponzin D., Pellegrini G., De Luca M. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc. Natl. Acad. Sci. USA. 2005;102:9523–9528. doi: 10.1073/pnas.0503437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koster M.I., Roop D.R. Transgenic mouse models provide new insights into the role of p63 in epidermal development. Cell Cycle. 2004;3:411–413. [PubMed] [Google Scholar]
- 15.McKeon F. p63 and the epithelial stem cell: more than status quo? Genes Dev. 2004;18:465–469. doi: 10.1101/gad.1190504. [DOI] [PubMed] [Google Scholar]
- 16.Ghioni P., Bolognese F., Duijf P.H., Van Bokhoven H., Mantovani R., Guerrini L. Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol. Cell. Biol. 2002;22:8659–8668. doi: 10.1128/MCB.22.24.8659-8668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishi H., Senoo M., Nishi K.H., Murphy B.A., Rikyiama T., Matsumara Y., Habu S., Johnson A.C. p53 homologue p63 represses epidermal growth factor receptor gene expression. J. Biol. Chem. 2001;276:41717–41724. doi: 10.1074/jbc.M101241200. [DOI] [PubMed] [Google Scholar]
- 18.Westfall M.D., Mays D.J., Sniezek J.C., Pietenpol J.A. The ΔNp63α phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay–Wells syndrome-derived mutations. Mol. Cell. Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu G., Nomoto S., Hoque M.O., Dracheva T., Osada M., Lee C.C., Dong S.M., Guo Z., Benoit N., Cohen Y., et al. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 2003;63:2351–2357. [PubMed] [Google Scholar]
- 20.Imbriano C., Gurtner A., Cocchiarella F., Di Agostino S., Basile V., Gostissa M., Dobbelstein M., Del Sal G., Piaggio G., Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol Cell. Biol. 2005;25:3737–51. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi A., A, Wu Z., Reed R.F., Suttle D.P. Nuclear factor-Y binding to the topoisomerase II alpha promoter is inhibited by both the p53 tumor suppressor and anticancer drugs. Mol. Pharmacol. 2003;63:359–367. doi: 10.1124/mol.63.2.359. [DOI] [PubMed] [Google Scholar]
- 22.Jung M.S., Yun J., Chae H.D., Kim J.M., Kim S.C., Choi T.S., Shin D.Y. p53 and its homologues, p63 and p73, induce a replicative senescence through inactivation of NF-Y transcription factor. Oncogene. 2001;20:5818–5825. doi: 10.1038/sj.onc.1204748. [DOI] [PubMed] [Google Scholar]
- 23.Krause K., Wasner M., Reinhard W., Haugwitz U., Dohna C.L., Mossner J., Engeland K. The tumour suppressor protein p53 can repress transcription of cyclin B. Nucleic Acids Res. 2000;28:4410–4418. doi: 10.1093/nar/28.22.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krause K., Haugwitz U., Wasner M., Wiedmann M., Mossner J., Engeland K. Expression of the cell cycle phosphatase cdc25C is down-regulated by the tumor suppressor protein p53 but not by p73. Biochem. Biophys. Res. Commun. 2001;284:743–750. doi: 10.1006/bbrc.2001.5040. [DOI] [PubMed] [Google Scholar]
- 25.Manni I., Mazzero G., Gurtner A., Mantovani R., Haugwitz U., Krause K., Engeland K., Sacchi A., Soddu S., Piaggio G. NF-Y mediates the transcriptional inhibition of the cyclin B1, cyclin B2, and Cdc25C promoters upon induced G2 arrest. J. Biol. Chem. 2001;276:5570–5576. doi: 10.1074/jbc.M006052200. [DOI] [PubMed] [Google Scholar]
- 26.Matsui T., Katsuno Y., Inoue T., Fujita F., Joh T., Niida H., Murakami H., Itoh M., Nakanishi M. Negative regulation of Chk2 expression by p53 is dependent on the CCAAT-binding transcription factor NF-Y. J. Biol. Chem. 2004;279:25093–25100. doi: 10.1074/jbc.M403232200. [DOI] [PubMed] [Google Scholar]
- 27.St Clair S., Giono L., Varmeh-Ziaie S., Resnick-Silverman L., Liu W.J., Padi A., Dastidar J., DaCosta A., Mattia M., Manfredi J.J. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: one involves direct binding to the cdc25C promoter. Mol. Cell. 2004;16:725–736. doi: 10.1016/j.molcel.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Yun J., Chae H.D., Choy H.E., Chung J., Yoo H.S., Han M.H., Shin D.Y. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J. Biol. Chem. 1999;274:29677–29682. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y., Mehta K.R., Choi A.P., Scolavino S., Zhang X. DNA damage-induced inhibition of securin expression is mediated by p53. J. Biol. Chem. 2003;278:462–470. doi: 10.1074/jbc.M203793200. [DOI] [PubMed] [Google Scholar]
- 30.Innocente S.A., Lee J.M. p73 is a p53-independent, Sp1-dependent repressor of cyclin B1 transcription. Biochem. Biophys. Res. Commun. 2005;329:713–718. doi: 10.1016/j.bbrc.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 31.Wu G., Osada M., Guo Z., Fomenkov A., Begum S., Zhao M., Upadhyay S., Xing M., Wu F., Moon C., et al. DeltaNp63alpha up-regulates the Hsp70 gene in human cancer. Cancer Res. 2005;65:758–766. [PubMed] [Google Scholar]
- 32.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 33.Kim I.S., Sinha S., de Crombrugghe B., Maity S.N. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol. Cell. Biol. 1996;16:4003–4013. doi: 10.1128/mcb.16.8.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caretti G., Salsi V., Vecchi C., Imbriano C., Mantovani R. Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J. Biol. Chem. 2003;278:30435–30440. doi: 10.1074/jbc.M304606200. [DOI] [PubMed] [Google Scholar]
- 36.Hackzell A., Uramoto H., Izumi H., Kohno K., Funa K. p73 independent of c-Myc represses transcription of platelet-derived growth factor beta-receptor through interaction with NF-Y. J. Biol. Chem. 2002;277:39769–39776. doi: 10.1074/jbc.M204483200. [DOI] [PubMed] [Google Scholar]
- 37.Romier C., Cocchiarella F., Mantovani R., Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- 38.Waltermann A., Kartasheva N.N., Dobbelstein M. Differential regulation of p63 and p73 expression. Oncogene. 2003;22:5686–5693. doi: 10.1038/sj.onc.1206859. [DOI] [PubMed] [Google Scholar]
- 39.Breitkreutz D., Stark H.J., Plein P., Baur M., Fusenig N.E. Differential modulation of epidermal keratinization in immortalized (HaCaT) and tumorigenic human skin keratinocytes (HaCaT-ras) by retinoic acid and extracellular Ca2+ Differentiation. 1993;54:201–217. doi: 10.1111/j.1432-0436.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 40.Chaturvedi V., Qin J.Z., Denning M.F., Choubey D., Diaz M.O., Nickoloff B.J. Apoptosis in proliferating, senescent, and immortalized keratinocytes. J. Biol Chem. 1999;274:23358–23367. doi: 10.1074/jbc.274.33.23358. [DOI] [PubMed] [Google Scholar]
- 41.Brauninger A., Strebhardt K., Rubsamen-Waigmann H. Identification and functional characterization of the human and murine polo-like kinase (Plk) promoter. Oncogene. 1995;11:1793–1800. [PubMed] [Google Scholar]
- 42.Bergamaschi D., Samuels Y., Jin B., Duraisingham S., Crook T., Lu X. ASPP1 and ASPP2: common activators of p53 family members. Mol. Cell. Biol. 2004;24:1341–1350. doi: 10.1128/MCB.24.3.1341-1350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uramoto H., Wetterskog D., Hackzell A., Matsumoto Y., Funa K. p73 competes with co-activators and recruits histone deacetylase to NF-Y in the repression of PDGF beta-receptor. J. Cell Sci. 2004;117:5323–5331. doi: 10.1242/jcs.01384. [DOI] [PubMed] [Google Scholar]
- 44.Serber Z., Lai H.C., Yang A., Ou H.D., Sigal M.S., Kelly A., E, Darimont B.D., Duijf P.H., Van Bokhoven H., McKeon F., et al. A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol. Cell. Biol. 2002;22:8601–8611. doi: 10.1128/MCB.22.24.8601-8611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liefer K.M., Koster M.I., Wang X.J., Yang A., McKeon F., Roop D.R. Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res. 2000;60:4016–4020. [PubMed] [Google Scholar]
- 46.Flores E.R., Tsai K.Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 47.Flores E.R., Sengupta S., Miller J.B., Newman J.J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 48.Ellisen L.W., Ramsayer K.D., Johannessen C.M., Yang A., Beppu H., Minda K., Oliner J.D., McKeon F., Haber D.A. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 49.Gressner O., Schilling T., Lorenz K., Schulze Schleithoff E., Koch A., Schulze-Bergkamen H., Maria Lena A., Candi E., Terrinoni A., Catani M., et al. TAp63alpha induces apoptosis by activating signaling via death receptors and mitochondria. EMBO J. 2005;24:2458–2471. doi: 10.1038/sj.emboj.7600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ihrie R.A., Marques M.R., Nguyen B.T., Horner J.S., Papazoglu C., Bronson R.T., Mills A-A., Attardi L-D. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120:843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 51.Barbieri C.E., Perez C.A., Johnson K.N., Ely K.A., Billheimer D., Pietenpol J.A. IGFBP-3 is a direct target of transcriptional regulation by DeltaNp63alpha in squamous epithelium. Cancer Res. 2005;65:2314–2320. doi: 10.1158/0008-5472.CAN-04-3449. [DOI] [PubMed] [Google Scholar]