Abstract
The envelope protein (Env) of human immunodeficiency virus type 1 forms homo-oligomers in the endoplasmic reticulum. The oligomeric structure of Env is maintained after cleavage in a Golgi compartment and transport to the surfaces of infected cells, where incorporation into budding virions takes place. Here, we use biophysical techniques to assess the oligomeric valency of virion-associated Env prior to fusion activation. Virion-associated Env oligomers were stabilized by chemical cross-linking prior to detergent extraction and were purified by immunoaffinity chromatography. Gel filtration revealed a single predominant oligomeric species, and sedimentation equilibrium analysis-derived mass values indicated a trimeric structure. Determination of the masses of individual Env molecules by scanning transmission electron microscopy demonstrated that virion-associated Env was trimeric, and a triangular morphology was observed in 20 to 30% of the molecules. These results, which firmly establish the oligomeric structure of human immunodeficiency virus virion-associated Env, parallel those of our previous analysis of the simian immunodeficiency virus Env.
The envelope protein (Env) of human immunodeficiency virus type 1 (HIV-1) mediates functions that are critical to the viral life cycle, including viral attachment to target cells and the fusion of viral and cellular membranes. The Env precursor gp160 is cotranslationally translocated into the luminal endoplasmic reticulum, where essential maturation steps, such as N-linked glycosylation, disulfide-bond formation, and oligomerization, occur (11). Cleavage in the Golgi complex by a cellular protease prior to egress to the cell surface is also requisite for viral infectivity (23). The products of cleavage, the surface subunit gp120 and the transmembrane (TM) subunit gp41, remain associated through noncovalent interactions. A TM domain within gp41 anchors an oligomeric complex of gp41 and gp120 to the surfaces of infected cells and, after budding takes place, to the surfaces of virions. Thus, the functional form of Env is oligomeric. Furthermore, oligomeric structure influences the antigenicity of Env, which is the main target of the neutralizing humoral immune response of the host. The virus-neutralizing capacity of monoclonal antibodies (MAb) derived from infected humans is predicted by binding to cell-associated oligomeric Env but not by binding to monomeric Env (15, 25).
Env displays considerable conformational flexibility. Sequential interactions between gp120 and the cell surface receptors CD4 and one of the members of the chemokine receptor family trigger conformational changes in both gp120 and gp41. These changes are believed to lead to the insertion of the gp41 N-terminal fusion peptides into the target cell membrane and ultimately to membrane fusion between the infected cell or virion and the target cell (reviewed in references 12 and 35). Crystallographic analysis revealed that upon mixing of peptides corresponding to predicted gp41 N- and C-terminal α-helical regions, a triple-stranded coiled-coil core of N-terminal peptides with three C-terminal peptides packed into grooves on the outer surface of the coiled-coil in an antiparallel orientation was formed (7, 32). The antiparallel packing places the N termini of the N-terminal helices and the C terminal of the C-terminal helices at the same end of a rod-like structure. This arrangement implies that the N-terminally located fusion peptides and C-terminally located TM domains (neither of which was present in the crystallized fragments) are in close proximity. This structure is believed to represent the conformation of gp41 after the fusion process (postfusion conformation), with the fusion peptides and TM domains being located in the same membrane.
The nature of the oligomeric structure of HIV-1 Env in the native conformation prior to fusion activation is uncertain. Biochemical approaches employing chemical cross-linking and sucrose gradient sedimentation revealed that recombinant gp160 forms dimers and higher-order oligomers (8, 10). Soluble uncleaved recombinant Env lacking the TM domain and cytoplasmic tail (gp140) has also been suggested to form dimers and tetramers (9) or dimers and trimers (13, 27). In contrast, gp140 devoid of complex carbohydrate groups was reported to form a relatively homogeneous population of trimers (37). Recombinant gp120 dimers have been described previously (5), while gp120 derived from recombinant gp160 has been shown to be dimeric under some conditions (24) but monomeric under others (10, 29). Virion-derived gp120 trimers have also been reported (30). The differences in the oligomeric states reported may be due to the difficulties in resolving large cross-linked glycoproteins by electrophoresis or, in the case of recombinant proteins, may reflect actual differences in oligomeric states resulting from the analysis of truncated or mutated species and the use of different expression systems. We have recently shown that virion-associated Env of another primate immunodeficiency virus, simian immunodeficiency virus (SIV), is trimeric (6). In the present study, we have used parallel biochemical and biophysical strategies to determine the oligomeric state of Env on the surfaces of HIV-1 virions.
Virion-associated HIV-1 Env purifies as one predominant oligomeric species.
HIV-1MN virions were purified by sucrose density gradient sedimentation and treated with 2,2′-dithiodipyridine, which blocks infectivity but does not compromise the structural or functional integrity of Env (26). In order to assess the quaternary structure of gp120/gp41 complexes on the virion surface, chemical cross-linking was performed prior to virion lysis to preserve the oligomeric contacts, which are otherwise readily disrupted by detergent (10). Virions (33.3 mg of total virion protein in a 46-ml volume of phosphate-buffered saline [PBS]) were cross-linked with a final concentration of 1 mM 3,3′-dithiobis(sulfosuccinimidyl propionate) (DTSSP; Pierce), a thiol-cleavable, amine-reactive cross-linker, as previously described (6). Virions were then lysed by the addition of hydrogenated Triton X-100 (TX-100h) (Calbiochem) at a final concentration of 1%. Env was purified from the lysate by immunoaffinity chromatography with anti-gp41 cytoplasmic domain MAb C8 (1) which had been coupled to CNBr-activated Sepharose (Amersham Pharmacia Biotech AB) according to the manufacturer's recommendations. After the lysate and Sepharose-coupled C8 were mixed overnight at 4°C and for 2 h at room temperature and then washed extensively with PBS-0.1% TX-100h, bound protein was eluted with 100 mM glycine-0.1% TX-100h-0.02% sodium azide (pH 2.8), followed by immediate neutralization. Concentrated eluate was subjected to gel filtration chromatography using a Superdex 200 column (Amersham Pharmacia Biotech AB) with PBS-0.1% TX-100h-0.02% sodium azide as the buffer (6).
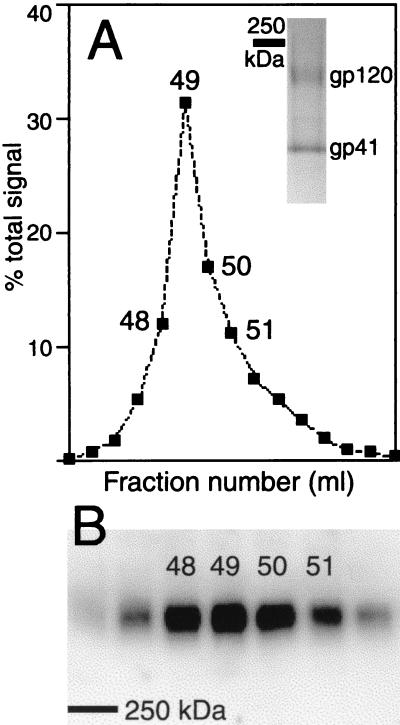
Aliquots of individual gel filtration fractions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in the presence of reducing agent (DTSSP cross-links broken) and immunoblotted with the gp120-specific MAb D19 (9), and the results were expressed for each fraction as a percentage of the total Env-specific signal. Env resolved as a symmetrical peak, with most protein eluting at 48 to 51 ml (Fig. 1A). SDS-PAGE analysis of a pool of fractions 48 to 51 under reducing conditions followed by Coomassie blue staining revealed the presence of bands corresponding to gp120 and gp41 (Fig. 1A, inset). A faint band of contaminating protein (between the gp120 and gp41 bands) constituting less than 5% of the total protein was also detected. Immunoblotting of nonreduced, DTSSP-cross-linked proteins (Fig. 1B) showed that Env had a lower electrophoretic mobility than a 250-kDa mass standard, in contrast to monomeric gp120, which migrated more rapidly than the 250-kDa mass standard (based on SDS-PAGE analysis of reduced Env) (Fig. 1A, inset), suggesting that Env was oligomeric. Furthermore, all cross-linked Env migrated to the same position, indicating that Env occurs as one predominant oligomeric species. Analysis of concentrated gel filtration fractions (data not shown) indicated the presence of a small amount of Env in fractions 60 to 62, consistent with dissociation or partial dissociation of some oligomers.
FIG. 1.
Gel filtration analysis of virion-derived HIV-1 Env. (A) Aliquots of gel filtration fractions were subjected to SDS-8% PAGE in the presence of reducing agent (cross-links broken) and sequentially immunoblotted with an HIV-1 gp120-specific MAb, a mouse immunoglobulin G-specific rabbit polyclonal serum, and iodinated protein A. gp120 was quantified by phosphor-screen autoradiography. Blue Dextran 2000 (giving the void volume) had an elution peak in fraction 43. (Inset) Pool of fractions 48 to 51 analyzed by SDS-4 to 20% PAGE in the presence of reducing agent and Coomassie blue staining. (B) Aliquots of gel filtration fractions analyzed by SDS-5% PAGE in the absence of reducing agent (cross-links maintained) and immunoblotted as described above. The bars in panels A (inset) and B indicate the electrophoretic mobility of a 250-kDa marker protein.
HIV-1 Env in the prefusion activated conformation is trimeric.
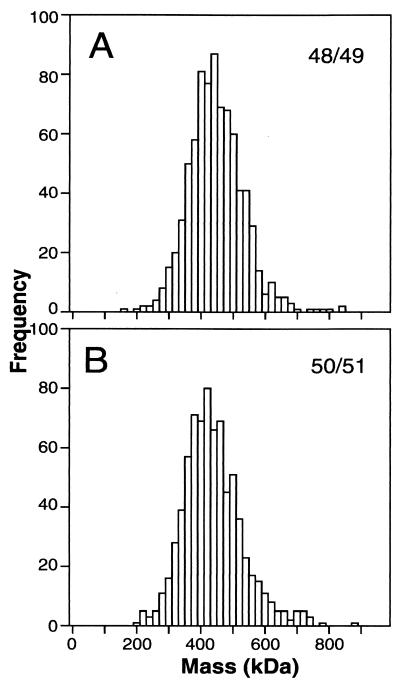
Gel filtration analysis has been used to estimate the mass of globular proteins by comparison with calibration standards; however, our previous studies with HIV-1 gp120 and SIV Env suggest that Env is nonglobular, precluding mass determination by this method (5, 6). Scanning transmission electron microscopy (STEM) (19) and sedimentation equilibrium analysis are biophysical methods that allow shape-independent mass determination. Env and tobacco mosaic virus were sequentially absorbed onto copper-mesh-supported carbon grids in 5-μl aliquots at a concentration of approximately 0.02 to 0.05 and 0.4 mg/ml, respectively, and STEM determination of Env mass was performed as previously described (6). Mass values were calibrated using tobacco mosaic virus particles contained in the same field as the Env molecules. Examination of a histogram of the measured masses of 795 Env molecules within a pool of fractions 48 and 49 revealed a peak with a Gaussian distribution (Fig. 2A). The mean of this distribution was 457 kDa with a standard deviation of 86 kDa (Table 1). Similarly, a histogram of the measured masses of 753 Env molecules within a pool of fractions 50 and 51 demonstrated a Gaussian distribution with a mean of 448 kDa and a standard deviation of 92 kDa (Fig. 2B; Table 1). These data are highly consistent with the expected mass of an Env trimer (441.9 kDa). The heterogeneity of glycosylation and random experimental error due to the low dosage used to generate STEM images may have contributed to the standard deviation. The bound cross-linker DTSSP (molecular weight, 608.51) is not expected to greatly add to the total mass values obtained. STEM analysis of a second independently purified preparation of virion-associated HIV-1 Env gave a mass value of 469 kDa for a pool of fractions 48 to 50 (data not shown). In approximately 20 to 30% of the STEM images of individual Env molecules, an apparent triangular morphology was observed (Fig. 3). This morphology is similar to that observed previously for the SIV Env of intact virions, purified-virion-derived SIV Env trimers, and SIV gp140 trimers (6, 16).
FIG. 2.
Mass distribution of virion-derived HIV-1 Env determined by quantitative analysis of STEM images. Shown are the results of an analysis of 795 molecules within a pool of gel filtration fractions 48 and 49 (A) and 753 molecules within a pool of fractions 50 and 51 (B).
TABLE 1.
Masses of virion-derived HIV-1 Env fractions
| Env fraction | Mass [kDa (no. of subunitsa)] measured by:
|
|
|---|---|---|
| Sedimentation equilibrium (wt avg) | STEM (no. avg) | |
| 48 | 488.5 (3.32) | 457 (3.10) |
| 49 | 525.7 (3.57) | |
| 50 | 459.9 (3.12) | 448 (3.04) |
| 51 | 471.9 (3.20) | |
The mass of one subunit (147.3 kDa) was estimated by summing the protein mass deduced from the amino acid sequence and a value of 1.739 kDa for each potential N-linked glycosylation site (see the text regarding sedimentation equilibrium analysis).
FIG. 3.
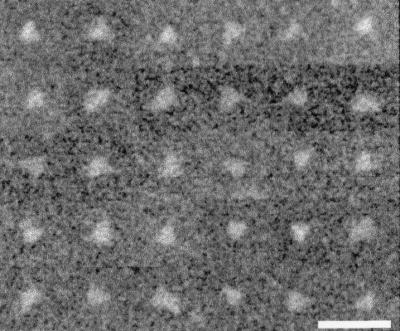
Montage of STEM images of virion-derived HIV-1 Env molecules exhibiting triangular morphology. Depicted are images from pooled gel filtration fractions 48 and 49. Smoothing was used to reduce pixelation. Bar = 50 nm.
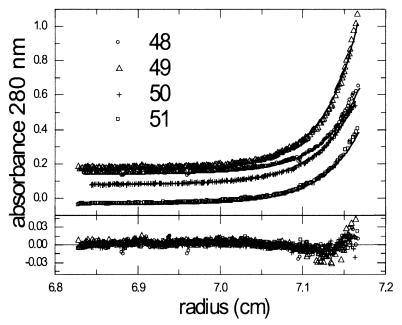
Weight-average molecular weights of Env within individual gel filtration fractions were determined by sedimentation equilibrium with an Optima XL-A/I analytical ultracentrifuge (Beckman). Cells were loaded with 120- to 135-μl sample volumes at a final concentration of 0.1 to 0.5 mg/ml. Sedimentation equilibrium absorbance data sets (wavelength, 280 nm) were measured at four different rotor speeds (7,000, 8,500, 10,000, and 12,000 rpm) at a rotor temperature of 10°C. The potential contribution of Env-associated TX-100h to the mass of Env was eliminated by matching the solvent density to the density of TX-100h by using a final concentration of 8.43% (wt/vol) sucrose, as previously described (6). Mass spectral analysis of HIV-1JRFL gp120 (cleaved from recombinant gp140; D. Sheeley and R. Center, unpublished data) allowed for the determination of the average carbohydrate molar mass contribution of each potential N-linked glycosylation site (1.739 kDa). This value is in close agreement with the average carbohydrate molar mass values for each potential site that were calculated using the mass spectral analyses of recombinantly derived, mammalian-cell-expressed gp120s of the BH8 (P. Earl and K. Parker, unpublished data), SF2 (3), and GB8 (17) strains (range, 1.68 to 1.78 kDa per site). The 1.739-kDa value was used to estimate the carbohydrate molar mass of virion-derived HIV-1 Env and therefore the weight fractions of protein (0.634) and carbohydrate (0.366). A partial specific volume (vbar) value of 0.622 ml/g was used for the carbohydrate weight fraction (20), and the protein vbar was calculated from the amino acid sequence to be 0.732 ml/g. The vbar of Env was determined by the formula 0.634 (0.732 ml/g) + 0.366 (0.622 ml/g) = 0.692 ml/g. Sedimentation equilibrium absorbance versus radial-position data sets (Fig. 4) were analyzed by global nonlinear regression analysis with the data analysis software package provided by Beckman-Coulter Instruments (version 4.0 and Microcal version 4.1) to give the mass values and calculated numbers of subunits shown in Table 1. The sedimentation equilibrium analysis-derived mean number of subunits for the four fractions is 3.30, slightly higher than both the STEM-derived mean number of subunits for the same fractions (3.07) and the expected value for trimers. This result could be explained by the presence of traces of oligomers with more than three subunits, as such molecules would contribute more significantly to the weight-average, molar mass measured at sedimentation equilibrium than to the number-average mass values obtained by STEM. In fact, a sedimentation coefficient distribution analysis of the sedimentation velocity boundary for each fraction showed a predominant peak at approximately 9S and a small peak (less than 10% of the total) in the 14S region (data not shown). The latter minor component is consistent with a low level of intertrimer cross-linking. Sedimentation equilibrium analysis performed on a second independently purified preparation of virion-associated HIV-1 Env gave a mass value for a pool of fractions 48 to 50 of 441.1 kDa (2.99 subunits [data not shown]).
FIG. 4.
Sedimentation equilibrium concentration profiles of individual gel filtration fractions of virion-derived HIV-1 Env. Solid lines show the best-fit distributions after global modeling of data obtained at four different rotor speeds. For clarity, only data obtained at 7,000 rpm are shown. The lower panel shows the residuals of the fitted lines to the experimental data.
In summary, the data from two independent biophysical techniques clearly indicated that HIV-1 gp120/gp41 complexes on the virion surface exist as trimers. Trimeric structure was also seen with SIV virion-associated Env prior to fusion activation (6) and in the crystal-derived structures of N- and C-terminal helical domains of HIV-1 gp41 and the SIV TM subunit, which are believed to represent the postfusion conformation (7, 21, 32, 36). Indeed, trimeric structure is a recurring theme in viral envelope protein biology. Crystal-derived structures of the Env TM subunits of human T-cell leukemia virus type 1 (18), Moloney murine leukemia virus (14), visna virus (22), Ebola virus (31), simian parainfluenza virus 5 (2), human respiratory syncytial virus (38), and influenza virus (4) are remarkably similar, all having a trimeric coiled-coil core. In the case of influenza virus hemagglutinin, a crystal-derived structure of the prefusion activated conformation also demonstrated a trimeric structure (34). It seems likely that functionally conserved aspects of Env-mediated membrane fusion underlie a common trimeric structural plan. However, the maintenance of a single Env oligomeric valency throughout the fusion process is not universal among enveloped viruses. For example, tick-borne encephalitis virus Env is a homodimer in the prefusion conformation and undergoes a transition to a homotrimer during fusion activation (28).
The homogeneous nature of the quaternary state of virion-associated HIV-1 Env that we observed contrasts with the oligomeric heterogeneity reported for recombinant, uncleaved forms of Env, such as membrane-anchored gp160 (8, 10) or soluble, secreted gp140 (9, 13, 27). High expression levels associated with recombinant systems and lack of membrane anchoring in the case of gp140 may induce aberrant quaternary forms not seen in natural infection. Alternatively, it is possible that heterogeneous quaternary structure may be a normal consequence of Env expression and that cellular mechanisms, which normally prevent nontrimeric forms from being exported to the cell surface, fail in the context of recombinant expression. Consistent with this suggestion, it has been reported that most of the gp160 expressed in infected cells is routed to lysosomes rather than the cell surface (33). Regardless of whether the uniformity of virion-associated envelope quaternary structure is controlled at the level of initial oligomerization in the endoplasmic reticulum or by subsequent cellular quality control mechanisms, it appears certain that trimerization is critical for Env function.
Acknowledgments
We thank G. Stubbs of Vanderbilt University for providing tobacco mosaic virus, D. Sheeley for mass spectral analysis, and P. Schuck for helpful discussions and manuscript review.
This work was supported in part by a National Institutes of Health Intramural AIDS Targeted Antiviral Program grant.
REFERENCES
- 1.Abacioglu, Y. H., T. R. Fouts, J. D. Laman, E. Claassen, S. H. Pincus, J. P. Moore, C. A. Roby, R. Kamin-Lewis, and G. K. Lewis. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retrovir. 10:371-381. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Borchers, C., and K. B. Tomer. 1999. Characterization of the noncovalent complex of human immunodeficiency virus glycoprotein 120 with its cellular receptor CD4 by matrix-assisted laser desorption/ionization mass spectrometry. Biochemistry 38:11734-11740. [DOI] [PubMed] [Google Scholar]
- 4.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 5.Center, R. J., P. L. Earl, J. Lebowitz, P. Schuck, and B. Moss. 2000. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J. Virol. 74:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Center, R. J., P. Schuck, R. D. Leapman, L. O. Arthur, P. L. Earl, B. Moss, and J. Lebowitz. 2001. Oligomeric structure of virion-associated and soluble forms of the simian immunodeficiency virus envelope protein in the prefusion activated conformation. Proc. Natl. Acad. Sci. USA 98:14877-14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Doms, R. W., P. L. Earl, and B. Moss. 1991. The assembly of the HIV-1 env glycoprotein into dimers and tetramers. Adv. Exp. Med. Biol. 300:203-219. [DOI] [PubMed] [Google Scholar]
- 9.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earl, P. L., B. Moss, and R. W. Doms. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 65:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 13.Farzan, M., H. Choe, E. Desjardins, Y. Sun, J. Kuhn, J. Cao, D. Archambault, P. Kolchinsky, M. Koch, R. Wyatt, and J. Sodroski. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 Å resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 15.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grief, C., D. J. Hockley, C. E. Fromholc, and P. A. Kitchin. 1989. The morphology of simian immunodeficiency virus as shown by negative staining electron microscopy. J. Gen. Virol. 70:2215-2219. [DOI] [PubMed] [Google Scholar]
- 17.Jones, D. H., B. W. McBride, M. A. Roff, and G. H. Farrar. 1995. Efficient purification and rigorous characterisation of a recombinant gp120 for HIV vaccine studies. Vaccine 13:991-999. [DOI] [PubMed] [Google Scholar]
- 18.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leapman, R. D., and S. B. Andrews. 1992. Characterization of biological macromolecules by combined mass mapping and electron energy-loss spectroscopy. J. Microsc. 165:225-238. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, M. S., and R. P. Junghans. 2000. Ultracentrifugal analysis of molecular mass of glycoproteins of unknown or ill-defined carbohydrate composition. Methods Enzymol. 321:136-149. [DOI] [PubMed] [Google Scholar]
- 21.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95:9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malashkevich, V. N., M. Singh, and P. S. Kim. 2001. The trimer-of-hairpins motif in membrane fusion: visna virus. Proc. Natl. Acad. Sci. USA 98:8502-8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCune, J. M., L. B. Rabin, M. B. Feinberg, M. Lieberman, J. C. Kosek, G. R. Reyes, and I. L. Weissman. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53:55-67. [DOI] [PubMed] [Google Scholar]
- 24.Owens, R. J., and R. W. Compans. 1990. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology 179:827-833. [DOI] [PubMed] [Google Scholar]
- 25.Parren, P. W. H. I., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossio, J. L., M. T. Esser, K. Suryanarayana, D. K. Schneider, J. W. Bess, Jr., G. M. Vasquez, T. A. Wiltrout, E. Chertova, M. K. Grimes, Q. Sattentau, L. O. Arthur, L. E. Henderson, and J. D. Lifson. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72:7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staropoli, I., C. Chanel, M. Girard, and R. Altmeyer. 2000. Processing, stability, and receptor binding properties of oligomeric envelope glycoprotein from a primary HIV-1 isolate. J. Biol. Chem. 275:35137-35145. [DOI] [PubMed] [Google Scholar]
- 28.Stiasny, K., S. L. Allison, A. Marchler-Bauer, C. Kunz, and F. X. Heinz. 1996. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J. Virol. 70:8142-8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas, D. J., J. S. Wall, J. F. Hainfeld, M. Kaczorek, F. P. Booy, B. L. Trus, F. A. Eiserling, and A. C. Steven. 1991. gp160, the envelope glycoprotein of human immunodeficiency virus type 1, is a dimer of 125-kilodalton subunits stabilized through interactions between their gp41 domains. J. Virol. 65:3797-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss, C. D., J. A. Levy, and J. M. White. 1990. Oligomeric organization of gp120 on infectious human immunodeficiency virus type 1 particles. J. Virol. 64:5674-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weissenhorn, W., A. Carfı, K.-H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 32.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 33.Willey, R. L., J. S. Bonifacino, B. J. Potts, M. A. Martin, and R. D. Klausner. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. USA 85:9580-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 35.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 36.Yang, Z.-N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct. Biol. 126:131-144. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, C. W.-H., Y. Chishti, R. E. Hussey, and E. L. Reinherz. 2001. Expression, purification and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J. Biol. Chem. 276:39577-39585. [DOI] [PubMed] [Google Scholar]
- 38.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]