Abstract
Here we present the first molecular characterization of the defect associated with an avian sarcoma and leukosis virus (ASLV) receptor resistance allele, tvbr. We show that resistance to infection by subgroups B, D, and E ASLV is explained by the presence of a single base pair mutation that distinguishes this allele from tvbs1, an allele which encodes a receptor for all three viral subgroups. This mutation generates an in-frame stop codon that is predicted to lead to the production of a severely truncated protein.
Functionally distinct alleles of the autosomal tva, tvb, and tvc loci that encode avian sarcoma and leukosis virus (ASLV) receptors have been defined for chickens (reviewed in references 12 and 22). Two classes of tva and tvc alleles are associated with either susceptibility (tvas and tvcs) or resistance (tvar and tvcr) to infection by subgroup A ASLV or subgroup C ASLV, respectively. The tvb locus is more complex since two chicken alleles of tvb that encode distinct ASLV receptors have been defined: tvbs1 for viral subgroups B, D, and E and tvbs3 for subgroups B and D (1, 2, 9, 20). In addition, there is another type of allele (tvbr) that cannot support entry by any of these ASLV subgroups (12). For each ASLV receptor gene, alleles that confer susceptibility to viral infection are dominant over those associated with resistance. Therefore, the only lines of chickens that are resistant to infection by subgroups A through D ASLVs are those that are homozygous for the resistance alleles of the cognate receptor gene. Resistance to subgroup E viral infection is more complicated because of the existence of endogenous ASLV elements in the chicken germ line which encode subgroup E ASLV-specific envelope (Env) proteins that interfere with the function of the TVBS1 receptor (1, 7). The recessive nature of the tvar, tvbr, and tvcr alleles rules out the possibility that their products interfere, in a dominant-negative manner, with those encoded by the corresponding susceptibility alleles. Instead, the defect(s) associated with these resistance alleles may be due either to their lack of expression or to the existence of specific amino acid substitutions in the corresponding proteins that abolish viral receptor function.
The tvas allele encodes a low-density lipoprotein (LDL) receptor-related protein that is a cellular receptor for ASLV subgroup A (5, 6, 26). The TVA protein contains an approximately 40-amino-acid long LDL-A module that harbors the major viral interaction determinants (16, 17, 27, 28). The molecular defect that is associated with the tvar allele has not yet been defined, but a preliminary characterization of this allele has documented no obviously debilitating defects within the LDL-A domain (5).
TVB is a tumor necrosis factor receptor (TNFR)-related death receptor that is most similar to the mammalian TNF-related apoptosis-inducing ligand (TRAIL) receptors DR4 and DR5 (1). TVB contains three extracellular cysteine-rich domains and a cytoplasmic death domain which can activate apoptosis (8, 9). Several ASLV subgroups that utilize TVB exhibit an acute cytopathic effect. The death-promoting activity of TVB may contribute to the cell death that is observed in cultures that have been infected by subgroups B and D of ASLV (8, 9, 23, 24).
In order to understand the molecular basis for the resistance to viral infection that is associated with the tvbr allele, we have characterized the nature of the defect. This analysis has revealed that the tvbr mRNA harbors a premature stop codon that is predicted to lead to the production of a severely truncated protein.
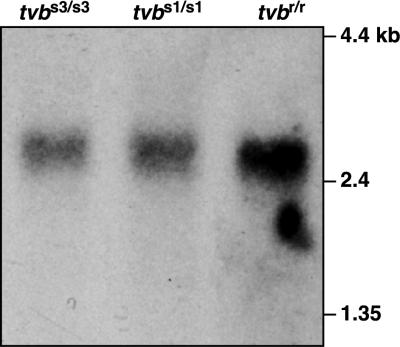
The tvbr allele is expressed as a normal-sized mRNA transcript.
Samples of mRNA were prepared from chicken embryo fibroblasts (CEFs) with distinct tvb genotypes: line 0 (tvbs3/tvbs3), line 15B1 (tvbs1/tvbs1), and line 72 (tvbr/tvbr) (provided by the Avian Disease and Oncology Laboratory, East Lansing, Mich.) (4). Approximately 1 μg of each mRNA sample, along with 5 μg of the 0.24- to 9.5-kb RNA ladder (Gibco/BRL), was subjected to electrophoresis in a 1.2% agarose gel containing 1.12% formaldehyde and morpholinepropanesulfonic acid (MOPS) buffer. The samples were then transferred to a nylon membrane (Amersham), and tvb mRNA was detected by probing with a 32P-labeled 2.5-kb HindIII-HindIII DNA fragment derived from the tvb genomic clone pBK-9 as described previously (9).
This analysis revealed that the tvbr mRNA transcript is similar in size to those of the functional tvbs3 and tvbs1 alleles (Fig. 1). Therefore, tvbr does not display any obvious defects in gene expression, mRNA stability, or mRNA splicing.
FIG. 1.
The tvbr allele is expressed as a normal-sized mRNA transcript. Northern blot analysis was performed with mRNA samples taken from CEFs with the indicated tvb genotypes. The nylon membrane was probed with a 32P-labeled tvb-specific probe derived from the BK-9 tvb genomic DNA clone as described previously (9). Autoradiography was performed at −80°C with intensifying screens.
The tvbr allele encodes a prematurely terminated protein product.
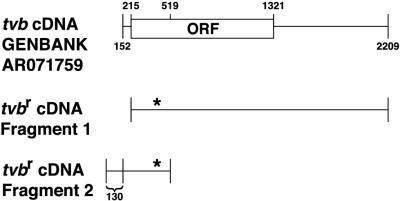
A reverse transcription-PCR (RT-PCR) amplification was used to further characterize the tvbr mRNA from line 72 (tvbr/tvbr) CEFs. Two sets of oligonucleotide primers were used in this procedure, generating two overlapping cDNA fragments that spanned the entire length of the mRNA transcript (Fig. 2). Fragment 1 was generated by subjecting 500 ng of total RNA to an RT-PCR amplification protocol with the ProStar Ultra HF RT-PCR kit (Stratagene). RT was performed with an oligo(dT) primer, and the subsequent PCR amplification was performed with the sense primer oSK2 (5′-GCTAGCTAGCCGATGCGCTCAGCTGCGCTCCGG-3′)and the antisense primer oSK73 (5′-GGGGTACCCCGCTGGTATTTGGCACAGGGG-3′). The oSK2 primer corresponds to nucleotides 215 to 235 and the oSK73 primer corresponds to nucleotides 2190 to 2209 of the tvb cDNA (GenBank accession number AR071759) (Fig. 2). To facilitate subsequent cloning of the PCR-amplified products, NheI and KpnI sites (underlined in the sequences) were incorporated at the 5′ ends of the oSK2 and oSK73 primers, respectively. A 1/10 fraction of the RT products was subjected to PCR amplification with Pfu Turbo DNA polymerase (Stratagene) for 40 cycles (94°C for 1 min, 65°C for 1 min, and 72°C for 3 min). The approximately 2-kb cDNA fragment 1, predicted to contain all of the putative tvbr coding sequence, was subcloned into the pCI mammalian expression vector (Promega).
FIG. 2.
Generation of two overlapping cDNA fragments derived from tvbr mRNA. A schematic drawing of the tvb cDNA clone (GenBank accession number AR071759) is shown along with the location of two overlapping tvbr cDNA fragments generated during this study. tvbr cDNA fragment 2 contains a unique 5′ sequence derived by rapid amplification of cDNA ends which extends 130 bp beyond that reported previously. The point mutation is in the region of overlap and is indicated by an asterisk.
The SMART RACE cDNA amplification kit (Clontech) was used to generate cDNA fragment 2. The 5′ rapid amplification of cDNA ends-ready cDNA was synthesized from 500 ng of line 72 (tvbr/tvbr) RNA, and PCR was performed utilizing the Clontech SMART universal primer and the gene-specific antisense primer oSK20 (5′-GAAGGAAGGAAGCGGCCGCGCACTGCGTGTTCCTGGTGGGG-3′; the NotI site is underlined) corresponding to nucleotides 496 to 519 of the tvb cDNA (Fig. 2). cDNA fragment 2 was T/A cloned into the pT-Adv vector (Clontech). The overlapping cDNA fragments 1 and 2 spanned the entire length of the tvbr mRNA, allowing us to analyze the full cDNA for any mutations (Fig. 2).
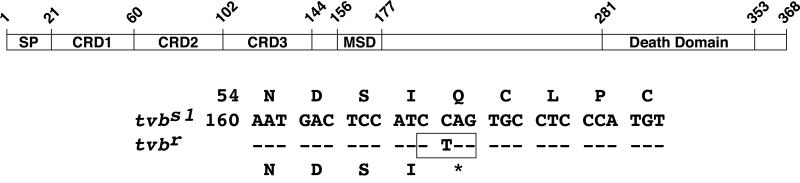
DNA sequence analysis of three independent clones of cDNA fragment 1 and nine independent clones of cDNA fragment 2 revealed only a single nucleotide difference between the open reading frames (ORFs) of tvbs1 and tvbr. A cytosine residue located 172 nucleotides downstream of the start methionine codon in tvbs1 is replaced by a thymidine residue in tvbr (Fig. 3). This change generates an in-frame stop codon (CAG→UAG). The presence of this mutation indicates that the tvbr allele encodes a severely truncated protein product.
FIG. 3.
The tvbr allele contains a premature stop codon. A schematic drawing of TVB is shown depicting the signal peptide (SP), cysteine-rich domains (CRDs), membrane spanning domain (MSD), and cytoplasmic death domain. The nucleotide and amino acid sequences are also shown surrounding the single nucleotide change that distinguishes tvbr from tvbs1 (numbered from the start methionine codon). The premature stop codon in tvbr is indicated by an asterisk and a BfaI site unique to tvbr is shown in a box.
The point mutation in tvbr generates a BfaI restriction enzyme site that can be used diagnostically.
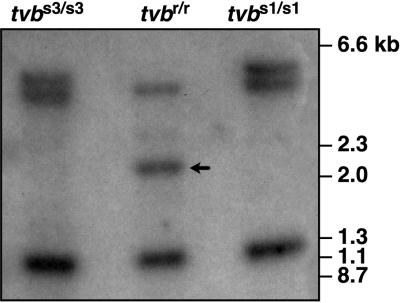
The nucleotide difference between the ORFs of tvbr and tvbs1 generated a BfaI restriction enzyme site that is specific to the resistance allele (Fig. 3). To test whether the presence of this site could be used as a diagnostic marker for the presence of this allele, Southern blot analysis was performed with BfaI-digested genomic DNA samples that were prepared from CEFs with different tvb genotypes. Using a panel of more than 50 independent restriction enzymes (not including BfaI), Smith et al. had previously failed to identify any restriction fragment length polymorphism that could be used to distinguish between the resistance and susceptibility alleles of tvb (20).
For the present studies, 25-μg samples of genomic DNA from line 72 (tvbr/tvbr), line 0 (tvbs3/tvbs3), and line15B1 (tvbs1/tvbs1) CEFs were digested overnight with 15 U of BfaI (New England Biolabs, Inc.). Approximately 10 μg of each sample was then subjected to electrophoresis on a 1% agarose gel, and the samples were then transferred to a nylon membrane (Amersham). These samples were hybridized with a 32P-labeled tvb-specific probe that was derived by PCR amplification with the tvbs3 cDNA clone pBK7.6-2 as the template DNA (9). The primers used for PCR amplification were the sense primer oSK2 (described above) and the antisense primer oSK65 (5′-GGCCAGCTGGTATTTGGCACAGGGG-3′), corresponding to nucleotides 2190 to 2209 of the tvb cDNA (GenBank accession number AR071759). The nylon membrane was incubated with the radiolabeled probe, washed at 65°C under standard conditions (10), and exposed to Kodak XAR-5 X-ray film with intensifying screens at −80°C. This analysis revealed the presence of a 2.1-kb DNA fragment that is diagnostic for the tvbr allele (Fig. 4). This DNA fragment serves as a useful marker for the tvbr allele and supports the presence of the thymidine mutation in tvbr DNA.
FIG. 4.
A 2.1-kb BfaI restriction fragment is diagnostic of the tvbr allele. BfaI-digested genomic DNA samples prepared from CEFs with the tvb genotypes indicated were subjected to Southern blot analysis with a 32P-labeled tvb-specific DNA probe under standard conditions. Autoradiography was performed for 10 days at −80°C with intensifying screens. The position of the 2.1-kb DNA fragment that is unique to tvbr is indicated with an arrow.
In this report we demonstrate the molecular defect associated with a resistance allele of an ASLV receptor gene. Resistance is due to the existence of a premature stop codon. This feature of tvbr presumably either completely abolishes protein expression or leads instead to the generation of a severely truncated protein product that consists of only the 57 N-terminal amino acids (including the N-terminal signal peptide). Although mechanisms exist for translating in-frame stop codons (i.e., nonsense suppression), this is unlikely to apply in the case of tvbr since the production of the full-length protein we predict would have ASLV receptor activity. The fact that tvbr encodes an aberrant protein product most probably explains why this allele is recessive in nature. Indeed, we have not observed a TVBR protein product of any size when the tvbr cDNA fragment 1 cloned into the plasmid vector pCI (Promega) was transfected into 293 cells (data not shown). We conclude that the premature stop codon results in a lack of TVB expression and is a null allele of TVB.
Resistance to human immunodeficiency virus (HIV) infection has also been ascribed to a mutation at the level of the receptor (in this case a coreceptor) that leads to an aberrant protein product. Several naturally occurring mutations in the human CCR5 coreceptor gene have been shown to confer resistance to R5-tropic strains of HIV, including the CCR5delta32 allele, which harbors a 32-bp deletion leading to the production of a severely truncated protein product that does not support viral entry (13, 18). Individuals who lack CCR5 are less susceptible to infection by HIV, indicating an important role for this receptor at the bottleneck of transmission. Amino acid substitutions and/or posttranslational modifications that abolish the function of other retroviral receptors have also been described as barriers to virus transmission both among and between species (11, 15, 21, 25).
The ORFs of tvbs1 and tvbs3 were previously characterized, and they were found to differ only at nucleotide residue 184 (a thymidine or an adenosine, respectively). As a consequence, residue Cys-62 of TVBS1 is replaced by a serine in TVBS3 (Fig. 3). The corresponding residue of tvbr is a thymidine, leading us to propose that this resistance allele evolved from tvbs1 during selective breeding among the commercial chicken population to confer resistance to subgroup B ASLV-induced lymphoid leukosis (4). As a consequence, the frequency of this allele and its homozygosity in the commercial chicken population is widespread.
The simple interpretation of the fact that chickens homozygous for tvbr are viable is that this receptor (and by inference its ligand) is not essential for the life of these birds. However, TVB is most closely related to the mammalian TRAIL receptors DR4 and DR5. TRAIL activity has been linked to lymphocyte death and activation, antiviral immune defenses, and tumor surveillance (reviewed in reference 14). TRAIL has a complex relationship with its receptors, which is underscored by the presence of multiple signaling-competent and decoy receptors in humans and mice (3). Although the loss or dysregulation of TNFR-related proteins and/or their cognate ligands is typically associated with severe pathological consequences, it may be that the loss of TVB can be compensated for by the expression of another TNFR-related receptor that can bind to the putative TVB ligand. Two naturally occurring mutations in human Fas involving prematurely terminated Fas polypeptides (one with only the first 57 and another with only the first 62 amino acids of the mature Fas protein translated) contain a ligand-independent assembly domain and have been shown to dominantly interfere with normal Fas function (19). Because the effect of tvbr on virus entry is recessive, we conclude that this truncated TVBR polypeptide, if it is expressed, cannot dominantly interfere with virus entry via full-length TVB receptors. However, it is not clear whether the short putative TVBR polypeptide would exert any negative effect on the natural function of the receptor, a possibility that can only be addressed when the TVB ligand has been identified and the normal physiological function of the receptor is known.
Nucleotide sequence accession number.
The sequences for cDNA fragment 1 and cDNA fragment 2, which were generated during this study, have been submitted to GenBank and assigned accession numbers AF507016 and AF507017, respectively.
Acknowledgments
We thank past and present members of the Young laboratory for many helpful discussions. We especially thank John Naughton for assistance in preparing the figures, Ken Bradley for critical reading of the manuscript, and the McArdle Laboratory Core DNA Sequencing Facility.
This work was supported by the National Institutes of Heath grant CA62000.
REFERENCES
- 1.Adkins, H. B., S. C. Blacklow, and J. A. Young. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J. Virol. 75:3520-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, H. B., J. Brojatsch, and J. A. Young. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J. Virol. 74:3572-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashkenazi, A., and V. M. Dixit. 1999. Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11:255-260. [DOI] [PubMed] [Google Scholar]
- 4.Bacon, L. D., H. D. Hunt, and H. H. Cheng. 2000. A review of the development of chicken lines to resolve genes determining resistance to diseases. Poult. Sci. 79:1082-1093. [DOI] [PubMed] [Google Scholar]
- 5.Bates, P., L. Rong, H. E. Varmus, J. A. Young, and L. B. Crittenden. 1998. Genetic mapping of the cloned subgroup A avian sarcoma and leukosis virus receptor gene to the TVA locus. J. Virol. 72:2505-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bates, P., J. A. Young, and H. E. Varmus. 1993. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell 74:1043-1051. [DOI] [PubMed] [Google Scholar]
- 7.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-435. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 8.Brojatsch, J., J. Naughton, H. B. Adkins, and J. A. Young. 2000. TVB receptors for cytopathic and noncytopathic subgroups of avian leukosis viruses are functional death receptors. J. Virol. 74:11490-11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845-855. [DOI] [PubMed] [Google Scholar]
- 10.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eiden, M. V., K. Farrell, and C. A. Wilson. 1994. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J. Virol. 68:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 13.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 14.Locksley, R. M., N. Killeen, and M. J. Lenardo. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104:487-501. [DOI] [PubMed] [Google Scholar]
- 15.Miller, D. G., and A. D. Miller. 1992. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J. Virol. 66:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rong, L., and P. Bates. 1995. Analysis of the subgroup A avian sarcoma and leukosis virus receptor: the 40-residue, cysteine-rich, low-density lipoprotein receptor repeat motif of Tva is sufficient to mediate viral entry. J. Virol. 69:4847-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rong, L., K. Gendron, B. Strohl, R. Shenoy, R. J. Wool-Lewis, and P. Bates. 1998. Characterization of determinants for envelope binding and infection in Tva, the subgroup A avian sarcoma and leukosis virus receptor. J. Virol. 72:4552-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 19.Siegel, R. M., J. K. Frederiksen, D. A. Zacharias, F. K.-M. Chan, M. Johnson, D. Lynch, R. Y. Tsien, and M. J. Lenardo. 2000. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science 288:2354-2357. [DOI] [PubMed] [Google Scholar]
- 20.Smith, E. J., J. Brojatsch, J. Naughton, and J. A. Young. 1998. The CAR1 gene encoding a cellular receptor specific for subgroup B and D avian leukosis viruses maps to the chicken tvb locus. J. Virol. 72:3501-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tailor, C. S., A. Nouri, and D. Kabat. 2000. Cellular and species resistance to murine amphotropic, gibbon ape, and feline subgroup C leukemia viruses is strongly influenced by receptor expression levels and by receptor masking mechanisms. J. Virol. 74:9797-9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss, R. A. 1993. Cellular receptors and viral glycoproteins involved in retrovirus entry, p. 1-108. In J. A. Levy (ed.), The Retroviridae, vol. 2. Plenum Press, New York, N.Y.
- 23.Weller, S. K., A. E. Joy, and H. M. Temin. 1980. Correlation between cell killing and massive second-round superinfection by members of some subgroups of avian leukosis virus. J. Virol. 33:494-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weller, S. K., and H. M. Temin. 1981. Cell killing by avian leukosis viruses. J. Virol. 39:713-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson, C. A., and M. V. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 65:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young, J. A., P. Bates, and H. E. Varmus. 1993. Isolation of a chicken gene that confers susceptibility to infection by subgroup A avian leukosis and sarcoma viruses. J. Virol. 67:1811-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zingler, K., C. Belanger, R. Peters, E. Agard, and J. A. Young. 1995. Identification and characterization of the viral interaction determinant of the subgroup A avian leukosis virus receptor. J. Virol. 69:4261-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zingler, K., and J. A. Young. 1996. Residue Trp-48 of Tva is critical for viral entry but not for high-affinity binding to the SU glycoprotein of subgroup A avian leukosis and sarcoma viruses. J. Virol. 70:7510-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]