Abstract
The mammalian gammaretroviruses gibbon ape leukemia virus (GALV) and feline leukemia virus subgroup B (FeLV-B) can use the same receptor, Pit1, to infect human cells. A highly polymorphic nine-residue sequence within Pit1, designated region A, has been proposed as the virus binding site, because mutations in this region abolish Pit1-mediated cellular infection by GALV and FeLV-B. However, a direct correlation between region A mutations deleterious for infection and loss of virus binding has not been established. We report that cells expressing a Pit1 protein harboring mutations in region A that abolish receptor function retain the ability to bind virus, indicating that Pit1 region A is not the virus binding site. Furthermore, we have now identified a second region in Pit1, comprising residues 232 to 260 (region B), that is required for both viral entry and virus binding. Epitope-tagged Pit1 proteins were used to demonstrate that mutations in region B result in improper orientation of Pit1 in the cell membrane. Compensatory mutations in region A can restore proper orientation and full receptor function to these region B mutants. Based on these results, we propose that region A of Pit1 confers competence for viral entry by influencing the topology of the authentic binding site in the membrane and hence its accessibility to a viral envelope protein. Based on glycosylation studies and results obtained by using N- and C-terminal epitope-tagged Pit1, region A and region B mutants, and the transmembrane helices predicted with the PHD PredictProtein algorithm, we propose a new Pit1 topology model.
Pit1 is the human ortholog of a ubiquitous multiple-membrane-spanning protein that functions as the type III sodium phosphate cotransporter (14, 19) and as the receptor for feline leukemia virus type B (FeLV-B) (33), woolly monkey virus (33), gibbon ape leukemia virus (GALV) (18), and 10A1 murine leukemia virus (10A1 MLV) (17, 38). Pit2, another type III sodium phosphate cotransporter, is the human ortholog of a highly related protein (approximately 60% amino acid identity), which functions as a receptor for amphotropic murine leukemia virus (17, 34), 10A1 MLV, and FeLV-B molecular recombinants (30), but not for GALV or naturally occurring FeLV-B isolates (21). The ability of Pit1 and Pit2 to function as discrete viral receptors with unique properties presumably is reflected in critical residue differences between these two proteins. Early efforts to identify regions within Pit1 important for virus receptor function implicated residues 550 to 558 for both GALV and FeLV-B entry. It has been proposed that this nine-Pit1-residue stretch, designated region A, is the binding site for both of these viruses (6, 11, 35) based on the following observations: (i) mutations in Pit1 region A render Pit1 nonfunctional for GALV or FeLV-B entry (3, 9, 22, 27, 31, 32); (ii) Pit2, MusPit1 (the murine ortholog of Pit1), and Pho-4 (a distantly related Neurospora crassa phosphate transporter), which are not GALV or FeLV-B receptors, become functional receptors when region A residues are substituted for the corresponding residues of these proteins (12, 13, 33); and (iii) region A is predicted by Kyte-Doolittle hydropathy plots to reside in an extracellular domain (12). It should be noted, however, that all published reports supporting the role of region A as a binding site have been based on functional assays of viral entry rather than actual receptor-binding studies (reviewed in reference 20). Therefore we sought to determine if mutations in region A that abolish virus receptor function do so by abolishing virus binding.
We have used data derived from the analyses of epitope-tagged Pit proteins, glycosylation studies, and transmembrane (TM) structure predictions performed by using the PHD PredictProtein algorithm (23, 24) to derive a new topology model for Pit1. Our proposed model differs from the current model in that it predicts that the N and C termini of Pit1 are extracellular, that Pit1 is an N-linked glycosylated protein, and that region A resides in a cytoplasmic region of Pit1. Finally, we provide evidence that intramolecular interactions between region A (residues 550 to 558) and region B (residues 232 to 260) are critical determinants in Pit1 receptor topogenesis that drive the accessibility or steric hindrance of the viral binding site. Based on these data and the lack of experimental evidence supporting direct binding of either GALV or FeLV-B to Pit1 region A, we have assigned a different role in virus entry to region A. We propose that the role of Pit1 region A in Pit1 virus receptor function is to direct the orientations of multiple viral binding sites relative to the extracellular space, such that the mature Pit1 protein, once embedded in the lipid bilayer, is competent for virus binding and entry.
MATERIALS AND METHODS
Cells and cell lines.
The following cell lines were used in this study: Mus dunni tail fibroblasts (MDTF), obtained from Olivier Danos, Institut Pasteur (Paris, France), and human embryonic kidney 293T cells, obtained from Cell Genesys Inc. (Foster City, Calif.). All cells were maintained in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, Calif.) supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 4 mM glutamine.
Production of retrovirus vectors and stable cell lines.
293T cells were seeded at a density of 106 cells/10-cm-diameter dish 2 days prior to transfection. Transfection by the calcium phosphate precipitation method (Promega, Madison, Wis.) was performed as described previously (10) using plasmids expressing the GALV or FeLV-B 90Z (1) envelope. Retrovirus vector supernatants were harvested 48 to 72 h posttransfection. MDTF cell lines stably expressing Pit1, mutant Pit1, or tagged Pit1 receptors were made by transducing cells with vesicular stomatitis virus (VSV) G protein-enveloped vectors with genomes expressing the appropriate receptor cDNA in retrovirus vector pLNSX (16). Forty-eight hours posttransduction the medium was changed to Dulbecco's modified Eagle medium containing 650 μg of Geneticin (G-418 sulfate; Invitrogen)/ml, and cells were selected for G-418 resistance for 7 to 10 days. Assessment of receptor function for MDTF cells expressing various receptor proteins was carried out by exposing cells to retrovirus vector-containing supernatant that had been passed through a 0.45 μm-pore-size Millipore (Bedford, Mass.) filter and then adjusted to contain 10 μg of Polybrene/ml. Twenty-four hours postinoculation the medium was changed, and cells were cultured for an additional 24 to 48 h before analysis for expression of β-galactosidase by histochemical staining with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as previously described (37). Titers were determined by averaging the number of blue foci obtained with vectors for each cell line tested in three or more independent experiments.
Construction of chimeric and mutant receptor cDNAs.
Chimeric receptor Pit1-2A was constructed by replacing residues 550 through 558 of Pit1 with the corresponding residues (residues 522 to 530) of Pit2. Complementary oligonucleotides 5′-GGTTTATAAGCAAGGAGGTGTTACTCAAGAAGCGGCAACACC (sense) and 5′-GGTGTTGCCGCTTCTTGAGTAACACCTCCTTGCTTATAAACCC (antisense), in which nucleotides encoding Pit1 residues were mutated to nucleotides encoding Pit2 residues (underlined), were designed; two segments of Pit1 cDNA were amplified by PCR mutagenesis as previously described (10) by using these primers plus upstream primer 5′-CCATGGCAATATGTGGCATGCC (sense) and downstream primer 5′-GAAGGCCAATATTTGATGCAATCACCACC (antisense) flanking the mutated region. These segments were annealed, after which a second round of PCR was carried out using only the outer primers. The final product was directly ligated into TA cloning vector pCR2.1 (Invitrogen, San Diego, Calif.) and sequenced with the Thermo Sequenase fluorescence-labeled primer cycle sequencing kit with CY-5-labeled primers (Integrated DNA Technologies, Inc., Coralville, Iowa) on an AlfExpress automated sequencer (Amersham Biosciences, Piscataway, N.J.). After the presence of the desired mutations and the absence of any unscheduled mutations were confirmed, the mutated DNA fragment was subcloned into the pLNSPit1 retrovirus expression vector between the HindIII and PflM1 restriction sites. Pit1-D550K and Pit1-D550T were created by mutating the aspartate at position 550 to a lysine or a threonine by PCR mutagenesis; the PCR product was subcloned into the pCR2.1 vector, sequenced, and then subcloned into pLNSPit1, as described above. Pit1-2B was constructed by first introducing an MfeI site at the codon for residue 230 and a BglII site at the codon for residue 391 of Pit1 by PCR mutagenesis and then substituting codons for residues 214 to 356 of Pit2, containing analogous PCR-introduced MfeI and BglII sites, for codons for Pit1 residues 230 to 391. Pit1-N-2B was made by replacing codons for residues 232 to 260 of Pit1 with codons for residues 216 to 244 of Pit2 between the KpnI site and a PCR-introduced AgeI site. Pit1-C-2B was made by substituting codons for residues 243 to 356 of Pit2 for codons for Pit1 residues 259 to 391 at the AgeI and BglII sites. Constructs containing PCR-induced changes were sequenced, as described above, in order to verify that only the desired mutations were introduced.
Tagging of receptors.
Pit1 was fused to a double-hemagglutinin (HA) epitope tag (YPYDVPDYA), derived from influenza virus HA, at the carboxy terminus by subcloning a DNA fragment encoding Pit1 into the pCS2-HA plasmid (5). By PCR mutagenesis, a C-terminal segment of Pit1 cDNA was amplified with primers 5′-CCATGGCAATATGTGGCATGCC (sense) and 5′-CAAACAGAGCTCCATTCTGAGGATGACC (antisense), resulting in the removal of the stop codon and the introduction of a SacI site (underlined). After the C-terminal portion of Pit1 was replaced with the PCR product, the modified Pit1 was subcloned into the pCS2-HA plasmid between HindIII and SacI sites in frame with the coding sequence for the tandem HA tag, resulting in plasmid pCS2Pit1-HA. This plasmid was then digested with HindIII and StuI, and the fragment containing the tagged Pit1 was subcloned into retrovirus vector pLNSX between HindIII and ClaI sites (the ClaI subcloning site was filled in with T4 DNA polymerase) to create the pLNSPit1-HA plasmid. HA-tagged mutant and chimeric Pit1 receptors were made by substitution of the appropriate DNA fragments in pLNSPit1-HA.
The N terminus of Pit1 was fused to a single copy of the HA epitope tag by mutation of sequences preceding the start of the Pit1 open reading frame in plasmid pLNSPit1; a start codon was also introduced just before the coding sequence for the HA epitope. PCR mutagenesis with sense primer 5′-GAGGCCGAGGCTTTTGCAAAAAGCTTCAATGTACCCCTATGATGTTCCTGATACGCACCACTAGTCGACAGAATGGCGACGCTG (coding sequence for the HA epitope is underlined) and antisense primer 5′-GATAAGGGCACAGAACACTGCACATCCCACCGAGATGAGGATGGTACC was used to synthesize a portion of Pit1 containing the tag. The product was ligated into the TA vector, and plasmids were screened by sequencing, as described above. The mutated fragment was then subcloned into the pLNSPit1 plasmid between HindIII and KpnI to create plasmid pLNSPit1-NHA.
Western blot analysis of cell lysates and cell membranes.
Whole-cell lysates were made from MDTF cells expressing various receptors, which had been grown to confluence in 10-cm-diameter dishes. The cells were washed in cold phosphate-buffered saline (PBS) and then incubated for 30 min on ice in 1 ml of lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1.0% Igepal CA-630 [NP-40], 10 mM phenylmethylsulfonyl fluoride [PMSF], 20 μM leupeptin, 20 μM aprotinin) and then were scraped from the dish and centrifuged at 16,000 × g for 10 min at 4°C. Supernatants were collected and analyzed by Western blotting. Cell membranes were obtained from confluent cells harvested from a 15-cm-diameter dish. Cells were resuspended, on ice, in 3 ml of membrane lysis buffer (20 mM Tris [pH 7.4], 5 mM EGTA, 10 mM PMSF, 20 mM aprotinin, 20 mM leupeptin) after being washed with PBS. The cells were scraped from the dish and disrupted with a Dounce homogenizer. Lysates were fractionated by centrifugation at 1,000 × g for 20 min at 4°C to pellet nuclei, followed by centrifugation of the nucleus-free supernatant at 30,000 rpm for 1 h at 4°C in a Beckman SW41 rotor to collect crude cell membrane pellets. Membrane pellets were solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample loading buffer and electrophoresed on a 4 to 20% polyacrylamide gel (Invitrogen). Proteins were transferred to polyvinylidene difluoride membrane (NEN Life Science Products, Boston, Mass.) and analyzed by Western blotting. HA-tagged receptor proteins were detected by incubation with 5 μg of monoclonal antibody HA.11 (Covance/Babco, Richmond, Calif.)/ml in TBST buffer (10 mM Tris-HCl [pH 8.0], 15 mM NaCl, 0.1% Tween 20), followed by incubation with the second antibody, goat anti-mouse immunoglobulin G (IgG) conjugated to horseradish peroxidase (1:25,000) (Pierce, Rockford, Ill.). Signals were detected with Renaissance Western blot chemiluminescence reagent (NEN Life Science Products) followed by exposure to Kodak X-Omat Blue XB-1 film.
Glycosylation assays.
Whole-cell lysates were prepared as described above from confluent 10-cm-diameter dishes of MDTF-Pit1-HA, MDTF-Pit1-2A-HA, or positive control 293T cells transiently expressing FeLV-B Z2-HA (14). One milliliter of lysate was immunoprecipitated overnight at 4°C with anti-HA affinity matrix (Roche, Indianapolis, Ind.) agarose beads covalently coupled to a rat monoclonal anti-HA antibody (clone 3F-10). The beads were washed in cold lysis buffer, and one-half of each lysate was set aside as the undigested control. The lysates were then digested with N-glycosidase F enzyme PNGaseF (New England Biolabs, Beverly, Mass.) at 37°C for 1 h. Digested and undigested samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting using monoclonal antibody HA.11, as described above.
Soluble-protein binding assays.
Binding assays using HA-tagged soluble FeLV-B or GALV (10, 15) surface subunit (SU) glycoproteins were performed by harvesting supernatants from 293T cells transiently transfected, as described above, with pCS2 expressing the soluble-protein cDNAs. Target MDTF cells expressing various receptor cDNAs were detached from tissue culture flasks with Cellstripper cell dissociation solution (Mediatech, Reston, Va.); 106 cells for each receptor cell line were suspended in 0.5 ml of supernatant containing soluble HA-tagged SU protein and incubated at 35°C for 45 min. Cells were washed twice with Hanks buffered saline solution (HBSS) containing 1% fetal bovine serum and then incubated in the presence of 5 μg of monoclonal antibody HA.11/ml in HBSS for 90 min at 4°C; cells were then washed with HBSS and incubated for 1 h at 4°C in the presence of HBSS containing 2 μg of goat anti-mouse IgG conjugated to fluorescein (Pierce)/100 μl. The cells were fixed in 1% paraformaldehyde and then analyzed by flow cytometry. HA-tagged receptors were detected on the cell surface by incubation of MDTF cells expressing receptors with monoclonal antibody HA.11, followed by incubation with a fluorescein-conjugated second antibody, as described above. Surface detection results are shown as histograms comparing MDTF cells expressing untagged Pit1 (negative control) and MDTF cells expressing HA-tagged Pit1 receptors exposed to the anti-HA antibody.
Immunofluorescence microscopy.
MDTF cells stably expressing HA-tagged receptors were seeded at 3 × 104 cells per chamber on four-chamber glass coverslips (Nalge Nunc International Corp., Naperville, Ill.) 2 days prior to analysis. For nonpermeabilized-cell analysis, cells were washed with PBS, blocked for 30 min at 25°C in 4 mg of normal goat serum (Pierce)/ml, and then incubated at 4°C for 1 h with 10 mg of monoclonal antibody HA.11 (Covance)/ml. Cells were then washed three times with cold PBS and incubated with a 1:50 dilution of 1.5 mg of fluorescein-conjugated goat anti-mouse IgG (Pierce)/ml at 4°C for 1 h. After being washed with PBS, cells were fixed for 10 min at 4°C with 4% formaldehyde. For permeabilization, cells were treated with 4% formaldehyde in PBS containing 0.1% Tween 20 for 30 min at 25°C before being subjected to the immunofluorescence protocol described above. Images were collected on either a Zeiss (Thornwood, N.Y.) LN410 laser scanning confocal microscope with a 25×, 0.8-numerical-aperture (NA) oil immersion objective or a Nikon (Tokyo, Japan) Diaphot-TMD inverted microscope with a 60×, 1.40-NA oil immersion objective.
Computational analysis.
The entire Pit1 sequence was submitted to the European Molecular Biology Laboratory (Heidelberg, Germany) PHD PredictProtein website (http://www.embl-heidelberg.de/Services/sander/predictprotein) for TM structure predictions. The expected accuracy of Pit1 TM structure prediction by using input domain sequences provided by the Prodom domain base (7) (http://prodes.toulouse.inra.fr/prodom/doc/prodom.html) was 72%. Input domains used for multiple sequence alignments are as follows (with GenBank accession numbers in parentheses): residues 1 to 34 and 296 to 345 of Felis silvestris catus Pit1 (AF074085); residues 35 to 169 of a phosphate transporter of Rhizobium meliloti (AF008187); residues 74 to 168, 527 to 552, and 564 to 673 of the Caenorhabditis elegans phosphate transporter (Z99102); residues 1 to 34 of Mus musculus Pit1 (Q61609); residues 152 to 199 of the Pyrococcus horikoshii phosphate transporter (AP00003); residues 35 to 169 of the Sinorhizobium meliloti phosphate transporter (AF008187); and residues 526 to 545 and 564 to 634 of the Halobacterium halobium phosphate transporter (AF016485).
RESULTS AND DISCUSSION
Mutations in Pit1 region A affect virus infectivity and binding.
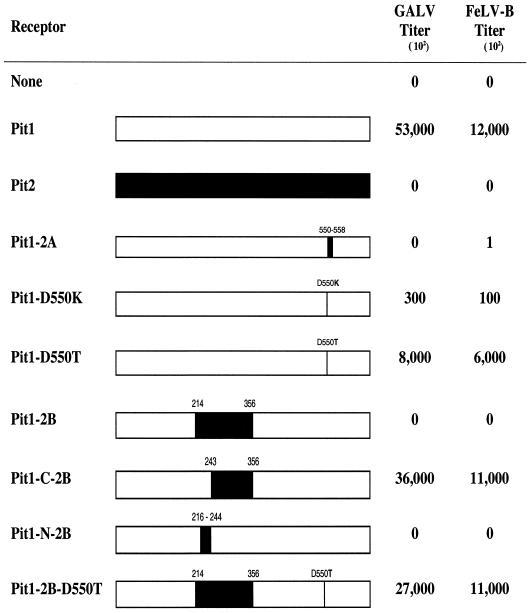
MDTF cells are resistant to GALV and FeLV-B, but MDTF cells expressing human Pit1 are rendered susceptible to these viruses (18). It has been previously reported that MDTF cells expressing a chimeric Pit1 protein in which Pit2 region A (residues 522 to 530) replaces Pit1 region A (residues 550 to 558) are not susceptible to either GALV or FeLV-B, suggesting an important role for region A in Pit1-mediated viral entry (21). It has not, so far, been determined whether this chimeric Pit1 protein is a nonfunctional virus receptor because it is not expressed, is inappropriately processed or transported, fails to bind virus, or prevents virus entry at a postbinding stage of infection.
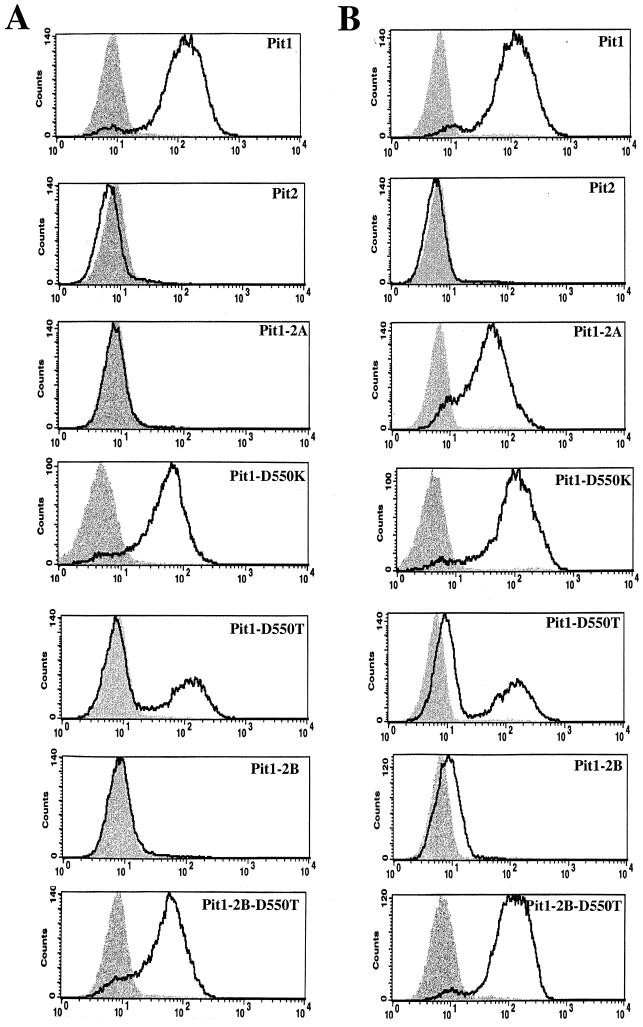
To determine the stage at which Pit2 region A blocks GALV or FeLV-B entry, we constructed a mutant Pit1 protein, Pit1-2A, in which Pit1 region A (DTGDVSSKV) was replaced with the corresponding region (KQGGVTQEA) of Pit2. We first verified that Pit1-2A, like Pit2, does not efficiently function as a GALV or FeLV-B receptor (Fig. 1). We then used flow-cytometric analysis to detect HA epitope-tagged soluble GALV or FeLV-B SU envelope proteins bound to MDTF cells expressing Pit1, Pit2, or Pit1-2A. As expected, both FeLV-B and GALV HA-tagged SUs bound MDTF-Pit1 cells (Fig. 2), whereas neither SU bound MDTF or MDTF-Pit2. Surprisingly, even though the Pit1-2A protein does not function efficiently as a receptor for FeLV-B, as evidenced by the substantially lower FeLV-B titers observed with MDTF-Pit1-2A than with MDTF-Pit1, it does efficiently bind FeLV-B SU (Fig. 1 and 2, respectively).
FIG. 1.
Infection results with GALV- and FeLV-B-enveloped retrovirus vectors on MDTF cells stably expressing Pit1, Pit2, or Pit1 chimeric or mutant receptors. Bars, schematic representations of receptors (white, Pit1; black, Pit2). Residue positions of mutations or substitutions are indicated. The titers from at least three independent experiments were averaged and are expressed as mean numbers of β-galactosidase-expressing cells.
FIG. 2.
Histograms from flow-cytometric analysis of cells stained with fluorescein-conjugated monoclonal antibody HA.11, recognizing soluble HA-tagged GALV SU (A) and FeLV-B SU (B) glycoproteins. The x axis represents fluorescence intensity (log scale), and the y axis represents cell number. Shaded areas correspond to negative-control MDTF cells exposed to HA-tagged glycoproteins; areas beneath boldface lines correspond to MDTF cells stably expressing receptors exposed to HA-tagged glycoproteins.
We reasoned that, as neither Pit2 nor the MDTF ortholog of Pit1 binds GALV or FeLV-B (Fig. 2) and as both proteins contain a lysine residue at the first position of region A (13, 34), the presence of a lysine residue at this position in region A may be deleterious for virus binding. Therefore, the substitution of a lysine for the aspartate at the first position of Pit1 region A would abolish Pit1 virus binding. To test this hypothesis, we replaced the aspartate residue at position 550 of region A in Pit1 with a lysine residue, corresponding to that found at position 522 of Pit2. This receptor (Pit1-D550K) retained both GALV and FeLV-B receptor function, albeit at an approximately 100-fold-reduced efficiency (Fig. 1). In contrast, the binding of both GALV and FeLV-B SUs to MDTF-Pit1-D550K cells was similar to that observed with MDTF-Pit1 cells (Fig. 2). These findings demonstrate that the presence of a basic residue at the first position of region A in Pit1 is not sufficient to disrupt the ability of either GALV or FeLV-B SU to bind the Pit1-D550K mutant receptor (Fig. 2), indicating that the absence of virus binding observed with Pit2 is not solely attributable to the basic residue at position 522.
We had previously reported that expression of a Pit1 mutant designated Pit1-D550T (4), wherein the aspartate at position 550 of Pit1 was replaced with a threonine residue, failed to render MDTF-Pit1-D550T cells susceptible to GALV. This cell line was made by exposing MDTF cells to PA317-produced vectors containing the Pit1-D550T cDNA. We have now developed improved methodology for assessing virus receptor function by establishing stable MDTF receptor-expressing cell lines using VSV G protein-enveloped retrovirus vectors produced in 293T cells (as described in Materials and Methods). MDTF-Pit1 and MDTF-Pit1-D550T cell lines generated with PA317 vectors express little or no receptor, respectively, as assessed by flow-cytometric analysis of envelope binding (data not shown). In contrast, MDTF-Pit1 cells generated by transduction with vectors bearing VSV envelopes, produced in 293T cells, express higher levels of receptor. Infection studies using VSV vector-derived MDTF-Pit1-D550T cells, in contrast to those derived with PA317 vectors (4), show that these cells bind both GALV and FeLV-B SUs (Fig. 2) and are susceptible to infection by these viruses (Fig. 1). We have also observed that VSV vector-transduced MDTF-Pit1-D550T populations are less stable than MDTF-Pit1 cells. In contrast to MDTF-Pit1 cells, MDTF-Pit1-D550T cells exhibit a bimodal shift in fluorescence intensity with two populations of cells: one population efficiently binds GALV and FeLV-B SU proteins, while the other does not (Fig. 2). Pit1, Pit1-D550T, and Pit1-D550K are functional receptors for GALV and FeLV-B; hence the receptors are expressed and localize to the plasma membrane.
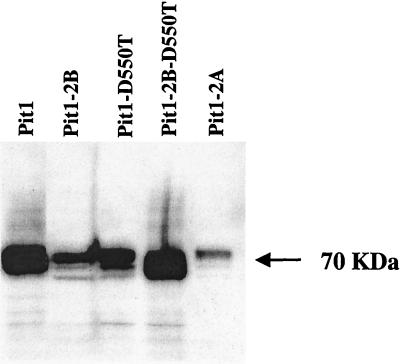
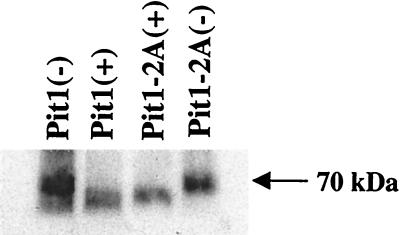
As reported above, neither GALV nor FeLV-B can use the Pit1-2A chimeric receptor to efficiently infect cells. To evaluate why Pit1-2A is not a functional receptor, we tagged this protein with an HA epitope so that expression and cellular location could be determined. Pit1, Pit1-D550T, and Pit1-D550K were similarly tagged to compare results to those obtained with Pit1-2A. Infection efficiency of the receptors was not altered by the addition of the epitope tag, as shown in Table 1. Western blot analysis of whole-cell lysates (data not shown) and membrane preparations prepared from MDTF cells expressing Pit1-HA, Pit1-D550T-HA, or Pit1-2A-HA receptor proteins revealed the presence of a 70-kDa protein in all transduced cells (Fig. 3), indicating that all these proteins are transported to the cell membrane. Pit1-2A, therefore, fails to function as a virus receptor due to inherent differences between the membrane-associated form of Pit1-2A and that of either Pit1 or Pit1-D550T.
TABLE 1.
Infection properties of HA-tagged receptors
| Receptora | Titer (bfu/ml)b of:
|
|
|---|---|---|
| GALV | FeLV-B | |
| None | <10 | <10 |
| Pit1 | 4.0 × 106 | 1.0 × 106 |
| Pit1-HA | 5.2 × 106 | 1.7 × 106 |
| Pit1-2A-HA | <10 | 7.6 × 102 |
| Pit1-D550T-HA | 1.8 × 106 | 0.3 × 106 |
| Pit1-D550K-HA | 0.7 × 106 | 2.7 × 104 |
| Pit1-2B-HA | <10 | <10 |
| Pit1-2B-D550T-HA | 2.4 × 106 | 0.3 × 106 |
Receptors are stably expressed in MDTF cells transduced by VSV G-enveloped vectors expressing receptor cDNA genomes, as described in Materials and Methods.
Titers are expressed as the mean numbers of blue foci (bfu) on target MDTF cells or MDTF cells expressing various receptors per milliliter from three experiments.
FIG. 3.
Western blot detection of HA-tagged mutant Pit1 receptor proteins using fluorescein-conjugated monoclonal antibody HA.11. Cell membranes were prepared from equivalent quantities of MDTF cells stably expressing HA-tagged receptors, as described in Materials and Methods.
A second region within Pit1 is also important in receptor function.
While it is clear that region A functions at a postbinding step in the entry of FeLV-B, its role in supporting GALV entry is more complex. We hypothesized that the failure of GALV to bind Pit1-2A was due to conformational changes induced by the substitution of Pit2 residues that block access to the authentic Pit1 GALV binding site. We therefore sought to identify other regions that are important in Pit1 GALV binding and constructed a series of Pit1 receptor chimeras that retained authentic Pit1 region A but contained Pit2-derived residues in other regions of the Pit1 backbone. Of the 17 chimeric receptors tested, only one receptor, Pit1-2B, failed to function as either a GALV or FeLV-B receptor (Fig. 1). Pit1-2B contains 143 residues from Pit2 (residues 214 to 356) in place of the corresponding residues of Pit1 (residues 230 to 391).
To determine whether the inability of Pit1-2B to function as a virus receptor was due to the absence of protein expression, we tagged Pit1-2B with an HA epitope. Western blot analysis of cell membranes prepared from MDTF cells (see Materials and Methods) showed that the Pit1-2B-HA protein is expressed (Fig. 3), even though it does not function for GALV or FeLV-B entry (Table 1). To further localize the specific Pit2 residues that were deleterious to Pit1-2B GALV and FeLV-B receptor function, two additional Pit1 chimeric receptors were made and tested. Pit1-N-2B, containing Pit2 residues 216 to 244 in place of Pit1 residues 232 to 260, failed to facilitate either FeLV-B or GALV entry (Fig. 1) or efficiently bind GALV or FeLV-B (Fig. 2). Pit1-C-2B, containing Pit2 residues 243 to 356 in place of Pit1 residues 259 to 391, retained viral receptor function comparable to that of wild-type Pit1 (Fig. 1). Therefore, only 28 of the 143 Pit2 residues present in Pit1-2B are required to abolish Pit1 receptor function.
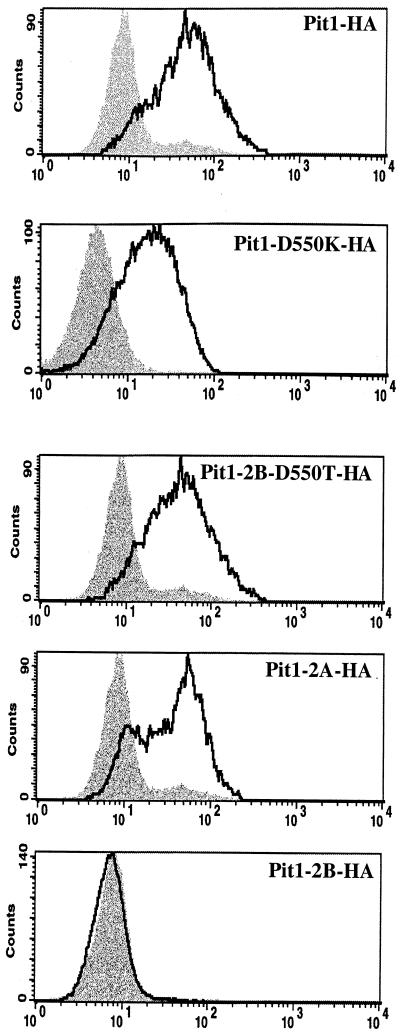
After establishing that the Pit1-2B protein is expressed, we next used flow-cytometric analysis to detect the Pit1-2B-HA protein, as well as other Pit1 HA-tagged receptors, on the cell surface. MDTF cells expressing Pit1-HA, Pit1-2A-HA, Pit1-D550K-HA, and Pit1-D550T-HA bound to the anti-HA antibody, suggesting that the C termini of these receptors are extracellular (Fig. 4). In contrast, the Pit1-2B-HA protein is undetectable (Fig. 4), indicating that its C terminus is located either inside the cell or in the cell membrane. To further authenticate the cellular location of the C-terminal HA tag attached to Pit1 and the other chimeric receptors, we performed immunofluorescence assays using an anti-HA antibody on both permeabilized and nonpermeabilized live MDTF cells expressing HA-tagged receptor proteins Pit1, Pit1-2A, and Pit1-2B; untagged MDTF-Pit1 cells were used as a negative control. We then used fluorescence scanning confocal microscopy to provide optical sectioning of cells. As expected, neither nonpermeabilized nor permeabilized MDTF-Pit1 cells immunoreacted with the fluorescein-conjugated HA antibody (Fig. 5), whereas both nonpermeabilized and permeabilized MDTF-Pit1-HA were highly immunoreactive (Fig. 5). The ability to detect the HA-tagged Pit1 receptor on nonpermeabilized cells clearly shows that the HA tag localizes to the cell membrane, confirming that the C terminus of Pit1-HA is extracellular. The same observation was made for Pit1-D550T-HA and Pit1-2A-HA. In contrast, the Pit1-2B-HA protein was detected only after cell membrane permeabilization (Fig. 5), an observation consistent with the C terminus of this membrane protein being positioned inside the cell membrane. Results obtained with Pit1-2B-HA suggest that this region of Pit2 is a topogenic determinant that controls the conformation of the receptor, thereby blocking access to the authentic binding site. It is possible that both Pit1 and Pit2 contain FeLV-B and/or GALV binding sites but that they are obscured in receptors containing Pit2 region B (e.g., Pit1-2B and Pit2).
FIG. 4.
Histograms of flow-cytometric analyses, showing detection of Pit1 receptors fused to C-terminal HA epitope tags by monoclonal antibody HA.11 stained with a fluorescein-conjugated second antibody. Shaded areas represent negative-control MDTF-Pit1 cells; areas beneath boldface lines represent MDTF cells expressing HA-tagged receptor proteins. The x axis represents fluorescence intensity (log scale), and the y axis represents cell number.
FIG. 5.
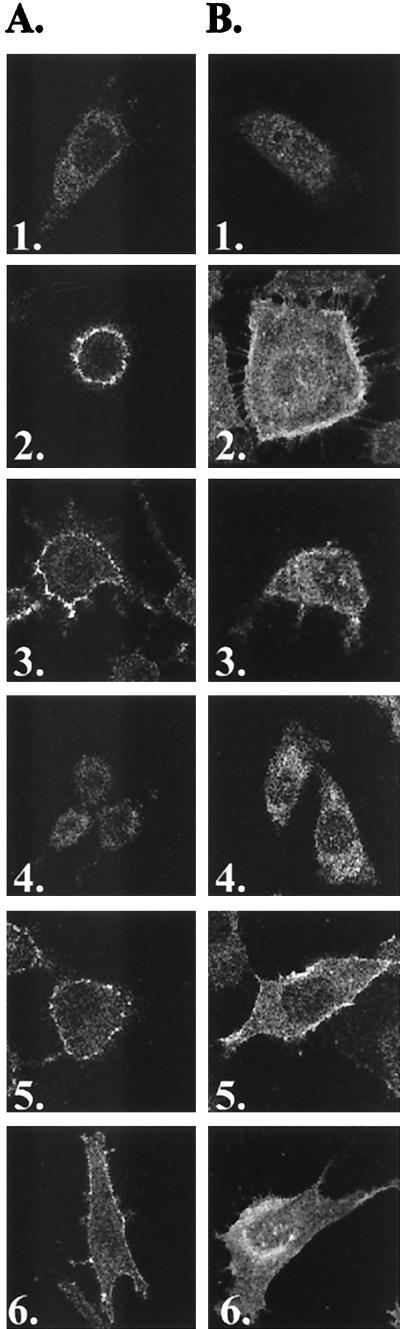
Confocal images of individual live nonpermeabilized (A) and permeabilized (B) cells from immunofluorescence studies. MDTF cells expressing receptors bearing C-terminal HA epitope tags were incubated with monoclonal antibody HA.11 and then stained with a fluorescein-conjugated second antibody. 1, MDTF-Pit1, untagged negative control; 2, MDTF-Pit1; 3, MDTF-Pit1-2A; 4, MDTF-Pit1-2B; 5, MDTF-Pit1-D550T; 6, MDTF-Pit1-2B-D550T.
Pit1 membrane orientation is regulated by at least two discrete determinants.
We have previously shown that, in Pit2, substitution of a single residue at the first position in region A of Pit2 (Pit2-K522E) is sufficient to confer GALV and FeLV-B receptor function to Pit2 (9). This finding suggests that mutations in Pit2 region A can compensate for the deleterious effects of Pit2 region B on GALV and FeLV-B receptor function. To determine if it might be possible to restore Pit1-2B GALV and FeLV-B virus binding and entry functions by making mutations in region A, we constructed mutant receptor Pit1-2B-D550T, in which a threonine was substituted for the aspartate at the first position of Pit1-2B region A. MDTF cells expressing this protein proved to efficiently bind GALV and FeLV-B SUs (Fig. 2) and to be susceptible to both viruses, as shown by infectivity assays (Fig. 1). We attached an HA epitope tag to the Pit1-2B-D550T protein; Western blot analysis of MDTF-Pit2B-D550T-HA whole-cell lysates (data not shown) and membrane preparations showed that the Pit1-2B-D550T-HA protein is expressed and localized to the cell membrane (Fig. 3). As expected from results obtained with the other HA-tagged Pit proteins, Pit1-2B-D550T-HA functions as a virus receptor as efficiently as Pit1-2B-D550T (Table 1 and Fig. 1). We then immunoreacted both permeabilized and nonpermeabilized live MDTF cells expressing Pit1-2B-D550T-HA with the fluorescein-conjugated HA antibody and used fluorescence scanning confocal microscopy to determine the membrane position of the HA tag. Both nonpermeabilized and permeabilized MDTF-Pit1-2B-D550T-HA cells were detected with the anti-HA antibody (Fig. 5). The ability to detect the HA-tagged receptor on nonpermeabilized cells indicates that the C terminus of Pit1-2B-D550T-HA is extracellular. Thus, a single residue change in region A of Pit1-2B can functionally compensate for the loss of GALV or FeLV-B receptor function associated with Pit2 region B residues, and replacement of the aspartate with a threonine at residue 550 causes a reorientation of the Pit1-2B protein in the cell membrane.
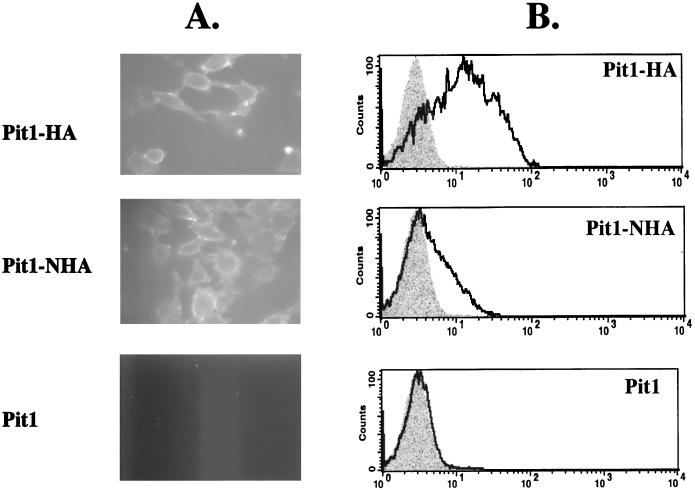
The current model for Pit1 topology predicts intracellular N- and C-terminal extremities (12). We have now determined, using Pit1-HA, that the C terminus of this receptor is extracellular. To resolve the location of the N terminus of Pit1, we constructed Pit1-NHA by attaching a single HA epitope tag in frame at the N terminus of Pit1. When expressed in MDTF cells, Pit1-NHA behaved like wild-type Pit1 in its ability to confer susceptibility to GALV and FeLV-B (data not shown), illustrating that the tag does not affect Pit1 receptor function. Flow-cytometric analysis and fluorescence microscopy with live nonpermeabilized cells were used to determine if the HA epitope could be detected on the surfaces of MDTF-Pit1-NHA cells. As shown in Fig. 6, flow cytometry demonstrates that the HA tag can be detected on the surfaces of MDTF-Pit1-NHA cells. In agreement with this finding, immunofluorescence analysis of nonpermeabilized cells showed immunoreactivity with the anti-HA antibody, supporting an extracellular positioning of the N terminus of Pit1-NHA (Fig. 6). Thus, both the C and N termini of epitope-tagged Pit1 are extracellular (Fig. 5 and 6, respectively). The current model also predicts that all potential N-linked glycosylation sites (residues 96, 371, 415, and 497) are intracellular. To further experimentally investigate the organization of the Pit1 protein, we performed glycosylation studies. Whole-cell lysates prepared from MDTF-Pit1-HA and MDTF-Pit1-2A-HA cells were analyzed by Western blotting using the anti-HA antibody. As expected, signals corresponding to a 70-kDa protein were detected in both cell lysates (Fig. 7). Treatment of both lysates with N-glycosidase F, which removes N-linked oligosaccharides, induced a shift in the molecular mass of the protein, observed as a lower band on the Western blot (Fig. 7). This observation suggests that Pit1 and Pit1-2A are both glycosylated proteins.
FIG. 6.
. Fluorescence images (A) and flow-cytometric histograms (B) detect the presence of both the N and C termini of the Pit1 receptor on the surfaces of live nonpermeabilized MDTF cells expressing Pit1 receptors bearing either C- or N-terminal HA epitope tags (Pit1-HA and Pit1-NHA, respectively). Pit1 bearing no epitope tag (Pit1) was used as a negative control. Cells were incubated with monoclonal antibody HA.11, washed, and then incubated with a fluorescein-conjugated second antibody. Shaded areas on histograms represent negative-control MDTF-Pit1 cells; areas beneath the boldface lines represent MDTF cells expressing HA-tagged receptors. The x and y axes are as defined for Fig. 2.
FIG. 7.
Glycosylation studies of Pit1-HA and Pit1-2A-HA; whole-cell lysates from MDTF cells expressing the receptors were immunoprecipitated with agarose beads coupled to a rat monoclonal anti-HA antibody (see Materials and Methods); one-half of each lysate was treated with N-glycosidase F enzyme, and then undigested (−) and digested (+) samples were analyzed by Western blotting. Signals were detected by using fluorescein-conjugated monoclonal antibody HA.11.
Proposed topology of Pit1.
A recently proposed topology model for Pit2 is based on broad database analysis of related sequences and experimental assessment involving protein glycosylation studies, epitope tagging of the N and C termini, and in vitro translation of truncated Pit2 mutants (26). In this model Pit2 has extracellular N and C termini and 12 TM domains. Salaün and coworkers used the ProDom database to identify a homology domain, named PD1131 (25); this domain is highly conserved among plant, bacterial, and eukaryotic representatives of the Pit family of proteins (24) and is contained in both the N- and C-terminal regions of Pit1 and Pit2. When the entire Pit2 sequence was analyzed using the PHD PredictProtein algorithm (23, 24), 11 TM domains were predicted, with N-PD1131 sequences harboring 3 TM domains and C-PD1131 harboring only 2. However, when the C-PD1131 homology domain was analyzed as an isolated sequence, PHD PredictProtein proposed that it contains three rather than two TM domains. The prediction of 11 TM domains for Pit2 is clearly inconsistent with the observation of Salaün et al. that both Pit2 termini are extracellular. To reconcile this discrepancy, Salaün et al. hypothesized that the C-PD1131 sequences, like the N-PD1131 sequences, contain 3 TM domains, and therefore their topological model of Pit2 contains a total of 12, not 11, TM domains.
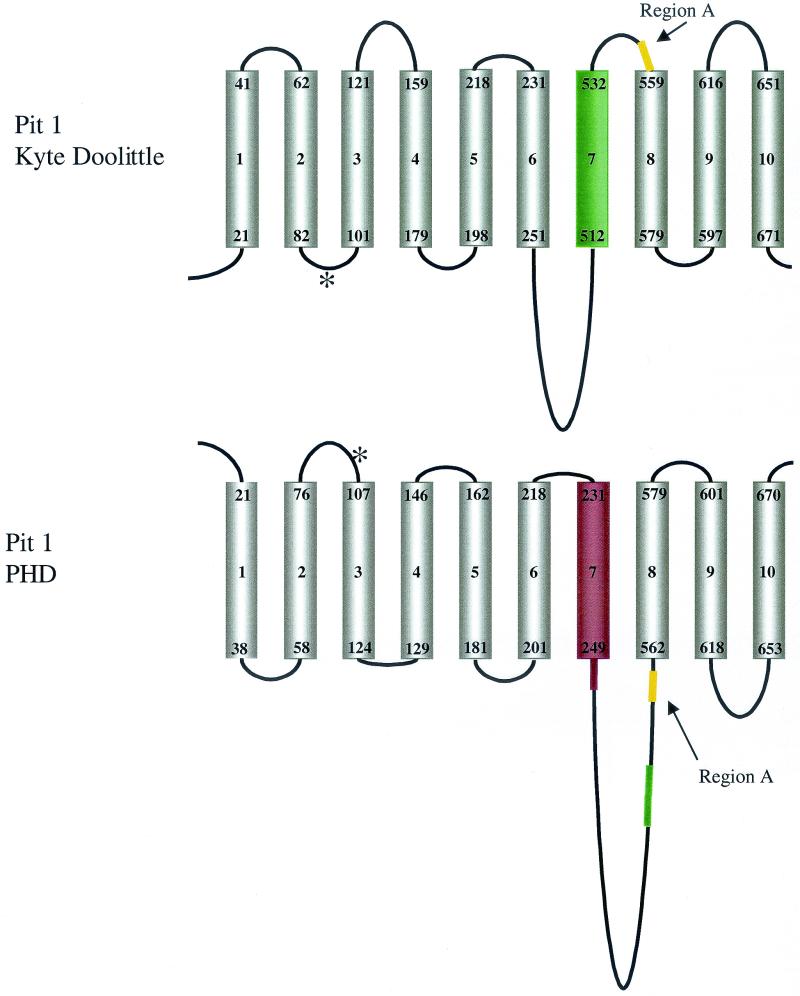
We propose a new model of Pit1 topology based on our combined experimental observations and a prediction of TM domain organization made by using the PHD PredictProtein algorithm (23, 24). PHD PredictProtein estimates that Pit1 has 10 TM domains, 9 of which are similar to those previously proposed using hydropathy plot analyses (12) (Fig. 8). This algorithm estimates a low probability that residues 512 to 532 comprise a TM domain and instead predicts that residues 129 to 146 form TM domain 4 (TM4) (Fig. 8). PHD PredictProtein also places region A in the large intracellular loop directly adjacent to TM8 (Fig. 8). The Pit1 topology predicted by PHD PredictProtein agrees with our experimental findings that the N and C termini of Pit1 are extracellular (Fig. 4 and 6). We have also determined that, like Pit2, Pit1 is a glycoprotein. In our topology model, both Pit1 and Pit2 have potential N-linked glycosylation sites within the predicted extracellular loop 1 (Fig. 8). All other asparagines that are found to constitute potential glycosylation sites are located in the large cytoplasmic loop between TM7 and TM8. Salaün and coworkers showed by site-directed mutagenesis and glycosylation studies that the asparagine present in the first extracellular loop of Pit2 carries an N-linked oligosaccharide (26). It is therefore probable that Pit1 asparagine 96 within the corresponding Pit1 extracellular loop also carries an N-linked oligosaccharide. The Pit2 PD7717 homology domain that extends from N-PD1131 to the large cytoplasmic loop region has a residue identity of 92% with Pit1. PHD PredictProtein invariably estimates three TM domains in aligned sequences containing the PD7717 domain. Based on these observations, it is reasonable to assume that Pit1 and Pit2 form similar structures up to and including TM7. The proposed 8th, 11th, and 12th TM domains of Pit2 do not correspond to any of the C-terminal TM regions assigned to Pit1, suggesting that the C-terminal regions of Pit1 and Pit2 vary in their topologies.
FIG. 8.
Schematic representations of Pit1 receptor protein based on Kyte-Doolittle (12) or PHD PredictProtein (23) systems. Cylinders, TM domains; lines above and below, extracellular and cytoplasmic domains, respectively. The residue numbers that represent the boundaries of the TM domains are shown at the opposing ends of each cylinder. Asterisk, potential N-glycosylation site at position 96. TM4 of Pit1 (residues 129 to 146) is present in the PHD-predicted model but absent from the Kyte-Doolittle system-based model. TM7 (residues 512 to 532; green cylinder) becomes part of the large cytoplasmic loop (green line) in the PHD model. Yellow lines, relative locations of region A; red line and cylinder (TM7 and flanking residues of PHD model), region B (residues 232 to 260).
Regions A and B are topogenic determinants.
Region A has been proposed to play a critical role in virus entry because the exchange or mutation of region A in Pit1 and Pit2 dramatically alters their abilities to support GALV or FeLV-B infection (reviewed in reference 20). The argument supporting region A as a viral binding site is weakened by two points: (i) the absence of a consensus binding site among functional Pit1 and Pit2 receptors and (ii) the failure to demonstrate that mutations in region A that abolish virus receptor function do so because they abolish virus binding. Our present findings support the hypothesis that region A is not a binding site for FeLV-B but instead controls virus entry at a postbinding step, since mutations in region A that abolish efficient receptor function do not affect FeLV-B binding (e.g., Pit1-2A; Fig. 1 and 2).
We have identified a second domain, region B, comprising Pit1 TM7 and the flanking region (Fig. 8), that plays a critical role in both virus binding and entry. Substitution of Pit2 region B residues for the corresponding region of Pit1 results in a chimeric protein (Pit1-2B) that fails to bind virus. The observation that Pit1-2B fails to bind GALV or FeLV-B (Fig. 2) even though it contains an intact region A suggests that sequences outside of region A can block Pit1 virus binding. We have ascertained that region A functions as a determinant that regulates Pit1 organization in that it directs proper membrane orientation. Mutations in region A (e.g., Pit1-D550T) ameliorate the effects of Pit2 region B on Pit1 virus binding and simultaneously reorient the C terminus to a position outside the membrane (Pit1-D550T-2B; Fig. 2 and 5).
Polytopic membrane assembly involves the precise orientation and integration of TM helices into the endoplasmic reticulum membrane, correct folding of cytoplasmic loop regions, and final assembly into a mature tertiary structure. For certain polytopic proteins it has been demonstrated that TM segments and/or their flanking regions contain topogenic determinants that direct membrane assembly in a cooperative manner (2, 8). For example, the Saccharomyces cerevisiae Sec61p protein requires intramolecular cooperation between sequences located in the C terminus and upstream sequences in TM5 for correct membrane topogenesis (36). Similar signal sequences have been identified within the human P glycoprotein (29) and the cystic fibrosis transmembrane conductance regulator protein (28). These determinants can act either synergistically or independently to direct both topogenesis and integration of the nascent chain into the membrane. The arrangement of topogenic information is critical to proper membrane integration. Region A and region B act similarly in that either each can independently abolish receptor function (e.g., Pit1-2A and Pit1-2B) or they can interact in a cooperative manner to direct appropriate Pit1 membrane orientation and render nonfunctional Pit1- or Pit2-derived receptors competent for GALV or FeLV-B entry (e.g., Pit1-2B-D550T). Stringent cellular processes have been demonstrated to recognize misfolded proteins and direct their subsequent degradation (28). A similar process may account for the diminished representation of the Pit1-2B protein in the cell membrane compared to Pit1-2B-D550T (Fig. 3). The arrangement of two distinct topogenic segments, located in regions A and B, serves to regulate the ability of Pit1 or Pit2 to bind or facilitate GALV or FeLV-B entry. In summary, we have determined that region A is not the Pit1 virus binding site but instead functions cooperatively with region B to facilitate Pit1-mediated viral entry.
Acknowledgments
We are grateful to Carolyn L. Smith (NINDS Light Imaging Facility) for confocal microscopy, Howard Mostowski (CBER, FDA) for flow cytometry binding analysis, and Julie Overbaugh for helpful discussions and the HA-tagged viral envelope plasmids. We also thank Carolyn Wilson, Lee Green, Steven Feldman, and Peggy Faix for critical comments on the manuscript.
REFERENCES
- 1.Boomer, S., M. V. Eiden, C. C. Burns, and J. Overbaugh. 1997. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J. Virol. 71:8116-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borel, A. C., and S. M. Simon. 1996. Biogenesis of polytopic membrane proteins: membrane segments assemble within translocation channels prior to membrane integration. Cell 85:379-389. [DOI] [PubMed] [Google Scholar]
- 3.Chaudry, G. J., and M. V. Eiden. 1997. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J. Virol. 71:8078-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudry, G. J., C. Schmitz, Y.-T. Ting, K. B. Farrell, C. J. Petropoulos, Y. S. Lie, and M. V. Eiden. 1999. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J. Virol. 73:2916-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C.-M. A., N. Kraut, M. Groudine, and H. Weintraub. 1996. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell 86:731-741. [DOI] [PubMed] [Google Scholar]
- 6.Coffin, J. 1996. Retroviridae: the viruses and their replication, p. 1767-1848. In B. Fields, D. M. Knipe, and P. M. Howley (ed.), Field's virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 7.Corpet, F., J. Gouzy, and D. Kahn. 1998. The ProDom database of protein domain families. Nucleic Acids Res. 26:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Do, H., D. Falcone, J. Lin, D. W. Andrews, and A. E. Johnson. 1996. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell 85:369-378. [DOI] [PubMed] [Google Scholar]
- 9.Eiden, M. V., K. B. Farrell, and C. A. Wilson. 1996. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to function as a gibbon ape leukemia virus receptor. J. Virol. 70:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrell, K. B., Y.-T. Ting, and M. V. Eiden. 2002. Fusion-defective gibbon ape leukemia virus vectors can be rescued by homologous but not heterologous soluble envelope proteins. J. Virol. 76:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Press, Cold Spring Harbor, N.Y. [PubMed]
- 12.Johann, S. V., J. J. Gibbons, and B. O'Hara. 1992. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J. Virol. 66:1635-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johann, S. V., M. van Zeijl, J. Cekleniak, and B. O'Hara. 1993. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J. Virol. 67:6733-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kavanaugh, M. P., D. G. Miller, W. Zhang, W. Law, S. L. Kozak, D. Kabat, and A. D. Miller. 1994. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc. Natl. Acad. Sci. USA 91:7071-7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauring, A. S., M. M. Anderson, and J. Overbaugh. 2001. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 75:8888-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. Bio/Technology 7:982-989. [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, D. G., and A. D. Miller. 1994. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 68:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Hara, B., S. V. Johann, H. P. Klinger, D. G. Blair, H. Rubinson, K. J. Dunne, P. Sass, S. M. Vitek, and T. Robins. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1:119-127. [PubMed] [Google Scholar]
- 19.Olah, Z., C. Lehel, W. B. Anderson, M. V. Eiden, and C. A. Wilson. 1994. The cellular receptor for gibbon ape leukemia virus is a novel high affinity phosphate transporter. J. Biol. Chem. 269:25426-25431. [PubMed] [Google Scholar]
- 20.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen, L., S. V. Johann, M. van Zeijl, F. S. Pedersen, and B. O'Hara. 1995. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J. Virol. 69:2401-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen, L., M. van Zeijl, S. V. Johann, and B. O'Hara. 1997. Fungal phosphate transporter serves as a receptor backbone for gibbon ape leukemia virus. J. Virol. 71:7619-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rost, B., and C. Sanders. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 24.Rost, B., and C. Sanders. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 25.Saier, M., B. Eng, S. Fard, J. Garg, D. Haggerty, W. Hutchinson, D. Jack, E. Lai, H. Lui, D. Nusinew, A. Omar, S. Pao, I. Paulsen, J. Quan, M. Sliwinski, T. Tseng, S. Wachi, and G. Young. 1999. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 1422:1-56. [DOI] [PubMed] [Google Scholar]
- 26.Salaun, C., P. Rodrigues, and J. M. Heard. 2001. Transmembrane topology of PiT-2, a phosphate transporter-retrovirus receptor. J. Virol. 75:5584-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneiderman, R. D., K. B. Farrell, C. A. Wilson, and M. V. Eiden. 1996. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J. Virol. 70:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skach, W. R. 2000. Defects in processing and trafficking of the cystic fibrosis transmembrane conductance regulator. Kidney Int. 57:825-831. [DOI] [PubMed] [Google Scholar]
- 29.Skach, W. R., and V. R. Lingappa. 1993. Amino-terminal assembly of human P-glycoprotein at the endoplasmic reticulum is directed by cooperative actions of two internal sequences. J. Biol. Chem. 268:23552-23561. [PubMed] [Google Scholar]
- 30.Sugai, J., M. Eiden, M. M. Anderson, N. Van Hoeven, C. D. Meiering, and J. Overbaugh. 2001. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or Pit2. J. Virol. 75:6841-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tailor, C., A. Nouri, and D. Kabat. 2000. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J. Virol. 74:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tailor, C. S., Y. Takeuchi, B. O'Hara, S. V. Johann, R. A. Weiss, and M. K. Collins. 1993. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J. Virol. 67:6737-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi, Y., R. G. Vile, G. Simpson, B. O'Hara, M. K. L. Collins, and R. A. Weiss. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Zeijl, M., S. V. Johann, E. Cross, J. Cunningham, R. Eddy, T. B. Shows, and B. O'Hara. 1994. An amphotropic virus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA 91:1168-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss, R. A., and C. Tailor. 1995. Retrovirus receptors. Cell 82:531-533. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson, B. M., A. J. Critchley, and C. J. Stirling. 1996. Determination of the transmembrane topology of yeast Sec61p, an essential component of the endoplasmic reticulum translocation complex. J. Biol. Chem. 271:25590-25597. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, C. A., M. V. Eiden, W. B. Anderson, C. Lehel, and Z. Olah. 1995. The dual-function hamster receptor for amphotropic murine leukemia virus (MuLV), 10A1 MuLV, and gibbon ape leukemia virus is a phosphate symporter. J. Virol. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, C. A., K. B. Farrell, and M. V. Eiden. 1994. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J. Virol. 68:7697-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]