Abstract
We have used X-ray crystallography to determine the structure of a decay accelerating factor (DAF)-binding, clinic-derived isolate of echovirus 11 (EV11-207). The structures of the capsid proteins closely resemble those of capsid proteins of other picornaviruses. The structure allows us to interpret a series of amino acid changes produced by passaging EV11-207 in different cell lines as highlighting the locations of multiple receptor-binding sites on the virion surface. We suggest that a DAF-binding site is located at the fivefold axes of the virion, while the binding site for a distinct but as yet unidentified receptor is located within the canyon surrounding the virion fivefold axes.
The echoviruses (enteric cytopathic human orphan viruses), a group of human virus isolates that could not be grown in suckling mice, are grouped with coxsackie B viruses to form a subgenus within the Enterovirus genus of the family Picornaviridae. While most echovirus infections are either asymptomatic or result in only mild symptoms, they can also cause meningitis, encephalitis, myocarditis, hemorrhagic conjunctivitis, and severe generalized neonatal infections. These syndromes are, however, not associated with single serotypes (30).
The molecular basis of enterovirus pathogenesis has been studied in detail for the polioviruses (11, 40, 44) and to a lesser degree for the group B coxsackieviruses (14, 19, 32). Studies of poliovirus have emphasized a role for the 5′ untranslated region in determining neurovirulence phenotypes (15, 22, 36), but the role of poliovirus receptor (PVR) distribution in pathogenesis has also been studied in some detail (39). Receptor usage patterns are also implicated in determining pathogenesis of cardiovirulent coxsackie B viruses (26). The molecular pathogenesis of other enterovirus infections has not been studied in detail; however, the use of different receptors by closely related viruses could in principle influence the pathogenesis directly by effecting tissue tropism to produce different clinical outcomes (10). Variation in receptor usage is found when different isolates of a single serotype are compared and following limited passage of clinical isolates in cell culture. Thus different clinical syndromes may result from preexisting viruses present within the infecting population or from the evolution of receptor or coreceptor use within the host during an infection.
For many picornaviruses, a single cell surface molecule appears to be sufficient for virus entry (binding and productive uncoating) in the experimental systems employed. For example, the PVR is used by all three poliovirus serotypes (28) while intercellular adhesion molecule 1 (ICAM-1) is the receptor for the major rhinovirus subgroup (45). In other cases, for example, a number of echoviruses and coxsackieviruses, the situation is more complex. A substantial number of enteroviruses that bind to the essentially ubiquitous complement regulatory protein decay accelerating factor (DAF; CD55) have been identified, and the DAF domains involved in infection for some viruses have been studied (4, 9, 46). These viruses are frequently capable of causing hemagglutination (HA) of human red blood cells, and this ability correlates with DAF binding (34, 38). In many cases monoclonal and polyclonal anti-DAF sera will efficiently inhibit binding, entry, and infection; the removal of DAF from cell surfaces by phospholipase (phosphatidylinositol-specific phospholipase C) treatment also inhibits these processes. However DAF binding alone is not sufficient to permit infection under most circumstances, and interaction with soluble DAF does not trigger formation of an “altered” (A) particle (4, 37), a structurally distinct form of the viral capsid (thought to be a cell entry intermediate (3, 25) where portions of the capsid proteins that were previously within the particle are externalized. However, it seems inherently unlikely that DAF binding by enteroviruses, a common event, is irrelevant to entry. Indeed DAF's ubiquity within the organism makes it ideal for sequestering viruses at cell surfaces. Thus the identification and analysis of the mechanisms by which DAF and its elusive coreceptors/cofactors act to promote infection are relevant to a large number of pathogenic viruses. The subtlety of the requirements for additional factors is emphasized by the plasticity of receptor usage in DAF-binding viruses; variant viruses with altered receptor usage are readily selected by passage.
The choice of receptor system is thus likely to be dependent on both the capsid structure of the particular virus and the disposition and quantity of binding/receptor molecules on the particular cell surface (which may be modulated by molecules which themselves make no contact with the virus capsid).
Picornaviruses possess a common capsid architecture, with the outer surface of the virus being made up of contributions from viral protein 1 (VP1), VP2, and VP3. Structures of rhinoviruses and enteroviruses have revealed a large depression running around the fivefold symmetry axis termed the canyon. Rossmann (41) proposed that this canyon could provide the site of interaction between these picornaviruses and their cellular receptors. Recent low-resolution structures have demonstrated that, while the details of the receptor-canyon interaction differ greatly for poliovirus (3, 15, 48) and ICAM-binding rhinoviruses (20) and coxsackieviruses (47), these viruses do indeed bind their cellular receptors within this surface depression.
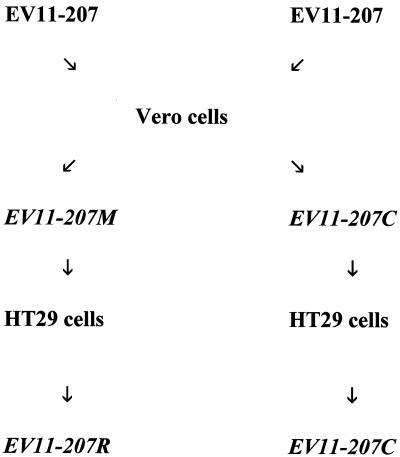
It is against this background that we have further characterized a DAF-binding clinical isolate of echovirus 11, EV11-207. We have previously reported a surface plasmon resonance study of EV11-207's DAF-binding properties (23), which mapped the site of interaction between DAF and EV11-207 to the third short consensus repeat (SCR) domain of DAF, which lies in the center of the four extracellular SCR domains. In light of this result it is difficult to envisage an organization of DAF SCR domains that would allow SCR3 to penetrate the canyon in the mode of a “classical” enterovirus receptor. We have now determined the structure of this virus isolate to nearly atomic resolution. We have also generated variants of EV11-207 in two independent experiments using serial blind passage of the virus on nonpermissive Vero (African green monkey kidney cells, which are human DAF negative) cells (Fig. 1), followed by passage on HT29 cells (the cell line used to propagate the parental virus) in an attempt to produce a revertant virus. We have characterized these variants and shown that they possess altered DAF-binding and utilization properties. These data are presented below.
FIG. 1.
Scheme for generation of variant viruses. See Materials and Methods for details.
MATERIALS AND METHODS
Cells and viruses.
The HT29 and Vero cells used in this study were obtained from the American Type Culture Collection. HT29 cells were passaged in RPMI medium supplemented with 10% fetal calf serum (FCS), 100 mM glutamine, 100 U of penicillin, and 10 μg of streptomycin/ml, and Vero cells were passaged in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% FCS, 100 mM glutamine, 100 U of penicillin, and 10 μg of streptomycin/ml. Virus wv207 (EV11-207) was isolated on HT29 cells from a clinical sample as previously described (35).
Antibodies and reagents.
Polyclonal anti-DAF (CD55) was a kind gift from Paul Morgan (Cardiff, United Kingdom. Monoclonal anti-DAF (monoclonal antibody 854) was a kind gift from Andrew Macadam (National Institute of Biological Standards and Control). Pischia pastoris expressing soluble DAF containing SCR domains 1 to 4 was a kind gift from David Evans (Glasgow, United Kingdom).
Generation and plaque purification of virus mutants EV11-207M, EV11-207R, and EV11-207C.
Confluent monolayers of Vero cells in 2-cm-diameter dishes were infected with purified EV11-207 at a multiplicity of infection (MOI) of 10. Infection was allowed to proceed for 3 days at 37°C. Adherent cells were scraped into the culture medium, which was then subjected to freezing and thawing three times followed by centrifugation at 2,000 × g for 5 min. A 10-fold dilution of this supernatant in culture medium was carried out, and a sample of this diluted material was used to infect a fresh plate of confluent Vero cells. This procedure was carried out seven times, and virus from the final passage was plaque purified on Vero cells three times. Five independent clones were isolated in this way; all proved to have identical characteristics on preliminary analysis, and one (EV11-207M) was chosen for further study. The same blind-passage procedure, using HT29 cells and infection with the EV11-207M variant, was carried out. This resulted in the isolation and plaque purification of a further variant, virus strain EV11-207R. In an independent experiment EV11-207 was passaged on Vero cells (as described above) to generate variant EV11-207C (passage of this variant back onto HT29 cells resulted in a virus with an identical sequence; hence only EV11-207C was used in further studies).
One-step growth curves.
One-step growth curves were obtained in the following manner. Confluent monolayers of cells in 24-well plates (Falcon) were pretreated with either medium alone or medium containing a polyclonal anti-DAF antibody (at 1:500) for 30 min at 37°C. These cells were then infected with virus at 5 PFU per cell (in the absence or presence of polyclonal anti-DAF). Virus was allowed to attach by incubation at room temperature for 60 min. The cells were then washed three times with phosphate-buffered saline, and the medium was replaced. Infection was allowed to continue by incubation at 37°C. At various time points the cells were scraped off and placed into 1.5-ml tubes. The tubes were frozen and thawed three times, and the medium was harvested and clarified by centrifugation at 2,000 × g for 5 min. Titers of virus were then assessed by plaque assay using HT29 cells for EV11-207 and EV11-207R and Vero cells for EV11-207M and EV11-207C.
Inhibition of virus replication by anti-DAF antibodies.
Cells in 96-well plates were pretreated with 50 μl of either medium alone or medium containing a 1:500 dilution of polyclonal anti-DAF for 30 min. Equal volumes of virus from a 10-fold dilution series were then added. Three replicate plates were produced. The level of virus cytopathic effect (CPE) was assessed 24, 48, and 72 h after the cells were fixed with formal saline and stained with 1% toluidine blue.
HA assay.
Enterovirus samples were serially diluted twofold. Fifty microliters of a virus dilution was added to an equal volume of 1% (vol/vol) type O human red blood cells in phosphate-buffered saline in round-bottom 96-well plates (Falcon). Samples were mixed and incubated at 4°C for 1 h. Controls of red blood cells incubated with mock-infected cell supernatants were routinely included. One HA unit was considered to be present in the first well in which HA was observed.
HA inhibition (HAI) assay.
Four HA units of virus was incubated with twofold serial dilutions of soluble DAF (starting concentration of 8 μM) for 1 h at room temperature. Fifty microliters of this mixture was added to an equal volume of 1% red blood cells and incubated for 1 h at 4°C. Twofold serial dilutions of anti-DAF monoclonal antibody 854 (starting concentration, 0.001×) were incubated with 1% red blood cells for 1 h at room temperature. Four HA units of virus was added, and the mixture was incubated for 1 h at 4°C.
Viral genome sequencing.
Virus RNA was purified by a method derived from that described by Chomczynski and Mackey (8). Concentrated virus particles (approximately 1011 PFU) were mixed with 1 ml of denaturing solution (4 M guanidinium isothiocyanate, 25 mM sodium citrate [pH 7], 0.5% sarcosyl, 100 mM 2-mercaptoethanol) and vortexed briefly. Sequential addition of 0.1 ml of 2 M sodium acetate, pH 4, 1 ml of phenol saturated with H2O, and 0.2 ml of a chloroform-isoamyl alcohol mixture (49:1 [vol/vol]) was followed by mixing and chilling on ice prior to centrifugation at 13,000 × g for 20 min at 4°C. The aqueous phase was mixed with 1 ml of isopropanol in a fresh tube and placed at −70°C for 1 h. The precipitated RNA was recovered by centrifugation at 13,000 × g for 20 min at 4°C. The pellet was dissolved in 0.3 ml of denaturing solution and precipitated once more with 1 volume of isopropanol. After recentrifugation the pellet was washed in 75% ethanol and dried under vacuum for 5 min
Sequence was obtained by direct, automated sequencing of PCR products. First-strand cDNA was synthesized from RNA prepared as described above by using Expand reverse transcriptase (Roche). PCR of the bulk of the genome was carried out with primer E9 (TGGCTGCTTATGGTGACAAT) and the NotI (dT)18 primer using a long-PCR kit from Roche. Cycling conditions were as follows: 3 min at 94°C; 10 cycles of 94°C for 30 s, 56°C for 1 min, and 68°C for 6 min; 20 cycles of 94°C for 30 s, 56°C for 1 min, and 68°C for 10 min; and finally 7 min at 72°C. The 5′-most 600 nucleotides were amplified with 5′ primer TTAAAACAGCCTGTGGGTTGATCCCA (24) and 3′ primer EV2 (CACCGGATGGCCAATCCA). Cycling conditions for this reaction were as follows: 3 min at 94°C; 30 cycles of 94°C for 30 s, 56°C for 1 min, and 68°C for 6 min; and finally 7 min at 72°C. PCR products were gel purified with Qiaex II (Qiagen) and sequenced with primers based on the previously determined virus sequence. DNA sequencing reactions were carried out with an Applied Biosystems Big Dye Terminator Ready Reaction kit and analyzed on either a PE/ABI Prism 377 or an ABI Prism 3700 sequence analyzer.
Purification of EV11-207.
Purification of virus was carried out using standard protocols. Briefly, confluent HT29 monolayers were infected with virus at a multiplicity of infection of 5 PFU/cell. Incubation was continued until complete CPE had developed (normally 16 h). Intracellular virus was released by freezing and thawing cells twice, and supernatants were clarified by centrifugation for 5 min at 1,000 × g. Initial purification of the virus was carried out by centrifugation of supernatants through a 30% (wt/vol) sucrose cushion at 100,000 × g for 2.5 h. The virus was then resuspended, and 10,000 cpm of radioactively labeled purified EV11-207 was added. The virus was then purified by rate zonal centrifugation on 15 to 45% sucrose gradients in a Beckman SW28 rotor for 4 h at 100,000 × g. Samples of 1.5 ml were collected, and 10 μl was subjected to scintillation counting. Fractions containing the radioactivity peak were selected; virus was pelleted by centrifugation at 100,000 × g for 2.5 h, and the rate zonal centrifugation step on 15 to 45% sucrose gradients was repeated after resuspension of virus. All sucrose solutions contained 10 mM Tris-Cl, pH 7.4. Purity of the virus preparation was monitored by electron microscopy.
Crystallization.
EV11-207 stocks were concentrated to approximately 10 mg/ml (as judged by optical densities at 260 and 280 nm) with a Centricon 30s (Amicon) and crystallized by equilibrating 2-μl sitting drops (1 μl of virus plus 1 μl of mother liquor) with microbridges (Crystal Microsystems, Oxford, United Kingdom) against 0.5 ml of mother liquor sealed in Linbro plates. Initial screens were made using the Hampton Research Crystal Screen 1 kit. Poor crystals grew in several conditions, and crystals suitable for collection of X-ray diffraction data grew in condition 32 (2.0 M ammonium sulfate). Crystal growth was further optimized by dilution of condition 32.
Structure solution and refinement.
Data were collected at the Synchrotron Radiation Station Daresbury (station 7.2) and the European Synchrotron Radiation Facility (ESRF) Grenoble (station ID2) at room temperature using crystals mounted in sealed quartz capillary tubes. This method of data collection was chosen as it allowed containment of the potentially infectious virus at all times during data collection. Data were collected as 0.5 to 1.5o oscillation images on MarResearch imaging plates (diameters, 18 [Daresbury] and 30 cm [ESRF]). Collection of data to 2.9 Å at the ESRF required swinging the detector away from the straight-through position by 15o.
RESULTS
Generation of EV11-207 variants by passage in cell culture.
Isolate EV11-207 (EBI accession no. AJ276224) is strikingly deficient in its capacity to replicate on Vero cells compared with highly passaged EV11 type strain Gregory (data not shown). However, EV11-207 RNA transfected into Vero cells replicates efficiently. This suggests that the inability of EV11-207 to replicate in Vero cells is a result of a defect in entry. Previous observations on the adaptation of isolates to growth in cell cultures of simian origin suggested that the ability to grow on Vero cells would be readily acquired by passage. The isolation, under controlled conditions, of such variants which are phenotypically similar to the EV11 type strain (Gregory) would enable analysis of the genetic and structural basis of this change in entry phenotype against the background of our EV11-207 sequence and structure.
An attempt to alter the cell tropism by blind passage in cultures of Vero cells was therefore made. Signs of CPE were observed after the third passage, and a growth rate equivalent to that of the parental strain was observed after the sixth passage. Initial analysis of the virus strains that resulted from plaque purification of the product of the seventh passage revealed that all the viruses showed an identical growth phenotype: they formed plaques efficiently on Vero cells, but their plaque-forming efficiency on HT29 cells was severely compromised. One of these isolates, EV11-207M, was chosen for further study. In an independent experiment, carried out by an identical method, the resulting virus was termed EV11-207C.
In an effort to obtain revertants or pseudorevertants of EV11-207M and EV11-207C, the viruses were subjected to seven passages on HT29 cells. Signs of CPE were observed after the second round of passage. Three rounds of plaque purification were carried out after the seventh passage. Once more all of the viruses isolated in this way showed identical phenotypes. The virus resulting from passage of EV11-207M was termed EV11-207R. However, subsequent studies showed that the virus generated by passage of EV11-207C on HT29 cells was identical to its parent and so will not be discussed further.
Characterization of virus mutants EV11-207M and EV11-207R. (i)Analysis of DAF-binding properties of EV11-207M and EV11-207R.
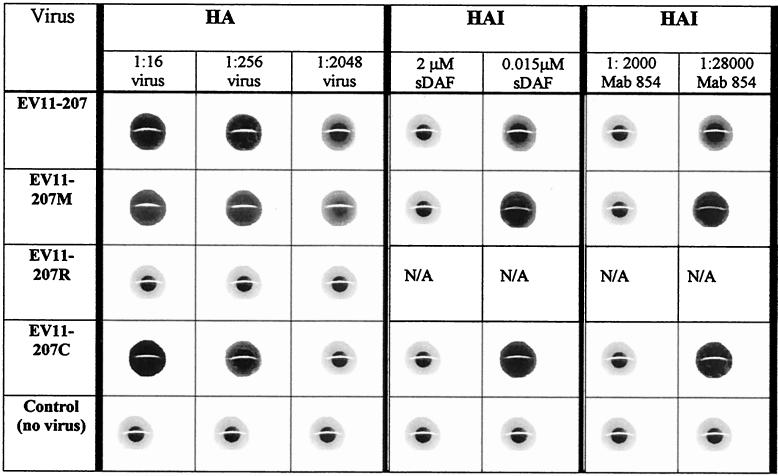
We have shown previously that EV11-207 is able to interact with DAF (23). To examine the DAF-binding ability of mutants EV11-207M, EV11-207R, and EV11-207C, we used a HA assay with human red blood cells. The results of the HA assay showed that EV11-207, EV11-207M, and EV11-207C could hemagglutinate red blood cells but that EV11-207R could not (Table 1). The HA titers for EV11-207, EV11-207M, and EV11-207C were each approximately 104 HA units/ml. The DAF-dependent nature of the HA was examined by using a HAI assay with anti-DAF monoclonal antibody 854 and soluble DAF. Both the monoclonal antibody and the soluble DAF could inhibit the HA induced by EV11-207, EV11-207M, and EV11-207C (Table 1), but a control antibody against CD59 had no effect (data not shown), indicating that all three viruses bind DAF. The results from the HA assay show that EV11-207R does not bind DAF at levels detectable in this assay.
TABLE 1.
DAF-specific HA of human red blood cells by EV11-207, EV11-207M, and EV11-207Ca
Columns HA show the HA resulting from incubation of human red blood cells with dilutions of EV11-207, EV11-207M, EV11-207R, or EV11-207C. The DAF specificity of this reaction was studied by using a HAI assay (columns HAI) with dilutions of soluble DAF (sDAF) or monoclonal antibody (Mab) 854 as shown. The control used was mock-infected cell supernatant.
(ii) Growth phenotypes.
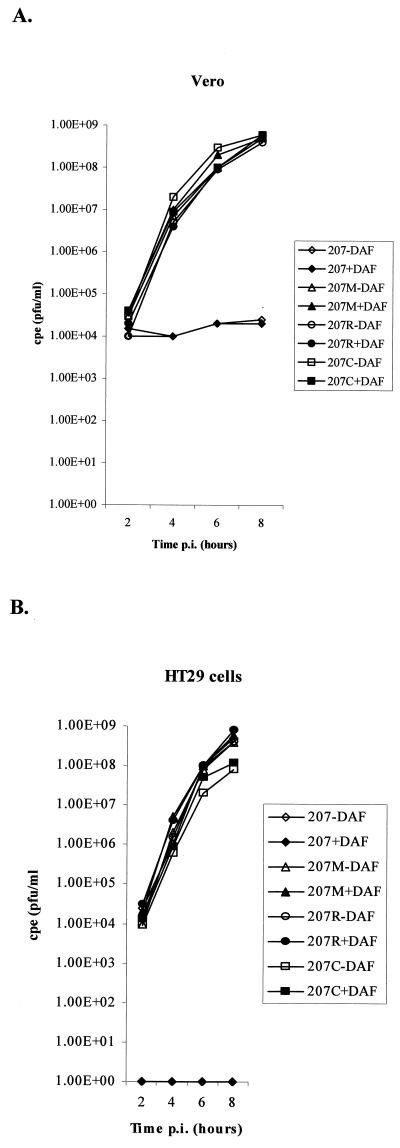
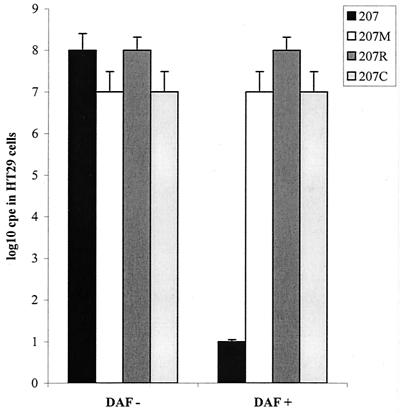
The capacity of viruses EV11-207, EV11-207M, EV11-207R, and EV11-207C to replicate in HT29 and Vero cells was investigated by measuring their one-step growth in these cell types in the absence or presence of polyclonal anti-DAF antibodies. These data demonstrated that EV11-207 was strikingly deficient in its ability to infect Vero cells (Fig. 2A) but could successfully infect HT29 cells (Fig. 2B). The three mutants (EV11-207M, EV11-207R, and EV11-207C) were able to infect both Vero cells (Fig. 2A) and HT29 cells (Fig. 2B).
FIG. 2.
One-step growth curves of EV11-207 and variants EV11-207M, EV11-207R, and EV11-207C on Vero and HT29 cells. (A) Titers resulting from infections of Vero cells by the four viruses in the presence and absence of polyclonal anti-DAF sera. (B) Titers from infection of HT29 cells, once again in the presence and absence of polyclonal anti-DAF sera.
The presence of the anti-DAF antibodies showed that infection of HT29 cells by EV11-207 is DAF dependent; however infection of Vero and HT29 cells by the mutants is DAF independent.
These data also reveal that, although EV11-207 is unable to replicate in Vero cells, it is able to adhere to the cells (both in the presence and absence of anti-DAF antibodies), as the virus added to the cells is not removed by the wash step (Fig. 2A). This is in contrast to what is found when EV11-207 is preincubated with polyclonal anti-DAF sera prior to addition to HT29 cells: the data presented in Fig. 2B clearly show that no virus remains adherent on the cell surface after washing.
(iii) Antibody inhibition of growth.
The effect of antibodies specific for DAF on the replication of viruses EV11-207, EV11-207M, EV11-207R, and EV11-207C was investigated. Results from antibody blocking assays shown in Fig. 3 demonstrate that DAF is necessary for infection by EV11-207 but is not required by EV11-207M, EV11-207R, or EV11-207C. Preincubation of cells with polyclonal anti-DAF sera resulted in a 106- to 107-fold reduction in the CPE resulting from infection by EV11-207 of HT29 cells (Fig. 3) but not Vero cells (data not shown). The antibodies were unable to block infection of either of the cell lines with EV11-207M, EV11-207R, or EV11-207C. Antibodies to receptors used by other enteroviruses (CD59, CAR, ICAM-1, and CD46) were tested to see if they had any effect on the EV11 viruses. None of these sera effected the growth of these EV11 viruses in HT29 or Vero cell lines (data not shown).
FIG. 3.
Infection by EV11-207 is inhibited by anti-DAF antibodies. HT29 cells were preincubated with a rabbit polyclonal sera (at a 1/500 dilution). Inhibition of CPE resulting from antibody treatment was scored 48 to 72 h after infection with EV11-207, EV11-207M, EV11-207R, or EV11-207C.
(iv) Sequencing of variant viruses.
To determine the genetic basis of these changes in cell tropism, the complete genomic sequences of viruses EV11-207M, EV11-207R, and EV11-207C were derived. The amino acid changes from the original EV11-207 sequence are presented in Table 2. There were no amino acid changes in VP4, VP3, or the nonstructural proteins.
TABLE 2.
Mutations present in cell tropism variants EV11-207M, EV11-207R, and EV11-207Ca
| Position | AA in:
|
|||
|---|---|---|---|---|
| EV11-207 | EV11-207M | EV11-207R | EV11-207C | |
| VP1 AA78 | E | Q | Q | E |
| VP1 AA83 | E | E | D | E |
| VP1 AA88 | K | K | K | E |
| VP1 AA132 | Q | K | K | K |
| VP1 AA240 | F | F | F | L |
| VP1 AA259 | K | E | E | K |
| VP2 AA139 | G | R | R | G |
| VP2 AA164 | N | N | D | N |
The amino acids (AA) present in the parent virus and variant viruses are shown for VP1 and VP2.
Determination of the structure of EV11.
Analysis of the symmetry of Bragg diffraction revealed that the crystals belong to space group R32 with unit cell dimensions of a = b = 300.9 Å and c = 1476.6 Å (all dimensions and coordinates described below use the hexagonal indexing scheme). Packing considerations suggested that one-third of a particle constitutes the crystallographic asymmetric unit, indicating that the center of the virus lay on the crystallographic threefold axis. X-ray data were indexed, integrated, and scaled with DENZO and SCALEPACK (33). The final data set consists of 167 images recorded from 75 crystals yielding 411,040 unique reflections (70% complete in the range of 40 to 2.9 Å) obtained by merging 1,648,270 reflections with a merging R factor of 22% (35% in the outer shell). Calculation of a native Patterson map by using data between 50 and 10 Å showed a peak with coordinates (1/3, 2/3, 1/6), equivalent to a Patterson vector with length (0, 0, [1/2]). This peak arises from two particles related by a crystallographic twofold axis (at Z = 0); the center of the virus therefore lies on the threefold axis along Z with a displacement of one-fourth of the cell edge (369.2 Å). A complete alpha-carbon model of coxsackievirus A9 (CAV9) (17) was then positioned at the origin of the unit cell with a threefold axis aligned along Z and a twofold axis aligned along X and then rotated around Z to search for the best orientation of the particle. Calculation of a Patterson correlation (PC) function at 0.2o intervals using data in the range of 12 to 8 Å revealed a maximum (PC = 20%) for a 1.8o rotation about the threefold axis. Rotation of the alpha-carbon model by 1.8o and translation to one-fourth in Z gave a correlation coefficient of 42% for data between 10 and 6 Å. Fifty steps of rigid-body minimization using data in the same range refined the rotation to be 1.88o and the translation to be 369.13 Å along z. These transformations were used as a skew matrix and applied to the standard 20 icosahedral operators required to generate the portion of the virion in the asymmetric unit from the icosahedral protomer. By using data to 3.5 Å, a full-atom model of CAV9 (17) was subjected to simple B-factor and positional refinement, giving an R factor of 33% (X-PLOR [5]), and a set of model amplitudes and phases was calculated. These were scaled to the observed amplitudes in resolution shells (program SHELL_SCALE) and used as the start point for 20 cycles of noncrystallographic symmetry averaging (over the 20 copies of the viral protomer in the asymmetric unit) and solvent flattening (with program GAP) to generate a 3.5-Å map into which the EV11 model was built. The phases generated by averaging had a mean phase difference of 33o from those generated from the CAV9 model. As expected, a 2Fobserved − Faveraged, αaveraged map showed essentially no bias toward the starting model, with the electron density map clearly showing side chains and loops differing significantly from those in the starting model. Cycles of refinement and rebuilding (using programs XPLOR [5], CNS [6], and O [18]) and incorporation of the data to 2.9 Å have led to a model with R = 24% for all data to 2.9 Å and 82% of residues in the most-favored regions of the Ramachandran plot (Table 3). Coordinates (identifier, 1 h8t) and structure factors (identifier, r1 h8tsf) have been deposited in the Protein Data Bank
TABLE 3.
Refinement statistics and model quality
| Parameter | Value |
|---|---|
| Refinement statistics | |
| No. of reflections used in refinement | 402,349 |
| Completeness in range 500-2.9 Å (%) | 71 |
| No. of nonhydrogen atoms in refinement | |
| Protein | 6,534 |
| Water | 209 |
| Cofactors | 30 |
| Crystallographic R factor (%) | 24 |
| Avg B factor (Å2) | |
| Protein | 17 |
| Water | 20 |
| Cofactors | 35 |
| Root mean square deviation between B factors for bonded atoms (Å) | 5 |
| Root mean square deviation between B factors for angle-related atoms (Å>) | 7 |
| Model chemistry | |
| Root mean square deviation from ideal values | |
| Bond lengths (Å) | 0.017 |
| Bond angles (°) | 2 |
| Ramachandran plot statistics (%) | |
| Core regions | 82 |
| Disallowed | 1 |
Structural Analysis. (i) Comparison to other enteroviruses.
As expected the overall structure of EV11 is similar to those of other picornaviruses and overall most similar to those of other enteroviruses; the main differences in the protein backbone structure occur in the surface-exposed antigenic loops (data not shown). Table 4 presents the root mean square difference in alpha-carbon position between the individual viral proteins and those of representatives of the main human picornavirus families for which structures are known. As for other enteroviruses the pocket in the center of the VP1 β-barrel is occupied by an extended aliphatic chain, here modeled on the basis of the electron density as palmitic acid (C16O2) as found in EV1 (13).
TABLE 4.
RMSD in alpha-carbon positions for comparisons with some other human picornavirusesa
| EV11 protein (no. of residues) | RMSD in alpha-carbon position (Å) versus that for:
|
||||
|---|---|---|---|---|---|
| EV1 (1EV1 [13]) (VP1, 281 residues; VP2, 254 residues; VP3, 239 residues) | CAV9 (1D4M [17]) (VP1, 299 residues; VP2, 261 residues; VP3, 238 residues) | Coxsackievirus B3 (1COV [31]) (VP1, 281 residues; VP2, 263 residues; VP3, 238 residues) | Type 1 poliovirus (2PLV [12]) (VP1, 302 residues; VP2, 272 residues; VP3, 238 residues) | Human rhinovirus 14 (4RHV [1]) (VP1, 289 residues; VP2, 262 residues; VP3, 238 residues) | |
| VP1 (289) | 0.8 (278) | 1.0 (277) | 0.9 (266) | 1.5 (266) | 1.8 (264) |
| VP2 (254) | 0.6 (250) | 0.6 (250) | 0.7 (250) | 1.7 (250) | 1.2 (249) |
| VP3 (240) | 0.8 (237) | 0.8 (234) | 0.7 (237) | 0.8 (234) | 1.0 (235) |
The root-mean square deviation (RMSD) in alpha-carbon positions is presented for the major capsid proteins of EV11 compared with proteins from some other human picornaviruses. The numbers of equivalent alpha carbons used to calculate the RMSD are shown in parentheses after the RMSD. The names and protein data bank codes for the other viruses are shown.
(ii) Mapping of variant virus changes onto structure.
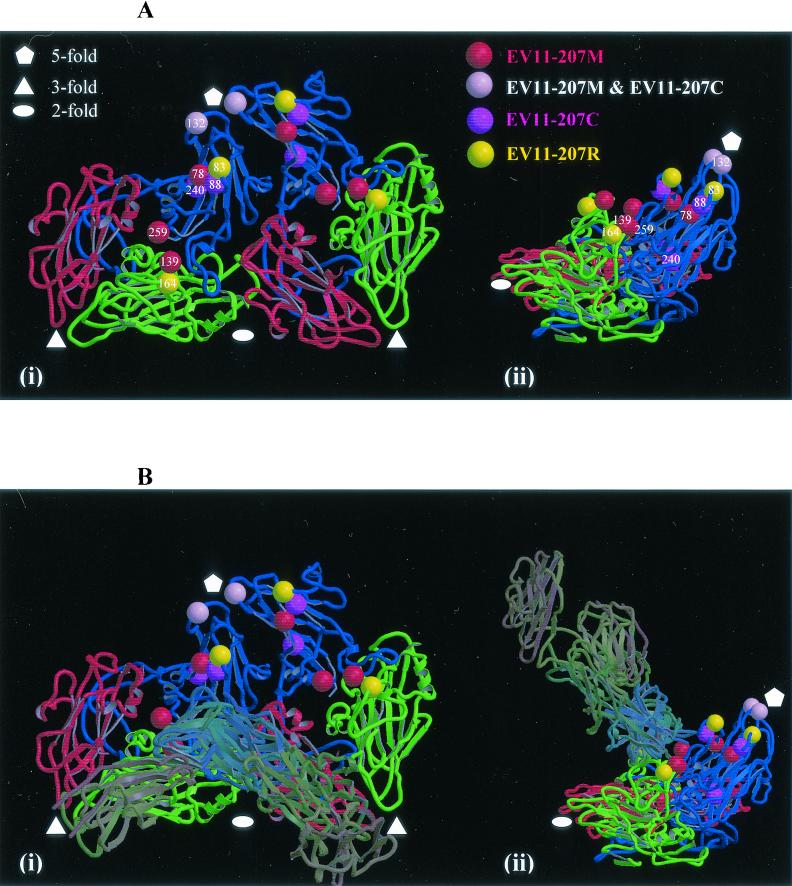
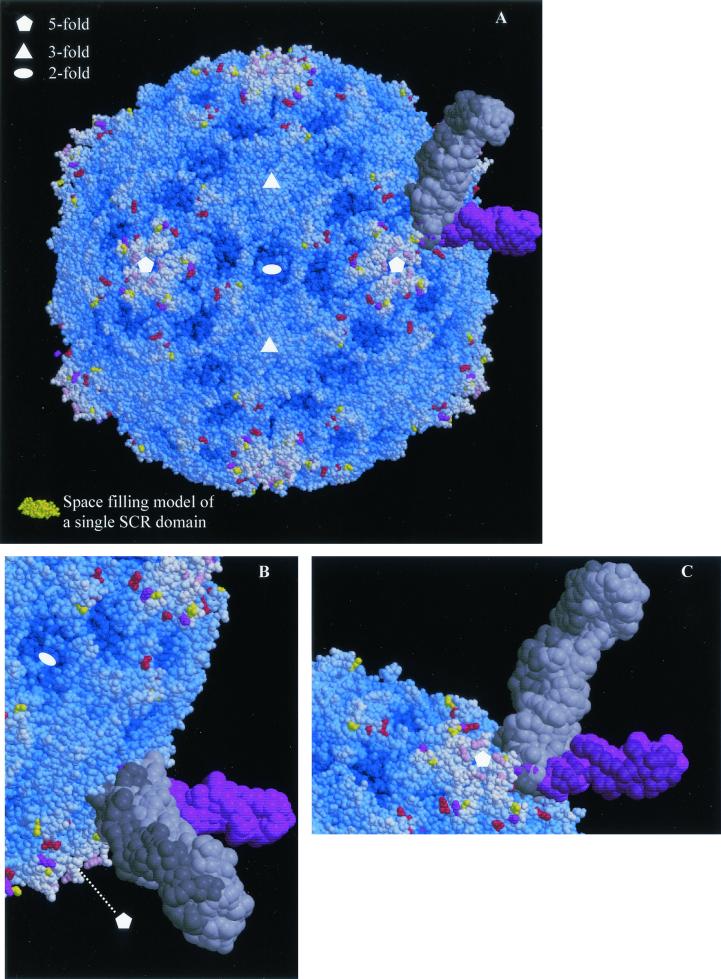
The structure shows that with the exception of conservative EV11-207C change of VP1 F240L the locations of all of the amino acid changes are on the surface of the virion (Fig. 4 and 5). VP2 G139R and VP2 N164D are both in the VP2 EF loop, VP1 E78Q, VP1 N83D, and VP1 K88E are in the BC loop, VP1 Q132K is in the DE loop, and VP1 K259E is at the C terminus of VP1. All of these changes are found in regions of considerable variation among published enterovirus sequences. Residues VP2 139R, VP2 164D, VP1 78Q, and VP1 132K are novel amino acids, while VP1 83D and VP1 159E are present among other sequenced enteroviruses. All of the changes represent significant alterations in size and/or charge. As indicated in Fig. 5 these changes map out an extended surface area and as such perturb a larger region that would seem consistent with the footprint of a single DAF domain (a single SCR domain is shown to allow the scale to be judged) and may therefore be interpreted as defining more than one receptor binding site. Single copies of the receptor molecules from those picornavirus-receptor complexes for which coordinates were available (HRV14-ICAM-1 and HRV16-ICAM-1 (21) and polio-PVR [3, 15]) have been overlaid on the EV11 structure. It may be seen that, while the changed residues in EV11-207M (with the exception of that at position 132 of VP1) located around the periphery of the canyon might be able to interact with a canyon-binding receptor of the same general class as the receptors shown, EV11-207M change VP1 Q132K seems to suggest a receptor interacting with the fivefold axes of the virion although it is not possible to rule out a side chain conformation which would allow interaction with a molecule binding in the canyon.
FIG. 4.
The structure of the viral protomer. (A) The structure of viral protomer is shown as a ribbon diagram to highlight secondary structural elements. Blue, VP1; green, VP2; red, VP3. Two neighboring protomers within a pentamer are shown, and the two views are related by a 90° rotation about the vertical axis (applicable also to panel B). The locations of the amino acid changes seen in variants are indicated (as shown in the key). (B) The same views as in panel A are shown with the structures of ICAM-1 and the PVR overlaid. These coordinates were obtained from the low-resolution structures of these receptors in complex with human rhinovirus 16 and human rhinovirus 14 (20) and poliovirus (3, 16). The coordinates were superposed on the EV11-207 structure by superposition of the picornavirus proteins. Both panels were drawn with Molscript (21) and rendered with Raster3D (29).
FIG. 5.
Mapping of EV11-207M, -C, and -R amino acid changes onto the surface of EV11-207. (A) Space-filling model of the complete EV11-207 virion. The majority of residues are blue, with the depth of color relating to their distance from the center of the virion. Those residues that are altered in the variants are colored as in Fig. 4. As in Fig. 4B the overlaid structures of PVR and ICAM-1 are shown to indicate the range of interactions with the viral capsid made possible by different modes of canyon-binding receptors. A single SCR domain from factor H (2) is shown in gold so that the small size of an SCR domain compared to those of the immunoglobulin (Ig) domains contained in ICAM-1 (two Ig domains contained in each coordinate set) and PVR (three Ig domains in each coordinate set) may be judged. (B and C) The canyon-binding receptors are shown more closely with views from opposite sides shown in the two panels. All panels were drawn with Molscript (21) and rendered with Raster3D (29).
The EV11-207R changes seem to lie at the extreme borders of the canyon; the change at position 83 of VP1 is most probably connected with altering the affinity for a molecule binding at the fivefold axes, although it is not possible to rule out altogether a conformation for the side chain of 83 that would enable it to contact a canyon-binding receptor. Residue 164 of VP2 is at the lower periphery of the canyon, and substitutions here are likely to affect the binding of molecules in the canyon. The two unique changes seen in EV11-207C (VP1 K88E and VP1 F240L) both occur within the canyon with VP1 88E exposed on the upper borders of the canyon. The EV11-207M change is therefore as likely to affect the binding of a molecule within the canyon as the EV11-207C changes. In contrast to this the change shared with the other two isolates (VP1 Q132K), as previously noted, is most likely to perturb the binding of a molecule at the fivefold axes of the virion. VP1 F240L (the other unique change seen in EV11-207C) lies in the pocket under the canyon and may either have no effect on receptor binding, due to its conservative nature, or exert some subtle effect due to alteration in the precise geometry of the floor of the pocket.
DISCUSSION
The structures of the three major capsid proteins, VP1 to VP3, closely resemble those of the other enteroviruses of known structure, with VP1 being the most structurally variant and VP3 being the most structurally conserved at the level of alpha carbons (Table 3). Our structure of EV11-207 allows the mapping of the changes present in this series of EV11 strains (Fig. 5) and shows that they all cluster on the virion surface. This is consistent with an interpretation of the biological phenotypes in terms of direct effects on cellular interactions. Other aspects of the structure are beyond the scope of this paper and will not be discussed further.
Considering the viruses one by one it can be seen that infection of HT29 cells by EV11-207 is dependent on DAF binding and that the virus is not able to infect cells in the absence of DAF although, as noted above, it is able to adhere to Vero cells in a DAF-independent fashion (Fig. 2A). By contrast the changes present in EV11-207M, while leaving it able to bind DAF, apparently allow infection of cells in a DAF-independent fashion. The assays used do not allow us to determine if the affinity of EV11-207M for DAF is subtly altered (use of surface plasmon resonance as previously described [22] was not possible due to difficulties in preparing large amounts of purified virus). We propose that this phenotype arises because the changes in VP1 and VP2, which cluster on the surface of the virion, act to increase the affinity of the virus for another cell surface molecule, which EV11-207M then utilizes as the receptor for cell entry. There has been much discussion in the literature about the role of DAF as a picornavirus receptor (37, 42, 43). DAF is thought by these groups to act as a primary receptor, sequestering virus at the cell surface, and so enabling interaction with another protein, which acts as the true receptor, leading to entry of the cell.
However, we have demonstrated previously that the interaction between EV11-207 and DAF plays an active role during infection and that DAF does not simply sequester virus at the cell surface. We have shown that binding to DAF directs internalization of EV11-207 through membrane domains known as lipid rafts and that it is during entry via this pathway that the virus uncoats and releases its RNA (A. D. Stuart, H. E. Eustace, T. A. McKee, and T. D. K. Brown, unpublished data). Work by Powell et al. (37) supports this hypothesis that some DAF-binding echoviruses, in contrast to other enteroviruses such as poliovirus, do not form A particles by interacting with a DAF at the cell surface but instead interact with an unknown factor(s) inside the cell during the entry process to uncoat the virus RNA.
In this light, we propose that the four changes between EV11-207 and EV11-207M, which have no discernible effect on the ability of EV11-207M to bind DAF, act by increasing the affinity of the virus for another cell surface molecule. This second molecule may not be used by EV11-207 (presumably due to insufficient affinity) since EV11-207 and EV11-207M enter the cell through different pathways (unpublished results). Since we are looking at the effects of only a few amino acid changes, increasing the affinity of a preexisting receptor-binding site seems more plausible than creation of a novel binding site on the virion surface. This hypothesis is supported by the ability of EV11-207 to adhere to Vero cells in a DAF-independent fashion (Fig. 2A). The changes present in EV11-207C must also act to increase the affinity for this alternate receptor.
The further changes present in EV11-207R result in the loss of DAF binding. These observations would be explained if the changes unique to EV11-207R leave binding to the second receptor unaffected but eliminate DAF binding.
In summary our interpretation is that the EV11-207R changes act to abolish the DAF-binding site while the EV11-207M changes act to increase the affinity for binding to a currently unidentified receptor. Mapping these changes onto the structure of EV11-207 might therefore point to those regions of the virion surface which are involved in binding DAF and the EV11-207M, -C, and -R receptor. This mapping is presented in Fig. 4 and 5 and leads us to propose that the EV11-207M, -C, and -R receptor is a classical, canyon-binding, picornavirus receptor in the mode of poliovirus-PVR (3, 15, 48), rhinovirus 16- and rhinovirus 14-ICAM-1 (20), and coxsackievirus A21-ICAM-1 interactions (47). The DAF-binding sites, in contrast, appear to lie at the fivefold axes. The mapping of DAF binding to the fivefold axes contrasts with work on other DAF-binding enteroviruses (42, 43), which has suggested that a depression at the twofold axis is important in DAF binding. This inconsistency can only be resolved by use of reverse genetics to map DAF binding precisely.
Binding in an exposed site (either at the two- or fivefold sites) rather than within the canyon may relate to the low affinity of EV11-207 for DAF (KD, ∼3 μM [23]) compared with that of poliovirus for PVR (estimates for KD range between 80 [48] and 100 to 700 nM [27]) or that of rhinovirus for ICAM-1 (KD, ∼190 nM [7]), since there is likely to be a smaller contact area between the virus and DAF leading to a weaker interaction overall.
Our data therefore support the idea that EV11 (and other DAF SCR3-binding viruses) infect cells via an infection pathway that involves interactions with a minimum of two proteins. The primary interaction is with the numerous DAF molecules present in the extracellular membrane; these interactions are characterized by a low affinity and a rapid off rate. By binding to DAF, the virus becomes sequestered in lipid raft domains, leading to the internalization and uncoating of the virus RNA by the interaction with an additional unidentified intracellular factor(s). The non-DAF-using EV11 mutants, however, appear to use a more classical picornavirus infection route, interacting with a cell surface receptor which is likely to be an extended molecule that binds with high affinity to the canyon surrounding the fivefold axis. Interaction with this receptor provides sufficient energy to allow A particle formation and hence cellular infection.
The use of the DAF SCR3 domain for entry by a substantial number of echovirus and coxsackie B virus clinical isolates is well documented. It is our view that this interaction initiates internalization of these viruses, thus permitting interactions with a further molecule (or molecules) that promote uncoating. However it is also clear that only a limited number of amino acid changes on the virion surface are required to permit productive recognition of alternative receptors. Given the high error rate in replication, such changes might occur during natural infections. The extent to which selection of receptor use variants actually occurs during natural infections and the specific identities of the alternate receptors are currently being investigated
Acknowledgments
Thanks to Bob Posse (IVEM, Oxford) for space in his category 2 virus laboratory; to Jonathan Grimes, Liz Fry, John Newman, Pete Mertens, and Bjarne Rasmussen for assistance in data collection at the ESRF; to David Goodridge and Stuart Williams in their role as disease security officers while X-ray data were collected; to Jim Gray (PHLS, Cambridge) for assistance with electron microscopy; to Paul Morgan (Cardiff) for anti-CD55 antibodies; and to David Evans (Glasgow) for soluble recombinant CD55. John Tate is thanked for assistance with preparation of figures.
S.M.L. was supported by a Royal Society Dorothy Hodgkin Fellowship. P.A.W. and A.D.S. were supported by the Wellcome Trust, and A.D.S. was also supported by Action Research. S.S. was supported by the PHLS central fund.
REFERENCES
- 1.Arnold, E., and M. G. Rossmann. 1990. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 angstroms. J. Mol. Biol. 211:763-801. [DOI] [PubMed] [Google Scholar]
- 2.Barlow, P. N., A. Steinkasserer, D. G. Norman, B. Kieffer, A. P. Wiles, R. B. Sim, and I. D. Campbell. 1993. Solution structure of a pair of complement modules by nuclear magnetic resonance. J. Mol. Biol. 232:268-284. [DOI] [PubMed] [Google Scholar]
- 3.Belnap, D. M., B. M. McDermott, D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergelson, J. M., M. Chan, K. R. Solomon, N. F. St. John, H. Lin, and R. W. Finberg. 1994. Decay accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc. Natl. Acad. Sci. USA 91:6245-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunger, A. T. 1992. X-Plor version 3.0. Yale University, New Haven, Conn.
- 6.Brunger, A. T., P. D. Adams, M. G. Clore, W. L. Delano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, N. Nilges, N.-S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Casasnovas, J. M., and T. A. Springer. 1995. Kinetics and thermodynamics of virus binding to receptor. Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J. Biol. Chem. 270:13216-13224. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and K. Mackey. 1995. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. BioTechniques 19:942-945. [PubMed] [Google Scholar]
- 9.Clarkson, N. A., R. Kaufman, D. M. Lublin, T. Ward, P. A. Pipkin, P. D. Minor, D. J. Evans, and J. W. Almond. 1995. Characterization of the echovirus 7 receptor: domains of CD55 critical for virus binding. J. Virol. 69:5497-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, D. J., and J. W. Almond. 1998. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 6:198-202. [DOI] [PubMed] [Google Scholar]
- 11.Evans, D. M., G. Dunn, P. D. Minor, G. C. Schild, A. J. Cann, G. Stanway, J. W. Almond, K. Currey, and J. V. Maizel, Jr. 1985. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature 314:548-550. [DOI] [PubMed] [Google Scholar]
- 12.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filman, D. J., M. W. Wien, J. A. Cunningham, J. M. Bergelson, and J. M. Hogle. 1998. Structure determination of echovirus 1. Acta Crystallogr. D Biol. Crystallogr. 54:1261-1272. [DOI] [PubMed] [Google Scholar]
- 14.Godeny, E. K., and C. J. Gauntt. 1987. In situ immune autoradiographic identification of cells in heart tissues of mice with coxsackievirus B3-induced myocarditis. Am. J. Pathol. 129:267-276. [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez, A. L., M. Denova Ocampo, V. R. Racaniello, and R. M. Del Angel. 1997. Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J. Virol. 71:3826-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, Y., V. Bowman, S. Mueller, C. Bator, J. Bella, X. Peng, T. Baker, E. Wimmer, R. Kuhn, and M. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendry, E., H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, J. Santti, M. Maaronen, T. Hyypia, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 15:1527-1538. [DOI] [PubMed] [Google Scholar]
- 18.Jones, T., Y. Zou, S. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 19.Klingel, K., C. Hohenadl, A. Canu, M. Albrecht, M. Seemann, G. Mall, and R. Kandolf. 1992. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc. Natl. Acad. Sci. USA 89:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 22.La Monica, N., J. W. Almond, and V. R. Racaniello. 1987. A mouse model for poliovirus neurovirulence identifies mutations that attenuate the virus for humans. J. Virol. 61:2917-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lea, S. M., R. M. Powell, T. McKee, D. J. Evans, D. Brown, D. I. Stuart, and P. A. van-der-Merwe. 1998. Determination of the affinity and kinetic constants for the interaction between the human virus echovirus 11 and its cellular receptor, CD55. J. Biol. Chem. 273:30443-30447. [DOI] [PubMed] [Google Scholar]
- 24.Lindberg, A. M., C. Polacek, and S. Johansson. 1997. Amplification and cloning of complete enterovirus genomes by long distance PCR. J. Virol. Methods 65:191-199. [DOI] [PubMed] [Google Scholar]
- 25.Lornberg-Holm, K., L. B. Gosser, and J. C. Kauer. 1975. Early alteration of poliovirus in infected cells and its specific inhibition. J. Gen. Virol. 27:329-342. [DOI] [PubMed] [Google Scholar]
- 26.Martino, T. A., M. Petric, M. Brown, K. Aitken, C. J. Gauntt, C. D. Richardson, L. H. Chow, and P. P. Liu. 1998. Cardiovirulent coxsackieviruses and the decay-accelerating factor (CD55) receptor. Virology 244:302-314. [DOI] [PubMed] [Google Scholar]
- 27.McDermott, B. M., Jr., A. H. Rux, R. J. Eisenberg, G. H. Cohen, and V. R. Racaniello. 2000. Two distinct binding affinities of poliovirus for its cellular receptor. J. Biol. Chem. 275:23089-23096. [DOI] [PubMed] [Google Scholar]
- 28.Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 29.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 30.Minor, P. D., and E. J. Bell, ed. 1990. Topley and Wilson's principles of bacteriology, virology and immunity, 8th ed, vol. 4. Edward Arnold, London, United Kingdom.
- 31.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5 angstrom resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 32.Neumann, D. A., N. R. Rose, A. A. Ansari, and A. Herskowitz. 1994. Induction of multiple heart autoantibodies in mice with coxsackievirus B3- and cardiac myosin-induced autoimmune myocarditis. J. Immunol. 152:343-350. [PubMed] [Google Scholar]
- 33.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 34.Pasch, A., J. H. Kupper, A. Wolde, R. Kandolf, and H. C. Selinka. 1999. Comparative analysis of virus-host cell interactions of haemagglutinating and non-haemagglutinating strains of coxsackievirus B3. J. Gen. Virol. 80:3153-3158. [DOI] [PubMed] [Google Scholar]
- 35.Patel, J. R., J. Daniel, and V. I. Mathan. 1985. An epidemic of acute diarrhoea in rural southern India associated with echovirus type 11 infection. J. Hyg. 95:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilipenko, E. V., V. M. Blinov, L. I. Romanova, A. N. Sinyakov, S. V. Maslova, and V. I. Agol. 1989. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology 168:201-209. [DOI] [PubMed] [Google Scholar]
- 37.Powell, R. M., T. Ward, D. J. Evans, and J. W. Almond. 1997. Interaction between echovirus 7 and its receptor, decay-accelerating factor (CD55): evidence for a secondary cellular factor in A-particle formation. J. Virol. 71:9306-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powell, R. M., V. Schmitt, T. Ward, I. Goodfellow, D. J. Evans, and J. W. Almond. 1998. Characterization of echoviruses that bind decay accelerating factor (CD55): evidence that some haemagglutinating strains use more than one cellular receptor. J. Gen. Virol. 79:1707-1713. [DOI] [PubMed] [Google Scholar]
- 39.Racaniello, V. R. 1996. Early events in poliovirus infection: virus-receptor interactions. Proc. Natl. Acad. Sci. USA 93:11378-11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren, R. B., E. G. Moss, and V. R. Racaniello. 1991. Identification of two determinants that attenuate vaccine-related type 2 poliovirus. J. Virol. 65:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossmann, M. G. 1989. The canyon hypothesis. Viral Immunol. 2:143-161. [DOI] [PubMed] [Google Scholar]
- 42.Schmidtke, M., H. Selinka, A. Heim, B. Jahn, M. Tonew, R. Kandolf, A. Stelzner, and R. Zell. 2000. Attachment of coxsackievirus B3 variants to various cell lines: mapping of phenotypic differences to capsid protein VP1. Virology 275:77-88. [DOI] [PubMed] [Google Scholar]
- 43.Shafren, D. R., D. T. Williams, and R. D. Barry. 1997. A decay-accelerating factor-binding strain of coxsackievirus B3 requires the coxsackievirus-adenovirus receptor protein to mediate lytic infection of rhabdomyosarcoma cells. J. Virol. 71:9844-9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svitkin, Y. V., N. Cammack, P. D. Minor, and J. W. Almond. 1990. Translation deficiency of the Sabin type 3 poliovirus genome: association with an attenuating mutation C472—U. Virology 175:103-109. [DOI] [PubMed] [Google Scholar]
- 45.Tomassini, J. E., D. Graham, C. M. DeWitt, D. W. Lineberger, J. A. Rodkey, and R. J. Colonno. 1989. cDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc. Natl. Acad. Sci. USA 86:4907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, T., P. A. Pipkin, N. A. Clarkson, D. M. Stone, P. D. Minor, and J. W. Almond. 1994. Decay-accelerating factor CD55 is identified as the receptor for echovirus 7 using CELICS, a rapid immuno-focal cloning method. EMBO J. 13:5070-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, C., C. M. Bator, V. D. Bowman, E. Rieder, Y. He, B. Hebert, J. Bella, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2001. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J. Virol. 75:2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xing, L., K. Tjarnlund, B. Lindqvist, G. G. Kaplan, D. Feigelstock, R. H. Cheng, and J. M. Casasnovas. 2000. Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J. 19:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]