Abstract
Herpesvirus gene expression is divided into immediate-early (IE) or α genes, early (E) or β genes, and late (L) or γ genes on the basis of temporal expression and dependency on other gene products. By using real-time PCR, we have investigated the expression of 35 human herpesvirus 6B (HHV-6B) genes in T cells infected by strain PL-1. Kinetic analysis and dependency on de novo protein synthesis and viral DNA polymerase activity suggest that the HHV-6B genes segregate into six separate kinetic groups. The genes expressed early (groups I and II) and late (groups V and VI) corresponded well with IE and L genes, whereas the intermediate groups III and IV contained E and L genes. Although HHV-6B has characteristics similar to those of other roseoloviruses in its overall gene regulation, we detected three B-variant-specific IE genes. Moreover, genes that were independent of de novo protein synthesis clustered in an area of the viral genome that has the lowest identity to the HHV-6A variant. The organization of IE genes in an area of the genome that differs from that of HHV-6A underscores the distinct differences between HHV-6B and HHV-6A and may provide a basis for further molecular and immunological analyses to elucidate their different biological behaviors.
In the mid- and late 1980s human herpesvirus 6A (HHV-6A) and HHV-6B were discovered and subsequently named as variants of HHV-6 (1, 20, 32, 34). Although they share approximately 90% nucleotide sequence identity (13, 18), the accumulating biological, genetic, and epidemiological data suggest that these viruses have independent biological functions (13, 18, 34, 37, 38). Whereas little is known about the disease potential of HHV-6A, HHV-6B is the causative agent of the childhood disease exanthem subitum, characterized by a few days of high fever followed by a rash appearing with fever subsidence (34, 39). Thus, the frequency of HHV-6B seropositive individuals reaches almost 100% in early childhood (7, 10), but late primary infection may provoke a mononucleosis-like disease similar to human cytomegalovirus (HCMV) mononucleosis. In addition, HHV-6B is thought to be involved in a number of other conditions, including encephalitis and neoplasia (2, 3).
HHV-6B displays primarily tropism for T cells and monocytes (22, 24). The virus enters the cells in part via binding to the ubiquitously expressed glycoprotein CD46 (33), but additional cellular proteins involved in virus entry still remain to be discovered (6, 33). As with other herpesviruses, HHV-6 establishes latency following the primary infection. In addition to its tropism for mononuclear cells, HHV-6 is highly neurotropic and may also reside in astrocytes within the central nervous system (8, 15), and it remains to be determined which cells harbor the majority of the latent virus (21, 23). Of clinical significance, HHV-6 is an important pathogen when reactivated in immunocompromised hosts, such as patients with AIDS, leukemia, lymphoma, and solid organ or bone marrow transplants (3).
The entire genomes of HHV-6B strains Z29 and HST have recently been sequenced (13, 18). Besides its homology to HHV-6A, HHV-6B shares significant homology to HHV-7 as well as a general sequence conservation in the overall genetic colinearity with HCMV (13). Thus, HHV-6B belongs to the β-herpesvirinae subfamily of herpesviruses together with HHV-6A, HHV-7, and HCMV. However, in contrast to HCMV and the gammaherpesviruses, HHV-6A, -6B, and -7 encode homologs of the origin binding protein from alphaherpesviruses. The relationship to both the alpha- and betaherpesviruses is unique for the Roseolovirus genus (17).
Traditionally, lytic herpesvirus genes are divided into immediate-early (IE) or α genes, early (E) or β genes, and late (L) or γ genes on the basis of their temporal expression and their dependency on other gene products. First, IE genes are expressed independent of de novo protein synthesis. The products from these genes are often transcription factors and other regulatory proteins. IE gene products are important regulators of E gene transcription, which in turn are involved in DNA metabolism and replication. Last, L genes are transcribed, encoding structural and other proteins involved in virion assembly. L genes are partially or completely dependent on the viral DNA replication.
The segregation of genes into just three kinetic classes may be an oversimplification. In the case of HCMV, the temporal gene expression has been extended to five instead of three kinetic classes, with E genes subdivided into β1 (early) and β2 (early late) genes, and L genes separated into γ1 (leaky late) and γ2 (true late) genes (27). In HCMV infection, transcription of α genes begins 1 h and peaks 4 to 8 h after infection, whereas transcription of β1 and β2 genes begins 4 to 24 h after infection. Differences in the regulation of maximal protein expression, changes in expression levels later in infection, and different responses to inhibitors of viral DNA synthesis separate β1 from β2 genes. The γ genes are transcribed more than 24 h after infection. Transcription of γ1 genes takes place in the presence of inhibitors of DNA replication at reduced levels, whereas transcription of γ2 genes is completely inhibited in the presence of DNA replication inhibitors (27).
Resolving the pattern of expression of herpesvirus genes is important for the molecular and immunological analysis of the virus. The genetic relationship with HCMV suggests that HHV-6B may display a similar pattern of gene expression, but experimental evidence has only been provided for a few of the genes (26). By using real-time PCR, we here describe the temporal expression of 35 genes from HHV-6B, including B-variant-specific genes (13). Our findings suggest the segregation of HHV-6B genes into six separate classes. Although HHV-6B has characteristics similar to those of HHV-6A in its overall gene regulation, it expressed B-variant-specific genes within the IE group. Moreover, IE genes were from a region of the genome that differed from that of HHV-6A, lending structural support to the notion that these viruses are biologically distinct.
MATERIALS AND METHODS
Virus, cell culture, and T-cell infection.
HHV-6B, strain PL-1 (generated by P. Lusso), was propagated in the T-cell line Molt-3. T cells were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, glutamine (0.292 g/liter), 10 mM HEPES, streptomycin (0.2 g/liter), and penicillin (0.2 g/liter). Virus was collected from supernatant of HHV-6B-infected T cells. Supernatant was cleared from cell debris by centrifugation at 4,500 × g for 1 h and was subsequently centrifuged at 100,000 × g for 1 h. Infection was performed with 8 × 106 Molt-3 or Sup-T1 cells per ml of 20-fold-concentrated virus supernatant.
RNA purification.
T cells (2 × 106) were washed in phosphate-buffered saline (PBS), and total RNA was isolated at room temperature by using a High Pure RNA Isolation kit (Roche Diagnostics Scandinavia AB, Hvidovre, Denmark). In essence, this kit is based on lysing the T cells in 4.5 M guanidine hydrochloride, 50 mM Tris-HCl, and 30% (wt/vol) Triton X-100, pH 6.6, followed by the collection of nucleotides on a filter by high-speed centrifugation. Nucleotides were subsequently treated with 200 U of DNase for 15 min and were washed once in a buffer consisting of 5 M guanidine hydrochloride and 20 mM Tris HCl (pH 6.6) in ethanol and were washed twice in a buffer consisting of 20 mM NaCl and 2 mM Tris-HCl (pH 7.5) in ethanol. Finally, RNA was eluted in nuclease-free, sterile, double-distilled H2O in a total volume of 60 μl. Total RNA concentration was determined in triplicate by absorbance measurements (GeneQuant II RNA/DNA Calculator; Pharmacia Biotech, Hørsholm, Denmark).
Reverse transcription (RT)-PCR.
First-strand cDNA synthesis was performed on 1 μg of total RNA with 100 pmol of p(dT)15 primer (Roche) for cDNA synthesis followed by a denaturation step at 65°C for 10 min. The RNA was reverse transcribed for 45 min at 42°C by using the method described by the manufacturer of the Expand Reverse Transcriptase kit for first-strand synthesis (Roche). In brief, 1× Expand Reverse Transcriptase buffer (first strand) was mixed with dithiothreitol in a final concentration of 10 mM, deoxynucleoside triphosphate mix, disodium salt solution with dATP, dCTP, dGTP, and dTTP at a final concentration of 1 mM, 20 U of RNase inhibitor, and 50 U of Expand Reverse Transcriptase enzyme. The reaction was stopped by raising the temperature to 95°C for 2 min. The cDNA was diluted to 40 μl with double-distilled H2O. To avoid sample-to-sample contamination, separate laboratories were used for RNA purification, PCR setup, and template addition.
PCR amplification.
Primers for PCR amplification of 35 genes from HHV-6B are listed in Table 1. The primers were derived from the published sequence of HHV-6B strain Z29 (GenBank accession number AF157706) by using the Primer Picking3 program (www-genome.wi.mit.edu). Amplification of cDNA was performed by real-time PCR on a LightCycler instrument (Roche) in the presence of 5 μl of cDNA, 2.5 mM MgCl2, 0.5 μM primers, and 2 μl of LightCycler-FastStart DNA Master SYBR Green I in a final volume of 20 μl. Each run included a positive control sample for the β2-microglobulin gene and negative controls for each primer pair with no cDNA and uninfected Molt-3 T-cell cDNA. All the reactions were done under the same conditions, with an initial denaturation step at 95°C for 10 min followed by 45 cycles of amplification at 95°C for 15 s, 62°C for 10 s, and 72°C for 15 s. These conditions of MgCl2 concentration and annealing temperature were optimized in initial experiments. Melting curves were performed with 1 cycle at 95°C for 0 s, 72°C for 15 s, and 99°C for 0 s. Run-to-run variation was minimized by ensuring that all PCRs with each test material were performed with the same batch of cDNA. To make sure that the amplified product originated from de novo synthesized mRNA and not from carry-over DNA from the viral infection, RT-PCR was performed on RNA purified from T cells mixed briefly with virus in the same amounts and under the same conditions as those used in all the infections. Furthermore, RNA that was not retrotranscribed was tested with real-time PCR to ensure that no contaminating DNA was present in the purified RNA solution. To ensure that the amplified fragments had the correct size, all the products from the temporal screening of the 35 genes were run in a 2% agarose gel containing ethidium bromide and visualized under UV illumination. As a marker of the DNA fragment size, DNA Molecular Weight Marker IX (Roche) was included in each run.
TABLE 1.
Primer pairs used for analysis of HHV-6B gene expression and sequencing
| Name | Primer coordinatesa | Nucleotide sequenceb (5′-3′)
|
Size of amplified product (bp) | |
|---|---|---|---|---|
| Forward | Reverse | |||
| β2mc | CAA GCA GAG AAT GGA AAG TC | GAT GCT GCT TAC ATG TCT CG | 269 | |
| B3 | 7351-7485 | GTC TAT CCG CCT GCG TGT AT | AGT AAC ACG GCA CGT AAG CA | 135 |
| B4 | 8972-9231 | GCG AGA GAG AAA GAG AGG CA | GGG TCG GGG GTC TAT AAA AA | 260 |
| B5 | 9555-9733 | AGA CAA CCA CGA AAA CGA CC | ACA TGG TAG AGC GGG AAA TG | 179 |
| U2 | 10519-10712 | GAG ACG AAG TCC CTC ACG TC | TTT TGT CGC TCT CCG TTT CT | 194 |
| U7 | 15666-15956 | ACG ACA AAC CTG CTG GTA GC | GGA TGT TGA ATG GGG AGT TG | 291/200d |
| U8 | 17687-17882 | TCC GTC TCT AGA TCG TGC AA | GGA ACT GCT GGC TGA GAT TC | 196 |
| U9 | 18225-18426 | ATT TCC TGC GAA TGT TGG AC | ATA CAG AAG CCG GAA CGA TG | 202 |
| U11 | 21476-21671 | ATC GAC GCT ATC CGA CTC TG | TGG GCC TCA CTC TTT AAT GG | 196 |
| U12 | 22652-22915 | AGC TGT CCA AAC TCC TGC AC | GCG TTA TGT TTT GCG ACT CTG | 187e |
| U14 | 25813-26010 | TCA CAG GAA GAC AGC AAT CG | GGG AGC ACG TCT TCT TTG TC | 198 |
| U21 | 34591-34800 | CCA TTT TAC CGC TTC TCC AA | TGG AGT TAG TGG GCG CTA GT | 210 |
| U22 | 34875-35085 | ATC CAA AGC AAA CCA GCA AG | TCA ATC GGT GAT GGA GTC AA | 211 |
| U23 | 35607-35855 | GAA TGC CCC GAC AAA ACT AA | GAG GCG CAG GAT ATT GAG AC | 249 |
| U27 | 38971-39167 | GAA TCG TCG CTC TCG CTA TC | GAG GAA TCT CGC TTT GAA CG | 197 |
| U29 | 42950-43155 | CGT AAG CCC GTG AAT GTT TT | GCG TGA GAA CCG CTT TAA CT | 206 |
| U30 | 44406-44602 | CCT CAA GAA CGG GAG AAT GA | GCC ATG TGG TTT GAG AGG AT | 197 |
| U35 | 55133-55324 | CCA TTT TCT GAG CCA TGA CC | AAG TCT GTG AGC AGG GAA GG | 192 |
| U36 | 55854-56109 | TCA CAT CCA AAG CGA AAA CA | GCG GAT AGT TAG GTC CGT GA | 256 |
| U37 | 57056-57255 | GCA ATC ACC ATC AAC ACA CC | GCG TAT CAG CTC CAT GTC CT | 200 |
| U38 | 58157-58356 | TGT GTT TGC GTC ATG TGA GA | TGT CCG CTC ATT TTG ATT TG | 200 |
| U39 | 62061-62501 | AAG AGG CTG CCA AAT CAA AA | CAT TTT GCA GAG CCG TTA GA | 441 |
| U41 | 65450-65645 | GTC ATA GAC CGG AGC ATC GT | TGA GGT GAT GAG GGA TAG GG | 196 |
| U47 | 78067-78261 | CGG TCG AGG AAA ATC TGA AC | TCG ACG TCA GCG TCT ATC AC | 195 |
| U51 | 83839-84034 | CAG CCA CTG CGG AGT TTT AT | GGC AAA GTC CAA TGT CCA CT | 196 |
| U54 | 87563-87761 | CGA CCG TGG TTA GAC TTG GT | CGC ATC AAT TAC TCG CAA GA | 199 |
| U58 | 96103-96310 | TAA GGC GGT TCC TGC AAT AC | GTC CGG CAA ACT GTA ATC GT | 208 |
| U69 | 106289-106489 | CCT GGA TTT CGA CCA CTT GT | CGC AAA CAA CCT TTT CTC GT | 201 |
| U73 | 111144-111349 | TGG GAA AGT TCA GAG CGA CT | AAA ACT CTT CGG CAG CAC AT | 206 |
| B6 | 119207-119308 | GGG AAA CCT CAA AAA TGC AC | CTT AGG ACA CCA TCC AAG CC | 102 |
| B7 | 120962-121063 | CCT AAT ACT TCT GAC GGG CG | TCA TCC AAA CCG AAA ACC TT | 102 |
| U81 | 123527-123730 | AAT TCG GGA TGC TTC TTT CC | ATC TAA CCG TCC ACC CCT CT | 204 |
| U86 | 128815-129014 | AAA GGA CTG GAG TCG AGC TG | CCA ACA TAC CTT CCC CTC AA | 200 |
| B8 | 142340-142636 | TTA GGG TCC TCA CCC ACT TG | CCG CAG CTT CTG TTT TTC AT | 297 |
| U94 | 144100-144302 | CGC CAC CAT TTT TCT TTG TT | TGC AAA GTG GTA CGC TCA AG | 203 |
| U95 | 147023-147222 | CCA AGA AAG GCG AAA AGT TG | AAT GTC GGC AAG CAA TCT CT | 200 |
Coordinates refer to the HHV-6B genome sequence in strain Z29 (accession no. AF157706).
The annealing temperature was 62°C for all primer pairs used.
β2m, β2-microglobulin.
Unspliced/spliced.
Covers a splice site.
Inhibition of protein synthesis and polymerase activity.
To identify IE, E, and L genes, the cells were incubated in the presence or absence of cycloheximide (CHX), phosphonoacetic acid (PAA), and aphidicolin (APH) (all from Sigma, Vallensbæk Strand, Denmark). For CHX or APH treatment, cells were infected in the presence of 100 μg of CHX (dissolved in H2O)/ml or 5 μg of APH (dissolved in dimethyl sulfoxide)/ml, which were the optimal concentrations on the basis of titration experiments on U81, U41, and U22, which are established IE, E, and L genes (14, 18, 26). After 1 h of adsorption the cells were diluted to a final cell concentration of 0.5 × 106 cells/ml with growth medium containing 100 μg of CHX/ml or 5 μg of APH/ml. Cells treated with CHX were harvested 8 or 24 h after infection, and cells treated with APH were harvested 24 h after infection. For PAA treatment, T cells (0.5 × 106 cells/ml) were mixed with 500 μg of PAA (dissolved in DMEM and neutralized with 10 M NaOH)/ml 1 h before infection, which was an optimal concentration on the basis of titration experiments. T cells were infected in the presence of 500 μg of PAA/ml and were incubated for 1 h before dilution to a final cell concentration of 0.5 × 106 cells/ml with growth medium containing 500 μg of PAA/ml. Cells treated with PAA were harvested 24 h after infection. To interpret the results with inhibitors independent of the absolute cycle number, the percentage inhibition was calculated by the following formula: % inhibition = [1 − (2x/2xi)] ×100%, where x is the crossing point value (Cp) without inhibitor and xi is Cp with inhibitor. Cp is the number of PCR cycles demanded to amplify the initial template amount above background fluorescence. In other words, the Cp value is the point where extrapolation of the exponential part of the PCR amplification curve is crossing the noise band, which is the border between background and relevant fluorescence. This means that an increase in Cp corresponds to a decrease in the amount of target. To define what should be considered a significant deviation in cycle number, Cp values were obtained from three independent determinations of each of the 35 genes in the absence of inhibitor. For each gene the maximal deviation from the mean value of the three determinations was calculated. On the basis of the distribution of these values, the standard deviation (SD) considering all genes was 0.6 cycles. Thus, in experiments using inhibitors, changes in Cp values of less than 2 SD were considered insignificant.
Normalization.
To evaluate the relative expression of the genes during kinetics experiments, the PCR data were normalized by calculating a value for each time point relative to the sum of all the Cp values obtained for the particular gene with the following equation: Nv(Cpy) = [(Cp1h + Cp3h + Cp8h + Cp22h + Cp48h)/Cpy], where Nv is the normalization value, Cpy is the Cp at the different time points, and y indicates a time point of 1, 3, 8, 22, or 48 h.
PCR reproducibility.
As an internal control, β2-microglobulin was amplified in each run. The β2-microglobulin values were highly reproducible in that the mean value ± SD of 36 measurements of β2-microglobulin was 16.3 ± 0.6 cycles. In three independent but identical experiments, cDNA from HHV-6B-infected T cells was tested in three separate runs with the 35 different primer pairs. The SD was always below 10%, and in 32 of the 35 observations it was even below 6%. That is, the real-time PCR results were highly reproducible.
Sequencing.
DNA was prepared from HHV-6B-infected Molt-3 T cells as previously described (16). In brief, 106 T cells were washed twice in PBS and resuspended in 0.5 ml of 0.1× PBS. T cells were then incubated at 94°C for 10 min to complete cell lysis and to denature the DNA. To degrade the proteins, the lysate was incubated for 30 min at 55°C in the presence of 0.4 mg of proteinase K (Roche)/ml. Finally, proteinase K was inactivated by incubation at 94°C for 10 min. The DNA was amplified by real-time PCR with the same reaction conditions described above. The amplified products were separated in a 2% agarose gel, and bands were cut out for sequencing. The specific fragments were purified with the QIAEX II Agarose Gel Extraction kit (QIAGEN, Valencia, Calif.). Five genes were sequenced with both sense and anti-sense primers on an ABI PRISM 377 DNA Sequencer (Perkin Elmer, Foster City, Calif.). PCR was performed with fluorescence-stained deoxynucleotides with a denaturation step at 96°C for 5 min and 25 cycles of amplification at 96°C for 30 s, 45°C for 15 s, and 60° for 4 min with the enzyme AmpliTaq DNA Polymerase. Reaction products were separated on a 6% polyacrylamide gel. The sequences were analyzed automatically and checked manually before they were aligned in the ClustalW program (www.ebi.ac.uk/clustalw).
RESULTS
Gene sequences of HHV-6B strain PL-1.
While the entire genomes of HHV-6B strains Z29 and HST have been sequenced, only limited information is available on the PL-1 strain. To establish the relationship between PL-1 and the sequenced strains, genes in PL-1 homologous to the most divergent genes between strain HST and strain Z29 were sequenced (Table 2). Partial sequences from U7, U58, B5, and B6 were 100% identical between PL-1 and Z29 but only 97 to 99% identical to those of HST, whereas PL-1 U39 was 99 and 98% identical to those of Z29 and HST, respectively. This suggests that PL-1 is most similar to the Z29 strain of HHV-6B.
TABLE 2.
HHV-6B strain PL-1 is highly similar to strain Z29
| PL-1 gene | GenBank accession no. | Similarity (%) to corresponding gene of:
|
Sequenced size (bp) | Description or predicted function | |
|---|---|---|---|---|---|
| Z29 | HST | ||||
| U7 | AF451281 | 100 | 97 | 162 | HCMV US22 gene family |
| U39 | AF451282 | 99 | 98 | 401 | Glycoprotein B |
| U58 | AF451283 | 100 | 98 | 168 | Unknown |
| B5 | AF451284 | 100 | 98 | 187 | Unknown |
| B6 | AF451285 | 100 | 99 | 150 | Unknown |
Early expression of HHV-6B genes.
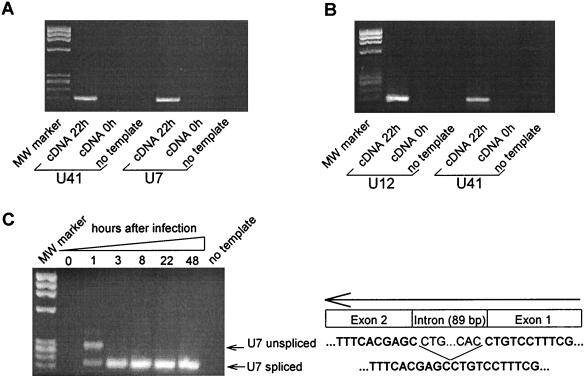
Initial experiments indicated that the expression of most HHV-6B genes could be detected as early as 1 h after infection with real-time PCR. To make sure that the products originated from de novo synthesis and not from contaminating RNA from the virus supernatant used during in vitro infection, cDNA derived from T cells briefly mixed with virus stock was amplified for 45 cycles by real-time PCR with primers for U41 and U7 (Table 1). None of these amplifications gave rise to detectable products (Fig. 1A). This indicated that there was no detectable contamination of RNA from the virus supernatant. To further ensure that no contaminating DNA was present in the RNA preparation, RNA was tested by PCR in the absence of retrotranscription with primers specific to U12 and U41. No products were detected after 45 cycles of real-time PCR, indicating that viral DNA did not contaminate the samples (Fig. 1B).
FIG. 1.
Early detected viral transcripts in HHV-6B-infected Molt-3 T cells do not originate from contaminating viral RNA or DNA. (A) Viral RNA is not detected in T cells after a few minutes of HHV-6B infection. Agarose gel electrophoresis of the products from 45 cycles of real-time PCR on cDNA derived from T cells infected by HHV-6B for 22 h (lane 1 and 4) or from T cells briefly mixed with virus supernatant (lane 2 and 5). Lanes 3 and 6, no template control. MW, DNA Molecular Weight Marker IX (Roche). Products in lanes 1, 2, and 3 were amplified with the primer pair for gene U41; products in lanes 4, 5, and 6 were amplified with the primer pair for gene U7. (B) RNA preparations from HHV-6B-infected T cells do not contain contaminating viral DNA. A cDNA reaction mixture not retrotranscribed was amplified for 45 cycles of real-time PCR by using primers for U12 (lanes 1, 2, and 3) and U41 (lanes 4, 5, and 6) and was separated by agarose gel electrophoresis. Lanes 1 and 4, HHV-6B-infected T cells (48 h). Lanes 2 and 5, RNA was purified from T cells briefly mixed with virus supernatant and cDNA reactions performed in the absence of retrotranscription. Lanes 3 and 6, no template control. MW, DNA Molecular Weight Marker IX. (C) Induction of a spliced form of U7 indicates de novo RNA synthesis. Agarose gel electrophoresis of the products from 45 cycles of real-time PCR on cDNA from uninfected (lane 1) and HHV-6B-infected T cells (lanes 2 to 6) is shown. Lane 7, no template control. All products have been amplified with the primer pair for gene U7. MW, DNA Molecular Weight Marker IX. The GenBank accession number for the U7 sequence is shown in Table 2.
Finally, on the basis of the sequencing data of HHV-6B, the U7 gene is predicted to be spliced. To further ensure that the products detected after 1 h originated from de novo synthesis and not from contaminating DNA, primers outside of the predicted intron in U7 was chosen so that unspliced and spliced amplimers could be distinguished. While infection for a few minutes (RNA preparation immediately after addition of virus) did not produce any U7-specific RNA (Fig. 1A), infection for 1 h produced two products (Fig. 1C) that were confirmed by sequencing to be a spliced and an unspliced form of U7. After 3, 8, 22, and 48 h of infection, only the spliced product was detected (Fig. 1C), indicating that the products detected in all the samples were derived from de novo synthesized mRNA. Taken together, these experiments demonstrated that our protocol was specific for de novo synthesized RNA, which could be detected from HHV-6B-infected T cells as early as 1 h after infection.
Temporal expression of HHV-6B genes.
Little is known about the temporal expression of the majority of HHV-6B genes. To further establish the kinetics of expression of HHV-6B genes, RNA from PL-1-infected Molt-3 T cells was purified 1, 3, 8, 22, and 48 h after infection. RT-PCR was performed by real-time PCR to establish the kinetic appearance of 35 HHV-6B gene transcripts. To emphasize the relative change in the level of product rather than the total amount of product, the PCR data were normalized as described in Materials and Methods and presented as a relative measure of the expressed product at different time points. That is, a steep curve would be expected between 1 and 3 h of infection in IE genes, between 3 and 8 h in E genes, and somewhere between 8 and 48 h in L genes.
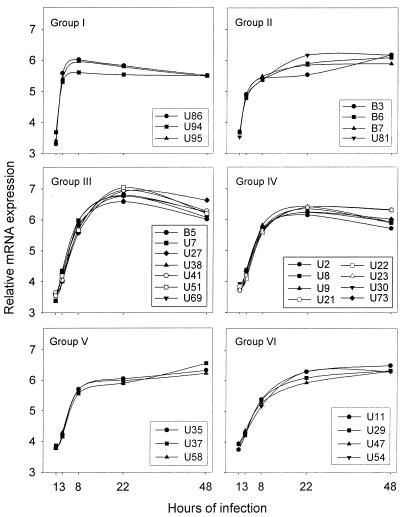
Superimposing the curves from the expression analysis of 35 HHV-6B genes suggested six patterns of expression (Fig. 2, groups I to VI). A major rise in product between 1 and 3 h of infection was characteristic for groups I and II. Likewise, two groups with a major rise in product between 3 and 8 h of infection could be distinguished and were termed groups III and IV. Finally, two groups, termed V and VI, with the highest level of expression after 48 h of infection were identified. The curves for groups IV and V were somewhat similar, since both of them display a major increase between 3 and 8 h of infection. Nevertheless, the relative amount of product after 48 h of infection distinguishes these patterns of expression. The average curves for these expression groups are shown in Fig. 3A.
FIG. 2.
Discrete groups of expression patterns among HHV-6B genes. The expression of the HHV-6B genes was determined by real-time PCR at different time points following infection of Molt-3 T cells. The temporal expression of the indicated gene is shown relative to its expression at the other time points. Thus, the curves reflect the changes in gene expression, as described in Materials and Methods, but do not indicate the total amount of expression.
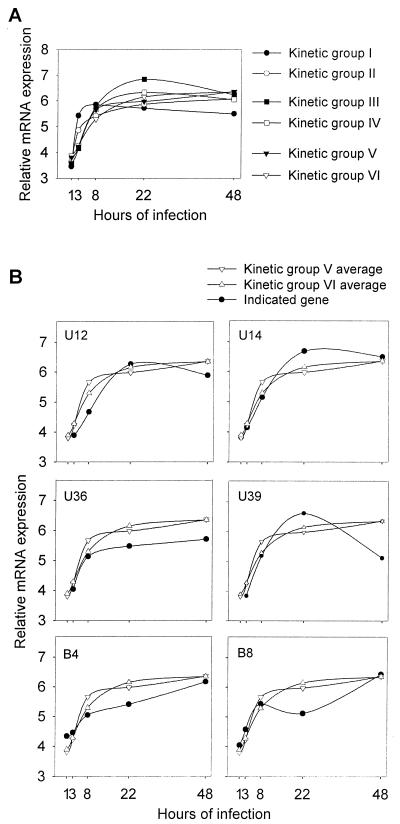
FIG. 3.
Kinetic expression of HHV-6B genes. (A) Average curves for kinetic groups I to VI, shown in Fig. 2. (B) Comparison of six genes that did not conform well to the established kinetic groups with the profile of kinetic groups V and VI.
RNA from all but 3 of the 35 genes (U12, U36, and U39) could be detected after 1 h of infection, and RNA from all 35 genes were detectable after 3 h of infection. Six of the examined genes had expression profiles that were highly reproducible but did not conform well to the defined kinetic groups (Fig. 3B). However, we found that these genes were most similar to genes of groups V and VI.
Expression of HHV-6B genes independent of de novo protein synthesis.
The classification of IE, E, and L genes cannot be made solely on the kinetics of expression. For instance, to fulfill the criteria for a bona fide IE gene, the expression should occur in a manner independent of de novo protein synthesis. To test this, cells were treated with CHX and were infected with HHV-6B. At the indicated time point, RNA was isolated and cDNA was analyzed by real-time PCR.
The degree of inhibition was virtually 100% in most of the examined genes following 8 h of CHX treatment (Fig. 4). Only 7 of the 35 examined genes (B3, B6, B7, U81, U86, U94, and U95) showed an inhibition of less than 80% after 8 h of CHX treatment. Indeed, these seven genes had been placed in groups I and II on the basis of their kinetics of expression. We therefore conclude that they are classic IE genes. U86 was the most promptly induced gene of all 35 HHV-6B genes examined and displayed a unique resistance to the inhibition by CHX even at 24 h of treatment.
FIG. 4.
Dependency on de novo protein synthesis for HHV-6B gene expression. Molt-3 T cells infected by HHV-6B were incubated in the presence or absence of the protein synthesis inhibitor CHX for 8 or 24 h. RNA was isolated at the indicated time point, and real-time PCR was performed on cDNA. The percentage of CHX-mediated inhibition of viral gene expression was calculated on the basis of the real-time PCR crossing points as described in Materials and Methods.
Viral DNA polymerase-dependent HHV-6B gene expression.
L genes, but not E genes, are dependent on the viral DNA polymerase for their transcription. Therefore, to identify true L genes transcription of HHV-6B genes was tested in the presence or absence of PAA, an inhibitor of viral DNA polymerase. In addition, we examined the inhibition by APH, an inhibitor of both cellular and viral DNA polymerases (5, 28).
Inhibition of HHV-6B gene expression by 24 h of treatment with PAA or APH varied between 0 and 99.8% and 97.4%, respectively (Fig. 5). Among the 10 genes with the highest inhibition by PAA (inhibited by more than 95%), 6 genes (U27, U29, U36, U37, U47, and U54) were also among the 10 most inhibited genes by APH (inhibited more than 80%). Conversely, 13 genes were not inhibited at all by APH. Twelve of these were also among the 13 genes with the lowest inhibition by PAA (less than 86% inhibition). Thus, PAA and APH had comparable effects, although inhibition by APH was less than that of PAA. Indeed, none of the 35 genes were inhibited more by APH than by PAA.
FIG. 5.
Dependency on DNA polymerase activity for HHV-6B gene expression. Molt-3 T cells infected by HHV-6B were incubated in the presence or absence of the DNA polymerase inhibitors PAA or APH for 24 h. RNA was isolated, and real-time PCR was performed on cDNA. The percentage of PAA- and APH-mediated inhibition of viral gene expression was calculated on the basis of the real-time PCR crossing points as described in Materials and Methods.
Despite these similarities, the two inhibitors had divergent effects on certain genes. B4, which was inhibited by 98.8% by PAA, was not inhibited at all by APH. Nevertheless, on the basis of PAA inhibition and the kinetics of expression, B4 and B8 behaved as L genes. Similarly, genes inhibited up to 85% were not inhibited by APH at all (e.g., U94, U69, U73, and U51). Of the seven genes in kinetic groups I and II and with incomplete CHX inhibition (IE genes), only U81 and B6 were inhibited by APH, whereas all of them except B3 and U95 were inhibited between 60 and 90% by PAA.
In concordance with the kinetic classification, seven of eight genes that were inhibited more than 97% by PAA were also in groups V and VI. Of the genes inhibited less than 97% by PAA, a combination of the temporal expression and the inhibition by PAA and APH suggests that U2, U11, U14, U21, U22, U23, U30, U35, U38, U39, U58, and possibly U12 may qualify as L genes. Similarly, on the basis of the above arguments and the CHX inhibition data, B5, U7, U8, U9, U41, U51, U69, and U73 appear to be E genes (Fig. 4 and 5).
DISCUSSION
Following a primary infection in childhood, HHV-6B establishes a latent infection that may give rise to severe clinical manifestations if reactivated during a state of immunosuppression (3, 10, 11). Nevertheless, viral gene expression is still poorly characterized. We have investigated the temporal expression of 35 viral genes by use of real-time PCR in HHV-6B (PL-1)-infected T cells. Understanding the kinetics of HHV-6B gene expression may be helpful in the functional characterization of the virus, since the temporal expression of herpesvirus genes is correlated with functional aspects. HHV-6B gene products can be divided into groups on the basis of the following functional properties: regulatory proteins (often encoded by IE genes), proteins involved in DNA replication (often E gene products), genes encoding structural proteins including proteins involved in virus assembly (often L genes), and genes encoding proteins modulating the host response. However, the majority of HHV-6B genes have not yet been functionally associated with any of these groups.
Surprisingly, we were able to detect transcripts by sensitive real-time PCR in 32 of 35 tested genes only 1 h after infection. This demonstrates that most of the genes are transcribed very early during the course of infection. Even genes which previously have been determined to be L genes, such as U22 (17), or genes with a known function suggesting late expression, such as the virion protein U11 (29), were detected at this early time point. This observation therefore suggests that the transcription from most of the HHV-6B genes is leaky or subject to a more complex mechanism of regulation than the one classically described for herpesviruses. Thus, it is likely that transcripts are detectable in cells infected by other herpesviruses if the same sensitive PCR assay is applied. However, the relative amounts of the transcripts between genes cannot be compared because the primer efficiencies may vary. Similarly, we cannot exclude the possibility that the genes not detected after 1 h of infection (U12, U36, and U39) could be transcribed at that time point.
While the initial classification of herpesviruses was based on three patterns of gene expression, HCMV is best described with the implementation of five classes (27). Our kinetics analysis of the HHV-6B genes conformed well to six patterns of expression (Fig. 2). However, the assignment of IE, E, and L genes also relies on their dependency on de novo protein synthesis as well as viral DNA polymerase activity for expression. That is, transcription from E or L genes, which are completely dependent on de novo protein synthesis, is shut down in the presence of CHX. Conversely, IE genes are expressed despite a block in protein synthesis. All but seven of the examined HHV-6B genes were virtually completely inhibited by 8 h of CHX treatment, indicating that they belong to the E or L genes. The seven genes that were expressed independently of de novo protein synthesis (U81, U86, U94, U95, B3, B6, and B7) belonged to the classes I and II, showing the most rapid onset of expression consistent with these genes being IE genes. For three of these genes (U81, U94, and U95), our analysis confirmed their previous classification as IE genes (18, 26, 36). Furthermore, U86 is homologous to IE-A in HHV-6A (13) and IE2 (UL122) in HCMV, and therefore it is expected to be an IE gene. U86 displayed the most immediate expression in the kinetic analysis and was unique in its complete resistance to CHX.
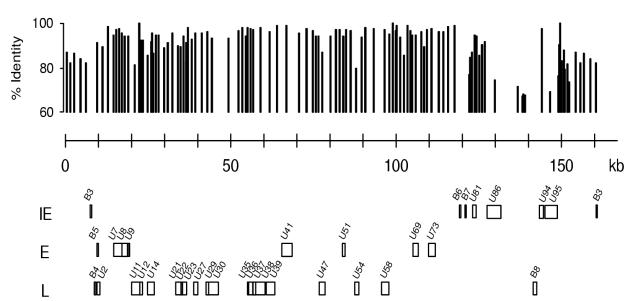
IE genes usually encode transcription factors, which may be critical in the establishment of the viral infection and thereby for the virus-host interactions. Interestingly, the IE genes clustered in a region of the genome (Fig. 6) that has the greatest sequence variation from the related viruses HHV-6A and HHV-7 (13). First of all, B3, B6, and B7 have no corresponding genes in HHV-6A or HHV-7 and are considered HHV-6B-specific genes within the herpesvirus family. The encoded protein from U94, a parvovirus rep homolog (31), is 97.6% identical to the homologous gene in HHV-6A but has no homolog in HHV-7. Moreover, the U86- and U95-encoded proteins have only 74.5 and 69.0% identity with the corresponding HHV-6A protein and just 29.5 and 26.7% identity with their HHV-7 counterparts (13). Thus, the genes characterized as IE genes appear in particular to distinguish HHV-6B from other roseoloviruses.
FIG. 6.
Classification of HHV-6B genes suggests a temporal organization in the expression from the viral genome. The HHV-6B genes examined in the present paper are shown as IE, E, or L genes in the order of their location in the HHV-6B genome. At the top of the figure an identity plot to homologous HHV-6A, based on a comparison of strain Z29 with U1102, is shown.
Genes inhibited to a significant extent after 24 h of CHX treatment but not after only 8 h (B3 and possibly also U81 and U95) are probably transcribed as IE genes in the beginning of the infection, but later accumulation of transcripts may be influenced by de novo protein synthesis. Further experiments are needed to determine whether CHX affects the transcription or the stability of the products. Toxic effects of CHX might also affect the results, although expression of U86 was completely unaffected by 24 h of CHX treatment.
In general we found that APH, a novel inhibitor affecting both cellular and viral DNA polymerases, inhibited the same genes, although to a less pronounced extent, as PAA. Experiments on purified herpes simplex virus DNA polymerase and cellular polymerases from B-lymphoblastoid cell lines and HeLa cells suggest that APH has comparable effects on viral and cellular DNA polymerases with a Km of approximately 1 μM (35). Moreover, concentrations above 6 μM inhibited cellular DNA synthesis with 97 to 98%. In the present studies we used 14.8 μM (5 μg/ml) APH, which is thus expected to inhibit cellular and viral DNA polymerases. Nevertheless, we observed that some genes were inhibited by PAA but not by APH. This was the case for B4, which was inhibited by more than 95% by PAA, and to a lesser degree for B5, U9, U51, U69, and U73, which were inhibited by more than 80% by PAA. We do not know the explanation for the divergent effect of the inhibitors on these genes. However, it may be an advantage for viral transcription that host-cell DNA synthesis is blocked, as suggested for HCMV expression (4, 12).
Combining the data on gene expression with inhibition in the presence of inhibitors of de novo protein synthesis and viral DNA polymerase activity allowed a classification of the examined genes (Fig. 6). The known or presumed function of U29, encoding a protein involved in capsid assembly and DNA maturation (18), and U36, encoding a protein involved in DNA packaging (30), supports their characterization as L genes. Moreover, our analysis of kinetics and sensitivity towards PAA and APH suggested that U11 might be an L gene, in agreement with its previous characterization as a de novo protein synthesis-dependent gene (31).
Some of the genes within the classes III and IV were significantly inhibited by PAA and APH and may thus be best characterized as L genes. These genes include U27, which has previously been suggested to be an E/L gene (9), and U30. The genes U21, U22, and U23 displayed virtually identical expression patterns (Fig. 2) and were inhibited by PAA and to a lesser extent by APH. We consider these genes as L genes despite the ambiguity in their classification. Likewise, we classify B5, U7, U8, U9, U41, U51, U69, and U73 as E genes, despite the borderline inhibition of the expression of some of these genes in the presence of PAA. This classification is supported by the previous finding of Menotti et al. (25) that U51 is not transcribed as an L gene, as well as by our finding that U51 is not an IE gene.
U8 has previously been categorized as an IE/regulatory gene (18), whereas our data suggest that this gene should be classified as an E gene. The use of different cell lines and different strains of HHV-6B may potentially explain this discrepancy between the two studies. U39 and U73 (26) have previously been characterized as IE genes. In contrast, our data suggest that U39 is an L gene and furthermore support the observation by Rapp et al. that U73 depends on de novo synthesis of protein (31). U41 has been characterized previously as an E gene (26), which would be in agreement with our data. Finally, U12 and U22 have been defined as L genes (14, 19). Our data support the characterization of U22 as an L gene, and although we failed to obtain convincing inhibition of U12 expression by PAA, the kinetic data suggest that U12 may be an L gene.
Nine open reading frames specific for HHV-6B have been predicted from the sequence data (nine B-variant-specific genes). We have detected transcripts from at least six of these (B3, B4, B5, B6, B7, and B8). Perhaps the most provocative finding in this regard is the grouping of three of these genes (B3, B6, and B7) as IE genes with potential impact on the establishment of infection. Although HHV-6A and -6B have 90% identity in their nucleotide sequence and both of them have tropism for T cells, they do not replicate in the same T-cell lines (37). Our finding of B-variant-specific IE gene expression and a clustering of HHV-6B IE genes in an area of the viral genome with the lowest identity to HHV-6A may be important for the further characterization of the biological differences between the two HHV-6 variants.
Acknowledgments
We thank P. Lusso for the PL-1 strain of HHV-6B and Z. Berneman and his laboratory for cell lines and advice. Helpful discussions with K. Poulsen and S. R. Paludan are appreciated. B. Bundgaard is thanked for excellent technical assistance.
This work was supported by grant 9903024 from the Danish Medical Research Council and by grants from the Novo Nordic Research Foundation, The Aarhus University Research Foundation, and the Foundation of 17.12.1981.
REFERENCES
- 1.Ablashi, D. V., S. Z. Salahuddin, S. F. Josephs, F. Imam, P. Lusso, R. C. Gallo, C. Hung, J. Lemp, and P. D. Markham. 1987. HBLV (or HHV-6) in human cell lines. Nature 329:207.. [DOI] [PubMed] [Google Scholar]
- 2.Akashi, K., Y. Eizuru, Y. Sumiyoshi, T. Minematsu, S. Hara, M. Harada, M. Kikuchi, Y. Niho, and Y. Minamishima. 1993. Brief report: severe infectious mononucleosis-like syndrome and primary human herpesvirus 6 infection in an adult. N. Engl. J. Med. 329:168-171. [DOI] [PubMed] [Google Scholar]
- 3.Braun, D. K., G. Dominguez, and P. E. Pellett. 1997. Human herpesvirus 6. Clin. Microbiol. Rev. 10:521-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 5.Bucknall, R. A., H. Moores, R. Simms, and B. Hesp. 1973. Antiviral effects of aphidicolin, a new antibiotic produced by Cephalosporium aphidicola. Antimicrob. Agents Chemother. 4:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume, G. 2000. Virus receptor arrays, CD46 and human herpesvirus 6. Trends Microbiol. 8:436-438. [DOI] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume, G., P. Mirandola, and L. Menotti. 1999. Human herpesvirus 6: an emerging pathogen. Emerg. Infect. Dis. 5:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caserta, M. T., C. B. Hall, K. Schnabel, K. McIntyre, C. Long, M. Costanzo, S. Dewhurst, R. Insel, and L. G. Epstein. 1994. Neuroinvasion and persistence of human herpesvirus 6 in children. J. Infect. Dis. 170:1586-1589. [DOI] [PubMed] [Google Scholar]
- 9.Chang, C. K., and N. Balachandran. 1991. Identification, characterization, and sequence analysis of a cDNA encoding a phosphoprotein of human herpesvirus 6. J. Virol. 65:2884-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, D. A. 2000. Human herpesvirus 6. Rev. Med. Virol. 10:155-173. [DOI] [PubMed] [Google Scholar]
- 11.Cone, R. W., M. L. Huang, L. Corey, J. Zeh, R. Ashley, and R. Bowden. 1999. Human herpesvirus 6 infections after bone marrow transplantation: clinical and virologic manifestations. J. Infect. Dis. 179:311-318. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French, C., P. Menegazzi, L. Nicholson, H. Macaulay, D. DiLuca, and U. A. Gompels. 1999. Novel, nonconsensus cellular splicing regulates expression of a gene encoding a chemokine-like protein that shows high variation and is specific for human herpesvirus 6. Virology 262:139-151. [DOI] [PubMed] [Google Scholar]
- 15.He, J., M. McCarthy, Y. Zhou, B. Chandran, and C. Wood. 1996. Infection of primary human fetal astrocytes by human herpesvirus 6. J. Virol. 70:1296-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Höllsberg, P., K. W. Wucherpfennig, L. J. Ausubel, V. Calvo, B. E. Bierer, and D. A. Hafler. 1992. Characterization of in vivo infected T cell clones. IL-2-independent growth of nontransformed T cells. J. Immunol. 148:3256-3263. [PubMed] [Google Scholar]
- 17.Inoue, N., T. R. Dambaugh, J. C. Rapp, and P. E. Pellett. 1994. Alphaherpesvirus origin-binding protein homolog encoded by human herpesvirus 6B, a betaherpesvirus, binds to nucleotide sequences that are similar to ori regions of alphaherpesviruses. J. Virol. 68:4126-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isegawa, Y., T. Mukai, K. Nakano, M. Kagawa, J. Chen, Y. Mori, T. Sunagawa, K. Kawanishi, J. Sashihara, A. Hata, P. Zou, H. Kosuge, and K. Yamanishi. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 73:8053-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isegawa, Y., Z. Ping, K. Nakano, N. Sugimoto, and K. Yamanishi. 1998. Human herpesvirus 6 open reading frame U12 encodes a functional beta-chemokine receptor. J. Virol. 72:6104-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Josephs, S. F., S. Z. Salahuddin, D. V. Ablashi, F. Schachter, F. Wong-Staal, and R. C. Gallo. 1986. Genomic analysis of the human B-lymphotropic virus (HBLV). Science 234:601-603. [DOI] [PubMed] [Google Scholar]
- 21.Kondo, K., Y. Hayakawa, H. Mori, S. Sato, T. Kondo, K. Takahashi, Y. Minamishima, M. Takahashi, and K. Yamanishi. 1990. Detection by polymerase chain reaction amplification of human herpesvirus 6 DNA in peripheral blood of patients with exanthem subitum. J. Clin. Microbiol. 28:970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo, K., T. Kondo, T. Okuno, M. Takahashi, and K. Yamanishi. 1991. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 72:1401-1408. [DOI] [PubMed] [Google Scholar]
- 23.Luppi, M., P. Barozzi, C. Morris, A. Maiorana, R. Garber, G. Bonacorsi, A. Donelli, R. Marasca, A. Tabilio, and G. Torelli. 1999. Human herpesvirus 6 latently infects early bone marrow progenitors in vivo. J. Virol. 73:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lusso, P., P. D. Markham, E. Tschachler, F. di Marzo Veronese, S. Z. Salahuddin, D. V. Ablashi, S. Pahwa, K. Krohn, and R. C. Gallo. 1988. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6). J. Exp. Med. 167:1659-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menotti, L., P. Mirandola, M. Locati, and G. Campadelli-Fiume. 1999. Trafficking to the plasma membrane of the seven-transmembrane protein encoded by human herpesvirus 6 U51 gene involves a cell-specific function present in T lymphocytes. J. Virol. 73:325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirandola, P., P. Menegazzi, S. Merighi, T. Ravaioli, E. Cassai, and D. Di Luca. 1998. Temporal mapping of transcripts in herpesvirus 6 variants. J. Virol. 72:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mocarski, E., and E. Tan Courcelle. 2001. Cytomegalovirus and their replication, p. 2629-2673. In David M. Knipe, Peter M. Howley, Diane E. Griffin, Robert A. Lamb, Malcolm A. Martin, Bernard Roizman, and Stephen E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Pedrali-Noy, G., and S. Spadari. 1979. Effect of aphidicolin on viral and human DNA polymerases. Biochem. Biophys. Res. Commun. 88:1194-1202. [DOI] [PubMed] [Google Scholar]
- 29.Pellett, P. E., D. Sanchez-Martinez, G. Dominguez, J. B. Black, E. Anton, C. Greenamoyer, and T. R. Dambaugh. 1993. A strongly immunoreactive virion protein of human herpesvirus 6 variant B strain Z29: identification and characterization of the gene and mapping of a variant-specific monoclonal antibody reactive epitope. Virology 195:521-531. [DOI] [PubMed] [Google Scholar]
- 30.Pellett, P., and G. Dominguez. 2001. Human herpesvirus 6A, 6B, and 7 and their replication, p. 2769-2784. In David M. Knipe, Peter M. Howley, Diane E. Griffin, Robert A. Lamb, Malcolm A. Martin, Bernard Roizman, and Stephen E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Rapp, J. C., L. T. Krug, N. Inoue, T. R. Dambaugh, and P. E. Pellett. 2000. U94, the human herpesvirus 6 homolog of the parvovirus nonstructural gene, is highly conserved among isolates and is expressed at low mRNA levels as a spliced transcript. Virology 268:504-516. [DOI] [PubMed] [Google Scholar]
- 32.Salahuddin, S. Z., D. V. Ablashi, P. D. Markham, S. F. Josephs, S. Sturzenegger, M. Kaplan, G. Halligan, P. Biberfeld, F. Wong-Staal, and B. Kramarsky. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596-601. [DOI] [PubMed] [Google Scholar]
- 33.Santoro, F., P. E. Kennedy, G. Locatelli, M. S. Malnati, E. A. Berger, and P. Lusso. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817-827. [DOI] [PubMed] [Google Scholar]
- 34.Schirmer, E. C., L. S. Wyatt, K. Yamanishi, W. J. Rodriguez, and N. Frenkel. 1991. Differentiation between two distinct classes of viruses now classified as human herpesvirus 6. Proc. Natl. Acad. Sci. USA 88:5922-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spadari, S., F. Focher, F. Sala, G. Ciarrocchi, G. Koch, A. Falaschi, and G. Pedrali-Noy. 1985. Control of cell division by aphidicolin without adverse effects upon resting cells. Drug Res. 35:1108-1116. [PubMed] [Google Scholar]
- 36.Takemoto, M., T. Shimamoto, Y. Isegawa, and K. Yamanishi. 2001. The R3 region, one of three major repetitive regions of human herpesvirus 6, is a strong enhancer of immediate-early gene U95. J. Virol. 75:10149-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyatt, L. S., N. Balachandran, and N. Frenkel. 1990. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J. Infect. Dis. 162:852-857. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto, T., T. Mukai, K. Kondo, and K. Yamanishi. 1994. Variation of DNA sequence in immediate-early gene of human herpesvirus 6 and variant identification by PCR. J. Clin. Microbiol. 32:473-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamanishi, K., T. Okuno, K. Shiraki, M. Takahashi, T. Kondo, Y. Asano, and T. Kurata. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065-1067. [DOI] [PubMed]