Abstract
We used a transient-transfection replication assay to identify two functional copies of the human herpesvirus 8 (HHV8) lytic origin of DNA replication (oriLyt). BCLB-1 cells were transfected with HHV8 subgenomic fragments containing the putative lytic origin along with a plasmid expressing viral transactivator open reading frame (ORF) 50. The HHV8 left-end oriLyt (oriLyt-L) lies between ORFs K4.2 and K5 and is composed of a region encoding various transcription factor binding sites and an A+T-rich region and a G+C repeat region. The right-end oriLyt (oriLyt-R) maps between ORF 69 and vFLIP, a region similar to the RRV oriLyt, and is an inverted duplication of oriLyt-L.
Human herpesvirus 8 (HHV8) is a gamma-2 class herpesvirus related to Epstein-Barr virus (EBV). HHV8 is the probable cause of Kaposi's sarcoma and is involved in the pathogenesis of primary effusion lymphoma and multicentric Castleman's disease (1-4). HHV8 appears to be distinct from EBV in that it encodes a number of genes of cellular origin (20-22, 27, 28).
HHV8 is typically latent in cultured B cells, and only a small number of cells undergo spontaneous lytic reactivation (18, 19, 26). In cell culture, HHV8 can be induced to enter the lytic cycle and produce infectious virus by treatment with tetradecanoyl phorbol acetate (TPA) and/or n-butyrate (26). In addition, transfection of a plasmid that expresses the viral transactivator open reading frame (ORF) 50 is sufficient for induction of the viral lytic cycle (9, 17, 33, 34). Induction of the viral lytic cycle consists of the expression of viral enzymes that participate in replication of the viral genome, resulting in an increase in viral DNA and production of infectious virus (9, 33). HHV8 ORF 50 appears to have a function similar to that of the EBV viral transactivator BZLF1 or Zta with respect to induction of the viral lytic cycle. Zta is a key viral protein that participates in reactivation and initiation of lytic EBV DNA replication (5, 6, 8, 12, 15, 29).
Recently, it was shown that the six core replication proteins for HHV8 are sufficient for replication of the cloned EBV origin of replication (36). In addition, a replication complex containing the HHV8 K8 protein was shown to colocalize with promyelocytic leukemia protein oncogenic domains (PODs) in lytic-cycle-induced or HHV8-infected dermal microvascular endothelial cells (11). Despite this recent elucidation of the core transacting factors, the discovery of a lytic origin for HHV8 has not been discovered.
The primarily latent nature of HHV8 in cell culture makes studies concerning the cis-acting lytic factors for this virus somewhat difficult. For this reason, we initially chose to examine the lytic replication machinery of the closely related virus rhesus rhadinovirus (RRV). In the RRV system, lytic replication occurs upon infection of rhesus fibroblasts (24, 32). The lytic origin for RRV is located at the right end of the genome and consists of a number of G+C- and A+T-rich repeat regions, along with various transcription factor binding sites, and maps between ORF 69 and vFLIP (24). This homologous region in HHV8, between vFLIP and ORF 69, has many of the features of the RRV lytic origin of DNA replication (oriLyt). In addition, Nicholas et al. identified this region, along with an inverted duplication of this region at the 5′ end of the genome, as a potential lytic origin (23). Within these regions, an approximately 1.0-kb DNA sequence is present that contains various transcription factor binding sites and A+T-rich palindromic sequences. This region is referred to as the oriLyt domains because of the strong resemblance to other herpesvirus oriLyt regions (23). The organization of the DNA sequence, with respect to the location of G+C- and A+T-rich DNA elements, of the 5′ (left-end) putative HHV8 oriLyt (oriLyt-L) closely resembles the RRV oriLyt. For this reason, we decided to initially subclone fragments from this region of the HHV8 genome for testing in a replication assay.
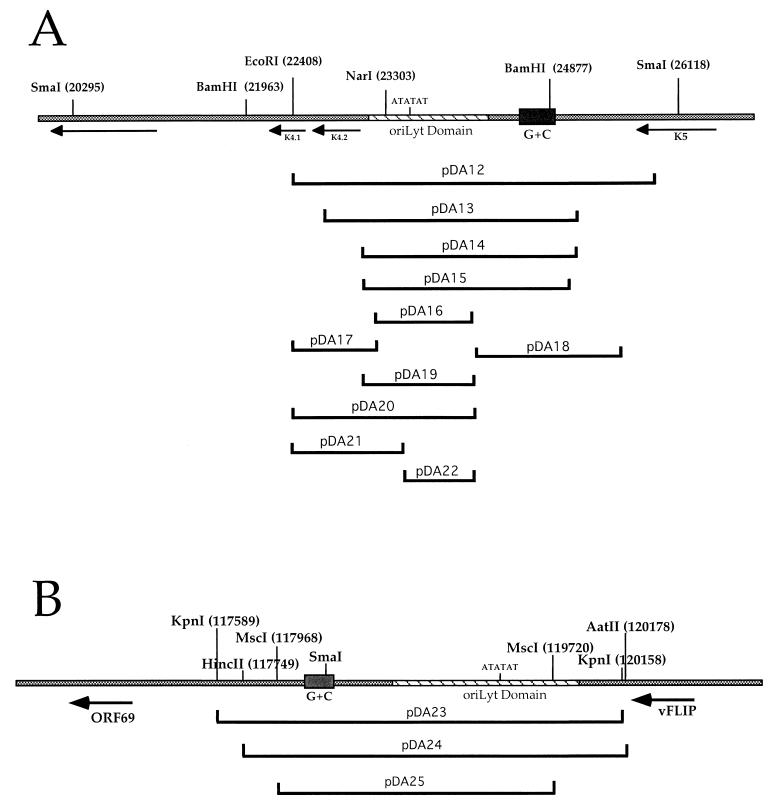
Table 1 shows the names, nucleotide coordinates, and restriction sites used for subclones that were made and subsequently transfected into BCBL-1 cells (AIDS Research and Reference Reagent Program). We initially subcloned a fragment encoding DNA elements that are consistent with a putative oriLyt. This 3.5-kb SacII-EcoRI fragment (Fig. 1A, pDA12), corresponding to nucleotides (nt) 22408 to 25913 of the genome, was transfected into BCBL-1 cells along with a plasmid expressing the HHV8 viral transactivator ORF 50 (pEXP50). Cotransfections included ORF 50 since this gene product was shown to induce the HHV8 lytic replication cycle in cells harboring latent virus. Total cellular DNA was harvested 4 days posttransfection and cleaved with EcoRI and DpnI. The restriction enzyme DpnI cleaves only methylated or unreplicated plasmid DNA. Therefore, replicated plasmid DNA is resistant to DpnI cleavage and migrates at a higher molecular weight than DpnI-sensitive cleavage products. Each transfection was repeated at least three times, and a replication signal more that 20% of that obtained with pDA12 was considered a valid replication signal.
TABLE 1.
HHV8 subclones used in transient-transfection replication assays
| Plasmid | Genomic location | Restriction enzymes | Insert size (kb) | Replication/assay resultc |
|---|---|---|---|---|
| OriLyt-L constructs | ||||
| pDA12 | 22408-25913 | SacII-EcoRI | 3,505 | + |
| pDA13 | 22705-25159 | ClaI-BsrGI | 2,454 | + |
| pDA14 | 23069-25159 | AgeI-BsrGI | 2,090 | + |
| pDA15 | 23069-25062 | AgeI-MluI | 1,993 | + |
| pDA16 | 23217-24165 | HincII-SmaI | 948 | − |
| pDA17 | 22408-23217 | HincIIa-SmaI | 836 | − |
| pDA18 | 24165-25552 | HincII-HincII | 1,387 | − |
| pDA19 | 23069-24165 | AgeI-HincII | 1,098 | − |
| pDA20 | 22408-24165 | HincIIa-HincII | 1,784 | − |
| pDA21 | 22408-23480 | HincIIaStuI | 1,099 | − |
| pDA22 | 23480-24165 | StuI-HincII | 685 | − |
| pDA12-Bb | 22408-25913 | SacII-EcoRI | 3,267 | + |
| pDA13-DEL11 | 23195-25159 | 1,964 | − | |
| pDA13-DEL12 | 23482-25159 | 1,677 | − | |
| pDA13-DELr1 | 22705-24619 | 1,914 | + | |
| pDA13-DELr2 | 22705-24303 | 1,598 | − | |
| pDA13-DELr3 | 22705-24043 | 1,338 | − | |
| pDA13-DELr4 | 22705-23884 | 1,179 | − | |
| pDA13-DELr5 | 22705-23719 | 1,014 | − | |
| OriLyt-R constructs | ||||
| pDA23 | 117589-120158 | KpnI-KpnI | 2,569 | + |
| pDA24 | 117749-120178 | HincII-AatII | 2,431 | + |
| pDA25 | 117968-119720 | MscI-MscI | 1,752 | + |
HincII site within the pBluescript multiple cloning site.
Internal BamHI fragment removed.
+, replication; −, no replication.
FIG. 1.
Schematic representation of HHV8 oriLyt regions and subclones used in the transient-transfection replication assay. Shown are the relative positions of ORFs. Hatched regions indicate the oriLyt domain sequences, and sold blocks indicate G+C repeat regions. Shown below are each oriLyt region and the locations of oriLyt subclones. Nucleotide coordinates for each subclone and other details are given in Table 1. (A) HHV8 oriLyt-L; (B) HHV8 oriLyt-R.
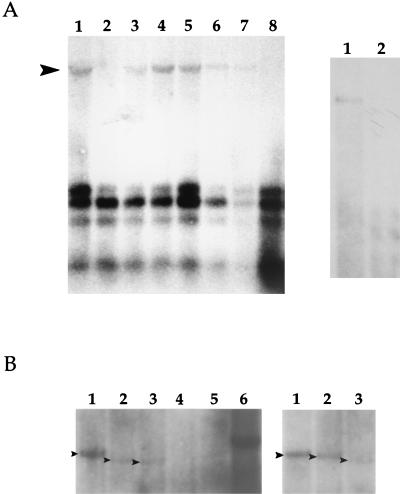
Transfection of pDA12 along with pEXP50 into BCBL-1 cells resulted in a DpnI-resistant band, indicating that the pDA12 plasmid was replicated (Fig. 2A, left side, lane 1). When pEXP50 was omitted from the cotransfection mixture, however, no replicated plasmid was detected (Fig. 2A, left side, lane 2). Increasing amounts of pEXP50 in the transfection mixture resulted in an increase in the amount of replicated pDA12, demonstrating the dose response effect of ORF 50 on oriLyt replication (Fig. 2A, left side, lanes 3 to 5). We also investigated whether incubation with TPA, another inducer of the virus lytic cycle, is sufficient to replicate the cloned HHV-8 oriLyt. Cells were pretreated with TPA for 24 h and then transfected with pDA12. Total cellular DNA was harvested 2 or 4 days posttransfection. Replication products were detected in samples where cells were treated with TPA, although the signal was not as strong as that of the replication product observed when pEXP50 was present in the transfection mixture (Fig. 2A, left side, compare lanes 3 and 4 to lanes 6 and 7). Replication products were not detected in samples cotransfected with pDA12 and pEXP50 and treated with phosphonoformic acid (PFA), an inhibitor of herpesvirus polymerase (Fig. 2A, left side, lane 8). This set of transfection experiments identified an HHV8 lytic origin in which replication was responsive to ORF 50 and inhibited by PFA.
FIG. 2.
Identification of HHV8 oriLyt. (A, left side) Autoradiogram of a Southern blot of EcoRI- and DpnI-cleaved total cellular DNA from BCBL-1 cells transfected by electroporation with pDA12 with or without a plasmid expressing HHV8 ORF 50 (pEXP50). Blots were hybridized with pGEM7zf(−). The arrowhead indicates the replicated product. Lanes: 1, transfection of pDA12 plus pEXP50 (5 μg); 2, pDA12 plus pGEM7zf(−) (5 μg); 3, pDA12 plus pEXP50 (0.5 μg); 4, pDA12 plus pEXP50 (2 μg); 5, pDA12 plus pEXP50 (5 μg); 6, pDA12 plus TPA treatment for 4 days; 7, pDA12 plus TPA treatment for 2 days; 8, pDA12 plus pEXP50 (5 μg) incubated with PFA (300 μg/ml). (A, right side) Replication products are susceptible to cleavage by the restriction enzyme MboI. Cells were transfected with pDA12 and pEXP50 (5 μg), and DNA was extracted and treated with EcoRI and DpnI (lane 1) or EcoRI, DpnI, and MboI (lane 2). (B) Both the oriLyt domain and the G+C repeat regions are required for replication and identification of the OriLyt-R sequence. Transfection of HHV8 oriLyt-L and oriLyt-R subclones was done to identify essential DNA elements. All transfections contained 20 μg of oriLyt-containing plasmids plus 5 μg of pEXP50. Arrowheads indicate replicated plasmids. Refer to Table 1 and Fig. 1 for details of subclones. (B, left side) Transfection of oriLyt-L subclones. Lanes: 1, pDA13; 2, pDA14; 3, pDA15; 4, pDA16; 5, pDA18; 6, pDA12. (B, right side) Transfection of oriLyt-R subclones. Lanes: 1, pDA23; 2, pDA24; 3, pDA25.
In order to show that DpnI-protected products were the result of DNA amplification and not incomplete cleavage by the DpnI restriction enzyme, samples were cleaved with the restriction enzyme MboI. MboI cleaves unmethylated DNA and is inhibited by methylated DNA. Treatment of pDA12 with EcoRI, DpnI, and MboI resulted in disappearance of the replicated DNA band in EcoRI- and DpnI-treated samples (Fig. 2A, right side, compare lanes 1 and 2).
To define the boundaries of the HHV8 oriLyt, we made several constructs in which DNA sequences were removed from the 5′ and 3′ ends of subclone pDA12 (Table 1 and Fig. 1A). Subclone pDA13, which lacks 754 bp from the right end and 303 bp from the left end of subclone pDA12, replicated when transfected into BCBL-1 cells (Fig. 2B, left side, lane 1). When an additional 364 bp was removed from the right end of pDA13 to make subclone pDA14, the replication efficiency was decreased to approximately 70% of that of the full-length oriLyt, indicating the possible existence of some ancillary element(s) (Fig. 2B, left side, lane 2). The replication efficiency was relatively unchanged when subclone pDA15, which has 97 bp deleted from the left end (Fig. 1A), was used in the transient-transfection replication assay (Fig. 2B, left side, lane 3). Subclone pDA16, which lacked all of the G+C repeat region but retained the putative oriLyt domains, failed to replicate, indicating that at least some of the G+C DNA sequence is required for replication (Fig. 2B, left side, lane 4). A subclone that contained the G+C repeat region plus some of the flanking DNA sequence (pDA18) also failed to replicate (Fig. 2B, left side lane 5). Therefore, neither the putative oriLyt domains nor the G+C repeat regions alone were sufficient for replication. Subclones shown in Fig. 1A that encoded either regions upstream of the oriLyt domains (pDA17 and pDA21) or those that contained portions of the oriLyt domains (pDA16, pDA19, and pDA22), along with a subclone in which most of the oriLyt domains but none of the G+C repeat region was present, did not replicate in the transient-transfection assays (data not shown). In addition, a construct that lacked some of the internal G+C repeat region from the BamHI site (nt 24637) to the BamHI site (nt 24877) did replicate, suggesting that some of the internal repeat elements within the G+C repeat region are dispensable (data not shown).
Examination of the HHV8 genome indicated that an almost exactly homologous region of oriLyt-L is located at the rightward end of the genome and is present as an inverted duplication of the 5′-end sequence (Fig. 1B). We subcloned this homologous region of the HHV8 genome and tested the resulting plasmid constructs in the transient-transfection replication assay. We initially subcloned a 2.5-kb fragment that contained the G+C repeat region and the putative lytic origin domains. This plasmid construct, pDA23, and pDA24, which is only 240 bp shorter at the 5′ end, efficiently replicated in BCBL-1 cells (Fig. 2B, right side, lanes 1 and 2). A smaller subclone, pDA25, lacks an additional 219 bp at the 5′ end and 458 bp from the 3′ end of pDA24. The pDA25 construct lacks a small portion (∼200 bp) of the oriLyt domain but includes the entire G+C repeat region. A replication signal was detected when pDA25 was used in the replication assay but at a severely decreased level (80% decrease in replication signal) compared to that obtained with pDA24, which included the complete oriLyt domain (Fig. 2B, right side, compare lanes 2 and 3). Smaller subclones further lacking the oriLyt-R domain failed to replicate (data not shown). Therefore, the boundaries of HHV8 oriLyt-R appear to be very similar to those for oriLyt-L. The smallest replicating fragment is a 1.7-kb region spanning nt 117968 to 119720.
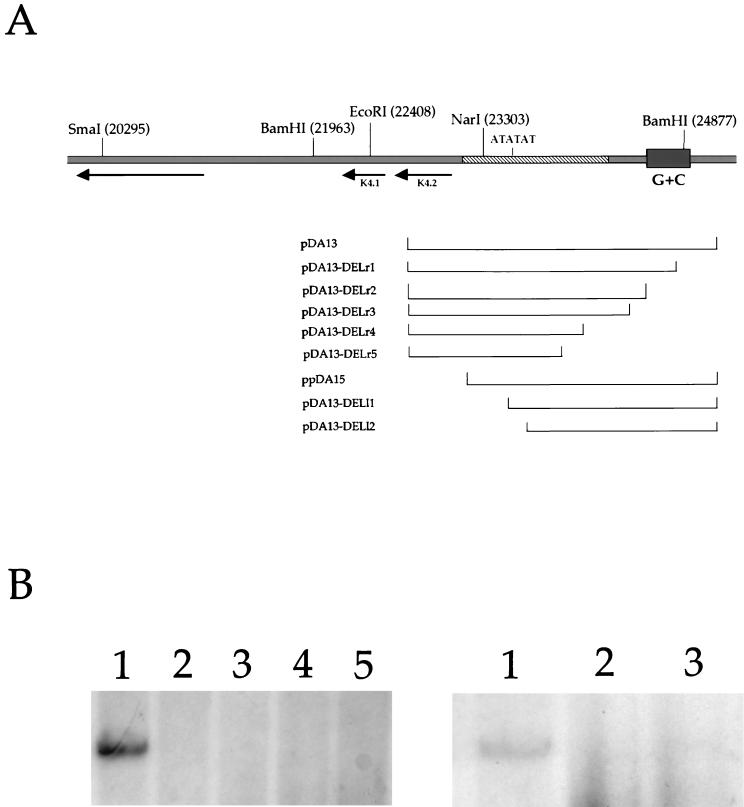
To further define the boundaries of the oriLyt, we generated a series of deletion constructs. We started with plasmid construct pDA13 and removed a DNA sequence from either the left (5′) or right (3′) end of the subclone (Fig. 3A). These constructs were then used in the transient-transfection replication assay. A construct lacking 540 bp at the 3′ end of pDA13 resulted in partial deletion of the G+C repeat region. This construct, pDA13-DELr1, replicated when transfected into BCLB-1 cells in the presence of ORF 50 (Fig. 3B, left side, lane 1). However, no other 3′-end deletion construct replicated in BCLB-1 cells. Plasmid construct pDA13-DELr2 lacked the entire G+C repeat region and failed to replicate in transfected cells (Fig. 3B, left side, lane 2). Other constructs lacking more of the intervening sequence between the G+C repeat region and the A+T-rich origin domain also did not replicate (Fig. 3B, left side, lanes 3 to 5). This series of deletion constructs revealed that at least half of the G+C-rich region within the HHV8 oriLyt is essential for efficient replication.
FIG. 3.
Mapping of the right- and left-hand boundaries of HHV8 oriLyt-R. (A) Exonuclease cleavage constructs used to define the 3′ and 5′ boundaries of the HHV8 oriLyt. Nucleotide coordinates for cleavage constructs are listed in Table 1. (B) 5′ and 3′ boundaries of the HHV8 oriLyt-R. BCBL-1 cells were transfected with a series of deletion constructs in which a DNA sequence from either the 5′ or the 3′ end of oriLyt subclone pDA13 was removed by exonuclease cleavage. An autoradiogram of a Southern blot of DNA from the transient-transfection replication assay is shown. (B, left side) 3′-end deletions. Lanes: 1, pDA13-DELr1; 2, pDA13-DELr2; 3, pDA13-DELr3; 4, pDA13-DELr4; 5, pDA13-DELr5. (B, right side) 5′-end deletions. Lanes: 1, pDA15; 2, pDA13-DELl1; 3, pDA13-DELl2.
Constructs that lacked a DNA sequence from the 5′ end of pDA13 revealed that none of the A+T repeat region can be removed and still allow efficient oriLyt replication. As shown previously, subclone pDA15, which lacked all of the K4.2 ORF, did efficiently replicate (Fig. 3B, right side, lane 1). However, when we tested a series of 5′-end deletions, it was shown that no other 5′-end deletion construct replicated upon transfection (Fig. 3B, right side, lanes 2 and 3). Plasmid construct pDA13-DELl1, which lacked some of the A+T-rich DNA sequence of subclone pDA15, did not replicate when transfected into BCLB-1 cells in the presence of the ORF 50 expression plasmid (Fig. 3B, lane 2). Likewise, construct pDA13-DELl2, lacking an additional 287 nt of pDA13-DELl1, also failed to replicate (Fig. 3B, right side, lane 3). This result indicates that an essential element lies between nt 23482 and 23591. Inspection of the HHV8 genomic sequence shows that the repeat element (AT)n was removed from construct pDA13-DELl2 (Fig. 3A) and could account for the loss of replication activity. On the basis of the transfection experiments involving deletion constructs, the boundaries of HHV8 oriLyt-L are nt 23069 and 24619, which corresponds to an approximately 1.6-kb region.
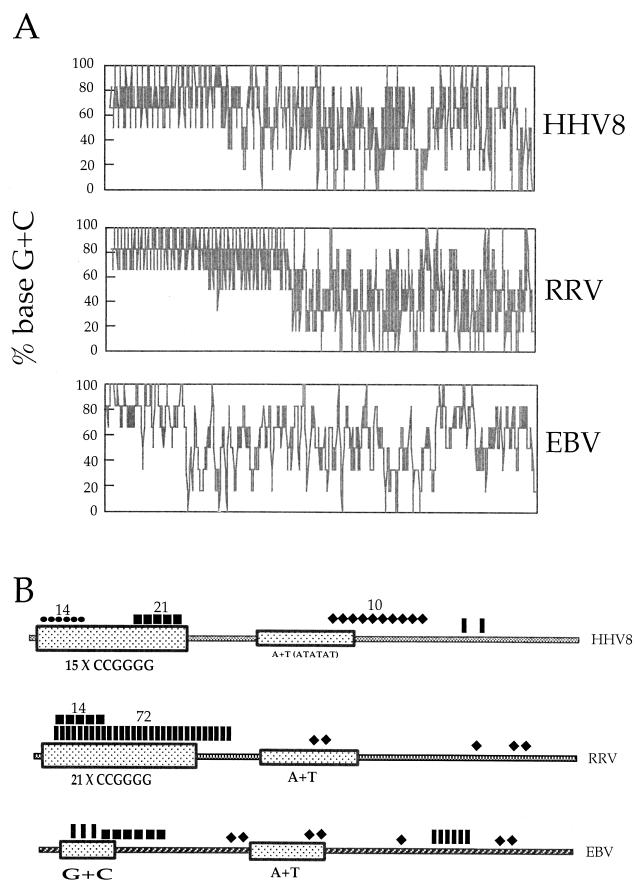
Comparison of the HHV8, RRV, and EBV oriLyt regions revealed that the HHV8 oriLyt more closely resembles the RRV origin than the EBV oriLyt. The G+C-rich regions of RRV and HHV8 are similarly arranged (Fig. 4A). The G+C-rich regions within HHV8 and RRV are much longer and have a higher G+C content than the EBV core origin (Fig. 4A). HHV8 and RRV both have multiple copies of the repeat CCGGGG, whereas EBV contains only one such motif (Fig. 4B). All three oriLyt regions encode A+T-rich regions; however, none of the viruses have the same motif within this region, with the exception of the basic helix-loop-helix transcription factor binding site. Several different transcription factor binding sites are present within all three core oriLyt regions. All three origins contain GC factor binding sites within the G+C-rich regions at the left end of the core sequence (Fig. 4B). The HHV8 oriLyt encodes 14 copies of the pentanucleotide repeat sequence (JCV), an element known to regulate JC virus DNA replication (Fig. 4B). This element appears to be unique to the HHV8 oriLyt, suggesting a distinct mode of regulation of DNA replication.
FIG. 4.
Sequence comparison of core oriLyt regions from HHV8, RRV, and EBV. (A) Comparison of the core oriLyt regions with respect to percent G+C content. (B) Identification of various transcription factor binding sites within HHV8, RRV, and EBV oriLyt regions. GCF, GC factor; JCV, pentanucleotide repeat sequence; bHLH, basic helix-loop-helix; AP2, activator protein 2.
We have described the identification of the HHV8 lytic origins of replication. Two functional lytic origins are present within the HHV8 genome. Replication products were sensitive to the herpesvirus DNA polymerase inhibitor PFA, indicating that the virus-encoded DNA polymerase is required for amplification of the cloned HHV8 oriLyt. oriLyt-L, located between nt 23069 and 24619, is composed of a small (365-nt) G+C repeat region, a central core encoding various transcription factor binding sites, and an ATATA repeat region. oriLyt-R is an almost exact inverted duplication of the oriLyt-L loci. oriLyt-R is located between vFLIP and ORF 69 in a region homologous to that of the closely related rhadinovirus RRV. The most robust replication signal was observed when an expression plasmid encoding the HHV8 viral transactivator ORF 50 was included in the transfection mixture. ORF 50 activates early gene promoters via direct binding and appears to require a cellular protein for transactivation function (16, 35). The EBV genome has two copies of oriLyt containing two essential core elements. One core contains Zta response elements (ZREs) for the transactivator protein Zta (10, 30). These regions are absolutely required for oriLyt replication in transient-transfection assays (30). The other core element contains two A+T-rich palindromes and an adjacent polypurine-polypyrimidine tract (10, 25, 31). Also present within the EBV oriLyt is a nonessential enhancer region containing the DNA binding sites for two virus-encoded transactivators, Rta and Zta (7).
HHV8 oriLyt-L lies just upstream of the K4.2 ORF. This arrangement is similar that of the EBV oriLyt, where one component of the origin is the BHLF1 promoter and leader sequence (10). The HHV8 oriLyt has two components; the first consists of an oriLyt domain that contains various transcription factor binding sites and a series of CAGCTG repeats. In addition, an A+T-rich palindromic region and the G+C repeat element are also present in both copies of the HHV8 oriLyt. Despite the similarities to the EBV oriLyt, the HHV8 oriLyt more closely resembles the lytic origin of RRV. The overall sequence homology between the two origin regions is 47% (24). The HHV8 oriLyt, like that of RRV, has an A+T-rich region adjacent to a G+C-rich region. Deletion studies of both the RRV and HHV8 origins revealed that, although a portion of the G+C-rich repeat sequence is dispensable, the A+T-rich sequence is absolutely required for oriLyt function. Within the A+T-rich sequence of the HHV8 oriLyt, a 16-base AT repeat sequence exists whereas the RRV A+T-rich sequence has several A and T repeat elements. These A+T-rich sequences are common in lytic origins and are thought to be sites where DNA melting and unwinding occur (13, 14). Future studies will define the role of the A+T and G+C repeat regions within the lytic origin.
The elucidation of the HHV8 oriLyt makes it possible to further define the cis- and trans-acting factors required for and contributing to the initiation of lytic replication.
Acknowledgments
These studies were funded by National Cancer Institute research grant RO1 CA85164 from the National Institutes of Health.
REFERENCES
- 1.Birley, H. D., and T. F. Schultz. 1997. Kaposi's sarcoma and the new herpesvirus. J. Med. Microbiol. 46:433-435. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 3.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 4.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 5.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, M. A., J. Leahy, and J. M. Hardwick. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammerschmidt, W., and B. Sugden. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427-433. [DOI] [PubMed] [Google Scholar]
- 11.Katano, H., K. Ogawa-Goto, H. Hasegawa, T. Kurata, and T. Sata. 2001. Human-herpesvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruits p53 to the PML bodies. Virology 286:446-455. [DOI] [PubMed] [Google Scholar]
- 12.Kenney, S., J. Kamine, E. Holley-Guthrie, J. C. Lin, E. C. Mar, and J. Pagano. 1989. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J. Virol. 63:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalski, D., and M. J. Eddy. 1989. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J 8:4335-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalski, D., D. A. Natale, and M. J. Eddy. 1988. Stable DNA unwinding, not “breathing,” accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc. Natl. Acad. Sci. USA 85:9464-9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 18.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, G., M. O. Rigsby, L. Heston, E. Grogan, R. Sun, C. Metroka, J. A. Levy, S. J. Gao, Y. Chang, and P. Moore. 1996. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 334:1292-1297. [DOI] [PubMed] [Google Scholar]
- 20.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholas, J., V. Ruvolo, J. Zong, D. Ciufo, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1997. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 71:1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholas, J., V. R. Ruvolo, W. H. Burns, G. Sandford, X. Wan, D. Ciufo, S. B. Hendrickson, H. G. Guo, G. S. Hayward, and M. S. Reitz. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287-292. [DOI] [PubMed] [Google Scholar]
- 23.Nicholas, J., J. C. Zong, D. J. Alcendor, D. M. Ciufo, L. J. Poole, R. T. Sarisky, C. J. Chiou, X. Zhang, X. Wan, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1998. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 23:79-88. [DOI] [PubMed] [Google Scholar]
- 24.Pari, G. S., D. AuCoin, K. Colletti, S. A. Cei, V. Kirchoff, and S. W. Wong. 2001. Identification of the rhesus macaque rhadinovirus lytic origin of DNA replication. J. Virol. 75:11401-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Portes-Sentis, S., A. Sergeant, and H. Gruffat. 1997. A particular DNA structure is required for the function of a cis-acting component of the Epstein-Barr virus OriLyt origin of replication. Nucleic Acids Res. 25:1347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 27.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarid, R., T. Sato, R. A. Bohenzky, J. J. Russo, and Y. Chang. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat. Med. 3:293-298. [DOI] [PubMed] [Google Scholar]
- 29.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schepers, A., D. Pich, and W. Hammerschmidt. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367-376. [DOI] [PubMed] [Google Scholar]
- 31.Schepers, A., D. Pich, J. Mankertz, and W. Hammerschmidt. 1993. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J. Virol. 67:4237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF 50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, F. Y., J.-H. Ahn, D. J. Alcendor, W.-J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]