Abstract
Foamy virus (FV) replication is resistant to most nucleoside analog reverse transcriptase (RT) inhibitors. In an attempt to create a 2′,3′-dideoxy-3′-thiacytidine (3TC)-sensitive virus, the second residue in the highly conserved YXDD motif of simian foamy virus-chimpanzee (human isolate) [SFVcpz(hu)] RT was changed from Val (V) to Met (M). Unexpectedly, the resultant virus, SFVcpz(hu) RT-V313M, replicated poorly, and Met rapidly reverted to Val. Despite the presence of approximately 50% of wild-type RT activity in RT-V313M virions, full-length DNA products were not detected in transfected cells. Using purified recombinant enzymes, we found that the wild-type FV RT is significantly more processive than human immunodeficiency virus type 1 RT. However, the V313M mutant has about 40% of the wild-type level of FV RT activity and has a lower processivity than the wild-type FV enzyme. The V313M mutant RT is also relatively resistant to 3TC. These results suggest that the decrease in RT activity and processivity of FV RT-V313M prevents completion of reverse transcription and greatly diminishes viral replication.
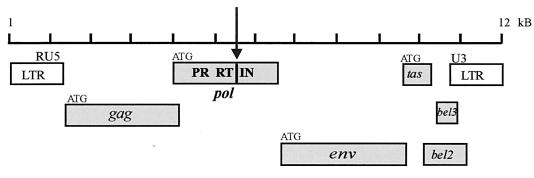
Foamy viruses (FVs), which are classified as retroviruses, have the classic retroviral genomic organization, including the hallmark genes gag, pol, and env (Fig. 1) (20). Despite this organization and classification, several aspects of the FV life cycle differ significantly from the life cycles of other retroviruses. Three of these differences involve the properties of the Pol protein and the reverse transcriptase (RT) contained within it. First, FV has an unusual mechanism for the expression of Pol. Typical retroviruses express Pol as part of a Gag-Pol fusion protein, and this fusion protein is incorporated into the virion through Gag-Pol-Gag interactions. In contrast, FV Pol is expressed from its own spliced message. Consequently, FV must employ a different strategy for incorporating Pol into viral particles (28). Second, relative to conventional retroviruses, reverse transcription occurs at a different stage in the FV life cycle. The virions of conventional retroviruses contain a single-stranded RNA genome; a significant portion of FV particles contain double-stranded DNA that is the functional genome when FV infects cells. (17, 30). This requires FV RT to be active before particles bud from an infected cell. Third, the FV Pol polyprotein undergoes limited processing. There is a single cleavage between RT and integrase (IN), which produces two mature viral enzymes, IN and a protease (PR)-RT fusion.
FIG. 1.
Schematic representation of PFV genome and encoded proteins. PFV is a complex retrovirus whose genome contains six open reading frames (shaded boxes). The Pol protein is synthesized independently of Gag and contains the enzymes PR, RT, and IN. The arrow indicates the single Pol cleavage site.
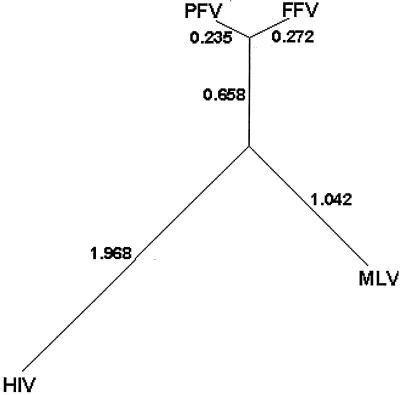
Because of its central role in the retroviral life cycle, RT has been a target of drugs designed to inhibit human immunodeficiency virus type 1 (HIV-1) replication and control infections. One major class of RT inhibitors is nucleoside analogs. Nucleoside analogs act as chain terminators and include the clinically important drugs 3′-azido-3′-deoxythymidine (AZT), 2′,3′-dideoxy-3′-thiacytidine (3TC), and 2′,3′-dideoxyinosine (ddI). In vitro, the triphosphate forms of these drugs are potent inhibitors of RT; the drugs can block the replication of a variety of retroviruses in cell culture. However, we have shown previously that while AZT specifically inhibits the replication of simian FV-chimpanzee (human isolate) (previously called human FV [HFV]; now referred to as prototype FV [PFV]), 3TC and ddI do not (30). In patients infected with HIV-1, these drugs inhibit viral replication; however, resistant strains of HIV-1 are selected rapidly. In the case of 3TC, resistance involves a mutation of the second residue in the HIV-1 catalytic motif YMDD to valine (M184V) (26). The YXDD catalytic motif is highly conserved among reverse transcriptases, and most wild-type retroviral RTs contain a methionine at the second residue. The exceptions are the murine leukemia viruses (MLVs) and FVs, which contain a valine at this position. Comparing the RT domains of PFV, feline FV (FFV), MLV, and HIV-1 in a neighbor-joining tree reveals that FVs are slightly more similar to MLV or HIV-1 than these two RTs are to each other, and that FVs are more similar to each other than they are to either MLV or HIV-1 (Fig. 2). Despite this relatively low level of similarity, there is considerable conservation of critical residues, including the YXDD motif (16). As might be expected, since both MLV and FV contain a valine in their catalytic YXDD motifs, both viruses are resistant to 3TC. Based on these observations, we wondered whether changing the YXDD motif of PFV from YVDD to YMDD would render it sensitive to 3TC.
FIG. 2.
Unrooted tree derived from the alignment of the core RTs from HIV, MLV, PFV, and FFV. The core RT sequences were derived from GenBank reference sequences NC_001802:2096.4174 (HIV), NC_001501:2337.4349 (MLV), NC_001736:3763.7194 (PFV), and NC_001871:1867.5337 (FFV). For the FV sequences, the PR and IN domains were removed using the consensus cleavage sequences defined by Pfrepper et al. (19). The amino- and carboxy-unaligned extensions of MLV and HIV, respectively, were removed after alignment using ClustalX (25). Numbers indicate relative differences between proteins. ClustalX was also used to calculate the distances for the tree.
A recent study found that MLV containing either wild-type YVDD or mutant YMDD RT was resistant to 3TC (8). This indicates that determinants other than the X position of the YXDD motif in MLV RT influence 3TC resistance and suggest that this may also be true for other RTs, including FV RT. To our surprise, changing YVDD to YMDD severely impairs the ability of PFV to replicate, and virus carrying this mutation rapidly reverts to the wild-type sequence, YVDD. We found that mutant virions retain approximately 50% of virion-associated RT activity yet are unable to complete reverse transcription, as judged by PCR and Southern analyses. In vitro studies using recombinant RT protein reveal that wild-type PFV RT is a highly processive enzyme and that the mutant RT with Met in the YXDD motif decreases processivity, which suggests that PFV requires a highly processive RT to replicate efficiently. We also found that the mutant enzyme is still resistant to 3TC despite the YMDD sequence.
MATERIALS AND METHODS
Construction of PFV RT-V313M mutant.
The shuttle vector pL2-Sub2 (1) was used to change the valine residue of the YXDD motif of PFV RT to a methionine (YVDD to YMDD). The mutagenic oligonucleotide polV313M was designed as a forward primer to change nucleotides 1682 to 1684 from GTT to ATG to create the V-to-M mutation and to change nucleotides 1694 to 1696 from TAT to TAC to create a unique AflII site to be used for screening (5′-CTAATGTACAAGTGTATATGGATGATATATACTTAAGCCATG-3′). The reverse primer Int(−) corresponds to nucleotides 2440 to 2459 of pL2-Sub2 (5′-CCCCAGGCTTTACACTTTATG-3′). Using these primers and the pL2-Sub2 template, PCR mutagenesis was used to generate a 794-bp fragment that was digested with BsrGI and XbaI and cloned back into the pL2-Sub2 vector. The clones were screened for the mutation in the insert with AflII, and two were identified. Appropriate segments from these two clones were moved back into the full-length PFV molecular clone pHFV13 by PacI/SwaI digestion and sequenced to verify the presence of the mutation. The resulting plasmids were named RT-V313M(A) and RT-V313M(B).
Tissue culture methods.
FAB cells (BHK cells containing PFV long terminal repeat [LTR]-β-galactosidase [β-Gal] DNA) (29) and 293T cells were grown in Dulbecco's modified Eagle medium with 10% fetal calf serum. Transfections of FAB cells were performed as previously described using Lipofectamine reagent (GIBCO-BRL, Gaithersburg, Md.) according to the manufacturer's instructions (30). Transfection of 293T cells was performed using a calcium phosphate method (4).
Viral supernatants were passed through a 0.2-μm-pore-size filter. Intracellular virus was released by scraping cells from plates followed by three freeze-thaw cycles, gentle sonication for 15 s, and centrifugation at 2,000 × g for 10 min. The virus was sedimented through a 20% sucrose cushion containing standard buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA [pH 8.0], 20% sucrose) by centrifugation at 24,000 rpm in a Beckman SW28 rotor at 4°C for 2 h. The pellets were resuspended in standard buffer with 10 mM MgCl2 and treated with RQ-1 RNase-free DNase (Promega, Madison, Wis.) (1 μl per 50-μl sample volume) at 37°C for 1 h. For polymerase assays, viral supernatants were concentrated to 1% of the original volume using Amicon Centriprep C-50 spin columns.
A panel of RT inhibitors (Table 1) were tested at a series of concentrations, and cell viability was analyzed for each at 24 h posttreatment by trypan blue staining. The inhibition experiments were performed with the highest inhibitor concentrations that did not affect cell viability. For pretreatment with RT inhibitors, FAB cells were treated with inhibitor for 4 h and then infected with cell-free virus stocks suspended in medium containing inhibitor. The virus was left on the cells for 24 h, and then the medium was removed and replaced with fresh medium containing inhibitor. The cells were fixed 48 h after infection and stained for β-Gal activity. For posttreatment with RT inhibitors, FAB cells were infected with cell-free virus stocks suspended in medium without inhibitor. Twenty-four hours after infection, the medium was removed and replaced with fresh medium containing inhibitor. Forty-eight hours postinfection, the virus was harvested and assayed for infectivity on fresh FAB cells (29).
TABLE 1.
Efficacies of RT inhibitors
| Inhibitora | concn (μM) | Titer relative to no-drug controlb
|
|
|---|---|---|---|
| Pretreated | Posttreated | ||
| No drug | 100 | 100 | |
| 3TC | 60 | 120 | 91 |
| AzddMeC | 25 | 70 | 13 |
| AzddU | 25 | 51 | 11 |
| AZT | 100 | 22 | <0.14 |
| D4T | 25 | 54 | 32 |
| ddC | 25 | 42 | 32 |
| FTC | 60 | 122 | 138 |
| PFA | 25 | 100 | 23 |
AzddMeC, 3′-azido-2′,3′-dideoxy-5-methylcytidine; AzddU, 3′-azido-2′,3′-dideoxyuridine; D4T, 2′,3′-didehydro-2′,3′-dideoxythymidine; ddC, 2′,3′-dideoxycytidine; FTC, beta-2′,3′-dideoxy-5-fluoro-thiacytidine; PFA, phosphonoformate.
Virus titers were determined on FAB cells as described in Materials and Methods.
Nucleic acid extractions.
DNA was extracted from viral particles as previously described (30). Briefly, resuspended pellets were DNase treated, lysed with sodium dodecyl sulfate (SDS), and extracted with phenol-chloroform-isoamyl alcohol (24:24:1), and nucleic acids were precipitated with ethanol. Pelleted nucleic acids were resuspended in the original volume of distilled H2O (dH2O) and treated with RNase A (Sigma) at 37°C for 1 h. The sample was then reextracted and resuspended in the original volume of dH2O.
Extraction of RNA from viral particles was performed by adding an equal mixture of phenol and 4 M guanidinium isothiocyanate at a 2:1 ratio to concentrated virus samples. The samples were extracted twice with chloroform-isoamyl alcohol (24:1), and nucleic acids were precipitated with ethanol in the presence of 10 μg of RNase-free carrier glycogen (Boehringer Mannheim, Indianapolis, Ind.). The pelleted nucleic acids were resuspended in the original volume of diethylpyrocarbonate-treated dH2O and treated with RQ-1 RNase-free DNase I (Promega) at 37°C for 1 h. The sample was then reextracted and resuspended in the original volume of diethylpyrocarbonate-treated dH2O.
Extraction of genomic DNA was performed by scraping transfected cells into PBS, centrifuging the cell suspension at 2,000 × g for 10 min, resuspending the cells in phosphate-buffered saline, and repeating the centrifugation. The cells were then resuspended in digestion buffer made with fresh proteinase K and RNase A (100 mM NaCl, 10 mM Tris-Cl [pH 8.0], 25 mM EDTA [pH 8.0], 0.5% SDS, 100 μg of proteinase K/ml, 1 μg of RNase A/ml) and incubated at 50°C overnight. Sodium perchlorate (1 M) was added at half the sample volume, and the sample was extracted twice with phenol-chloroform followed by one chloroform extraction, as described above. The DNA was ethanol precipitated and resuspended in dH2O.
RT-PCR, cloning, and sequencing.
Viral supernatants were pelleted by ultracentrifugation through a 20% sucrose cushion, and viral RNA was extracted as described above. RT-PCR was performed using the forward primer pol1441+ (5′-CCAACACTCTGCTGGTATTTTAGCTACTA-3′) and the reverse primer pol2351− (5′-CAGCTGACAAATTTGGACGTCCG-3′). PCR mixtures contained 2.5 U of avian myeloblastosis virus RT (U.S. Biochemicals, Cleveland, Ohio), 6 U of RNase inhibitor (Boerhinger Mannheim), 1× PCR buffer (Perkin-Elmer, Branchburg, N.J.), 1.5 mM MgCl2, 0.1 mM deoxynucleoside triphosphate (dNTP) mix (Gibco-BRL, Grand Island, N.Y.), 1 U of Platinum Pfx DNA Pol (Gibco-BRL), and 4 ng of each primer. Samples were incubated at 42°C for 45 min, followed by denaturation for 2 min at 95°C before thermal cycling was done. Temperatures and times for denaturing, annealing, and extension were 95, 50, and 72°C for 45 s, 45 s, and 1 min, respectively, each for 35 cycles. The final extension reaction was performed for 10 min at 72°C. The RT-PCR products were purified using Qiaex II (Qiagen), and A overhangs were added to the blunt-ended products using 5 U of Platinum Taq (Gibco-BRL), 1× PCR buffer (Perkin-Elmer), 1.5 mM MgCl2, and 0.1 mM dATP (Gibco-BRL) at 65°C for 30 min. The DNA products were cloned into the pGEM T Easy vector (Promega), and blue-white screening was used to identify clones that contained the insert. White colonies were picked and sequenced using the forward primer pol1548 (5′-GGTTAACAGCATTTACCTGGCAAG-3′).
Virion-associated RT assays.
Virion-associated RT assays were performed using viral supernatants collected 4 days posttransfection of FAB cells and concentrated with Centriprep 50 spin columns (Amicon). The substrate used in these assays was poly(A) · poly(dT)10 (Sigma). Concentrated viral supernatants were added to an RT cocktail containing final concentrations of 40 mM Tris-HCl (pH 8.0); 50 mM NaCl; 0.5 mM MnCl2; 15 mM dithiothreitol (DTT); 25 mM (each) dATP, dCTP, and dGTP; 0.1% NP-40; 2 μg of poly(A) · poly(dT)/ml; and 0.25 μl of [α-32P]dTTP/ml. The reaction mixtures were incubated at 37°C for 90 min; time points were taken at 30-min intervals. At each time point, 25 μl of the reaction mixture was spotted onto DE81 filters and allowed to dry. The filters were washed four times at room temperature with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min each, followed by two washes with 95% ethanol. The filters were then dried and counted in scintillation fluid.
Western blots.
Viral supernatants were concentrated as described above for the virion-associated RT assay. Concentrated virus was then added to SDS loading dye and fractionated on an SDS-10% polyacrylamide gel electrophoresis (PAGE) gel. Proteins were transferred to an Immobilon-P membrane (Millipore, Bedford, Mass.) and reacted with antibodies according to standard protocols. The membrane was developed using ECL reagents (Amersham Pharmacia) and exposed to film.
2LTR circle PCR.
100 ng of DNA was subjected to PCR using primers 350R (5′-AFAAGGGTCCATCTGAGTCAC-3′) and 546F (5′-GATTAAGGTATGAGGTGTGTGG-3′). The reaction conditions were 1× PCR buffer (Perkin-Elmer), 1.5 mM MgCl2, 0.2 mM dNTP mix (Gibco-BRL), and 1 U of Taq polymerase. Samples were denatured at 95°C for 5 min, followed by 30 cycles of denaturation, annealing, and extension at 95, 55, and 72°C for 30 s, 30 s, and 1 min, respectively. A final extension was carried out at 72°C for 10 min. The PCR products were fractionated on a 0.8% agarose gel, and the gel was subjected to Southern blotting as described below.
Southern blots.
Ten micrograms of DNA was digested with 40 U of the restriction enzyme NcoI (New England Biolabs) overnight. The digested DNA samples were fractionated on a 0.9% agarose gel. The gel was washed and transferred to Hybond membrane and hybridized following standard protocols. Radiolabeled probe to the LTR region was generated with the PrimeItII random-priming kit (Stratagene).
Construction of PFV RT expression clones.
The construct RT2 pET16b (13) contains part of the HFV RT coding region but is missing the protease coding region and most of the RNase H domain. The construct RTVM pET16b is similar except that the mutation V313M changes the polymerase active-site motif from YVDD to YMDD. PCR amplification was used to generate DNA fragments containing the protease coding region and the RNase H domain that could be linked to the HFV polymerase domain. The PCR amplification of the protease coding region used a 5′ primer that generated an NcoI site at the ATG initiation codon (5′-GCGGCGCCATGGCGAATCCTCTTCAGCTGTTACAGCCGCTTCCGGCGG-3′). The introduction of the NcoI site converts the start of the protease amino acid sequence from MNPLQ- to MANPLQ-. The 3′ primer in the PCR amplification spans a unique AflIII restriction enzyme recognition sequence in the segment encoding the polymerase domain of HFV RT (5′-GCGGCGCCTTGAGGAAGACGTGTCCAACAATACTGTTTACC-3′). This set of PCR primers was used for two separate PCR amplifications using different substrates: One amplification used pHFV13 (15a), which is a full-length wild-type HFV clone. The second PCR amplification used PFV D/A, which is similar to pHFV13 except that the protease coding region contains the active-site mutation D24A, which renders the HFV protease inactive (15). The PCR products from these two amplifications were digested with NcoI; the 3′ end remained blunt from the PCR amplification. The resulting 840-bp fragment was cloned as an NcoI blunt-end insert into NcoI/EcoRV-digested Litmus-29 (New England Biolabs), generating the constructs NcoI/AflIII PFV and NcoI/AflIII PFV D/A. The RNase H domain was also obtained by PCR amplification. The 5′ primer in the amplification spanned a unique PflMI restriction endonuclease recognition site in the PFV pol domain (5′-GCGGCGGGATCCGCTTTACCCATTAGTGGATAACATGGATGAC-3′). The primer also generated a BamHI site 5′ of the PflMI site. The 3′ primer added a TAG termination codon after the tyrosine codon that encodes the last amino acid in the RNase H domain and also generated an EcoRI site 3′ of the termination codon (5′-GCGGCGGAATTCGCGCTAATATTGTTTGGGATATCCTTTTATATAATGACCCTG-3′). The underlined sequence is a unique EcoRV restriction endonuclease recognition sequence present in the coding region. The PCR fragment was digested with BamHI/EcoRI and cloned into BamHI/EcoRI-digested Litmus-29. This clone, designated 3′ PFV, contains the normal C terminus of the PFV RNase H domain. To simplify protein purification, the C terminus was further modified by the addition of six histidine residues before the termination codon. The unique EcoRV site was used as an entry point. The clone 3′ PFV was digested with EcoRV and EcoRI and then ligated to synthetic DNA fragments to construct the clone 3′ PFV (His). The synthetic DNA fragments were generated by kinasing oligonucleotide 1 (5′-ATCCCAAACAATATTCTTCCCATCATCACCACCATCATTAGTAAGGTACCCG-3′) and oligonucleotide 2 (5′-AATTCGGGTACCTTACTAATGATGGTGGTGATGATGGGAAGAATATTGTTTGGGAT-3′) and then heating and slowly cooling the oligonucleotides to allow them to anneal. The C terminus of PFV RT is normally -PKQY. In PFV, the coding region was altered so that the C terminus was -PKQYPSSGHHHHHH.
Three fragments were ligated to generate clones containing all of the protease as well as the entire polymerase and RNase H domains. The fragments are the 840-bp NcoI/AflIII fragment from either NcoI/AflIII PFV or NcoI/AflIII D/A, which contains either the active or inactive protease coding region, respectively; the AflIII/PflMI fragment from either RT2 pET16b or RTVM pET16b, which contains the wild-type or V313M active site in the polymerase domain; and the PflMI/EcoRI fragment from 3′ PFV (His). The fragments were coligated into NcoI/EcoRI-digested Litmus-29 to generate four clones: PFV (His), which has a wild-type protease and a wild-type RT; D/A (His), which has an inactive protease and a wild-type RT; RTV313M (His), which has an active protease and the V313M mutation in the RT; and D/A RTV313M (His), which has an inactive protease and the V313M mutation in the RT. For protein expression, the inserts were cloned into NcoI/EcoRI-digested pT5m, which is similar in concept to the pET vectors. The clones were transformed into the Rosetta Escherichia coli strain (Novagen, Madison, Wis.), a BL21 derivative. Only the clones that had the inactive protease expressed significant levels of protein in this system.
Protein expression and purification.
Bacteria were grown at 37°C with agitation to an optical density at 600 nm of 0.5 to 0.6. Expression of the RT protein was induced by the addition of 0.2 μM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubation of the bacteria for an additional 3 h before harvesting them. Fifty grams of pelleted bacteria was extracted in 100 ml of 50 mM NaPO4 (pH 8.0), 50 mM NaCl, 1.5 mM phenylmethylsulfonyl fluoride, and 0.75 mg of lysozyme/ml. The sample was incubated on ice for 30 min; 10.75 ml of 4 M NaCl was added to the suspension, followed by three 30-s sonications at 90% power (maximum, 350 W) and 70% pulse. A 3/4-in probe was used with 5 min between sonications. The suspension was centrifuged at 85,000 × g for 90 min, and the clear portion of the supernatant was removed. The remaining, somewhat viscous portion of the supernatant was recentrifuged, and the clear supernatant was collected. The supernatants were diluted 1:1 with 66 mM NaPO4, pH 6.8, and 300 mM NaCl. A 15-ml Q-Sepharose column and a 15-ml nickel column (Qiagen) were poured and connected in series with the Q column first. The columns were equilibrated with 50 mM NaPO4, pH 7.0, and 300 mM NaCl. The diluted supernatants were loaded onto the columns at 1 ml/min. After being loaded, the columns were washed with equilibration buffer. After 100 ml, the Q column was removed and the nickel column was washed with an additional 150 ml of buffer. The Q column was next washed with 250 ml of 50 mM NaPO4 (pH 6.0), 10 mM imidazole, 300 mM NaCl, and 10% (wt/vol) glycerol. A 150- by 150-ml, 10 to 500 mM imidazole (in pH 6.0 buffer) gradient was used to elute the protein. Eight-milliliter fractions were collected. The fractions were pooled based on SDS-PAGE analysis. The resulting pool was between 60 and 70 ml. This pool was divided in half, and each pool was dialyzed against three 500-ml volumes of 25 mM Tris acid-25 mM Tris base. A sample (50% of the pool) was removed from dialysis and centrifuged at 12,000 × g for 30 min. The pellet was resuspended with 3 ml of 20 mM HEPES (pH 7.0), 100 mM imidazole, 300 mM NaCl, 1 mM EDTA, and 10 mM DTT. One milliliter of 2 M NaCl was added to the suspension, and the sample was stirred for 30 min and centrifuged at 12,000 × g for 30 min. The supernatant was loaded onto a 1.6- by 85-cm Sephacryl 200 column (Amersham-Pharmacia) equilibrated with 20 mM HEPES (pH 7.0), 100 mM imidazole, 300 mM NaCl, 1 mM EDTA, and 1 mM DTT. The column was run at 0.2 ml/min with 10-min fractions collected. After gel analysis, the fractions were pooled. The gel filtration runs were then combined, resulting in approximately 20 ml. The sample was then dialyzed versus 25 mM Tris acid-25 mM Tris base, 10% glycerol, 1 mM EDTA, 10 mM imidazole, and 1 mM DTT. A sample was removed from dialysis, centrifuged at 12,000 × g for 30 min, and loaded onto a 5-ml Q column equilibrated with the dialysis buffer. The flow rate was 1 ml/min, and 4-ml fractions of the flowthrough were collected and analyzed by SDS-PAGE. Samples were pooled, 1/10 volume of NaCl was added to the pool, and the samples were concentrated by centrifugal ultrafiltration using a 10-kDa cutoff membrane (Filtron). Sample analysis by SDS-PAGE and Coomassie staining showed purity of >97%. Gel filtration indicated the presence of a small amount of dimmers, which increased upon storage in the cold room. Yields were generally between 12 and 20 mg. All steps were done at 4°C.
In vitro polymerase assays.
The substrate for in vitro polymerase assays was the M13 −47 sequencing primer annealed to single-stranded M13mp18 DNA (New England Biolabs). The −47 primer was end labeled using T4 polynucleotide kinase and [γ-32P]ATP and then annealed to the single-stranded M13mp18 DNA. The substrate was resuspended in 90 μl of buffer lacking dNTPs to a final concentration of 2 nM (see below) and 1.0 μg of enzyme. The mixture was incubated at room temperature for 5 min to allow the enzyme to bind the template-primer. The reaction was initiated by the addition of 10 μl of 0.2 mM dATP, 0.2 mM dCTP, 0.2 mM dGTP, and 0.2 mM dTTP. In a processivity assay, 0.5 U of poly(rC) · oligo(dG) was also included as a cold trap. The final concentrations of buffer in the reactions were 25 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 10.0 mM CHAPS {3-[(-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 100 μg of acetylated bovine serum albumin/ml, and 10.0 μM (each) dNTP in a final volume of 100 μl. After 10 min at 37°C, the reaction was terminated by extraction with an equal volume of phenol-chloroform, and the mixture was precipitated with isopropyl alcohol. The sample was resuspended in 10 μl of loading dye, and 4 μl was loaded on a 6% sequencing gel. The products of the reaction were visualized by exposure to X-ray film. The dNTP curves were performed using the buffer described above and the indicated concentration of dNTPs in the absence of poly(rC) · oligo(dG). The reaction mixtures were incubated at 37°C for 15 min.
The 3TC triphosphate (3TCTP) inhibition assays were performed using the M13 template and the −47 primer. The −47 sequencing primer was annealed to the single-stranded M13mp18 DNA by heating it to 95°C and slowly cooling it to room temperature. The template-primer was extended by adding 1.0 μg of RT in 25 mM Tris (pH 8.0); 75 mM KCl; 8.0 mM MgCl2; 100 μg of bovine serum albumin/ml; 10 mM CHAPS; 10 μM (each) dATP, dGTP, and dTTP; 2.0 μM [α-32P]dCTP; and the appropriate concentrations of 3TCTP (Moravek Biochemicals, Brea, Calif.) in a 100-μl reaction volume. The mixture was incubated at 37°C for 30 min and then halted by the addition of 3 ml of ice-cold trichloroacetic acid, and the precipitated DNA was collected by suction filtration through Whatman GF/C glass filters. The amount of incorporated radioactivity was determined by liquid scintillation counting.
RESULTS
Efficacies of RT inhibitors.
The timing of PFV reverse transcription allows the virus to infect cells that are pretreated with the RT inhibitor AZT (since infectious PFV particles already contain viral DNA). However, PFV is unable to produce infectious particles from cells that are treated with AZT. Although AZT could block PFV replication, neither 3TC nor ddI was able to do so (30). To better understand the susceptibility of PFV, a more complete panel of HIV-1 RT inhibitors was tested.
In studies designed to measure the effect of adding RT inhibitors to target cells, FAB cells, which are BHK derived and contain β-Gal driven by the FV LTR (29), were treated with inhibitor 4 h prior to the addition of PFV stock. Forty-eight hours after infection, the cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining solution, and the viral titer was measured. The RT inhibitors 3TC, 3′-azido-2′,3′-dideoxy-5-methylcytidine, 3′-azido-2′,3′-dideoxyuridine, AZT, 2′,3′-didehydro-2′,3′-dideoxythymidine, 2′,3′-dideoxycytidine, beta-2′,3′-dideoxy-5-fluoro-3′-thiacytidine, and phosphonoformate were used in these experiments at the highest concentrations that showed no cellular toxicity. All concentrations used are known to inhibit HIV-1 replication. None of the inhibitors had a dramatic effect on PFV infectivity, as predicted by the FV life cycle (Table 1). To test the effects of the inhibitors on virus-producing cells, FAB cells were infected with PFV at a multiplicity of infection of 1, and inhibitor was added 24 h after infection. Forty-eight hours after infection, virus was harvested from these cells and titered on fresh FAB cells. Only AZT demonstrated a specific decrease in infectivity of >10-fold (Table 1).
Replication of RT-V313M.
Given the fact that the highly conserved YXDD sequence in PFV RT is YVDD, the sequence of the YXDD motif in 3TC-resistant HIV-1 RT, it is not surprising that 3TC was unable to inhibit PFV replication (16, 26). To determine whether mutating the RT sequence to YMDD might render PFV sensitive to 3TC, valine (V) was changed to methionine (M) in the PFV RT YXDD motif to create the mutant PFV RT-V313M. FAB cells were transfected with PFV RT-V313M(A) and -M(B), two independently derived clones. Two days after transfection, the supernatants were collected and titers were determined on fresh FAB cells. The titer of the mutant virus was extremely low (Table 2). In an attempt to select for second-site mutations or revertants, cells transfected with clones containing the mutant RT were passaged. Supernatants were collected at 2-day intervals, and titers were determined on fresh FAB cells; the cells were passaged normally. By day 12, the cells produced virus with titers similar to those of wild-type PFV (Table 2).
TABLE 2.
Replication of PFV RT-V313M mutant in FAB cells
| Days posttransfection | Virus titera
|
|||
|---|---|---|---|---|
| WT PFV | V313M clone A | V313M clone B | Mock | |
| 2 | 1.4 × 103 | 1 × 101 | 1 × 101 | <101 |
| 4 | 1.7 × 104 | 1 × 101 | 5 × 101 | <101 |
| 6 | 1.4 × 104 | 4 × 101 | 8 × 101 | <101 |
| 8 | 8.7 × 103 | 1 × 101 | 3 × 101 | <101 |
| 10 | 3.6 × 102 | 4.9 × 102 | <101 | |
| 12 | 5.6 × 103 | 8.8 × 103 | <101 | |
| 14 | 1.5 × 105 | 1.7 × 105 | <101 | |
| 16 | 1.5 × 105 | 1.2 × 105 | <101 | |
| 18 | 2.9 × 104 | 2.0 × 104 | <101 | |
Virus titers were determined on FAB cells as described in Materials and Methods. WT, wild-type.
Virus was isolated from the cell-free supernatants of day 14, when titers of the RT-V313M viruses were at the highest level. RNA was extracted from the virus and used in RT-PCRs designed to amplify the region of PFV pol encoding the V313M mutation. Individual clones were obtained from the RT-PCR products and were sequenced. All of the clones sequenced had a 2-nucleotide change at the ATG methionine codon to regenerate the valine codon present in the wild type. The clones also had nonspecific single-nucleotide changes; however, none of these individual mutations were found in any other clones, suggesting that they were not second-site mutations involved in reversion (data not shown). Thus, it appears that PFV does not replicate if the RT has a methionine in the second residue of the YXDD motif.
Virion-associated activity of RT-V313M.
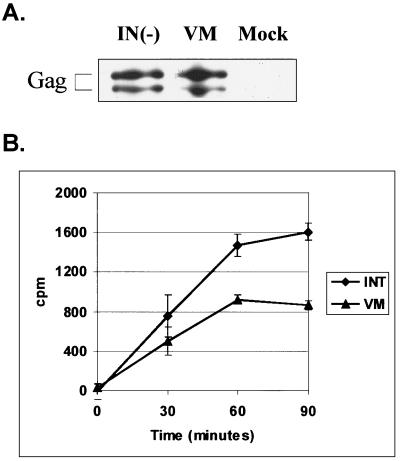
The fact that the V313M mutation is located near the polymerase active site suggests that this mutation might affect the catalytic activity of the enzyme. To test whether RT-V313M had any exogenous polymerase activity, supernatants were collected from transfected FAB cells. Concentrated virions were used in an RT assay which measures the incorporation of radiolabeled nucleotide onto a poly(A) · oligo(dT) primer-template over the course of 90 min. As a positive control for this assay, we used another mutant virus, PFV-IN(−), which contains a wild-type RT but has a mutation in the active site of IN, making it replication defective. A Western blot performed on the IN(−) and the V313M concentrated virus particles showed that enzyme from similar numbers of viral particles were used in the assays (Fig. 3A). The plasmid pNEB193, which contains no viral sequences, was used as a negative control for normalization of the assays. The results from the RT assays showed that the V313M mutant RT retains approximately 50% of wild-type polymerase activity (Fig. 3B). Given the dramatic effect this mutation has on replication, it was surprising to find a relatively modest decrease in RT-V313M activity.
FIG. 3.
Virion-associated RT activity of PFV RT-V313M on a homopolymeric template. FAB cells were transfected with viral or irrelevant (Mock) plasmid DNA, and extracellular virions were harvested 4 days posttransfection and concentrated. (A) Anti-Gag Western blot of concentrated cell supernatants. (B) RT activity was determined for each of the concentrated samples by measuring incorporation of a radiolabeled nucleotide on a poly(A) · poly(dT) template-primer over time. Each time point was normalized to the mock sample. The error bars indicate standard deviations.
RT-V313M reverse transcription products in cells.
Since the mutant RT retains substantial polymerase activity, we wanted to determine whether reverse transcription is completed in cells transfected with PFV RT-V313M. Because the RT-V313M mutant does not replicate well, it is difficult to obtain significant quantities of virus or viral products in cells that are transfected using Lipofectamine. To increase the production of mutant viruses in transient transfections, the viral genome was placed under the control of the cytomegalovirus (CMV) promoter. Calcium phosphate transfection was used to introduce the CMV-driven viral clones into 293T cells. This technique greatly increased viral expression, simplifying the analysis of replication-defective viruses.
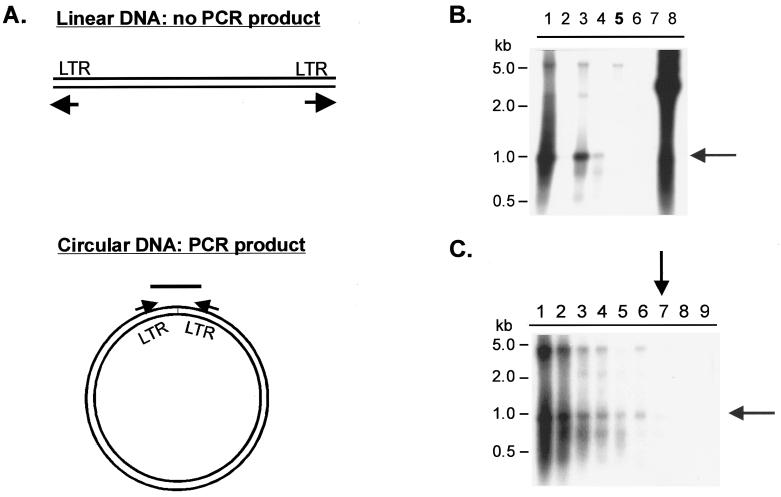
Two different approaches were used to address the question of whether reverse transcription is completed in cells transfected with PFV RT-V313M, the first of which was the detection of 2LTR circles using PCR combined with Southern blot analysis. 2LTR circles are a by-product of the completion of the full-length linear viral DNA. A fraction of the full-length linear DNAs are joined by blunt-end ligation to form 2LTR circles. Primers were designed to hybridize to the 5′ and 3′ LTR regions such that polymerization would extend the primers to the ends of unintegrated linear viral DNA. When 2LTR circles are present, a specific PCR product is formed (Fig. 3A). Two positive controls containing wild-type RTs are the IN(−) mutant, described above, and the FST4 mutant, which lacks the Env cleavage site. Neither mutant produces infectious virus, but both release particles that contain wild-type RT. The PolΔ5 mutant, which contains a large deletion in the RT coding region, was used as a negative control.
293T cells were transfected with these mutants, and genomic DNA was isolated 4 days posttransfection. PCR products from 500 ng of cellular DNA were separated on agarose gels and transferred to a nylon membrane for Southern hybridization. A radiolabeled fragment of the PFV LTR region was used as the probe. The Southern blot (Fig. 4B) showed that the PCR product specific for the 2LTR circle was present in all of the samples derived from transfections of viruses with wild-type RT (lanes 1, 3, and 4) but was absent in samples derived from virus with the RT deletion (lane 6) and the RT-V313M virus (lane 5). Since the positive-control virus FST4 had a weaker signal than the wild-type and IN(−) controls, we used it to determine the sensitivity of this assay. Genomic DNA from cells transfected with the FST4 virus was serially diluted, and the samples were subjected to the PCR and Southern blot procedure described above (Fig. 4C). 2LTR circles could be detected in a 100-fold dilution of the original sample (lane 7), indicating that the production of 2LTR circles is at least 100-fold lower with the V313M mutant RT than with the wild-type RT (Fig. 4B).
FIG. 4.
Detection of PFV 2LTR circles in transfected cells. 293T cells were transfected with CMV-driven viral plasmids or mock plasmid, and genomic DNA was isolated 4 days posttransfection and subjected to PCR. (A) Diagram of the PCR strategy used to detect 2LTR circles. The arrows indicate the positions and directions of primers. (B and C) Southern blots of PCR products using an LTR-specific radiolabeled probe. (B) Comparison of PCR products generated from genomic DNA of cells transfected with various plasmids: lane 1, wild type; lane 2, mock; lane 3, IN(−); lane 4, FST4; lane 5, V313M; lane 6, PolΔ5; lane 7, no DNA; lane 8, plasmid DNA positive control. (C) Detection of serial dilutions of PCR products generated from genomic DNA of cells transfected with the FST4 virus: lane 1, 500 ng; lane 2, 250 ng; lane 3, 100 ng; lane 4, 50 ng; lane 5, 25 ng; lane 6, 10 ng; lane 7, 5 ng; lane 8, 1 ng; lane 9, 0.5 ng. A 1-kb ladder (New England Biolabs) was run alongside the samples as indicated. The shaded arrows indicate the specific 2LTR PCR product; the solid arrows indicate the lowest dilution at which the specific PCR product is still detectable.
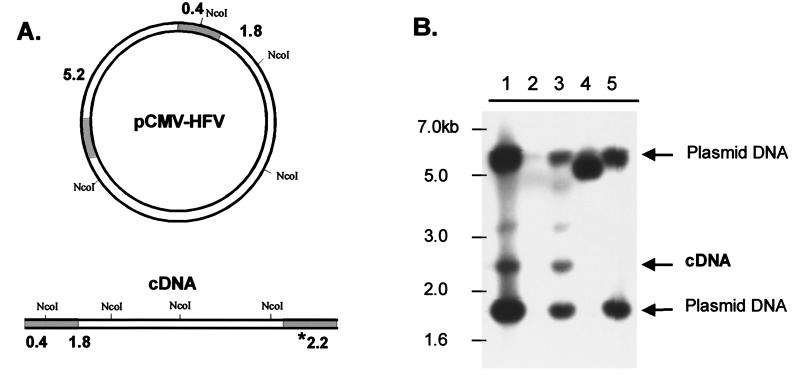
The second approach to address whether the RT-V313M mutant is able to complete reverse transcription involved Southern blotting of the DNA isolated from transfected 293T cells. The genomic-DNA samples used in the 2LTR PCR experiments were digested with the restriction enzyme NcoI. This enzyme produces DNA fragments that distinguish input plasmid DNA from linear viral cDNA (Fig. 5A). After digestion, the DNA samples were fractionated on an agarose gel, transferred to a nylon membrane, and probed with the same radiolabeled PFV LTR fragment described above. Detection of the 2.2-kb band corresponding to the 3′ LTR of the cDNA product shows that viral DNA is present in the transfected cells (Fig. 5A). The blot shows that cells transfected with either of the viruses that contain wild-type RT [wild type and IN(−)] generated detectable amounts of cDNA (Fig. 5B, lanes 1 and 3). However, cDNA could not be detected in cells transfected with the PolΔ5 or V313M viruses (lanes 4 and 5). These results agree with the results of the PCR assay and show that although the V313M RT has about 50% activity in an exogenous RT assay, it is unable to complete reverse transcription in an infected cell.
FIG. 5.
Detection of full-length PFV DNA in transfected cells. 293T cells were transfected with CMV-driven viral plasmids or empty (mock) plasmid, and genomic DNA was isolated 4 days posttransfection and digested with NcoI. (A) Diagram of NcoI sites in the viral plasmid used for transfection and predicted sizes of resulting products that hybridize to the probe. The shaded regions of the DNA indicate the locations of the LTRs. The asterisk indicates the size of the fragment corresponding to the unique cDNA fragment. (B) Southern blot of digested DNAs using an LTR-specific radiolabeled probe: lane 1, wild-type HFV; lane 2, mock; lane 3, IN(−); lane 4, PolΔ5; lane 5, RT-V313M. A 1-kb ladder (New England Biolabs) was run alongside the samples for size orientation.
RT-V313M activity in vitro.
In order to perform more detailed characterization of V313M RT activity, a His-tagged version of the protein was produced in E. coli and purified (see Materials and Methods). In the context of the virus, RT is expressed as part of a larger Pol polyprotein, which also contains IN and PR. Unlike those of most other retroviruses, the FV Pol undergoes a single cleavage event, which releases IN, leaving a PR-RT fusion protein. However, in some bacterial overexpression systems a second cleavage event between PR and RT has been reported (19). To avoid this cleavage and the toxicity of PR in bacteria, the PR-RT fusion protein expressed in bacteria had a point mutation in the PR active site that inactivated the protease (15). Expression plasmids with the mutation in the protease active site are named D/A.
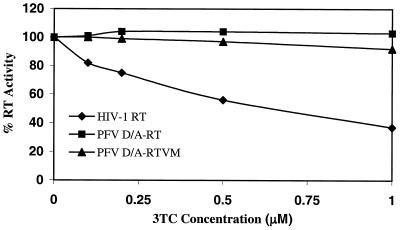
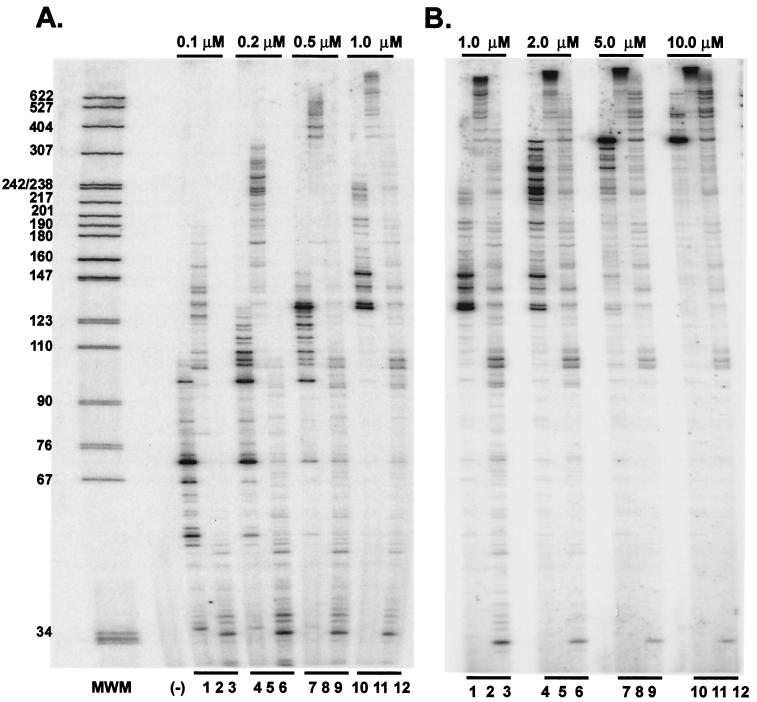
Using the purified recombinant PFV enzymes along with purified recombinant HIV-1 RT, we first wanted to determine whether the FV RT-V313M was sensitive to 3TC. The M13 template and primer were annealed and then incubated with either FV D/A-RT, FV D/A-RTVM, or HIV-1 RT and increasing concentrations of 3TCTP. Samples were trichloroacetic acid precipitated and bound to glass filters, and incorporation of radiolabeled dCTP was measured. In the absence of 3TCTP, we found that both the V313M RT and HIV-1 RT displayed about 35% of the polymerase activity of FV D/A RT. This is similar to the decrease in virion-associated RT activity observed for the V313M mutant virus (Fig. 3). These levels of RT activity in the absence of drug were set at 100% activity for each recombinant RT. HIV-1 RT demonstrated sensitivity to 3TC, retaining only 37% of its activity at 1.0 μM 3TCTP (Fig. 6). FV D/A-RT demonstrated complete resistance to 3TC, as seen in our tissue culture experiments. The mutant FV D/A-RTVM was relatively resistant to 3TC, retaining 92% of its activity at 1.0 μM 3TCTP, despite the presence of a valine in the second position of the YXDD motif (Fig. 6).
FIG. 6.
3TCTP inhibition curves for HIV-1, FV D/A-RT, and FV D/A-RTVM recombinant RTs. Polymerization by purified recombinant RTs using a heteropolymeric template was measured by incorporation of radiolabeled dCTP in the absence or presence of 3TCTP at concentrations of 0.1, 0.2, 0.5, and 1.0 μM. HIV-1 RT, FV D/A-RT, and FV D/A-RTVM were tested.
To compare the RT activities of the mutant and wild-type FV enzymes, we performed standard kinetic analyses using homopolymeric templates. However, the results obtained from these experiments suggest that a homopolymeric template may not be the appropriate substrate to study these enzymes (data not shown). As an alternative method to compare the mutant and wild-type FV RTs, we examined the effects of dNTP concentration on polymerization using a heteropolymeric template. The M13 single-stranded DNA template and radiolabeled primer were annealed and incubated with either HIV-1 RT, FV D/A-RT, or FV D/A-RTVM and a reaction mixture containing increasing concentrations of each dNTP. The reaction was carried out for 15 min, and then the reaction mixture was fractionated on a 6% sequencing gel. At concentrations above 1.0 μM, most of the DNA synthesized by FV D/A-RT is too large to be measured accurately on a sequencing gel (Fig. 7B, lanes 2, 5, 8, and 11). The mutant FV D/A-RTVM is severely impaired in its polymerization at the lowest dNTP concentrations (Fig. 7A, lane 3). However, the mutant does synthesize DNAs of 100 nt or longer at dNTP concentrations of 0.2 μM or greater and does not approach saturation until the highest dNTP concentrations are reached (Fig. 7A, lanes 6, 9, and 12, and 7B, lanes 3, 6, 9, and 12). These results suggest that the Km of the V313M mutant might be in the 5 to 10 μM range. HIV-1 RT is also approaching saturation at the highest dNTP concentrations, which suggests a Km in the 5 to 10 μM range, similar to that of the V313M RT (Fig. 7B, lanes 1, 4, 7, and 10). To measure the sizes of the DNA products synthesized by FV D/A-RT more accurately, another polymerization assay was performed using an M13 template, and the products were fractionated on a 1% alkaline agarose gel. DNAs up to 7 kb were synthesized in 20 min, but the rate of DNA synthesis was still increasing at dNTP concentrations of >20 μM (data not shown). This suggests that the Km of FV D/A-RT for dNTPs is higher than that of HIV-1 RT and that if anything the Km of the wild-type FV enzyme is higher than that of the mutant.
FIG. 7.
Primer extension at various dNTP concentrations by HIV-1, FV D/A-RT, and FV D/A-RTVM recombinant RTs. Extension by purified recombinant HIV-1 RT, FV D/A-RT, and FV D/A-RTVM using a heteropolymeric template in the presence of increasing concentrations of each dNTP is shown. Lanes 1, 4, 7, and 10, HIV-1 RT; lanes 2, 5, 8, and 11, FV D/A-RT; lanes 3, 6, 9, and 12, FV D/A-RTVM. (A) Lower concentrations of each dNTP. Lanes 1 to 3, 0.1 μM; lanes 4 to 6, 0.2 μM; lanes 7 to 9, 0.5 μM; lanes 10 to 12, 1.0 μM. (B) Higher concentrations of each dNTP. Lanes 1 to 3, 1.0 μM; lanes 4 to 6, 2.0 μM; lanes 7 to 9, 5.0 μM; lanes 10 to 12, 10.0 μM.
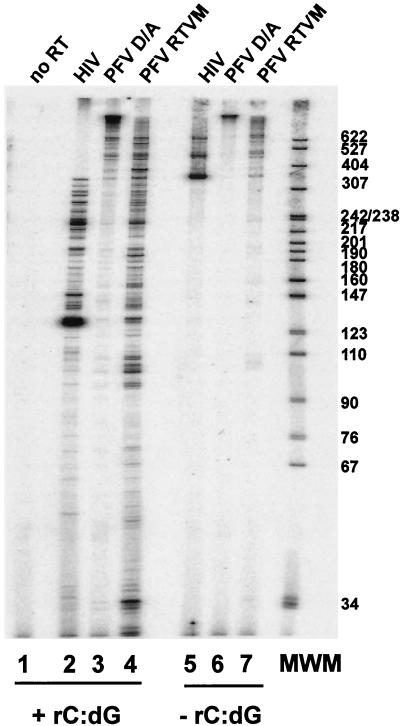
Because FV RT-V313 M does not support the synthesis of full-length cDNA (Fig. 3 and 4) and the recombinant FV D/A-RTVM protein showed a significant decrease in primer extension, particularly at low dNTP concentrations (Fig. 7A), we measured the processivity of the mutant FV enzyme and compared it to both wild-type FV D/A-RT and HIV-1 RT. The −47 primer was end labeled and then annealed to single-stranded M13mp18 DNA. This radiolabeled primer-template was incubated with the purified HIV-1 RT, FV D/A-RT, and FV D/A-RTVM proteins to allow the enzyme to bind the primer-template. The reaction was initiated by that addition of all four dNTPs. In some reactions, an unlabeled poly(rC) · oligo(dG) trap was also added. The trap was added in excess and bound any RT proteins that dissociated from their original primer-template. The RT extension products produced in the presence of a trap indicate how far the enzyme was able to extend before it dissociated from the template. The reactions were carried out for 10 min, and the samples were fractionated on a 6% sequencing gel. In the absence of the trap, the HIV-1 RT generated products 350 to 600 nt in length, while the FV D/A RT generated products well above 600 nt in length (Fig. 8, lanes 5 and 6). The mutant FV D/A-RTVM shows products of an intermediate length (Fig. 8, lane 7). In the presence of the trap, the HIV-1 RT showed predominantly shorter products; however, the FV D/A-RT still generated mostly products longer than 600 bp (Fig. 8, lanes 2 and 3). In contrast to the wild-type FV RT, the FV D/A-RTVM enzyme did not generate products longer than 600 nt but instead showed a range of smaller products (Fig. 8, lane 4). These results show that wild-type FV RT is a highly processive enzyme, producing large quantities of long products after only 10 min. The inability of the FV D/A-RTVM RT to generate longer products indicates that this enzyme is significantly less processive than the wild-type enzyme, although it is similar to HIV-1 RT. Substitution of methionine for valine in the the YXDD motif affected processivity; this could explain the inability of the mutant RT to complete reverse transcription.
FIG. 8.
Primer extension and processivity of HIV-1, FV D/A-RT, and FV D/A-RTVM recombinant RTs. Extension by purified recombinant HIV-1, FV D/A-RT, and FV D/A-RTVM RTs using a heteropolymeric template in the presence (lanes 1 to 4) or absence (lanes 5 to 7) of the poly(rC) · poly(dG) trap. Lane 1, no RT; lanes 2 and 5, HIV-1 RT; lanes 3 and 6, FV D/A-RT; lanes 4 and 7, FV D/A-RTVM. MWM, molecular weight markers.
DISCUSSION
The highly conserved YXDD motif contains two of the three aspartic acid residues that make up the RT active site (12, 14, 18, 23). The three-dimensional crystal structure of HIV-1 RT shows that the YXDD motif is located on the β9-β10 hairpin and is part of the dNTP binding site (10, 11). More specifically, the second variable residue (X) appears to interact with the ribose of the terminal primer nucleotide (6, 21, 23). It has been postulated that this interaction between the second (X) residue and the primer may be important for affinity to the template-primer, and different residues in the second position may alter the flexibility of the dNTP binding pocket, leading to changes in the behavior of the enzyme (3, 5, 6, 8, 21, 23). Therefore, it is likely that the size and/or the hydrophobicity of the amino acid side chain of the second residue in the YXDD motif influences the catalytic activity of the polymerase RT. Changes in the YXDD motif may also alter the active-site structure and/or lead to the repositioning of the template-primer in such a way that the RT-DNA complex incorporates normal dNTPs less efficiently. Such an altered structure could be less stable, and thus PFV RT-V313M could dissociate more easily from its nucleic acid substrate than the wild-type enzyme, reducing processivity.
In contrast to PFV, HIV-1 with the 3TC resistance mutation YVDD (M184V) replicates almost as well as the wild type (YMDD) (22). In vitro, HIV-1 M184V RT displays 75% of the activity of wild-type RT and has reduced processivity (3). Further, MLV with the mutant YMDD motif (V223M) retains 20% of wild-type RT activity, yet it is still able to replicate with only a 10-fold decrease in titer compared to the wild type (7). In vitro, recombinant wild-type and YMDD MLV RTs show similar polymerase activities and have similar extension abilities (2). Thus, although mutations in the YXDD motif affect the polymerase activities of both HIV-1 and MLV, these viruses are still able to replicate with either M or V present at the variable position of the YXDD motif; the architecture of the active site for both HIV-1 and MLV must be able to tolerate either amino acid. Our data show that the same is not true for PFV RT. A PFV RT with YMDD (V313M) could not support productive replication of the virus. The mutant virus rapidly reverted to YVDD despite the fact that the mutant RT retained approximately 50% of wild-type activity.
Conventional retroviruses generate both Gag and Gag-Pol fusion proteins at a ratio of about 20:1. The result is a virus particle that contains approximately 50 to 75 Pol proteins per particle (27). FV Pol is expressed from its own spliced message, and the mechanism for packaging the Pol protein is unclear. In FV-infected cells, the Pol protein is sufficiently abundant that it can be easily detected by Western blotting. However, we have been unable to detect any Pol in virus particles by Western blotting or by radioimmunoprecipitation (1), although in more recent work using a monoclonal antibody to Pol, it can be detected (M. Heinkelein, M. Rammling, K. Peters, and A. Rethwilm, submitted for publication. There is some evidence for a cis-acting RNA sequence at the 5′ end of the FV genome (CAS I) that is not involved in RNA packaging but is required for PR activity (9) and for Pol packaging (M. Heinkelein, M. Rammling, K. Peters, and A. Rethwilm, submitted for publication). If Pol must bind to a specific region of the RNA for encapsidation, this would limit the number of enzyme molecules that can be incorporated.
A low number of Pol molecules in a viral particle places a special burden on PFV RT relative to other retroviral RTs. One or two FV RTs would have to accomplish the same tasks carried out by the 50 to 75 RTs present in other retroviral particles (27). In order to overcome this disadvantage, FV RT would necessarily be a particularly efficient polymerase, and our results suggest that this is the case. It is likely that a moderate decrease in RT activity in viruses such as HIV-1 and MLV does not have a dramatic effect on replication because there are a relatively large number of RTs in the particle that can collaborate to carry out reverse transcription (12, 24). In the case of HFV, a minor decrease in RT activity could be much more detrimental if only one or two RTs were responsible for reverse transcription.
As might be expected if PFV requires a highly active RT, our results also show that FV RT is an extremely processive enzyme. In the processivity assay, wild-type PFV RT was able to generate products that were significantly longer than 600 nt in 10 min. HIV-1 RT is much less processive, generating products ranging from 100 to 400 nt in length. This high level of processivity for PFV should effectively compensate for the small number of Pol proteins in a virion. Our results support this idea, showing that the mutant V313M, which cannot replicate, has a decrease in processivity compared to wild-type PFV RT. Yet in vitro, the mutant enzyme has a processivity that is comparable to, or slightly better than, that of HIV-1 RT. Thus, FV is not able to replicate with a mutant RT that has a processivity similar to that of HIV-1 RT.
Acknowledgments
This investigation was supported by grant CA-18282 from the NCI to M.L.L. and grants from the NCI and NIGMS to S.H.H. C.S.R. was supported by NCI training grant CA-09229.
We thank Pat Clark and Peter Frank for purification of the proteins, Rolf Flugel (Deutsches Krebsforschungszentrum, Heidelberg, Germany) for the PFV RT protein expression clone RT2-pET16b, Ray Schinazi (Emory University) for the panel of RT inhibitors, and Michael Emerman for critical review of the manuscript.
REFERENCES
- 1.Baldwin, D. N., and M. L. Linial. 1999. Proteolytic activity, the carboxy terminus of Gag, and the primer binding site are not required for Pol incorporation into foamy virus particles. J. Virol. 73:6387-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer, P. L., H. Q. Gao, P. K. Clark, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. YADD mutants of human immunodeficiency virus type 1 and Moloney murine leukemia virus reverse transcriptase are resistant to lamivudine triphosphate (3TCTP) in vitro. J. Virol 75:6321-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer, P. L., and S. H. Hughes. 1995. Analysis of mutations at position 184 in reverse transcriptase of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 39:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding, J., S. H. Hughes, and E. Arnold. 1997. Protein-nucleic acid interactions and DNA conformation in a complex of human immunodeficiency virus type 1 reverse transcriptase with a double-stranded DNA template-primer. Biopolymers 44:125-138. [DOI] [PubMed] [Google Scholar]
- 6.Gao, H. Q., P. L. Boyer, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2000. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J. Mol. Biol. 300:403-418. [DOI] [PubMed] [Google Scholar]
- 7.Halvas, E. K., E. S. Svarovskaia, and V. K. Pathak. 2000. Development of an in vivo assay to identify structural determinants in murine leukemia virus reverse transcriptase important for fidelity. J. Virol. 74:312-319. [PMC free article] [PubMed] [Google Scholar]
- 8.Harris, D., N. Kaushik, P. K. Pandey, P. N. Yadav, and V. N. Pandey. 1998. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J. Biol. Chem. 273:3324-3334. [DOI] [PubMed] [Google Scholar]
- 9.Heinkelein, M., J. Thurow, M. Dressler, H. Imrich, D. Neumann-Haefelin, M. O. McClure, and A. Rethwilm. 2000. Complex effects of deletions in the 5′ untranslated region of primate foamy virus on viral gene expression and RNA packaging. J. Virol. 74:3141-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 11.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, and P. Clark. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kogel, D., M. Aboud, and R. M. Flügel. 1995. Molecular biological characterization of the human foamy virus reverse transcriptase and ribonuclease H domains. Virology 213:97-108. [DOI] [PubMed] [Google Scholar]
- 14.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 15.Konvalinka, J., M. Löchelt, H. Zentgraf, R. M. Flügel, and H.-G. Krausslich. 1995. Active spumavirus proteinase is essential for virus infectivity but not for formation of the Pol polyprotein. J. Virol. 69:7264-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Löchelt, M., H. Zentgraf, and R. M. Flügel. 1991. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel1 gene. Virology 184:43-54. [DOI] [PubMed] [Google Scholar]
- 16.Maurer, B., H. Bannert, G. Darai, and R. M. Flügel. 1988. Analysis of the primary structure of the long terminal repeat and the gag and pol genes of the human spumaretrovirus. J. Virol. 62:1590-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moebes, A., J. Enssle, P. D. Bieniasz, M. Heinkelein, D. Lindemann, D. Bock, M. O. McClure, and A. Rethwilm. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nanni, R. G., J. Ding, A. Jacobo-Molina, S. H. Hughes, and E. Arnold. 1993. Review of HIV-1 reverse transcriptase three-dimensional structure: implications for drug design. Perspect. Drug Discov. Des. 1:129-150. [Google Scholar]
- 19.Pfrepper, K. I., H. R. Rackwitz, M. Schnolzer, H. Heid, M. Löchelt, and R. M. Flügel. 1998. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J. Virol. 72:7648-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rethwilm, A. 1995. Regulation of foamy virus gene expression. Curr. Top. Microbiol. Immunol. 193:1-24. [DOI] [PubMed] [Google Scholar]
- 21.Sarafianos, S. G., K. Dasgupta, A. D. Clark, J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc. Natl. Acad. Sci. USA 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schinazi, R. F., R. M. Lloyd, Jr., M. H. Nguyen, D. L. Cannon, A. McMillan, N. Ilksoy, C. K. Chu, D. C. Liotta, H. Z. Bazmi, and J. W. Mellors. 1993. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob. Agents Chemother. 37:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tantillo, C., J. Ding, A. Jacobo-Molina, R. G. Nanni, P. L. Boyer, S. H. Hughes, R. Pauwels, K. Andries, P. A. Janssen, and E. Arnold. 1994. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J. Mol. Biol. 243:369-387. [DOI] [PubMed] [Google Scholar]
- 24.Telesnitsky, A., and S. P. Goff. 1993. Two defective forms of reverse transcriptase can complement to restore retroviral infectivity. EMBO J. 12:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication—a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 29.Yu, S. F., and M. L. Linial. 1993. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J. Virol. 67:6618-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, S. F., M. D. Sullivan, and M. L. Linial. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]