Abstract
The papain-like protease p29, derived from the N-terminal portion of the hypovirus CHV1-EP713-encoded open reading frame (ORF) A polyprotein, p69, was previously shown to contribute to reduced pigmentation and sporulation by the infected host, the chestnut blight fungus Cryphonectria parasitica, while being dispensable for virus replication and attenuation of fungal virulence (hypovirulence). We now report that deletion of the C-terminal portion of p69, which encodes the highly basic protein p40, resulted in replication-competent mutant viruses that were, however, significantly reduced in RNA accumulation. While the Δp40 mutants retained the ability to confer hypovirulence, Δp40-infected fungal strains produced more asexual spores than strains infected with either wild-type CHV1-EP713 or a Δp29 mutant virus. As observed for Δp29-infected colonies, pigment production was significantly increased in Δp40-infected fungal strains relative to that in CHV1-EP713-infected strains. Virus-mediated suppression of laccase production was not affected by p40 deletion. A gain-of-function analysis was employed to map the p40 symptom determinant to the N-terminal domain, encompassing p69 amino acid residues Thr(288) to Arg(312). Evidence that the gain of function was due to the encoded protein rather than the corresponding RNA sequence element was provided by introducing frameshift mutations on either side of the activity determinant domain. Moreover, restoration of symptoms correlated with increased accumulation of viral RNA. These results suggest that p40 indirectly contributes to virus-mediated suppression of fungal pigmentation and conidiation by providing an accessory function in hypovirus RNA amplification. A possible role for p40 in facilitating ORF B expression and the relationship between hypovirus RNA accumulation and symptom expression are discussed.
Members of the virus family Hypoviridae are distinguished by the ability to persistently attenuate virulence (hypovirulence) and stably alter complex biological processes upon infection of their fungal host, the chestnut blight fungus, Cryphonectria parasitica. Infection-related phenotypic changes can include reduced pigment production, suppressed asexual sporulation, altered colony morphology, loss of female fertility, and modified expression of specific host genes (1, 12, 22-25). Efforts to identify hypovirus symptom determinants have benefited from the complete nucleotide sequence determinations of four members of the family (7, 18, 28, 30) and construction of full-length infectious cDNA clones for two family members: the prototypic severe hypovirus isolate CHV1-EP713 (9) and the mild isolate CHV1-Euro7 (7).
Using the robust transformation protocol available for C. parasitica (10), Choi and Nuss (8) showed that the 5′-proximal CHV1-EP713 coding domain, open reading frame (ORF) A, when expressed in C. parasitica transformants in the absence of virus infection, caused a subset of phenotypic changes exhibited by CHV1-EP713-infected strains, e.g., a white phenotype (reduction in orange pigmentation), reduced asexual sporulation, and a slight reduction in the production of fungal laccase activity. ORF A encodes a 69-kDa polyprotein (p69) that is autocatalytically processed into p29 and p40 by the action of a papain-like cysteine protease domain located in the p29 coding region. Craven et al. (11) subsequently mapped the ORF A suppressive activity to p29, located within the N-terminal portion of p69. By deleting all but the first 24 codons of p29 in the context of the CHV1-EP713 infectious cDNA clone (mutant virus Δp29), these authors were also able to show that 88% of the p29 coding domain is dispensable for viral replication and that p29 contributes to the suppression in orange pigment production and in conidiation but not to virulence attenuation.
Suzuki et al. (31) subsequently devised a gain-of-function analysis involving progressive repair of the Δp29 mutant virus to map the p29 symptom determinant domain to a region extending from Phe(25) to Gln(73). These authors noted a moderate level of sequence similarity between this region of p29 and the N-terminal portion of the HC-Pro papain-like protease encoded by plant-infecting potyviruses, including four conserved cysteine residues, Cys(38), Cys(48), Cys(70), and Cys(72). While substitution of a glycine for Cys(38) and Cys(48) had no apparent phenotypic effect, replacement of Cys(72) resulted in reduction of symptoms approaching that observed for the Δp29 mutant. In contrast, replacement of Cys(70) resulted in a very severe phenotype that included significantly reduced mycelial growth and profoundly altered colony morphology. Given the proposed evolutionary relationship between hypoviruses and potyviruses (20), it will be instructive to compare p29- and HC-Pro-mediated modulation of specific cellular regulatory pathways in the respective fungal and plant hosts.
Additional insights into the functional role of viral coding regions were indirectly provided during recent efforts to develop hypoviruses as gene expression vectors (32). The nucleotide sequence corresponding to the first 24 codons of p29 were found to be required for viral replication, while the remaining 598 codons of ORF A, including all of the p40 coding region, was found to be dispensable. Substantial alterations were also tolerated in the pentanucleotide UAAUG that contains the ORF A termination codon and the overlapping putative ORF B initiation codon. For example, replication competence was maintained following either a frameshift mutation that caused a two-codon extension of ORF A or a modification that produced a single-ORF genomic organization.
The deduced p40 amino acid sequence is very basic (pKa = 11.96), indicative of a possible role in viral RNA binding or replication. Moreover, this protein is conserved between two major hypovirus species, CHV1 and CHV2 (30). Thus, the observation that p40 is not required for CHV1-EP713 replication (32) was surprising. We now report the consequences of p40 deletion for CHV1-EP713-mediated alteration of fungal phenotype, virulence, and viral RNA accumulation and also report the mapping of the functional domain of this highly basic protein to amino acid residues Thr(288) to Arg(312).
MATERIALS AND METHODS
Construction of mutated viral cDNAs.
Modifications of the ORF A coding domain within the context of the CHV1-EP713 infectious cDNA clone, plasmid pLDST (9), were facilitated by use of the pTNR4-based mutation-modification cassette system as described by Suzuki et al. (32) for construction of CHV1-EP713 gene expression vector candidates. Plasmid pTNR4 contains the 5′-terminal 3.7-kb XbaI-NheI fragment derived from pLDST in the background of modified plasmid pTZ19R (U.S. Biochemicals, Cleveland, Ohio). Sequence information for all the primers and probes used in this study is shown in Table 1. The coordinates for viral nucleotide and amino acid sequences were based on the CHV1-EP713 map presented by Shapira et al. (28). Since viral proteins p29 and p40 are derived from p69 (ORF A polyprotein product) by autocatalytic proteolysis, the p69 amino acid numbering system is used throughout this article. Thus, the p40 polypeptide extends from p69 Gly(249) to Phe(622).
TABLE 1.
Oligonucleotide primers and probes used in this study
| Name | Sequence | Map positiona | Comments |
|---|---|---|---|
| BR16 | GAAGCTAATCCGATGGTT | 364-381 | |
| NS7 | CCGAACGAGGTCCGAACA | 476-493 | |
| NS9 | CTTTCGCCTCTTCCATGGG | 3261-3279 | Natural NcoI site at position 3268 in bold |
| NS16 | AGCTTTGGCCCGCTCGGCG | 1851-1869 | |
| NS17 | CACGGATCCCGTGGCCGCCACGAAGGTCCAC | 2327-2349 | Added BamHI site in bold |
| NS18 | CATGGATCCAATGTATAAGGAAGCCGAACGACCTATTG | 2363-2391 | Added BamHI site in bold |
| NS19 | CACGGATCCTGCGTCGTCACGGGCCCGCCCGTC | 1573-1598 | Added BamHI site in bold |
| NS20 | CACGGATCCCGGCCGCGCGTTTGGTTACGTGC | 2128-2152 | Added BamHI site in bold |
| NS22 | CGCCACACTTCAATAGGTCG | 2382-2401 | |
| NS35 | CACAGATCTCGGCCGCCAATCCGGGCAAGGG | 1223-1245 | Added BglII site in bold |
| NS40 | CACGGATCCTCGTCGCCTTTGGGCGAGATACG | 1470-1492 | Added BamHI site in bold |
| NS41 | CACGGATCCAACCATTGTCTCCGTTGGCAAGG | 1354-1378 | Added BamHI site in bold |
| NS42 | CACGGATCCACAACGCCTTGCTGGTCTCTCC | 1645-1668 | Added BamHI site in bold |
| NS44 | CACGGATCCGCTCGCTGCCGAGTTTGCGCC | 1252-1274 | Added BamHI site in bold |
| NS45 | ACAGGATCCACAGGTTACCCGCCGTAAAGG | 1305-1328 | Added BamHI site in bold |
| NS46 | CACGGATCCCACCAGCATCCAGGATGCTCTTG | 1392-1414 | Added BamHI site in bold |
| NS47 | CACGGATCCGAACGTCAATTTCGGCCGTCGTG | 1431-1453 | Added BamHI site in bold |
| NS57M | TGACTGGTCGTGACCCCAACCATTGTCTCCGTTGG | 1340-1373 | Inserted C residue in bold |
| NS58M | CTTGTGGCTACGGAGT(t)GAGGAACGTCAATTTCGGC | 1411-1446 | Deleted T residue indicated by (t) |
| NS52PE | GCGCCGAAGGGTTTTGTA | 12471-12488 | Used in real-time PCR |
| NS53PE | GACCGAAGCAAGCACAGAGACT | 12341-12362 | Used in real-time PCR |
| NS52TP | ACAACGAAGCGCGTAGCCCATACTTTC | 12433-12459 | TaqMan probe used in real-time PCR |
| PDF1373F | ATAACAGGTCTGTGATGCCCTTAGA | 1373-1397 | Used in real-time PCR |
| PDF1439R | CTCGCTGGCTCTGTCAGTGTAG | 1418-1439 | Used in real-time PCR |
| PDF1400T | TTCTGGGCCGCACGCGC | 1400-1416 | TaqMan probe used in real-time PCR |
Two independent ORF A deletion mutants, Δp69a and Δp69b, were constructed by replacing the BamHI-NcoI fragment in pTNR4 (spanning CHV1-EP713 map positions 562 to 3263 [28]) (Fig. 1) with corresponding modified BamHI-NcoI PCR amplification fragments generated from the pLDST plasmid template with the thermostable Pfu DNA polymerase (Stratagene Cloning Systems, La Jolla, Calif.). The PCR fragment used to construct Δp69a was generated with a primer set consisting of NS17 and NS9 (Table 1), while the modified fragment used for construction of Δp69b was generated with the primer set NS18 and NS9. Substitution of the modified fragments for the BamHI-NcoI fragment in pTNR4 regenerated a BamHI site at the deletion boundary in the resulting intermediate plasmids pTNR4Δp69a and pTNR4Δp69b (Fig. 1). Mutant CHV1-EP713 viral cDNAs were reconstituted by insertion of the 3′-terminal 8.9-kb NheI-SpeI CHV1-EP713 cDNA fragment, also derived from pLDST.
FIG. 1.
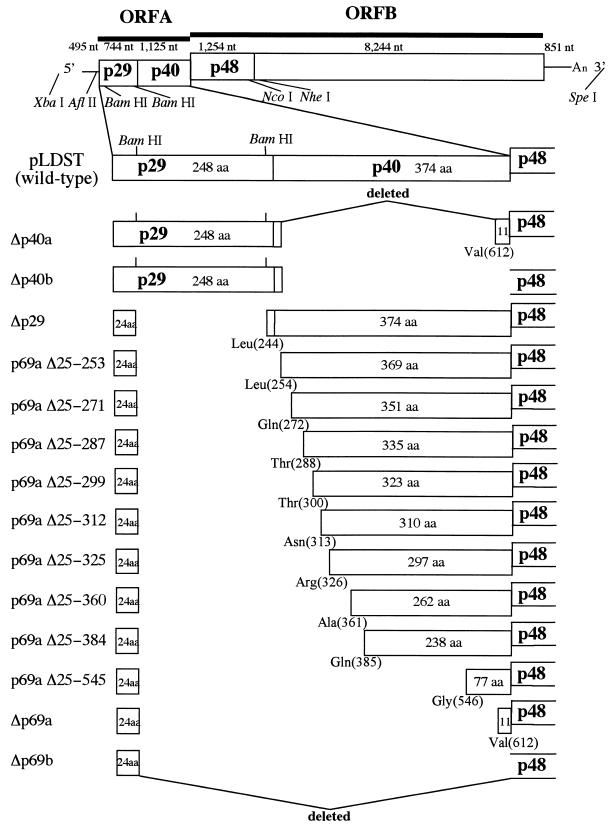
Diagram showing the organization of ORF A mutant viruses. The genetic organization of hypovirus CHV1-EP713 is shown at the top. The plus-sense strand of CHV1-EP713 hypovirus dsRNA is 12,712 nucleotides in length, excluding a poly(A) tract, and contains a 495-nucleotide (nt) 5′ noncoding leader sequence, two contiguous ORFs (1,869-nucleotide ORF A and 9,498-nucleotide ORF B) and an 851-nucleotide 3′ noncoding region. ORF A encodes a 69-kDa polyprotein (p69) that is autocatalytically processed into p29 and p40 by the action of a papain-like cysteine protease domain located within the p29 coding region. Key restriction enzyme recognition sites within the full-length CHV1-EP713 infectious cDNA clone, pLDST (9), used for mutant viral cDNA constructions included AflII (CHV1-EP713 map position 450 [28]), BamHI (CHV1-EP713 map positions 562 and 1219), PstI (CHV1-EP713 map position 1846), NcoI (CHV1-EP713 map position 3263), NheI (CHV1-EP713 map position 3705), SpeI (vector sequence), and XbaI (vector sequence). ORF A deletion mutant viruses are shown below the wild-type virus cDNA. Δp40a and Δp40b lack 96.5 and 99.5% of the p40 coding sequence within the context of di- and monocistronic genome organizations, respectively. For Δp40a, two BglII-derived amino acid residues (aspartic acid and proline) were added during the cloning procedure downstream of p40 Arg(2) and fused with p40 Val(612), while the added proline is fused with Met(1) of p48 encoded by ORF B in Δp40b. Δp29, described earlier (11), has a deletion of 87.5% of the p29 gene while retaining the N-terminal 24 codons necessary for virus viability (32). Extreme mutants Δp69a and Δp69b contained 94.4 and 96.1% deletions of the ORF A sequence, again, within the context of the di- and monocistronic genome organizations described for Δp40a and Δp40b, respectively. p29 residue Pro(24) is fused with p40 Val(612) for Δp69a or with p48 Met(1) for Δp69b. A series of nine mutants containing progressive extensions from p40 Val(612) towards the p40 N terminus Leu(254) are presented between Δp69a and Δp29. These extension mutant viruses were used in transfection (gain-of-function) assays to map the region of the p40 coding domain responsible for suppression of pustule formation.
For construction of p40 deletion mutants, a DNA fragment (bases 363 to 1245) containing a portion of the 5′ noncoding sequence plus the entire p29 coding sequence and the first two codons of p40 was amplified on pLDST by using a primer set consisting of BR16 and NS35. The fragment was cloned into pCRScript (Stratagene Cloning Systems), liberated by digestion with AflII and BglII, and substituted for the AflII-BamHI fragment in the intermediate plasmids pTNR4Δp69a and pTNR4Δp69b (Fig. 1). As in the case of Δp69a and Δp69b, the NheI-SpeI fragment of pLDST was inserted into these modified plasmids to generate mutant virus cDNAs Δp40a and Δp40b (Fig. 1).
A double frameshift mutation was introduced to change the amino acid sequence of the p40 activity domain [Thr(288) to Arg(312)] without significantly altering the RNA nucleotide sequence. This was accomplished in a stepwise manner by first introducing a C residue between map positions 1355 and 1356 with the mutagenic primer NS57M and then deleting a T residue at position 1427 with the mutagenic primer NS58M. Oligonucleotides NS7 and NS16 were included as common terminal primers in the sequential PCRs, with pLDST serving as the template. The amplified fragment generated in each step was cloned into pCRScript and subjected to sequence analysis to confirm the introduction of the desired mutation. The BamHI-PstI fragment in plasmid pTNR4 (pLDST map positions 562 to 1845) was replaced with the fragment (map positions 1219 to 1845) liberated from the doubly mutagenized amplification product. The 8.9-kb NheI-SpeI fragment from pLDST was subsequently ligated with the altered pTNR4 to give plasmid pDFS2, a p40 double frameshift mutant of Δp29.
A series of nine variants of Δp69a were constructed to contain progressive extensions of the p40 coding region from Val(612) toward the N terminus, Gly(249). These mutants were given designations beginning with p69aΔ25 followed by the p69 amino acid residues that remain deleted. Thus, repair of the Δp69a deletion mutant by extension from Val(612) to Gly(546) gave mutant p69aΔ25-545 (which still lacked p69 amino acid residues 25 to 545). DNA fragments used to construct mutants p69aΔ25-545 and p69aΔ25-384 were generated with primer pair NS20 and NS9 and primer pair NS42 and NS22, respectively. DNA fragments used to construct the remaining mutants were generated with the reverse primer NS16 and the following forward primers: NS19 for p69aΔ25-360, NS40 for p69aΔ25-325, NS47 for p69aΔ25-312, NS46 for p69aΔ25-299, NS41 for p69aΔ25-287, NS45 for p69aΔ25-271, and NS44 for p69aΔ25-253. The resulting fragments were first cloned into pCRScript and then digested with BamHI and PstI or (for the p69aΔ25-545 mutant) with BamHI and NcoI, and the liberated fragments were used to replace the corresponding fragments (BamHI-PstI [spanning positions 562 to 1846] or BamHI-NcoI [spanning positions 562 to 3263]) in pTRN4. Mutant viral cDNAs were generated as described above.
In vitro transcription and transfection.
Synthetic transcripts were synthesized in vitro from SpeI-linearized wild-type and mutant pLDST-based plasmids with T7 RNA polymerase and Promega (Madison, Wis.) reagents according to the manufacturer's instructions. The resulting transcripts were extracted with phenol, precipitated with ethanol, and electroporated into 5 × 105 spheroplasts, prepared from C. parasitica strain EP155 (ATCC 38755) by the method of Churchill et al. (10), using a Gene Pulser II System electroporator (Bio-Rad Laboratories, Hercules, Calif.) and settings developed by Chen et al. (5). Surviving spheroplasts were cultured on osmotic solid regeneration media for 7 to 9 days to allow cell wall regeneration, and then transferred to potato dextrose agar (PDA; Difco, Detroit, Mich.) plates for phenotypic characterization.
Analysis of viral RNAs recovered from fungal transfectants.
Mycelia grown in 20 ml of EP complete medium (26) for 5 days were harvested by filtration through Miracloth (Calbiochem, La Jolla, Calif.), ground to powder with a mortar and pestle in liquid nitrogen, and suspended in 4 ml of extraction buffer composed of 100 mM Tris (pH 8), 200 mM NaCl, 4 mM EDTA, and 2% sodium dodecyl sulfate. Total nucleic acid was extracted sequentially with phenol, phenol-chloroform, and chloroform-isoamyl alcohol (27) and precipitated with ethanol. The extracted nucleic acid samples were exposed to two rounds of RQ1 DNase (Promega) digestion (4 U each) in 0.5 ml of 20 mM Tris (pH 8.0)-20 mM MgCl2, in the presence of 40 U of RNase inhibitor (RNasin; Promega) for 1 h. at 37°C, and finally suspended in distilled water at a final optical density (OD) at 260 nm of 20. The quality and relative quantity of nucleic acid preparations were examined by electrophoresis through 0.7% NuSieve (Biowhittaker, Rockland, Maine) agarose as described previously (31).
The relative level of viral RNA accumulation in fungal colonies infected with wild-type and mutant viruses was also examined by semiquantitative real-time reverse transcriptase (RT)-PCR analysis. cDNA specific for viral plus- and minus-strand RNA was generated with primers NS52PE and NS53PE, respectively. Each cDNA reaction mixture also contained a primer complementary to C. parasitica 18S rRNA, PDF1439R. RT reaction mixtures were assembled by first mixing 2 μl of total RNA with 40 pmol of each primer (e.g., PDF1439R for rRNA priming and NS53PE for viral minus-sense RNA priming) in 90% dimethyl sulfoxide, heating the solutions at 65°C for 20 min to denature double-stranded RNA (dsRNA) species, and then precipitating the denatured nucleic acids and primers by addition of 1/10 volume of 3 M sodium acetate and 2.5 volumes of ethanol (2). The dried precipitates were suspended in 30 μl of preheated (42°C) RT reaction mixture containing 100 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 0.2 mM concentrations of each deoxynucleoside triphosphate, and 1.25 U of avian myeloblastosis virus RT (Invitrogen, Gaithersburg, Md.) and incubated for 45 min. The relative amounts of viral RNA were determined by amplifying the viral cDNA products with AmpliTaq Gold in TaqMan universal PCR master mix (Applied Biosystems, Foster City, Calif.), exploiting the 5′ nuclease assay in a GeneAmp 5700 sequence detection system (Applied Biosystems). Values were normalized against 18S rRNA cDNA. Primer sets used in the PCR were PDF1373F and PDF1439R for 18S rRNA and NS52PE and NS53PE for viral RNA. PDF1373F was designed to span the insertion site of a 547-bp intron found in the C. parasitica 18S gene (6) so that only rRNA cDNA is amplified. The terminal portion of the CHV1-EP713 genome (map positions 12341 to 12488) was chosen for amplification because this region is conserved in internal-deletion-containing RNA species that are frequently generated in hypovirus-infected fungal isolates (29). TaqMan probes designed for measuring rRNA cDNA and viral cDNA amplification were PDF-1400T and NS52TP.
The nucleotide extraction procedure used for the p40 gain-of-function study was modified to include a lithium chloride-mediated single-stranded RNA precipitation step prior to the RQ1 digestion step (17) in order to enrich dsRNA in the preparation analyzed by electrophoresis through 0.7% agarose. The genetic stability of viral RNA mutants was monitored by sequence analysis of ClampR (21) RT-PCR-amplified fragments generated from the same RNA samples following S1 nuclease treatment to remove remaining single-stranded RNAs according to Suzuki et al. (31, 32) using a single primer set consisting of NS7 and NS22. ClampR-generated fragments were cloned into pCRScript (Stratagene) and sequenced.
Phenotypic measurements.
Differences in colony morphologies for hypovirus-free C. parasitica strain EP155 (ATCC 38755) and corresponding wild-type- and mutant-hypovirus-transfected strains were examined following incubation for 1 week or 1 month on PDA (Difco) on the laboratory bench top at 22 to 24°C and a light intensity of 500 to 1,500 lx. Asexual spores (conidia) were quantified as described by Hillman et al. (17). Conidia were liberated from 1-month-old colonies with a glass rod in 15 ml of 0.15% Tween 80, filtered through three layers of Miracloth to remove mycelial fragments, and quantified with the aid of a hemacytometer. Laccase activity produced by virus-free and transfected fungal strains was measured by the method of Smart et al. (30). Colonies were cultured for 1 week on Bavendamm's (4) solid media containing 0.5% tannic acid (pH 4.5), 1.5% malt extract, and 2% Bacto-agar (Difco). Replicate uniform samples of medium were taken from the central portion of each plate, placed in a microtube, weighed, and dissolved in 10 volumes of 6 M sodium iodide. The absorbance at 405 nm was measured in a Beckman model DU-65 spectrophotometer. Virulence assays were performed with dormant American chestnut stems as previously described (9, 19), with a minimum of six duplicate inoculations per fungal strain. Inoculated stems were kept in an aquarium covered with Saran Wrap at room temperature to maintain humidity.
RESULTS
Construction and viability of p40 deletion mutants.
Four independent mutant CHV1-EP713 viruses were constructed to test the effect of p40 deletion on host phenotype in either the absence (Δp69a and Δp69b) or presence (Δp40a and Δp40b) of p29 (Fig. 1). Mutants Δp69a and Δp69b each contained the N-terminal 24 codons of p29 shown previously to be required for CHV1-EP713 replication (32). The Δp69a mutant retained the two-ORF genomic configuration of wild-type CHV1-EP713, in which the pentanucleotide UAAUG serves as both the translation termination codon (UAA) of ORF A and the 5′-proximal translation initiation codon (AUG) of ORF B. As a result, the 3′-terminal coding region of p40 (encoding 11 C-terminal codons), which is rich in A and U residues and thought to facilitate translation reinitiation at the UAAUG pentanucleotide separating ORF A and ORF B (32), was also retained, i.e., p29 Pro(24) was fused to p69 Val(612). The single-ORF configuration of mutant Δp69b was accomplished by fusing the p29 Pro(24) codon with the Met(1) codon of the ORF B p48 coding domain, resulting in deletion of the entire p40 coding domain. To construct mutants Δp40a and Δp40b, a fragment encoding all of p29 and the first two codons past the p29 cleavage site to ensure efficient cleavage, plus two BglII-derived codons (encoding Asp and Pro) added during the cloning procedure, was inserted into the Δp69a and Δp69b mutant backgrounds, respectively.
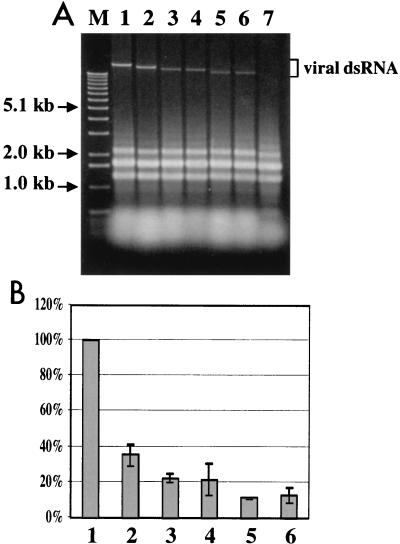
The replication competence of each mutant viral RNA was tested in parallel with synthetic transcripts derived from wild-type CHV1-EP713 cDNA and the Δp29 deletion mutant by transfecting spheroplasts prepared from virus-free C. parasitica strain EP155. As indicated by the ability to recover viral dsRNA from regenerated mycelium of each transfectant, all four mutant viruses were viable (Fig. 2A), confirming that p40 is dispensable for CHV1-EP713 replication. However, it is also apparent from Fig. 2A that the viral dsRNA of the different mutant classes accumulated to different levels.
FIG. 2.
(A) Agarose gel electrophoretic analysis of total RNA isolated from C. parasitica colonies transfected with mutant recombinant viruses. Equal amounts (OD, 0.25) of total RNA extracted from uninfected mycelia (lane 7) and mycelia infected with CHV1-EP713 wild-type virus (lane 1) or mutant viruses Δp29 (lane 2), Δp40a (lane 3), Δp40b (lane 4), Δp69a (lane 5), and Δp69b (lane 6) were electrophoresed through a 0.7% agarose gel in the TAE buffer system (40 mM Tris-acetate-1 mM EDTA, pH 7.8) and stained with ethidium bromide. Lane M was loaded in parallel with 1-kb ladder DNA size markers (Gibco-BRL, Gaithersburg, Md.). Relative mobilities of viral dsRNA are indicated. (B) Relative accumulation of viral minus-strand RNA in fungal colonies infected with wild-type CHV1-EP713 and mutant viruses. Total RNA was isolated from mycelia infected with CHV1-EP713 (bar 1) and mutant viruses Δp29 (bar 2), Δp40a (bar 3), Δp40b (bar 4), Δp69a (bar 5), and Δp69b (bar 6) and used for strand-specific cDNA synthesis after denaturation in 90% dimethyl sulfoxide at 65°C. The resulting cDNA was subjected to semiquantitative PCR analysis in a GeneAmp 5700 sequence detection system by using cDNA of 18S rRNA generated in the same RT reaction for normalization. The sequences of primers and TaqMan probes used in the quantification are described in Methods and Materials. Viral minus-strand RNA accumulation levels are reported as percentages of the value for CHV1-EP713-infected colonies, with standard deviations based on three independent measurements indicated by the error bars.
Quantification of the relative accumulation of viral RNA in the colonies infected with wild-type and mutant viruses was attempted with real-time PCR. The accumulation of Δp40 mutant minus-strand RNA was found to be reduced to approximately 20% of the level measured in colonies infected with wild-type CHV1-EP713 (Fig. 2B). A further reduction, to approximately 10% of the wild-type levels, was observed in Δp69-infected colonies. Thus, the relative values for viral minus-strand RNA accumulation measured by RT-PCR were consistent with the differences in viral dsRNA accumulation estimated by visual inspection of the agarose gel analysis presented in Fig. 2A. A similar level of reduction in relative accumulation was observed with an RT primer specific for viral plus-strand RNA (data not shown). Thus, there was no indication that there was a preferential strand-specific reduction in the RNA accumulation. Unexpectedly, Δp29 mutant RNA was also found to accumulate to a lower level (approximately 40%) than wild-type viral RNA, an observation not previously noted.
Phenotypic consequences of p40 deletion.
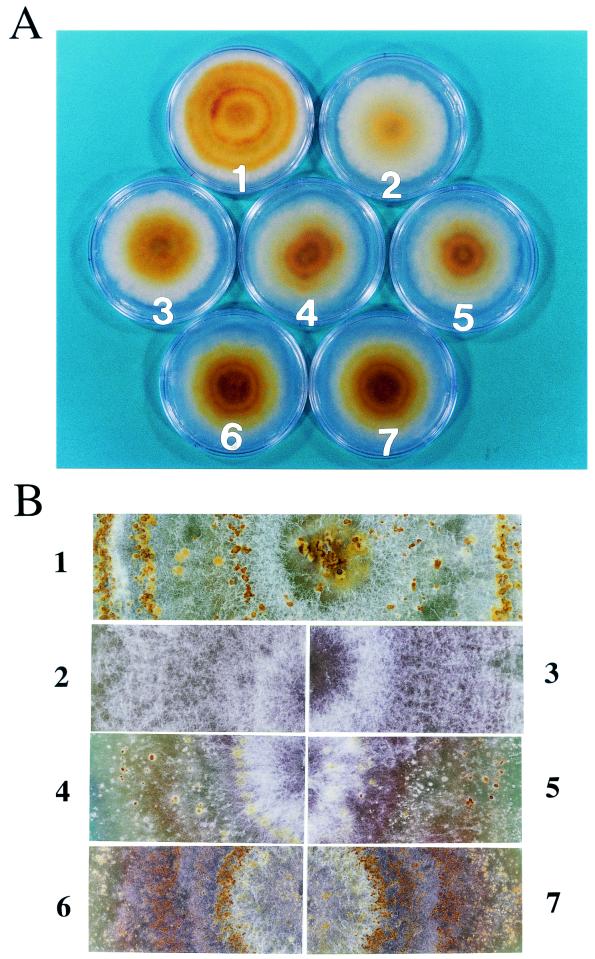
Deletion of the p40 coding domain had a significant effect on virus-mediated alteration of host colony morphology. As previously reported (17), infection of C. parasitica strain EP155 with CHV1-EP713 results in altered colony morphology characterized by reduced mycelial growth, irregular colony margins, and significantly reduced pigment production (Fig. 3A). Partial relief of each of these symptoms occurs upon deletion of the p29 coding domain (11, 31). Interestingly, colonies infected with the Δp40a and Δp40b deletion mutant viruses also exhibited increased growth rates and increased pigment production relative to colonies infected with the wild-type virus. However, these colonies still produced irregular margins. Deletion of p69 (Δp69a and Δp69b) resulted in further restoration in pigmentation relative to the Δp29- or Δp40-infected strains but still failed to produce the level observed for uninfected strain EP155 (Fig. 3A).
FIG. 3.
Phenotype of C. parasitica colonies infected with mutant viruses. Spheroplasts of C. parasitica strain EP155 were transfected with synthetic transcripts derived from each of the mutant virus cDNA plasmids, Δp29 (3), Δp40a (4), Δp40b (5), Δp69a (6), and Δp69b (7). Virus-free host strain EP155 (1) and wild-type virus (CHV1-EP713)-infected EP155 (2) are also shown. (A) Fungal colonies were photographed after culturing on PDA plates for 6 days. (B) One-month-old cultures of the same colonies were magnified to clearly show formation of conidia-containing pustules.
Prolonged incubation of the transfected colonies resulted in more pronounced morphological differences. A principal difference involved the formation of conidium-containing stromal pustules that normally cover the surface of virus-free C. parasitica colonies. As indicated in Fig. 3B, colonies infected with CHV1-EP713 and the Δp29 mutant failed to produce these pustules even after 1 month of incubation, while a substantial number of pustules, albeit smaller than those produced by virus-free colonies, formed on colonies transfected with the Δp40a and Δp40b mutants. Colonies infected with Δp69a and Δp69b produced even more (approximately 10-fold; data not shown), but smaller, pustules than the Δp40 infected colonies.
A description of the influence of ORF A deletions on conidium production must be prefaced by a discussion of the influence of light intensity on conidium production. Hillman et al. (17) confirmed that C. parasitica conidiation is light dependent (colonies do not form conidia in the dark) and reported that hypovirus-mediated suppression of conidiation can be partially overcome by exposure of infected colonies to high light intensity. Craven et al. (11) noted that Δp29-infected strains were more sensitive to increases in light intensity than CHV1-EP713-infected isogenic strains, i.e., the two strains were both severely suppressed in conidiation when cultured on the bench top while the Δp29-infected strain produced 100- to 1,000-fold more conidia than the CHV1-EP713-infected strain at moderate (2,000 to 3,000 lx) light intensity.
As shown in Table 2, conidiation levels exhibited by the different ORF A deletion mutant virus transfectants mirrored pustule formation. When incubated on the laboratory bench for 1 month, colonies infected with CHV1-EP713 produced 5 to 6 logs fewer conidia than virus-free strain EP155. Similar to the results reported by Craven et al. (11), colonies infected with the Δp29 mutant virus did not produce significantly more conidia than CHV1-EP713-infected colonies under these conditions. Rather than the 5- to 6-log reduction in conidiation caused by CHV1-EP713 infection, the Δp40 mutant viruses caused a 2- to 3-log reduction (Table 2). Deletion of p69 resulted in an additional 1-log increase in conidium production. While colonies infected with the Δp29 mutant responded to incubation under moderate light conditions with a 2- to 3-log increase in conidium production, as previously reported, no significant increase in conidiation was observed for colonies infected with CHV1-EP713 or the Δp40 and Δp69 mutants under these conditions (data not shown). This resulted in a narrowing in the difference between conidium production for the strains infected with the Δp29 and Δp40 mutants to 2 to 3 logs rather than the 4 to 5 logs observed at bench top.
TABLE 2.
Conidiation by C. parasitica strains transfected with hypovirus deletion mutants
| Fungal strain or transfecting virus | No. of conidia/mla |
|---|---|
| CHV1-EP713 | 7.8 × 103 ± 5.6 × 103 |
| Δp29 | 5.3 × 103 ± 2.9 × 103 |
| Δp40a | 2.0 × 107 ± 1.5 × 107 |
| Δp40b | 7.4 × 107 ± 8.1 × 107 |
| Δp69a | 3.4 × 108 ± 2.3 × 108 |
| Δp69b | 6.8 × 108 ± 4.1 × 108 |
| EP155 | 1.5 × 109 ± 2.8 × 108 |
Conidiation levels are the means and standard deviations for four cultures.
Deletion of the p29 coding domain was previously reported to cause a small but measurable relief of CHV1-EP713-mediated reduction in laccase accumulation while expression of p29 in the absence of virus infection caused a clear reduction in laccase accumulation (11). Thus, it was of interest to examine laccase accumulation in Δp40- and Δp69-infected colonies. Laccase activity can be quantified by dissolving agar plugs from tannic acid-containing Bavendamm's assay medium in 6 M sodium iodine and measuring the adsorbance at 405 nm (Table 3). As previously reported (8), CHV1-EP713 infection caused a significant reduction in laccase production (compare levels for EP155 and CHV1-EP713 in Table 3). The level of laccase activity produced by transfectants infected with the Δp40 mutant viruses was indistinguishable from that produced by the CHV1-EP713-infected strain (30) (Table 3). In contrast, transfectants infected with mutant viruses lacking the p29 coding domain (Δp29, Δp69a, and Δp69b) all produced three- to fivefold more laccase activity. Thus, deletion of p40 does not alter virus-mediated suppression of laccase production.
TABLE 3.
Laccase activities of fungal colonies
| Fungal strain or transfecting virus | Laccase activity ina:
|
|
|---|---|---|
| Expt I | Expt II | |
| CHV1-EP713 | 0.17 | 0.05 |
| Δp40a | 0.20 | 0.07 |
| Δp40b | 0.22 | 0.03 |
| Δp29 | 0.60 | 0.52 |
| Δp69a | 0.63 | 0.47 |
| Δp69b | 0.61 | 0.43 |
| EP155 | >2.0 | 1.5 |
For each colony laccase activity is shown as the mean OD at 405 nm for two Bavendamm plates melted as described in Materials and Methods.
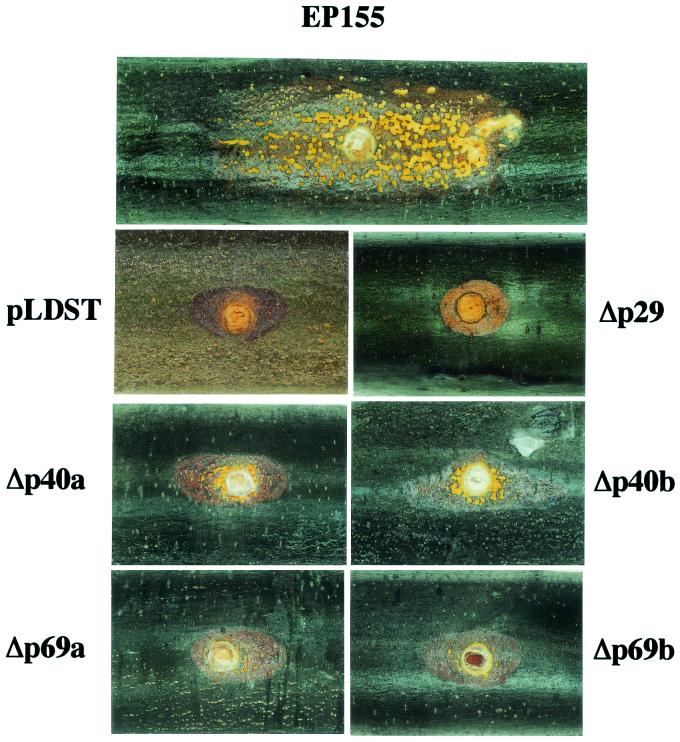
The effect of p40 deletion on CHV1-EP713-mediated attenuation of C. parasitica virulence was examined by testing the ability of the mutant virus-infected strains to cause cankers on dormant American chestnut stems. Virus-free C. parasitica strains produced large cankers that continued to expand until they completely girdled the stem. The surface of these cankers was densely packed with orange spore-containing stromal pustules that erupted through the bark as the canker expanded. Transfection with CHV1-EP713 severely reduced the ability of C. parasitica to expand on chestnut tissue, resulting in the formation of small, superficial cankers that produced very few stromal pustules (Fig. 4; Table 4). Strains infected with the Δp29 mutant virus produced cankers indistinguishable from those produced by CHV1-EP713-infected strains (Fig. 4). While strains infected with Δp40 mutant viruses also produced small cankers (approximately 10-fold smaller than cankers produced by EP155), they were consistently between two- and threefold larger and contained significantly more pustules than CHV1-EP713-infected or Δp29-infected colonies. Interestingly, deletion of p69 also resulted in a very slight increase in canker size and pustule production (Fig. 4).
FIG. 4.
Morphology of cankers induced on dormant chestnut stems by fungal colonies infected with mutant viruses. Dormant chestnut stems were inoculated with freshly grown fungal mycelia transfected with wild-type CHV1-EP713 (pLDST) or mutant viruses Δp29, Δp40a, Δp40b, Δp69a, and Δp69b and incubated in an aquarium under appropriate moisture and temperature conditions for 1 month. Virus-free C. parasitica strain EP155 was also used as an inoculum in parallel. A representative canker photograph is shown for each of the fungal strains.
TABLE 4.
Mean canker areas induced by virus-infected and virus-free fungal colonies
| Fungal strain or transfecting virus | Canker area (cm2)a
|
|
|---|---|---|
| 3 wk | 4 wk | |
| EP155 (virus free) | 20.12 ± 5.48 | 31.09 ± 5.92 |
| CHV1-EP713(pLDST) | 1.28 ± 0.28 | 1.36 ± 0.11 |
| Δp29 | 1.18 ± 0.63 | 1.34 ± 0.70 |
| Δp40a | 2.23 ± 0.73 | 3.27 ± 1.72 |
| Δp40b | 2.59 ± 0.90 | 2.96 ± 0.76 |
| Δp69a | 1.51 ± 0.45 | 1.69 ± 0.36 |
| Δp69b | 1.75 ± 0.28 | 1.89 ± 0.40 |
Values are means ± standard deviations for six replicates.
Fine mapping of the p40 functional domain.
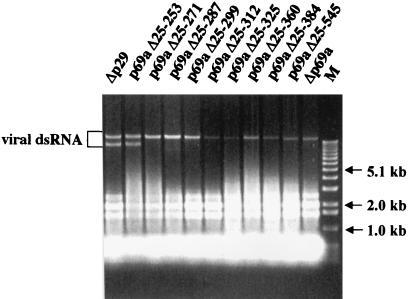
The gain-of-function strategy successfully used to map the p29 symptom determinant domain (31) was adopted to map the p40 functional domain. The Δp69a mutant was systematically repaired by progressive extension of the p40 coding domain from Val(612) toward the N terminus (Fig. 1). The resulting nine recombinant synthetic transcripts were then tested for replication competence and effect on fungal phenotype. As shown in Fig. 5, viral dsRNA was recovered from all nine transfectants, confirming the replication competence of each modified viral RNA.
FIG. 5.
Agarose gel electrophoresis pattern of p40 gain-of-function mutant viral dsRNA. Equal amounts (OD, 0.25) of enriched dsRNA fractions obtained from C. parasitica transfectants with each of the mutant viruses Δp29, p69aΔ25-253, p69aΔ25-271, p69aΔ25-287, p69aΔ25-299, p69aΔ25-312, p69aΔ25-325, p69aΔ25-360, p69aΔ25-384, p69aΔ25-545, and Δp69a were applied to the wells. Electrophoresis was performed in a 0.7% agarose gel in 1× TAE (40 mM Tris-acetate-1 mM EDTA, pH 7.8) as for Fig. 2. Migration positions of 1-kb ladder DNA size markers (lane M) (Gibco-BRL) and mutant viral dsRNA are shown. The dsRNA fractions isolated from Δp29 and p69aΔ25-253 transfectants also contain defective viral dsRNAs that migrate faster than the viral genome. These defective dsRNAs have been generated in many of the transformants examined in this study. However, they are most often observed upon continued passage of the infected mycelium and in the transfectants that accumulate the most dsRNA, e.g., wild-type and Δp29 transfectants. The presence of defective dsRNAs, first described by Shapira et al. (29), does not correlate with any alterations in host phenotype.
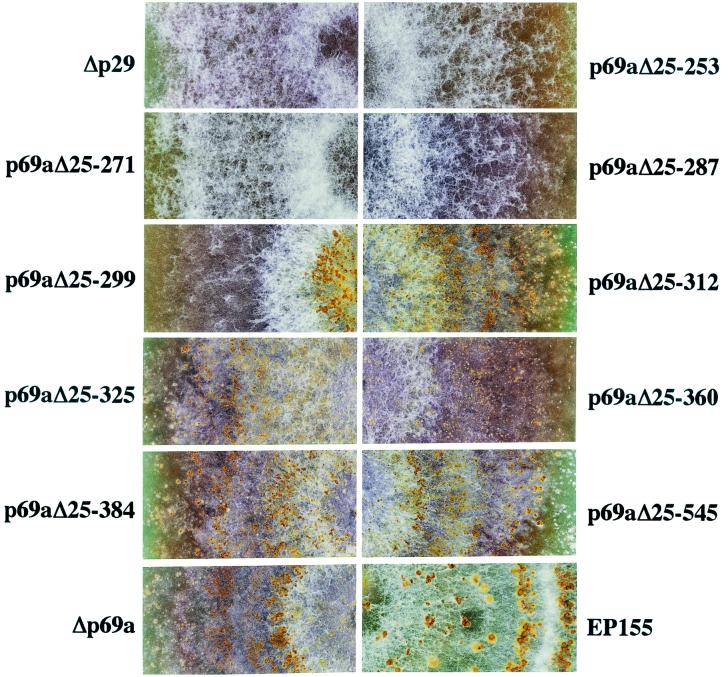
Fungal strains infected with recombinant viruses containing progressive extensions of the p40 coding domain from Val(612) toward the N terminus to Gly(546), Gln(385), Ala(361), Arg(326), and Asn(313) all resembled the Δp69b-infected strain in terms of pigmentation and colony morphology. The colonies all formed stromal pustules, although some (e.g., p69aΔ25-384 and p69aΔ25-312) produced slightly more pustules than the Δp69a-infected strain (Fig. 6). However, a significant change in morphology and pustule formation was observed after extension to Thr(300). Further extension of the p40 coding domain to Thr(288) and beyond to Gln(272) and Leu(254) resulted in colony morphology very similar to that of colonies transfected with the Δp29 mutant. Thus, the domain responsible for the p40-mediated contribution to alteration of colony morphology can be defined within the region extending from Thr(288) to Arg(312).
FIG. 6.
Enlargements of photographs of C. parasitica colonies infected with p69 extension mutant viruses. Colonies of a healthy C. parasitica EP155 colony and fungal colonies transfected with Δp29 or Δp69 extension mutant viruses p69aΔ25-253, p69aΔ25-271, p69aΔ25-287, p69aΔ25-299, p69aΔ25-312, p69aΔ25-325, p69aΔ25-360, p69aΔ25-384, p69aΔ25-545, and Δp69a were grown on PDA for 1 month at bench top and photographed.
The effect of progressive p40 repair on fungal asexual sporulation paralleled that observed for colony morphology (Table 5). No significant reduction in sporulation was observed after extension to Asn(313). However, a 2- to 3-log reduction resulted after extension to Thr(300). Further extension to Thr(288), Gln(272), or Leu(254) resulted in further reductions to the levels observed for colonies transfected with the Δp29 mutant. Thus, the determinant for p40-mediated alteration of colony morphology colocalizes with the determinant that contributes to the suppression of fungal asexual sporulation.
TABLE 5.
Conidiation by C. parasitica strains transfected with hypovirus deletion mutants
| Fungal strain or transfecting virus | No. of conidia/mla |
|---|---|
| Δp29 | 9.0 × 103 ± 2.8 × 103 |
| p69aΔ25-253 | 6.3 × 103 ± 1.5 × 103 |
| p69aΔ25-271 | 4.5 × 103 ± 3.4 × 103 |
| p69aΔ25-287 | 9.5 × 103 ± 1.0 × 104 |
| p69aΔ25-299 | 4.9 × 106 ± 6.2 × 106 |
| p69aΔ25-312 | 1.1 × 109 ± 5.8 × 108 |
| p69aΔ25-325 | 6.9 × 108 ± 5.0 × 108 |
| p69aΔ25-360 | 7.1 × 107 ± 9.5 × 107 |
| p69aΔ25-384 | 9.5 × 108 ± 1.4 × 108 |
| p69aΔ25-545 | 7.1 × 108 ± 4.7 × 107 |
| Δp69a | 6.2 × 108 ± 4.1 × 108 |
| EP155 | 1.9 × 109 ± 7.1 × 108 |
Conidiation levels are the means and standard deviations for four cultures.
As indicated in Fig. 5, gel analysis of viral RNA recovered from strains infected in the gain-of-function analysis indicated a direct correlation between the restoration of p40-related suppressive activity and an increased level of viral RNA accumulation. To examine whether these changes were dependent on the peptide sequence of the identified activity domain [Thr(288) to Arg(312)] or possibly mediated by the corresponding RNA sequence, a double frameshift mutation of the p40 region was constructed within the context of the Δp29 virus. This was accomplished by addition of a C residue between nucleotides 1355 and 1356 and the deletion of a T residue at position 1427, resulting in a +1 frameshift that replaced the natural amino acid sequence extending from Thr(288) to Leu(311) (TIVSVGKVGMAITSIQDALVATEL) with the unrelated sequence (NHCLRWQGWYGHHQHPGCSCGYGV). The double frameshift mutant (pDFS2) was viable, and infected colonies exhibited a phenotype indistinguishable from that exhibited by Δp69-infected strains (data not shown). These results provide strong evidence that it is the peptide sequence extending from Thr(288) to Arg(312), and not the corresponding RNA sequence, that is responsible for the p40-mediated amplification in RNA accumulation.
Stability of mutant viruses.
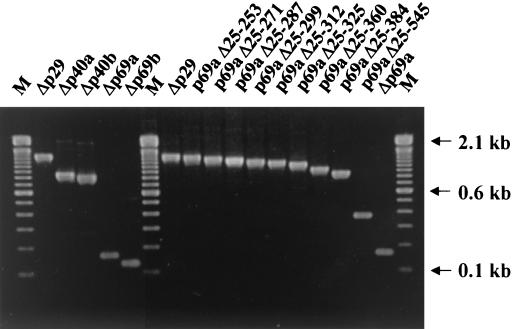
Although the dsRNA recovered from the strains transfected with the Δp29, Δp40, and Δp69 mutants exhibited relative migration rates consistent with the size of the deletions engineered (Fig. 2A and 5), a more sensitive confirmation of the stability of all of the engineered deletions was undertaken by ClampR analysis. As indicated in Fig. 7, the relative sizes of the ClampR-amplified fragments generated from each mutant dsRNA were consistent with the relative size of the deletion introduced into the mutant cDNA clone. Sequence analysis confirmed that mutant viruses Δp40b, Δp69a, and Δp69b all stably retained the altered sequences. However, an interesting modification was observed in five independent clones of ClampR fragments derived from RNA recovered from Δp40a-infected strains. This involved the insertion of an additional A in the cluster of four A residues located between 9 and 12 residues upstream of the UAAUG pentanucleotide at the boundary between ORF A and ORF B. This insertion resulted in conversion from a dicistronic to a monocistronic genome organization. Additional dicistronic-to-monocistronic conversions as a result of nucleotide insertions were observed for several of the p40 repair constructs used in the mapping study (Table 6). For example, ClampR fragments amplified from viral RNA isolated from strains infected with mutant viruses p69aΔ25-545 and p69aΔ25-312 were all found to contain the insertion of an A residue in the same area as observed for the Δp40a mutant. A similar A insertion was found in one of five and two of five cloned ClampR fragments generated from progeny RNA of mutants p69aΔ25-360 and p69Δ25-384, respectively. The viral RNA recovered from the strain infected with p69aΔ25-325 contained an additional U residue in the U cluster immediately adjacent to the pentanucleotide. To reduce the possibility that compensatory mutations distant from the ORF A/ORF B boundary contribute to the viability of the monocistronic gain-of-function progeny viruses, the PstI-SalI fragment (nucleotide coordinates 1846 to 2637) in p69aΔ25-312 and p69aΔ25-325 was replaced by the corresponding ClampR fragments amplified from their respective progeny virus genomic dsRNA. Each of the reconstructed monocistronic mutants was found to be replication competent, and the colony morphologies exhibited by fungal strains infected with these mutants were indistinguishable from that exhibited by the strains from which the original monocistronic progeny viruses were recovered.
FIG. 7.
ClampR analysis of dsRNA of virus mutants. ClampR was performed on dsRNA recovered from the fungal colonies transfected with mutant virus Δp29, Δp40a, Δp40b, Δp69a, Δp69b, p69aΔ25-253, p69aΔ25-271, p69aΔ25-287, p69aΔ25-299, p69aΔ25-312, p69aΔ25-325, p69aΔ25-360, p69aΔ25-384, and p69Δ25-545. The fragments covering the deletion sites were amplified for all transfectants with a single primer set consisting of NS7 and NS22 (see Table 1 for primer sequences). Amplified fragments were electrophoresed in a 2.0% agarose gel in TBE (89 mM Tris-89 mM boric acid-2.5 mM EDTA, pH 8.3). Lanes M contain 100-bp ladder DNA size standards (Gibco-BRL). Sequence results of ClampR fragments cloned into pCRScript are summarized in Table 6.
TABLE 6.
Characteristics of mutant viruses
| Mutant virus | Repression in pustule formation | Infectivitya | Genome organizationb
|
|
|---|---|---|---|---|
| cDNA | ClampR | |||
| Δp40a | No | 3/3 (P, P, P) | Dicistronic | Monocistronic |
| Δp40b | No | 3/3 (S, S, S) | Monocistronic | Monocistronic |
| Δp29 | Yes | 3/3 (S, S, S) | Dicistronic | Dicistronic |
| p69aΔ25-253 | Yes | 3/3 (S, S, S) | Dicistronic | Dicistronic |
| p69aΔ25-271 | Yes | 3/3 (S, S, S) | Dicistronic | Dicistronic |
| p69aΔ25-287 | Yes | 3/3 (S, S, S) | Dicistronic | Dicistronic |
| p69aΔ25-299 | Noc | 3/3 (S, S, S) | Dicistronic | Dicistronic |
| p69aΔ25-312 | No | 1/3 (P) | Dicistronic | Monocistronic |
| p69aΔ25-325 | No | 1/3 (P) | Dicistronic | Monocistronic |
| p69aΔ25-360 | No | 3/3 (S, S, S) | Dicistronic | Mono/dicistronicd |
| p69aΔ25-384 | No | 3/3 (S, S, P) | Dicistronic | Mono/dicistronic |
| p69aΔ25-545 | No | 1/3 (P) | Dicistronic | Monocistronic |
| Δp69a | No | 3/3 (S, S, S) | Dicistronic | Dicistronic |
| Δp69b | No | 3/3 (S, S, S) | Monocistronic | Monocistronic |
Infectivity is shown as the ratio of infected plates (colonies) to total plates (colonies) regenerated from transfected spheroplasts and by the letters P and S, indicating partial and systemic infection.
Genome organization of mutant viruses is based on sequences of cDNA originally designed for in vitro transcription and sequences of ClampR fragments that were amplified on dsRNA recovered from transfected fungal colonies.
Although only the center of an infected colony produced stromal pustules, the surrounding area was suppressed in pustule production.
Both monocistronic and dicistronic genomes were detected in the progeny virus population.
It is noteworthy that the dicistronic-to-monocistronic conversion observed for several of the gain-of-function mutants is predicted to result in the fusion of the partial p40 amino acid sequence to p48. In the case of mutant p69aΔ25-312, the p40 N-terminal portion of the fusion would extend for 310 amino acid residues. Sequence analysis of ClampR-amplified fragments generated from progeny viral RNA of the double frameshift mutant also revealed a conversion to a monocistronic organization (data not shown). In this case it is predicted that p48 would form an N-terminal fusion with a full-length modified p40 protein that would extend a total of 374 amino acids.
DISCUSSION
The detailed examination of the phenotypic and molecular consequences resulting from deletion of the CHV1-EP713 p40 coding domain described in this report was prompted by the surprising observation that mutants lacking the coding region for this highly basic and conserved viral protein were viable (32). While the results confirm the dispensability of p40 for viral replication, it is now clear that deletion of the p40 coding domain is accompanied by a significant reduction in the accumulation of viral RNA (Fig. 2). As successfully employed for mapping the p29 activity determinant domain, a gain-of-function analysis was used to map the p40 functional domain to a region extending from Thr(288) to Arg(312). Evidence that the gain of function was due to the encoded protein rather than the corresponding RNA sequence element was provided by introducing frameshift mutations on either side of the activity determinant domain. The severity of virus-mediated phenotypic suppression by p40 deletion and gain-of-function mutants was directly related to the accumulation of viral RNA. While deletion of p40 had little effect on virus-mediated hypovirulence, virus-mediated suppression of several other host processes, such as orange pigment production and asexual sporulation, was significantly affected (Fig. 3, 4, and 6; Tables 2 and 5).
The phenotypic consequences of p40 deletion are strikingly similar to those previously reported for deletion of p29, e.g., partial relief of virus-mediated suppression of conidiation, pigmentation, and hyphal growth rate (11). The similarities extend to a reduced level of RNA accumulation for both the Δp40 and Δp29 mutant viruses, an observation not previously noted for the latter mutant. However, there are several indications that the two proteins influence virus-mediated phenotypic alterations by distinct mechanisms. When independently transformed into virus-free C. parasitica, the p29 coding domain, but not the p40 coding domain, caused reduced pigmentation and conidiation (11; our unpublished observation). Thus, the p29 protein can alter fungal phenotype independent of virus infection. This characteristic could not be demonstrated for p40. An additional distinction is that deletion of p29 causes a reduction in the level of virus-mediated suppression of laccase activity (11) while deletion of p40 does not.
The combined results suggest that p29 alters host phenotype directly through action of the protein on host factors and indirectly by contributing to viral RNA accumulation. In contrast, p40 appears to act indirectly through its accessory role in amplifying viral RNA. However, one cannot rule out the possibility that p40 also acts directly in conjunction with another virus-encoded protein and consequently fails to cause any phenotypic alterations when expressed independently in the absence of virus infection.
The inability to experimentally regulate the accumulation of wild-type hypovirus RNA currently precludes a precise determination of the relative effect of reduced hypovirus RNA accumulation on symptom expression. However, several symptoms appear to be more sensitive to changes in viral RNA accumulation than others. For example, the 70 to 80% reduction in viral RNA accumulation observed for the p40 mutants correlated with a significant relief in virus-mediated suppression of pigmentation, conidiation, and mycelial growth. While a further reduction in RNA accumulation observed for the p69 mutants was accompanied by additional relief in suppression of pigmentation and conidiation, remarkably, this 90% reduction in viral RNA accumulation had only a minimal effect on virus-mediated virulence attenuation (Fig. 4; Table 4). The suggestion that the magnitude and range of hypovirus-mediated symptom expression can be influenced by the level of virus RNA accumulation raised by this study also reinforces the importance of measuring viral RNA accumulation for viral mutants used to identify viral symptom determinant domains. Differences in viral RNA accumulation levels could also contribute to the significant diversity in symptom expression observed for hypovirus-infected C. parasitica field isolates (13-15, 22).
Real-time PCR measurements of the relative accumulation of hypovirus positive- and negative-strand RNA in mutant- and wild-type-virus-infected fungal strains were entirely consistent with the relative accumulation of hypovirus dsRNA as judged by agarose gel electrophoresis (Fig. 2). Given the evolutionary relatedness between hypoviruses and positive-strand RNA potyviruses, it was of interest to examine the relative accumulation of positive- and negative-strand RNA. However, the confidence level for use of real-time PCR to determine absolute amounts of positive- and negative-strand-RNA accumulation was complicated by differences in the priming efficiency for the control positive and negative strands tested individually or as a duplex (data not shown). Nevertheless, even after these differences were factored in, there was no evidence for a large excess of positive-strand RNA. The ratio of CHV1-EP713 positive-strand RNAs to negative-strand RNAs ranged between 2 and 4 (data not shown). In this regard, Fahima et al. (16) reported that vesicles isolated from CHV1-EP713-infected mycelia produced full-length positive- and negative-strand RNAs at ratios between 2 and 8. Thus, the positive- to negative-strand-RNA ratios observed for hypoviruses both in vitro and in vivo differ from values generally accepted in the classic model for a productive infection by a positive-strand RNA virus, as exemplified by poliovirus (3). Poliovirus positive-strand RNA is reported to accumulate to a level 20- to 50-fold in excess of minus-strand RNA accumulation, serving primarily as mRNA early after infection and siphoned off into virus particles as the infection proceeds. The persistent nature of the hypovirus infection and the absence of a capsid protein may contribute to the lower positive- to negative-strand ratio observed for hypoviruses. For example, during the transition from acute to persistent coxsackievirus infection, the ratio of positive- to negative-strand RNA changed from 75/1 to nearly 1/1, with the viral genetic information then persisting in a double-stranded configuration (33). The absence of a capsid protein may result in greater exposure of single-stranded positive-strand RNA to nucleases, thereby increasing turnover. Certainly, ongoing efforts to engineer hypoviruses for enhanced biological control or for use in fundamental studies on fungal virulence (12, 24, 25) will benefit from a clearer understanding of the mechanisms that regulate hypovirus RNA accumulation.
The ClampR analysis of progeny of mutant viral RNAs revealed an interesting pattern involving the conversion from a dicistronic gene organization in several mutant virus constructs to a monocistronic genome organization in corresponding progeny viruses as the result of single nucleotide insertions in the AU-rich sequences just upstream of the UAAUG pentanucleotide separating ORF A and ORF B. With one exception, Δp69a, all of the dicistronic mutant viral RNAs that underwent this conversion lacked the p40 activity domain [Thr(288) to Arg(312)], including the double frameshift mutant. One interpretation of this striking observation is that the p40 activity domain in some way facilitates ORF B expression, perhaps by facilitating the proposed translational termination and reinitiation at the ORF A/ORF B boundary. That is, the conversion from the dicistronic to the monocistronic configuration may partially compensate for a deficiency in the level of ORF B expression caused by the absence of the p40 activity domain. In this scenario, replication of a dicistronic mutant lacking the p40 activity domain would be predicted to proceed with very low efficiency. Conversion to the monocistronic configuration at an early stage after transfection would increase ORF B expression and consequently promote replication efficiency, causing increased viral RNA accumulation and a further increase in the expression of ORF B gene products. Consistent with this possibility, these mutants all exhibited a lower level of transfection efficiency (ratio of successful infections to transfection attempts) and generally were able to only partially, rather than systemically, infect the regenerated mycelia present on regeneration plates (Table 6). Moreover, we have never recovered any monocistronic progeny viruses from dicistronic mutant viruses that contained the p40 activity domain. This could indicate that conversion to a monocistronic configuration provides no advantage, or is even deleterious, in the context of the p40 activity domain. For Δp69a, the one mutant lacking the p40 activity domain for which no conversion was observed, one could envision that conversion is not needed because the ORF B AUG initiation codon is sufficiently close to the 5′ noncoding leader to allow an adequate level of internal translation initiation to occur at this site.
The dicistronic-to-monocistronic conversion in the progeny of the different p40 viral mutants is also predicted to result in fusion of portions of p40 to the N terminus of p48. In the case of the double frameshift mutant, the N-terminal extension would include the entire modified p40 protein of 374 amino acids. It is surprising that the fusion of such a long extension would not compromise the function of the p48 coding domain. The combined observations and predictions presented in this report clearly warrant further investigation into the role of p40 in translational control of hypovirus gene expression and viral RNA amplification. Identification of the 25-amino-acid p40 functional domains should significantly facilitate such studies.
Acknowledgments
We are grateful to Brad Hillman, Todd Parsley, Angus Dawe, Gert Segers, and Lynn Geletka for helpful comments.
This work was supported in part by NIH grant GM55981 to D.L.N.
REFERENCES
- 1.Anagnostakis, S. L. 1982. Biological control of chestnut blight. Science 215:466-471. [DOI] [PubMed] [Google Scholar]
- 2.Asamizu, T., D. Summers, M. B. Motika, J. V. Anzola, and D. L. Nuss. 1985. Molecular cloning and characterization of the genome of wound tumor virus: a tumor-inducing plant reovirus. Virology 144:398-409. [DOI] [PubMed] [Google Scholar]
- 3.Baltimore, D. 1969. The replication of picornaviruses, p. 101-176. In H. B. Levy (ed.), The biochemistry of viruses. Marcel Dekker, New York, N.Y.
- 4.Bavendamm, W. 1928. Uber das vorkommen und den Nachweis von Oxydasen bei holzzerstorenden Pilzen. Z. Pflanzenkr. Pflanzenschutz 38:257-276. [Google Scholar]
- 5.Chen, B., G. H. Choi, and D. L. Nuss. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264:1762-1764. [DOI] [PubMed] [Google Scholar]
- 6.Chen, B., C.-H. Chen, B. H. Bowman, and D. L. Nuss. 1996. Phenotypic changes associated with wild-type and mutant hypovirus RNA transfection of plant pathogenic fungi phylogenetically related to Cryphonectria parasitica. Phytopathology 86:301-310. [Google Scholar]
- 7.Chen, B., and D. L. Nuss. 1999. Infectious cDNA clone of hypovirus CHV1-Euro7: a comparative virology approach to investigate virus-mediated hypovirulence of the chestnut blight fungus Cryphonectria parasitica. J. Virol. 73:985-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, G. H., and D. L. Nuss. 1992. A viral gene confers hypovirulence-associated traits to the chestnut blight fungus. EMBO J. 11:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, G. H., and D. L. Nuss. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800-803. [DOI] [PubMed] [Google Scholar]
- 10.Churchill, A. C. L., L. M. Ciufetti, D. R. Hansen, H. D. Van Etten, and N. K. Van Alfen. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17:25-31. [Google Scholar]
- 11.Craven, M. G., D. M. Pawlyk, G. H. Choi, and D. L. Nuss. 1993. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67:6513-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawe, A. L., and D. L. Nuss. 2001. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35:1-29. [DOI] [PubMed] [Google Scholar]
- 13.Elliston, J. E. 1978. Pathogenicity and sporulation in normal and diseased strains of Endothia parasitica in American chestnut, p. 95-100. In W. L. MacDonald, F. C. Cech, J. Luchok, and C. Smith (ed.), Proceedings of the American Chestnut Symposium. University Books, Morgantown, W.Va.
- 14.Elliston, J. E. 1985. Characterization of dsRNA-free and dsRNA-containing strains of Endothia parasitica in relation to hypovirulence. Phytopathology 75:151-158. [Google Scholar]
- 15.Enebak, S. A., W. L. MacDonald, and B. I. Hillman. 1994. Effect of dsRNA associated with isolates of Cryphonectria parasitica from the central Appalachians and their relatedness to other dsRNAs from North America and Europe. Phytopathology 84:528-534. [Google Scholar]
- 16.Fahima, T., P. Kazmierczak, D. R. Hansen, P. Pfeiffer, and N. K. Van Alfen. 1993. Membrane-associated replication of an unencapsidated double-stranded RNA of the fungus, Cryphonectria parasitica. Virology 195:81-89. [DOI] [PubMed] [Google Scholar]
- 17.Hillman, B. I., R. Shapira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80:950-956. [Google Scholar]
- 18.Hillman, B. I., B. T. Halpern, and M. P. Brown. 1994. A viral dsRNA element of the chestnut blight fungus with a distinct genetic organization. Virology 201:241-250. [DOI] [PubMed] [Google Scholar]
- 19.Jaynes, R. A., and J. E. Elliston. 1980. Pathogenicity and canker control by mixtures of hypovirulent strains of Endothia parasitica in American chestnut. Phytopathology 70:453-456. [Google Scholar]
- 20.Koonin, E. V., G. H. Choi, D. L. Nuss, R. Shapira, and J. C. Carrington. 1991. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl. Acad. Sci. USA 88:10647-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalik, T. F., Y.-Y. Yang, and J. K.-K. Li. 1990. Molecular cloning and comparative sequence analysis of bluetongue virus S1 by selective synthesis of specific full-length DNA copies of dsRNA genes. Virology 177:820-823. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald, W. L., and D. W. Fulbright. 1991. Biological control of chestnut blight: use and limitation of transmissible hypovirulence. Plant Dis. 75:656-661. [Google Scholar]
- 23.Nuss, D. L. 1992. Biological control of chestnut blight: an example of virus mediated attenuation of fungal pathogenesis. Microbiol. Rev. 56:561-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuss, D. L. 1996. Using hypoviruses to probe and perturb signal transduction processes underlying fungal pathogenesis. Plant Cell 8:1845-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuss, D. L. 2000. Hypovirulence and chestnut blight: from the field to the laboratory and back, p. 149-170. In J. W. Kronstad (ed.), Fungal pathology. Kluwer Press, Dordrecht, The Netherlands.
- 26.Puhalla, J. E., and S. L. Anagnostakis. 1971. Genetics and nutritional requirements of Endothia parasitica. Phytopathology 61:169-173. [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Shapira, R., G. H. Choi, and D. L. Nuss. 1991. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10:731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapira, R., G. H. Choi, B. I. Hillman, and D. L. Nuss. 1991. The contribution of defective RNAs to complexity of viral-encoded double-stranded RNA populations present in hypovirulent strains of the chestnut blight fungus Cryphonectria parasitica. EMBO J. 10:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smart, C. D., W. Yuan, R. Foglia, D. L. Nuss, D. W. Fulbright, and B. I. Hillman. 2000. Cryphonectria hypovirus 3, a virus species in the family Hypoviridae with a single open reading frame. Virology 265:66-73. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki, N., B. Chen, and D. L. Nuss. 1999. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J. Virol. 73:9478-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki, N., L. M. Geletka, and D. L. Nuss. 2000. Essential and dispensable virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J. Virol. 74:7568-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tam, P. M., and R. P. Messner. 1999. Molecular mechanisms of coxsackievirus persistence in chronic inflammatory myopathy: viral RNA persists through formation of a double-stranded complex without associated genomic mutations or evolution. J. Virol. 73:10113-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]