Abstract
The protease of the porcine endogenous retrovirus (PERV) subtypes A/B and C was recombinantly expressed in Escherichia coli as proteolytically active enzyme and characterized. The PERV Gag precursor was also recombinantly produced and used as the substrate in an in vitro enzyme assay in parallel with synthetic nonapeptide substrates designed according to cleavage site sequences identified in the PERV Gag precursor. The proteases of all PERV subtypes consist of 127 amino acid residues with an Mr of 14,000 as revealed by determining the protease N and C termini. The PERV proteases have a high specificity for PERV substrates and do not cleave human immunodeficiency virus (HIV)-specific substrates, nor are they inhibited by specific HIV protease inhibitors. Among the known retroviral proteases, the PERV proteases resemble most closely the protease of the murine leukemia retrovirus.
Organ xenotransplantation from pigs may be one possibility to solve the shortage of human organ donors. An unanswered question is, however, the risk of viral infection by porcine endogenous retroviruses (PERVs) upon xenografting (3, 5, 7, 10, 11, 16). One way to minimize this problem might be the option of designing an effective antiviral chemotherapy against PERV, comparable to that presently employed against human immunodeficiency virus (HIV) infection. Here we have characterized the PERV protease as a target for protease inhibitors (PIs) that might be employed as antiviral agents for chemotherapy.
Three main subtypes of PERV, A, B, and C, are known to date (14). All are classical C-type retroviruses (e.g., 1, 2, 15) and are genetically similar but not identical. We have revealed the coding sequences for the proteases of all three PERV subtypes, produced active proteases by recombinant methods in Escherichia coli, and established an in vitro assay system for PERV protease activity.
Coding sequence of the PERV protease.
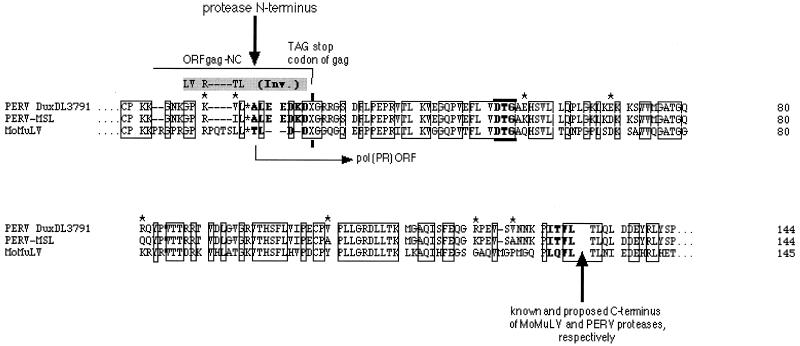
For recombinant expression of the PERV protease, the boundaries of its coding sequence had to be revealed. Possible N and C termini of the functional PERV protease coding sequence were predicted by multiple alignment of the translated sequences of gag and pol regions of PERV subtypes A/B (DuxDL3791 [J. Blusch et al., unpublished data]) and C (PERV-MSL [1]) with the protease sequence of Moloney murine leukemia virus (MMLV [8, 20]), the phylogenetically closest C-type retrovirus with a known protease protein sequence (Fig. 1).
FIG. 1.
Amino acid sequences for the protease of PERV subtypes A/B and C. The Pol ORF sequences of PERV subtype A/B (DuxDL3791), PERV subtype C (MSL), and MMLV (MoMuLV) were compared in a multiple alignment. Identical residues in the putative protease common to all three sequences are boxed. The suppressed gag stop codon, TAG, is indicated by an X. Asterisks indicate amino acid differences between PERV proteases of subtypes A/B and C.
The alignment indicated that the C terminus of the PERV protease seemed to be highly similar to that of the murine leukemia virus protease; the N terminus of PERV protease, however, showed significant differences (Fig. 1). The N terminus of MMLV protease is known to overlap into the C-terminal Gag open reading frame (ORF) by 4 amino acid residues (T L D D) (21). Thus, the gag stop (amber) codon TAG (X in Fig. 1) is part of the protease coding sequences and can be read through by a Gln suppressor tRNA. An alignment of the protease N terminus emerged only in the way shown in Fig. 1; the L*A was provided as the reasonable N-terminal PERV protease cleavage site corresponding to that of the murine L*T. Thus, the N terminus of PERV protease was proposed to have three more hydrophilic residues than the murine viral protease N terminus (Fig. 1).
The alignment in this form shows a high sequence similarity among the compared enzymes. Proteases of the PERV subtypes A/B and C have 59 and 60% sequence identity, respectively, to the MMLV protease. The two PERV proteases are up to 95% identical. The hydrophobicity plots of the PERV proteases are identical; the hydrophobicity profile of PERV protease is slightly less hydrophobic in the N and C termini than those of the murine virus enzyme (data not shown).
Expression of recombinant PERV protease and Gag precursor substrate in E. coli.
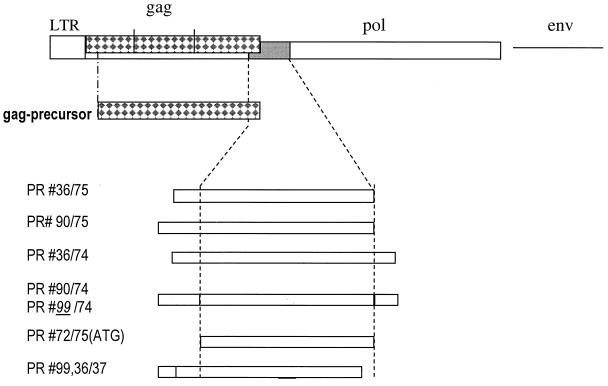
Recombinant constructs (Fig. 2) were made by inserting the proposed protease coding sequences of subtypes A/B and C into the His-tag expression vector pTrcHisA (Invitrogen). Constructs prepared with primers #36, #C36, #90, #C90, #99, and #C99 carry few adjacent nucleotides of the gag ORF upstream of the protease N terminus. Constructs #75 and #C75 terminated with the proposed C terminus boundary, while constructs with #37 were C-terminally truncated. Constructs based on #74 and #C74 extended over the proposed protease C terminus containing few adjacent reverse transcriptase coding sequences. Fragments for expression of PERV A/B protease were directly amplified from Du × DL crossbred 3791 pig genomic DNA.
FIG. 2.
Recombinant expression. All recombinant Gag and protease constructs are depicted. The dotted lines are the boundary of the coding region of the PERV protease. The construct numbers reflect the primer number. Primer sequences were #36 (GCGCGGATCCGAAGATAAAGATCAGGGGAGACG), #37 (CGCGAAGCTTTTAAAAAGAAATTTGAGCTCCCATC), #72 (GCGGTACCATG GAAGAAGATAAAGATCAGGGGAG), #C74 (CGCGAAGCTTAGTCATCTAATTGGAGGGTCAACAC), #74 (CGCGAAGCTTTTAGGGAGAATATAGTCGATATTCATC), #75 (CGCGAAGC TTTTACAACACAGTGATGGGTTTG), #90 (CGCGGATCCGGACCGAAGGTCCTAGCTCTAGAAGAAGATAAAGATCAGGGGAGAGGGGTTC), #C90 (CGCGGATCCGGACCAAGGATCCTAGCTCTAGAAGAAGATAAAGATCAG), #92 (GCGGT ACCATGGGACAGACAGTGACGACC), #93 (GCGGTACCTTAATCTTTATCT TCTTCTAGAGCTAGGAC), and #99 (CGCGGATCCTTGGTCCGGGTCCTAGCTCTAGAAGAAGATAAAGAT). LTR, long terminal repeat.
PERV C protease was expressed by first preparing a PERV C #36/#75 fragment by a heminested amplification strategy applying primers #36 and the PERV env C-specific reverse primer PL206 (14), which served as a template for the generation of additional PERV C-specific fragments with primers #36 and #C90 in combination with oligonucleotides #C74 and #75.
A direct (non-His-tagged) ATG/Met construct of the PERV A/B protease was made by switching the N-terminal amino acid, Ala, to Met (start codon) and switching the construct ends to the proposed C-terminal Leu. Fragments were generated with primer #72 in combination with the above reverse primers, digested with HindIII and NcoI, and ligated in two steps into pTrcHisA. This was necessary because the PERV protease gene contains an internal NcoI site and because NcoI is inevitably connected with ATG in common E. coli expression vectors.
In all constructs the gag TAG stop (X) codon was switched to CAG coding for Gln, as described for expression of the MMLV protease (8, 20). Recombinant PERV protease was produced in E. coli Top10 (Invitrogen); proteolytically active enzyme was in the supernatant. It was purified by using the cation-exchange column Fractogel EMB 650 S (Merck, Darmstadt, Germany) and was eluted by a linear gradient containing 1 M NaCl and was then concentrated by 50% saturated NH4SO4 precipitation. The pellet was dissolved and further purified by hydrophobic interaction chromatography (HIC) on a PhenylSuperose column (Pharmacia) eluted with a linear low-salt gradient. At this stage the protease was sufficiently pure to be tested for enzymatic activity (Fig. 3a).
FIG. 3.
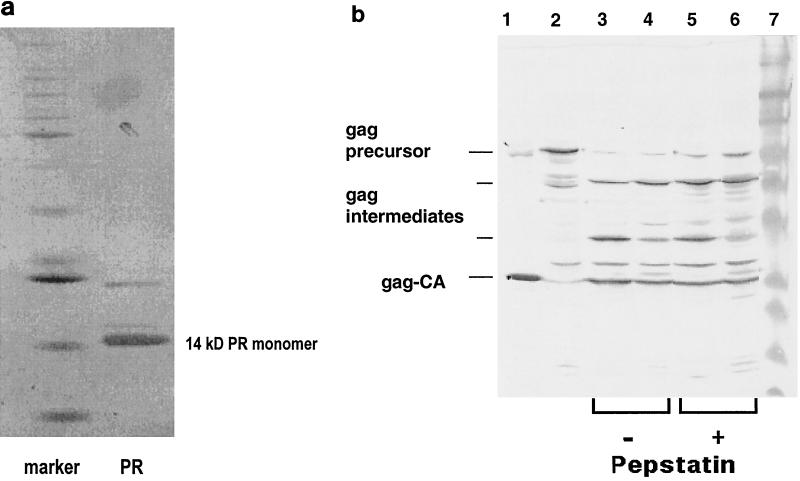
(a) Enzyme purification. After purification by cation and HIC column chromatography, the recombinant protease (PR) was analyzed by silver-stained SDS-PAGE. The marker lane contained commercial standard protein size markers (12 to 85 kDa). (b) Protease activity. Shown are the recombinant Gag precursor alone (lane 2) and its cleavage products after incubation at 37°C for 20 min with the recombinant PERV protease (lanes 3 to 6). The reaction mixture was separated by SDS-PAGE and was then Western blotted and reacted with antibody against PERV Gag (polyclonal rabbit anti-PERV GagCA serum [produced in collaboration with Charles River, Kisslegg, Germany] and a secondary goat anti-rabbit immunoglobulin G antiserum coupled to horseradish peroxidase [Dako, Hamburg, Germany]). Shown are cleavage of the recombinant Gag precursor by recombinant PERV protease subtype A/B (lane 3) and subtype C (lane 4) and cleavage in presence of 0.1 mM pepstatin A by PERV A/B (lane 5) or PERV C (lane 6). Purified PERV subtype A/B virus served as a marker (lane 1); lane 7 is a standard protein size marker.
All constructs indicated in Fig. 2, with the exception of the truncated #36/#37 and #99/#37, produced active enzyme (see below), however, at different rates and amounts. The highest yield of active enzyme was obtained with the protease construct of PERV subtype C, #C90/#C74. After purification of the protease, the bulk protein migrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at about 14 kDa (Fig. 3a) in agreement with the calculated Mr of a proposed protease monomer with 127 amino acid residues. This implied that, briefly after synthesis or even cotranslationally, the protease had been (auto)catalytically processed within the E. coli cells from its redundant overlapping (zymogen) sequences (i.e., #36, #90, #99, and #74 plus the vector fusion part) and had thus been enzymatically activated, presumably as a homodimer, as known from other retroviral proteases. Such activation cleavage appears to be, however, not a stringent prerequisite: a recombinant protease construct with an ATG start codon (Met), where no zymogen cleavage is required, was also enzymatically active (Fig. 2).
During control sequencing of the expression construct, the sequence directly upstream of the protease N terminus (i.e., translated GPKVL [Fig. 1]) in construct #C90/#C74 turned out to be altered to LVRTL, obviously due to an inversion of the PERV C-specific internal BamHI fragment in the three-fragment ligation strategy. Constructs containing the LVRTL sequence yielded a significantly higher production of recombinant protease than those containing the GPKV sequence. Both wild-type and inversion variant recombinant proteases were identical by N-terminal sequencing (see below). Hence, the LVRT construct was reconstructed by a new primer, #99, to obtain high-level expression of PERV A/B protease. It is not known why this inversion caused an increased expression and/or maturation rate of the protease. It is known, however, that sequences that do not encode proteases (p6*) and precede the N terminus of the HIV protease appear to be involved in the regulation of the expression and or maturation of the protease.
Sequencing of N and C termini of the PERV protease.
N-terminal amino acid sequencing of the purified recombinant protease confirmed our alignment prediction yielding the N-terminal ALEEDK for the protease of PERV subtypes A/B and C (Fig. 1). PERV protease thus contained one more Lys and two more Glu residues than did the MMLV protease N terminus.
Determination of the PERV protease C terminus was performed indirectly: a synthetic nonapeptide, TLQLPITVL, representing the 5 amino acid residues upstream and the 4 residues downstream of the predicted C-terminal cleavage site, L*P, was cleaved by the recombinant protease of all PERV subtypes into TLQL, confirming the predicted C-terminal Leu for the protease of PERV subtypes A/B and C (Fig. 1), and into PITVL, the presumable N terminus of the PERV reverse transcriptase. (The fragments had been N-terminally sequenced).
Protease activity and in vitro cleavage system.
The proteolytic activity of the purified recombinant enzyme was assayed using two different forms of the PERV Gag substrate: (i) a Gag precursor produced recombinantly in E. coli of PERV subtype A/B and (ii) synthetic nonapeptides representing the sequence of the cleavage sites of Gag MA*CA or CA*NC of PERV subtypes A/B and C.
(i) For the expression of the PERV Gag precursor protein, oligonucleotides #92 and #93 were used to amplify the entire gag sequence from pig genomic DNA and subcloned as a KpnI fragment in vector pTrcHisB (Invitrogen). Expression of recombinant Gag protein precursor was carried out in E. coli. The Gag precursor was in the supernatant; it was used without further purification.
The in vitro cleavage of this recombinant Gag precursor by recombinant PERV protease of all subtypes is shown in Fig. 3b: the PERV Gag precursor disappeared and three new bands appeared, the major one migrating exactly as the PERV GagCA protein from a virus preparation did. As this band reacted also with an anti-PERV Gag antiserum, we suppose that it is identical with the GagCA protein. This means that the recombinant proteases of subtypes A/B and C were proteolytically active toward the PERV Gag precursor protein. The other two visible fragments represent presumably partially cleaved intermediate forms of the Gag cleavage process as they react also with the Gag-specific antibody.
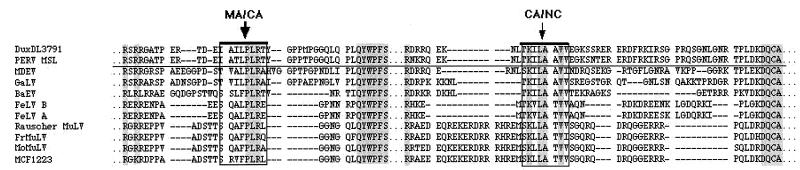
(ii) To confirm the PERV cleavage specificity, the protease was reacted with synthetic nonapeptide substrates representing the amino acid sequence around the Gag cleavage sites MA*CA and CA*NC. These cleavage sites had been derived by multiple sequence alignment (Fig. 4) with PERV subtypes A/B and C and various retroviruses genetically closely related with the pig retrovirus. The sequence of the MA*CA cleavage site had been experimentally confirmed by N-terminal sequencing of the PERV GagCA protein of a highly purified virus preparation from supernatant of the porcine PK15 cell line (11).
FIG. 4.
Gag precursor cleavage sites. Prediction of the sequence of cleavage sites between PERV Gag MA*CA and CA*NC proteins by multiple sequence alignment of PERV A/B and C and several other related retrovirus Gag sequences. Accession numbers are as follows: PERV DuxDL 3791 (AF147808), PERV MSL (AF038600), MMLV (AF033811 J02255 J02256 J02257 M76668); Fr-MuLV (X02794 J02192 M12528 M19209); GALV (M26927), Rauscher MuLV (U94692), and BaEV (D10032 D00088 N00088).
Nonapeptide I (EIAILPLRT) represents the cleavage site MA*CA of the PERV subtype A/B Gag precursor, nonapeptide II (EIATLPLRT) represents the same site of the PERV subtype C Gag precursor (both differ genetically in the 4th amino acid [T/I]), and nonapeptide III (LTKILAAVV) represents the amino acid sequence of the cleavage site of CA*NC of all PERV subtypes. The protease activity was assayed by measuring the amount of cleaved peptide fragments either by high-performance liquid chromatography or by fluorophotometric analysis. In the latter case Dansyl/Edans-labeled decapeptide IV (Dansyl-LVRTLALEED-Edans) was used as the substrate representing the (Gag)*protease cleavage site. All nonapeptides (I to IV) were cleaved by all PERV subtype proteases. The Km values for I and III were in the range of 0.1, comparable to those of other retroviral peptide substrates. Peptide II (EIATL*PLRT), representing the MA*CA site of PERV-C, had an eightfold-higher Km, implying lower cleavage and probably maturation rates of the subtype C virus. Interesting, in this context, may be the fact that PERV C has been shown to have replication disadvantages in human cells (18, 19).
As control an HIV-specific protease substrate (KARVLAEA) was tested. It was not cleaved by the PERV protease of either subgroup A/B or C (not shown).
The pH optimum for the proteolytic reaction using nonapeptide I substrate in different buffers was in the range of pH 6.0, which appears to be slightly higher than that of many other retroviral proteases. The optimum for the salt concentrations [NaCl] was about 2 M, which is comparable with those of other retroviral proteases (not shown).
Inhibition of the PERV protease.
The PERV protease was assayed in the presence of pepstatin A, as all known retroviral proteases are of the aspartic type and are (at least slightly) inhibited by this aspartic prototype inhibitor (17). Both proteases of PERV subtypes A/B and C were inhibited by about 40% at a concentration of 0.1 mM pepstatin A (Fig. 3b); this was also confirmed using the nonapeptide substrate assay (not shown).
PIs that inhibit specifically the HIV protease at nanomolar range are employed as chemotherapeutic drugs against HIV infection (saquinavir, ritornavir, indinavir, and nelfinavir). These drugs were also tested against the PERV protease. They had no inhibitory effect on the PERV protease, not even in the micromolar range. This is in agreement with recent data from reference 13 and is not surprising. PIs are specific for the structure of the active center, and differences in the structure between various retroviral proteases require distinct design for each individual retroviral protease, as has been done for HIV (4, 9, 17) and recently for the human endogenous retrovirus K10 (6).
The enzyme assay using fluorophotometric analysis established here is suitable as an inhibitor-screening assay for newly designed PERV protease-specific inhibitors, which might be an encouraging opportunity for antiviral chemotherapy against the risk of a PERV infection to human xenograph recipients. Those PIs should possibly be applied in combination with reverse transcriptase inhibitors, such as the existing reverse transcriptase substrate analogue, zidovudine. Reverse transcriptase inhibitors have a wide inhibitory spectrum, in contrast to PIs; zidovudine is employed against HIV reverse transcriptase and could be applied against other retroviral reverse transcriptases such as PERV reverse transcriptase (12, 13). Thus, a combination chemotherapy of existing reverse transcriptase inhibitors and novel PERV PIs might offer a most promising chance for antiviral protection against the potential risk of a human PERV infection.
Acknowledgments
We are grateful to the Bayrische Forschungsstiftung, Munich, Germany, for financial support.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, L. J. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus in miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czauderna, F., N. Fischer, K. Boller, R. Kurth, and R. R. Toenjes. 2000. Establishment and characterization of molecular clones of porcine retroviruses replicating on human cells. J. Virol. 74: 4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng, Y.-M., B. E. Tuch, and W. D. Rawlinson. 2000. Transmission of porcine endogenous retroviruses in severe combined immunodeficient mice xenotransplanted with fetal porcine pancreatic cells. Transplantation 70:1010-1016. [DOI] [PubMed] [Google Scholar]
- 4.Flexner, C. 1998. HIV protease inhibitors. N. Engl. J. Med. 338:1281-1292. [DOI] [PubMed] [Google Scholar]
- 5.Heneine, H., W. M. Switzer, M. Soucie, et al. 2001. Evidence of PERV in porcine factor VIII and evaluation of transmission to recipients with haemophilia. J. Infect. Dis. 183:648-652. [DOI] [PubMed] [Google Scholar]
- 6.Kuhelj, R., C. J. Rizzo, C.-H. Chang, P. K. Jadhav, E. Towler, and B. Korant. 2001. Inhibition of human endogenous retrovirus-K10 in cell-free and cell-based assays. J. Biol. Chem. 276:16674-16682. [DOI] [PubMed] [Google Scholar]
- 7.Martin, U., V. Kiessig, J. H. Blusch, A. Haverich, K. von der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692-694. [DOI] [PubMed] [Google Scholar]
- 8.Menendez-Arias, L., D. Gotte, and S. Oroszlan. 1993. Moloney murine leukemia virus protease: bacterial expression and characterization of the purified enzyme. Virology 196:557-563. [DOI] [PubMed] [Google Scholar]
- 9.Molla, A., and A. Japour. 1997. HIV protease inhibitors. Curr. Opin. Infect. Dis. 10:491-495. [Google Scholar]
- 10.Paradis, K., G. Langford, Z. Long, W. Heneine, P. Sandstrom, W. M. Switzer, L. E. Chapman, C. Lockey, D. Onions, E. Otto, and the XEN 111 Study Group. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. Science 285:1236-1241. [DOI] [PubMed] [Google Scholar]
- 11.Patience, C., Y. Takeuchi, and R. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 12.Powell, S. K., M. E. Gates, G. Langford, M. L. Gu, C. Lockey, Z. Long, and E. Otto. 2000. Antiretroviral agents inhibit infection of human cells by porcine endogenous retroviruses. Antimicrob. Agents Chemother. 44:3432-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quari, S. H., S. Magre, J. G. Garcia-Lerma, A. I. Hussein, Y. Takeuchi, C. Patience, R. Weiss, and W. Heneine. 2001. Susceptibility of the porcine endogenous retrovirus to reverse transcriptase and protease inhibitors. J. Virol. 75:1048-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. LeTissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retroviruses. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tönjes, R. 1999. Xenogene porcine endogene Retroviren: Genanalysen und Replicationsverhalten. Tx. Med. 11:211-215. [Google Scholar]
- 16.van der Laan, L. J. W., C. Lockey, B. C. Griffith, F. S. Frasier, C. A. Wilson, D. E. Onions, B. J. Hering, Z. Long, E. Otto, B. E. Torbett, and D. R. Salomon. 2000. Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature 407:90-94. [DOI] [PubMed] [Google Scholar]
- 17.von der Helm, K. 1996. Retroviral proteases: structure, function and inhibition from a non-anticipated viral enzyme to the target of a most promising HIV therapy. Biol. Chem. 377:765-774. [PubMed] [Google Scholar]
- 18.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson, C. A., S. Wong, M. VanBrocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshinaka, Y., I. Katoh, T. D. Copeland, and S. Oroszlan. 1985. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl. Acad. Sci. USA 82:1618-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshinaka, Y., I. Katoh, T. D. Copeland, and S. Oroszlan. 1985. Translational readthrough of an amber termination codon during synthesis of feline leukemia virus protease. J. Virol. 55:870-873. [DOI] [PMC free article] [PubMed] [Google Scholar]