Abstract
Human immunodeficiency virus type 1 uses ribosomal frameshifting for translation of the Gag-Pol polyprotein. Frameshift activities are thought to be tightly regulated. Analysis of gag p1 sequences from 270 plasma virions identified in 64% of the samples the occurrence of polymorphism that could lead to changes in thermodynamic stability of the stem-loop. Expression in Saccharomyces cerevisiae of p1-β-galactosidase fusion proteins from 10 representative natural stem-loop variants and three laboratory mutant constructs (predicted the thermodynamic stability [ΔG°] ranging from −23.0 to −4.3 kcal/mol) identified a reduction in frameshift activity of 13 to 67% compared with constructs with the wild-type stem-loop (ΔG°, −23.5 kcal/mol). Viruses carrying stem-loops associated with greater than 60% reductions in frameshift activity presented profound defects in viral replication. In contrast, viruses with stem-loop structures associated with 16 to 42% reductions in frameshift efficiency displayed no significant viral replication deficit.
In human immunodeficiency virus type 1 (HIV-1), the frameshift structure needed for translation of the Gag-Pol precursor polyprotein includes a slippery heptamer (UUUUUUA) region that allows ribosome repositioning in −1 frame and, seven nucleotides downstream, a 28-nucleotide stem-loop structure that putatively facilitates ribosomal pausing and shifting (11, 12, 18, 20). These elements are encoded by the conserved p1 region (1). HIV-1 polyproteins Gag and Gag-Pol are produced at estimated 9:1 to 19:1 stoichiometry ratios, depending on the in vitro or in vivo systems used to analyze the phenomenon (17, 18). Changes in frameshifting have the potential to dramatically alter viral assembly (8, 14, 17) and, thus, are thought to be tightly regulated (15). In the L-A double-stranded RNA virus of Saccharomyces cerevisiae, increasing or decreasing the frameshift efficiency more than twofold by alterations in the slippery site disrupts viral propagation (7).
The identification of frequent clinical viral isolates carrying variant frameshift structures by us and by others (4) led us to perform a detailed analysis of the functionality of polymorphic stem-loops and to explore the issue of the tight control of the frameshift frequency proposed as necessary for correct viral assembly. For this purpose, we determined the in vivo frameshifting activities of a series of natural and mutant variants of the stem-loop, as well as the phenotype (protein maturation, reverse transcriptase [RT] activity, infectivity, and replication) of recombinant viruses carrying variant stem-loops.
Circulating viruses frequently present stem-loop polymorphism.
The frequency of stem-loop variants was established in a series of p1 sequences obtained from plasma virions from 270 HIV-1-infected individuals (Table 1). The sequence obtained reflected the majority population in plasma. In 64% of the samples studied, we observed p1 polymorphism that could lead to changes in the thermodynamic stability of the stem-loop. The thermodynamic stability at 37°C of the stem-loop structures was calculated by using the mfold program (http://bioinfo.math.rpi.edu/∼mfold/rna/form1.cgi) (Table 1). The clinical characteristics of HIV-1 infection in individuals infected with stem-loop variants did not differ from those of individuals infected with wild-type viruses: viremia and CD4 count at initiation of antiretroviral therapy were (mean ± standard deviation) 4.23 ± 1.00 log RNA copies/ml and 218 ± 190 CD4 cells/μl for variant stem-loop strains versus 4.39 ± 1.12 log RNA copies/ml and 202 ± 194 CD4 cells/μl for the wild-type strains (P > 0.05). There was no relationship between predicted stem-loop thermodynamic stability and virological or immunological parameters.
TABLE 1.
Frequencies and estimated thermodynamic stability of variant stem-loops
| Straina | Stem-loop sequencec | Frequency (%) | ΔG° (kcal/mol at 37°C)c |
|---|---|---|---|
| NL4-3 | UCUGGCCUUCCCACAAGGGAAGGCCAGG | 98 (36.2) | −23.5 |
| Natural variants (wild type) | |||
| C15U/A26G/A33G | -U----------G------G-------- | 1 (0.4) | −24.3 |
| G41A | ---------------------------A | 1 (0.4) | −24.0 |
| A29G | ---------------G------------ | 1 (0.4) | −23.4 |
| U22G/C23U/A26C/G32A/A33C | --------GU--C-----AC-------- | 1 (0.4) | −23.1 |
| U21C/A26G | -------C----G--------------- | 1 (0.4) | −23.1 |
| A26C | ------------C--------------- | 1 (0.4) | −23.0 |
| A33Gd | -------------------G-------- | 66 (24.3) | −23.0 |
| C27G/A33G | -------------G-----G-------- | 1 (0.4) | −23.0 |
| A29G/A33G | ---------------G---G-------- | 2 (0.7) | −22.9 |
| C27G/A29G/A33G | -------------G-G---G-------- | 2 (0.7) | −22.9 |
| A39G | -------------------------G-- | 21 (4.1) | −22.8 |
| InsA29AGG/A33Gb | ----------------I--G-------- | 1 (0.4) | −22.6 |
| A33G/A39G | -------------------G-----G-- | 2 (0.7) | −22.3 |
| C25U | -----------U---------------- | 2 (0.7) | −21.3 |
| C15U | -U-------------------------- | 3 (1.1) | −21.1 |
| C25U/A26G/A33G | -----------UG------G-------- | 2 (0.7) | −21.1 |
| C25U/A26C | -----------UC--------------- | 7 (2.6) | −21.0 |
| C25U/A33G | -----------U-------G-------- | 3 (1.1) | −20.8 |
| C19U | -----U---------------------- | 4 (1.5) | −20.8 |
| C19U/C27G | -----U-------G-------------- | 1 (0.4) | −20.8 |
| C15A | -A-------------------------- | 2 (0.7) | −20.6 |
| C15U/A33G | -U----------------G-------- | 3 (1.1) | −20.6 |
| C25A/A26G/G30A | -----------AG---A----------- | 1 (0.4) | −20.4 |
| C24U/C27G/A28G/A29G | ----------U--GGG------------ | 1 (0.4) | −20.4 |
| C25A | -----------A---------------- | 4 (1.5) | −20.3 |
| C25A/A26G | -----------AG--------------- | 2 (0.7) | −20.3 |
| C25A/A29G | -----------A---G------------ | 1 (0.4) | −20.3 |
| C24U/C27A/A33G | ----------U--A-----G-------- | 1 (0.4) | −20.0 |
| A26C/G30A | ------------C---A----------- | 1 (0.4) | −19.9 |
| G30A | ----------------A----------- | 2 (0.7) | −19.9 |
| C15A/A39G | -A-----------------------G-- | 1 (0.4) | −19.9 |
| C25A/G30A/A33G | -----------A----A--G-------- | 1 (0.4) | −19.9 |
| A26G/G30C | ------------G---C----------- | 1 (0.4) | −19.8 |
| G30C | ----------------C----------- | 2 (0.7) | −19.8 |
| C25A/A26G/A33G | -----------AG------G-------- | 2 (0.7) | −19.8 |
| C25A/A33G | -----------A-------G-------- | 4 (1.5) | −19.8 |
| C15U/A33G/A39G | -U-----------------G-----G-- | 1 (0.4) | −19.8 |
| A34Cd | --------------------C------- | 3 (1.1) | −19.4 |
| U21C | -------C-------------------- | 1 (0.4) | −19.4 |
| U22A/A33G | --------A----------G-------- | 1 (0.4) | −19.1 |
| U22C | --------C------------------- | 1 (0.4) | −19.1 |
| U22C/C25U/A26G | --------C--UG--------------- | 1 (0.4) | −19.0 |
| C15U/C25A/G30A | -U---------A----A----------- | 1 (0.4) | −18.0 |
| C15A/C24U | -A--------U----------------- | 2 (0.7) | −17.6 |
| C15U/C25A/G30A/A33G | -U---------A----A--G-------- | 1 (0.4) | −17.5 |
| C15U/C25A/A26G/A33G | -U---------AG------G-------- | 7 (2.6) | −17.4 |
| C15U/C25A/A33G | -U---------A-------G-------- | 4 (1.5) | −17.4 |
| G17U | ---U------------------------ | 1 (0.4) | −17.0 |
| G32Ad | ------------------A--------- | 2 (0.7) | −16.7 |
| C25U/G32A | -----------U------A--------- | 1 (0.4) | −16.6 |
| C25U/G32A/A33C | -----------U------AC-------- | 1 (0.4) | −16.6 |
| G18C | ----C----------------------- | 2 (0.7) | −16.4 |
| Consensus | U-U---C--------A----AGGCC-G- | ||
| Laboratory mutants | |||
| mut2ad | -------AA------------------- | NAe | −17.6 |
| mut4ad | -------AAAA----------------- | NAe | −10.2 |
| mut6ad | -------AAAAAA--------------- | NAe | −4.3 |
In bold are the strains whose stem-loops were analyzed for frameshift activity.
Ins, insertion (AGG).
ΔG°, calculated thermodynamic stability.
Virions produced.
NA, not applicable.
p1 stem-loop variants selected to represent a range of predicted thermodynamic stabilities were introduced by site-directed mutagenesis (Quickchange; Stratagene, Basel, Switzerland) into the NL4-3 laboratory strain. In addition, three additional constructs, mut2a, mut4a, and mut6a, were created by replacement of two, four, and six stem-loop nucleotides in pNL4-3 with adenine by site-directed mutagenesis (Table 1). Recombinant viruses were obtained by HeLa cell transfection (Geneporter transfection reagent; Axon Labs, Baden, Switzerland).
Variant stem-loops from clinical isolates display reduced frameshift activity.
Construction of frameshift reporter vectors for in vivo expression in S. cerevisiae FY1679-28C (MATaura3-52 trp1 leu2-1 his3-200) (6) used the LacZ reporter vector pCDR1-p367lacZ (D. Sanglard, unpublished data). The different p1 variants were amplified by PCR with primers Y1 (5′AAAAGGTCGACTGGAAATGTGGNAAGGADGGACAC) and Y3 (5′ AACCTGAAGCTTTCTTCTGGTGGGGCTGTTGG) and cloned after the start codon of lacZ after digestion with SalI and HindIII so that the reporter was placed in −1 frame relative to the initiation codon. Minus one frame vectors have been previously used to demonstrate that frameshifting is performed in yeast in response to retroviral mRNA signals (15). An in-frame positive control was built by using primers Y1 and Y4 (5′AACCTGAAGCTTCTCTTCTGGTGGGGCTGTTGG). An out-of-frame negative control was built by insertion of an unrelated 241-bp fragment into pCDR1-p367lacZ. Transformation of S. cerevisiae was performed by the lithium acetate method (9). After transformation, colonies were grown on YNB medium (0.67% yeast nitrogen base, 2% glucose, 0.25% uracil, 1% histidine, 1% tryptophan, 2% agar) plates. Transformants were resuspended in 5 ml of YNB medium and grown to an optical density of 0.4 to 0.6. The expression levels of the various p1-LacZ fusion proteins were assessed by the β-galactosidase liquid assay. Reproducibility of results was ensured by simultaneous transformation of the same batch of competent S. cerevisiae FY1679-28C (to minimize intra-assay variation) and analysis in triplicate on pooled transformants for each clone (to minimize intertransformant variation). To assess interassay variation, experiments were performed thee times on separate days.
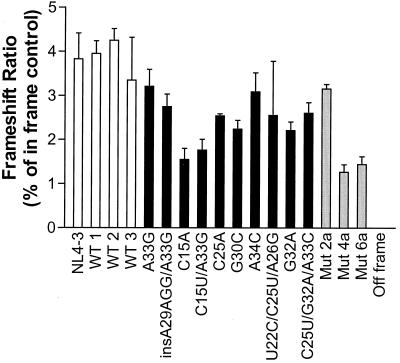
Frameshift analyses used the stem-loop of NL4-3, 13 stem-loops from clinical isolates (3 wild-type and 10 variant stem-loop sequences), and 3 mutant stem-loops derived from mutant clones (mut2a, mut4a, and mut6a). While cloning of the p1 regions from NL4-3 and three wild-type clinical isolates resulted in a mean (± the standard error of the mean [SEM]) frameshift activity in S. cerevisiae of 3.91% ± 0.23%, all variant stem-loops presented diminished expression of the p1-β-galactosidase fusion protein with frameshift ratios of 2.45% ± 0.17% (ranging from 1.55 to 3.21%; P < 0.001) (Fig. 1). These translate into an overall frameshift reduction of 18 to 60% in variant stem-loop constructs versus the wild type NL4-3 stem-loop. The stem-loop mutant mut2a (ΔG°, −17.6 kcal/mol) had a frameshift ratio of 3.17% ± 0.09%, while mut4a (ΔG°, −10.2 kcal/mol) and mut6a (ΔG°, −4.3 kcal/mol) displayed markedly diminished frameshift activities, 1.26% ± 0.17% and 1.38% ± 0.16%, compared to that of the wild-type NL4-3 stem-loop, 68 and 65% reductions, respectively. Deletion of the stem-loop has been reported to result in a 65 to 83% reduction in frameshifting in eukaryotic expression systems (S. cerevisiae, QT6, COS-7, and mouse NIH 3T3 cells) (2, 18). There was a correlation between the calculated thermodynamic stability of the stem-loop and the frameshift activity (r = −0.58, P = 0.01). The inter- and intra-assay coefficients of variation were 18.7 and 20.8%, respectively.
FIG. 1.
Frameshifting activity of the NL4-3 stem-loop and three wild-type stem-loop structures (white bars), 10 variant stem-loops (black bars), and three artificial stem-loop mutants (gray bars) as determined by a yeast frameshift reporter assay. Shown are the mean ± SEM of triplicate analyses from a representative experiment.
Recombinant clones with variant stem-loops display a range of infectivity, Pol incorporation into virions, and replication.
There are limited data on the behavior of molecular clones or of mutant HIV-1 with changes in the p1 region. Mutation of the slippage site resulting in overexpression of the Gag-Pol precursor results in a limited effect on overall viral gene expression, yet viral particle formation is inhibited (11, 17). Drastic mutation in the stem-loop, including deletion, has been shown to result in diminished synthesis of Gag-Pol upon transfection of QT6 or COS-7 cells (18), but no data on the phenotype of emerging viruses are available. To better understand the effect of variant stem-loops on the viral phenotype, we studied three natural variant p1 molecular clones and three mutants constructed by site-directed mutagenesis in pNL4-3. These were chosen to represent the spectrum of predicted free energy and in vitro frameshifting activities: stem-loop variants A33G (the most common variant stem-loop in clinical isolates; ΔG°, −23.0 kcal/mol; frameshift ratio, 3.20), A34C (ΔG°, −19.4 kcal/mol; frameshift ratio, 3.09), and G32A (ΔG°, −16.7 kcal/mol; frameshift ratio, 2.21) and mut2a (ΔG°, −17.6 kcal/mol; frameshift ratio, 3.17), mut4a (ΔG°, −10.2 kcal/mol; frameshift ratio, 1.26), and mut6a (ΔG°, −4.3 kcal/mol; frameshift ratio, 1.38).
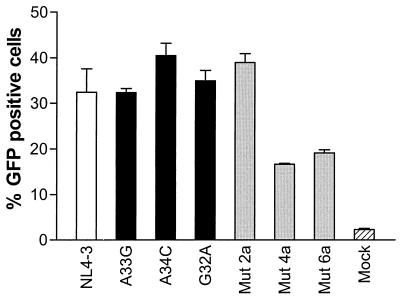
The single-cycle infectivity assay (3) used GHOST-CXCR4 cells obtained through the National Institutes of Health AIDS Research and Reference Reagent Program from D. Littman and V. K. Ramani. The infectious titer was determined by fluorescence-activated cell sorter analysis as the proportion of green fluorescent protein-positive cells. Viruses carrying natural variant stem-loops and mu2a viruses displayed no infectivity deficit compared to NL4-3: mean ± SEM percentage of infected cells of 36.7% ± 1.3% versus 32.4% ± 5.2%, respectively (P = 0.3; there was no significant difference for each of the one-to-one comparisons) (Fig. 2). In contrast, mut4a (16.6% ± 0.2% infected cells) and mut6a (19.1% ± 0.7% infected cells) presented impaired infectivity compared to that of wild-type NL4-3 (P = 0.02) (Fig. 2).
FIG. 2.
Single-cycle infectivity assay using GHOST-CXCR4 cells. Results for NL4-3 (white bar), molecular clones carrying 10 variant stem-loops (black bars), and three artificial stem-loop mutants (gray bars) are shown. Shown are the mean ± SEM of triplicate analysis. GFP, green fluorescent protein.
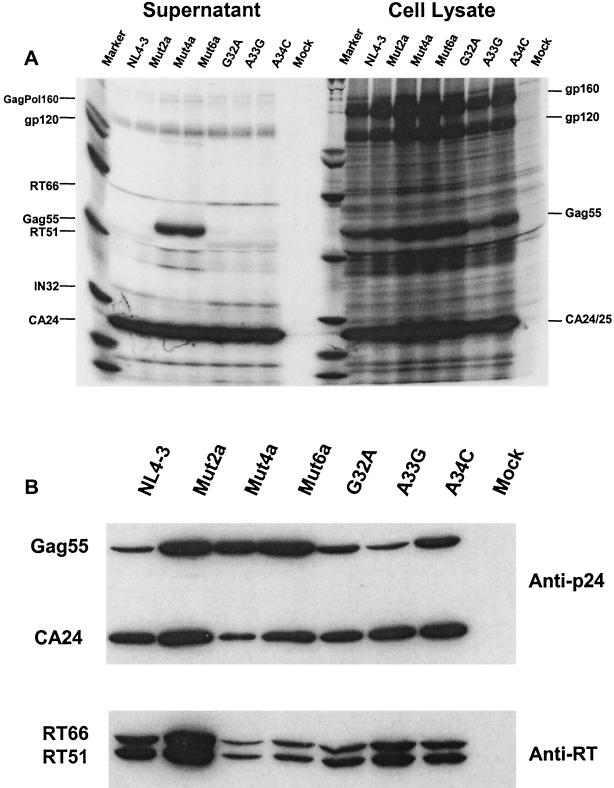
Analysis of particle-associated and cell-associated viral proteins was performed by transfecting COS-7 cells as previously described (3). On sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of immunoprecipitated proteins, mut4a and mut6a displayed a marked maturation impairment, as indicated by the presence in the virion of a large concentration of unprocessed Gag55, accumulation of a high-molecular-weight species (probably the p160 Gag-Pol precursor), and a markedly diminished content of the Pol products RT66, RT51, and integrase IN32 (Fig. 3A). Western blot analysis with a rabbit anti-p24 polyclonal antibody (Division of AIDS, National Institute of Allergy and Infectious Diseases, Rockville, Md.) diluted 1:50,000 in blocking solution (5% powdered milk, 0.05% Tween 20) and a mouse anti-RT monoclonal antibody (Intracell, Issaquah, Wash.) diluted 1:200 in blocking solution confirmed the impaired maturation of Gag and a profound deficit in RT incorporation for mut4a and mut6a: estimation of the RT-to-Gag (Gag55 plus p24) ratio indicated 73 and 77% reductions in the RT content of particles compared to that of NL4-3 (Fig. 3B). Western blot analysis also identified moderate maturation impairment and a reduction in the RT content of clones mut2a (33% reduction in RT content), G32A (46% reduction in RT content), A33G (25% reduction in RT content), and A34C (56% reduction in RT content) compared to that of NL4-3 (Fig. 3B).
FIG. 3.
Protein processing and maturation profiles in cell lysate and supernatant virions of NL4-3 and molecular clones carrying variant and mutant stem-loops. (A) For sodium dodecyl sulfate-polyacrylamide gel electrophoresis, COS-7 cells were transfected with recombinant clones and metabolically labeled with [35S]methionine-[35S]cysteine. Proteins were immunoprecipitated with anti-HIV human immunoglobulin G and analyzed by 5 to 15% gradient gel electrophoresis and fluorography. (B) For Western blotting, proteins from supernatant virions were reacted with anti-p24 polyclonal and anti-RT monoclonal antibodies. Shown is a representative blot from three separate experiments.
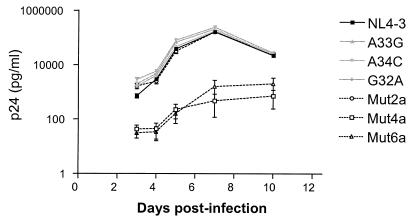
To assess the replication competence of the different molecular clones, peripheral blood lymphocytes (3 × 106 cells) were infected with recombinant virus (3,000 pg of p24 antigen) in 1 ml of supplemented RPMI 1640 medium for 3 h. Thereafter, the residual inoculum was removed by washing and cells were maintained in culture for 10 days at 106 cells/ml. Aliquots of culture supernatant were collected to monitor viral replication with an HIV-1 p24 antigen enzyme-linked immunosorbent assay (HIVAG-1 Monoclonal; Abbott) (3). The stability of the various mutations was confirmed by sequencing of the viruses at completion of the replication kinetics assay. Viruses carrying natural stem-loop variants and mutant mut2a replicated efficiently. In contrast, mut4a and mut6a showed severely impaired replication (Fig. 4). Thus, despite the diminished frameshift activities of the stem-loop variants and the mut2a clone and the deficit in Pol and Gag maturation observed by Western blotting, there is a conserved infectivity and replication efficiency of viruses. Possible explanations include (i) the production of sufficient numbers of correctly assembled and matured particles in culture (where, in any case, the majority of virions are noninfectious) and (ii) a progressive maturation of viral particles after budding that might allow recovery of infectivity over time (5, 13). The latter hypothesis was examined, and no postbudding maturation was observed (results not shown).
FIG. 4.
Replication kinetics of NL4-3 and molecular clones carrying variant and mutant stem-loops. Peripheral blood lymphocytes were infected with p24-normalized amounts of viral particles. Virus production was monitored by measuring the p24 antigen concentration in the culture supernatant. Bars represent the SEM of triplicate analysis.
We assume that the impairment of molecular clones mut4a and mut6a relates solely to the changes in stem-loop function and not to associated changes in the peptides encoded by the p1 region. There is no recognized function for the p1Gag peptide and limited information on the corresponding transframe peptide (16, 19). Most natural variants have amino acid polymorphism in either frame. The mutant clones presented S8T (mut2a), P7K and S8T (mut4a), and P7K and S8K (mut6a) substitutions in p1Gag and F8N (mut2a), F8K and P(L)9T (mut4a), and F8K and P(L)9K (mut6a) substitutions in the transframe peptide-p6Pol that would potentially alter the transframe octapeptide-p6Pol cleavage site. Processing of protease from Gag-Pol and optimal catalytic activity require native cleavage sites (16, 19), and it is conceivable that some of the processivity defects observed do reflect changes in protease activation rather than defects derived from changes in frameshift activity. Unfortunately, modification of the stem-loop by two, four, and six nucleotides to achieve progressive changes in stem-loop stability without altering one or both reading frames (p1Pol and p1Gag) was not feasible.
Thus, despite predicted changes in the thermodynamic stability of variant stem-loop structures, decreased frameshifting in reporter constructs, and impairment of Gag-Pol maturation, the phenotype of molecular clones carrying natural variants of the stem-loop is that of infectious viruses. This may reflect a tolerance in HIV-1 for a wider range of frameshifting rates than previously considered or the existence of adequate compensatory viral mechanisms (10). Constructs with more profound destabilization of the stem-loop and further reductions in frameshifting activity presented a disruption of viral propagation. Overall, the biological data in the present work are consistent with the frequent identification of polymorphic stem-loop variants in nature and the progressive nature of infection in humans carrying these variants.
Nucleotide sequence accession numbers.
Representative variant p1 nucleotide sequences have been submitted to the GenBank database and assigned accession numbers AF293420 to AF293427.
Acknowledgments
Support for this work was provided by the Swiss National Science Foundation (grants 3346-62092.99 and 33-61321.00) and by the Santos-Suarez Foundation.
REFERENCES
- 1.Bally, F., R. Martinez, S. Peters, P. Sudre, and A. Telenti. 2000. Polymorphism of HIV-1 Gag p7/p1 and p1/p6 cleavage sites. Clinical significance and implications for resistance to protease inhibitors. AIDS Res. Hum. Retrovir. 16:1209-1213. [DOI] [PubMed] [Google Scholar]
- 2.Bidou, L., G. Stahl, B. Grima, H. Liu, M. Cassan, and J.-P. Rousset. 1997. In vivo HIV-1 frameshift efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA 3:1153-1158. [PMC free article] [PubMed] [Google Scholar]
- 3.Bleiber, G., M. Munoz, A. Ciuffi, P. Meylan, and A. Telenti. 2001. Individual contribution of protease and reverse transcriptase to infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S.-Y., R. Sutthent, P. Auewarakul, C. Apichartpiyakul, M. Essex, and T.-H. Lee. 1999. Differential stability of the mRNA secondary structures in the frameshift site of various HIV type 1 viruses. AIDS Res. Hum. Retrovir. 15:1591-1596. [DOI] [PubMed] [Google Scholar]
- 5.Davis, D. A., K. Yusa, L. A. Gillim, F. M. Newcomb, H. Mitsuya, and R. Yarchoan. 1999. Conserved cysteines of the human immunodeficiency virus type 1 protease are involved in regulation of polyprotein processing and viral maturation of immature virions. J. Virol. 73:1156-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decottignies, A., A. M. Grant, J. W. Nichols, H. de Wet, D. B. McIntosh, and A. Goffeau. 1998. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273:12612-12622. [DOI] [PubMed] [Google Scholar]
- 7.Dinman, J. D., and R. B. Wickner. 1992. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66:3669-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, K. M., and S. P. Goff. 1988. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J. Virol. 62:2179-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda, A., T. Nakamura, and S. Nishimura. 1995. RNA signals for translation frameshift: influence of stem size and slippery sequence. Biochem. Biophys. Res. Commun. 213:575-582. [DOI] [PubMed] [Google Scholar]
- 11.Hung, M., P. Patel, S. Davis, and S. Green. 1998. Importance of ribosomal frameshift for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 72:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 13.Jardine, D. K., D. P. Tyssen, and C. J. Birch. 2000. Effect of protease inhibitors on HIV-1 maturation and infectivity. Antiviral Res. 45:59-68. [DOI] [PubMed] [Google Scholar]
- 14.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193:661-671. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. I., J. G. Umen, and H. E. Varmus. 1995. A genetic screen identifies cellular factors involved in retroviral −1 frameshifting. Proc. Natl. Acad. Sci. USA 92:6587-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louis, J. M., E. M. Wondrak, A. R. Kimmel, P. T. Wingfield, and N. T. Nashed. 1999. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 274:23437-23442. [DOI] [PubMed] [Google Scholar]
- 17.Park, J., and C. D. Morrow. 1991. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 65:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkin, N. T., M. Chamorro, and H. E. Varmus. 1992. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 66:5147-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulus, C., S. Hellebrand, U. Tessmer, H. Wolf, H.-G. Kräusslich, and R. Wagner. 1999. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. J. Biol. Chem. 274:21539-21543. [DOI] [PubMed] [Google Scholar]
- 20.Reil, H., H. Kollmus, U. H. Weidle, and H. Hauser. 1993. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J. Virol. 67:5579-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]