Abstract
In vitro studies have indicated that retroviral nucleocapsid (NC) protein facilitates both DNA synthesis by reverse transcriptase (RT) and annealing of the nascent DNA with acceptor template. Increasing the rate of DNA synthesis is expected to reduce the frequency of RT template switching, whereas annealing the nascent DNA with acceptor template promotes template switching. We performed a mutational analysis of the murine leukemia virus (MLV) NC zinc finger domain to study its effect on RT template switching in vivo and to explore the role of NC during reverse transcription. The effects of NC mutations on RT template switching were determined by using a previously described in vivo direct-repeat deletion assay. A trans-complementation assay was also developed in which replication-defective NC mutants were rescued by coexpression of replication-defective RT mutants that provided wild-type NC in trans. We found that mutations in the MLV NC zinc finger domain increased the frequency of template switching approximately twofold. When a predicted stem-loop RNA secondary structure was introduced into the template RNA, the template-switching frequency increased 5-fold for wild-type NC and further increased up to an additional 6-fold for NC zinc finger domain mutants, resulting in an overall increase of as much as 30-fold. Thus, wild-type NC increased the efficiency with which RT was able to reverse transcribe through regions of RNA secondary structure that might serve as RT pause sites. These results provide the first in vivo evidence that NC enhances the rate of DNA synthesis by RT in regions of the template possessing stable RNA secondary structure.
In order for retroviruses to synthesize a double-stranded viral DNA with complete long terminal repeats (LTRs) at both ends, two obligatory template-switching events from the ends of the retroviral genome—minus-strand DNA transfer and plus-strand DNA transfer—must take place during reverse transcription (9). Additionally, internal template switching occurs frequently due to low template affinity and low processivity of the viral reverse transcriptase (RT) (47). The additional template switches result in recombination, which is an important source for increasing retroviral genetic variation (32, 62). It was proposed that RT pausing during viral DNA synthesis triggers template switching from internal regions of RNA templates (5, 64). The RNase H activity of RT facilitates this event by degrading the RNA template as it is copied into DNA, so that the nascent DNA is released from the template and is then available to anneal onto the acceptor template (13, 17, 61).
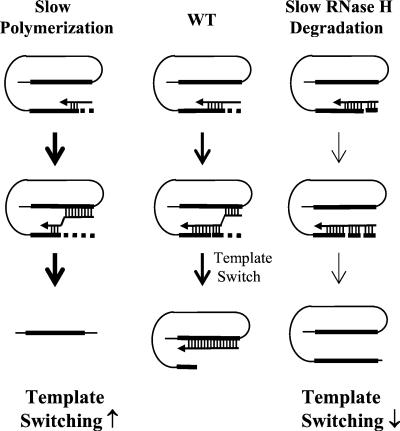
Retroviral vectors containing directly repeated sequences provide a powerful in vivo experimental system for elucidating the mechanism of RT template switching (47-49). The frequency of direct-repeat deletion appears to correlate with the size of the repeat and the distance between direct repeats (13, 47). We recently proposed the dynamic copy choice model for template switching, which suggests that a steady state between the rates of DNA polymerization and RNA template degradation by RNase H determines the amount of nascent DNA that is available for annealing to the acceptor RNA template and thus strongly influences the frequency of RT template switching (Fig. 1). Conditions that slow down the rate of DNA polymerization, such as mutations in the polymerase domain or depletion of intracellular nucleotide pools, result in a higher frequency of template switching by allowing more efficient degradation of the template RNA by RNase H and/or provide more time for annealing between the nascent DNA and the acceptor template. On the other hand, mutations in RNase H that reduce the ability of RNase H to degrade the template RNA result in a lower frequency of RT template switching, because less nascent DNA is available for annealing with the acceptor template. Several other factors could also influence template switching by affecting the balance between DNA synthesis and annealing between the nascent DNA and acceptor RNA. These factors include effective RT concentration, hydrogen bonding between the nascent DNA and the acceptor template, and the efficiency of reinitiation of DNA synthesis by RT on the acceptor template.
FIG. 1.
Dynamic copy choice model for RT template switching. The thick lines represent direct repeats in an RNA template. The horizontal arrows represent nascent DNA. The thick dashed lines represent RNA degraded by the RNase H activity of RT. Annealing between the RNA template and nascent DNA is designated by short vertical lines. Vertical arrows of various thicknesses indicate the relative efficiency of template switching. Annealing between the nascent DNA and acceptor template RNA (upper direct repeat) stimulates RT template switching. A higher frequency of template switching results in a higher rate of direct-repeat deletion, whereas a lower frequency of template switching results in a lower rate of direct-repeat deletion. Increased (↑) or decreased (↓) levels of template switching relative to wild-type RT are indicated.
The model predicts that the nature of the viral template will influence RT template switching. The secondary and tertiary structures in the template RNA or DNA, which might serve as RT pause sites, could slow down the rate of DNA synthesis and increase the frequency of template switching (34, 61). Another prediction of the model is that viral proteins other than RT might play a role in RT template switching if they directly or indirectly affect the process of reverse transcription. The nucleocapsid (NC) protein could be especially important in RT template switching because of its potential ability to interact with templates and nascent DNA, as well as its nucleic acid chaperone activity.
Retroviral NC protein is released from the C-terminal portion of the Gag precursor upon proteolytic processing during virion maturation. Murine leukemia virus (MLV) NC contains one zinc-binding motif (termed the zinc finger domain or CCHC motif) and numerous basic amino acids and is tightly associated with the genomic RNA dimer in the virion core (9). NC has a strong affinity for both DNA and RNA in vitro and exhibits both specific and nonspecific RNA-binding activity (3, 4, 20). Studies have indicated that the basic residues are important for nonspecific RNA binding and the zinc finger is indispensable for specific RNA recognition (3, 4, 8, 10, 24, 59). The basic residues and the zinc finger domains of NC both contribute to tight RNA binding in vitro (8, 12, 14, 60) and play a role in selecting and packaging viral genomic RNA (24, 54).
NC exhibits nucleic acid chaperone activity that catalyzes nucleic acid rearrangements to favor more thermodynamically stable conformations (11, 29, 63). It is able to transiently break base pairs in nucleic acid secondary structures and then reanneal the bases in alternative conformations to maximize the number of base pairs, which accelerates annealing of complementary sequences and directly contributes to viral RNA dimerization and maturation (55).
The chaperone activity of NC may contribute to the process of reverse transcription by several mechanisms. Human immunodeficiency virus type 1 (HIV-1) Gag polyprotein has been shown to be involved in tRNA primer unwinding and its placement on the primer-binding site (6, 19); other in vitro studies suggest that the NC domain of Gag is important for this activity (7, 33). HIV-1 NC also plays essential roles in suppression of nonspecific self-priming (18, 26, 44) and minus- and plus-strand DNA transfers (26, 27, 50). Furthermore, addition of HIV-1 NC to in vitro reverse transcription reactions reduces the total amounts of DNA synthesized, but the amounts of full-length DNA products relative to shorter products are increased in the presence of NC, especially when extensive RNA secondary structures are present in the template RNA (36, 42, 65). These in vitro studies have suggested that HIV-1 NC increases the processivity of DNA synthesis by reducing RT pausing at regions of stable secondary structures in the template RNA (36, 42). Studies have also shown that MLV NC contributes to the synthesis of full-length viral cDNA (21, 22, 31, 53) and stimulation of minus-strand DNA transfer (1, 53).
The role of HIV-1 NC in facilitating minus-strand DNA transfer is clearly demonstrated through in vitro assays. In vitro observations that RT pausing stimulates strand transfer and that HIV-1 NC reduces pausing but increases strand transfer seem to be a paradox. Based on previous in vitro studies, it has been postulated that NC might have both stimulatory and inhibitory effects on RT template switching (57). The ability of NC to reduce pausing should inhibit template switching (64, 65). On the other hand, NC might also promote template switching by stimulating base pairing of the nascent DNA with the homologous acceptor template upon degradation of the donor template by RNase H (15, 16, 50). In vitro studies using different assay conditions might have introduced some bias toward one or the other activity (46). In this report, we describe the first in vivo experiments to analyze the role of NC in RT template switching during reverse transcription, using templates with different lengths of homologous sequences and/or the presence of a predicted RNA secondary structure. Our results indicate that one of the in vivo roles of MLV NC is to enhance the rate of DNA synthesis, especially through regions of template containing RNA secondary structure.
MATERIALS AND METHODS
Definitions.
pES-GF250FP, pGF100FP, and pGFSLFP refer to plasmids, whereas vGF250FP, vGF100FP, and vGFSLFP denote the viruses derived from these plasmids. cGF250FP, cGF100FP, and cGFSLFP denote the cell lines containing a single copy of the corresponding proviruses and the MLV amphotropic envelope. cGF250FP(D150E), cGF100FP(D150E), and cGFSLFP(D150E) refer to pools of cells generated by transfection of the MLV RT mutant pD150E plasmid into corresponding cell lines.
Plasmids and retroviral vectors.
The construction of the MLV-based retroviral vector pES-GF250FP was previously described (61). The vector contains the selectable neomycin phosphotransferase gene (neo), which is expressed from an internal ribosomal entry site (IRES) of encephalomyocarditis virus and confers resistance to G418 (a neomycin analog) (35). The pES-GF250FP vector also contains overlapping fragments of the green fluorescent protein (GFP) gene that are 250 bp in length and are called the F portion.
A new pair of vectors named pGF100FP and pGFSLFP were constructed. The structures of the MLV-based retroviral vectors pGF100FP and pGFSLFP are similar to that of pES-GF250FP. However, the length of the F portion of GFP in vectors pGF100FP and pGFSLFP is 100 bp. In addition to the 100-bp repeated F portion, pGFSLFP contains a 41-bp sequence between the overlapping fragments that is predicted to form a stem-loop structure. The sequences of the GF and FP fragments and the intervening regions present were confirmed by PCR amplification and DNA sequencing of both strands (data not shown).
Plasmid pLGPS expresses MLV gag and pol from a truncated MLV LTR promoter (45). Plasmid pD150E, derived from pLGPS, expresses MLV gag-pol containing a replacement of one of the catalytic-site aspartic acid residues with a glutamic acid, which renders it replication defective (28). Plasmid pSV-A-MLV-env expresses the amphotropic MLV envelope gene from the LTR promoter and simian virus 40 enhancer (43). Plasmid pSVα3.6 encodes the α subunit of the murine Na+/K+ ATPase gene and confers resistance to ouabain (40). Plasmid pSVhygro encodes the hygro gene and thus confers resistance to hygromycin (25).
Mutagenesis and plasmid construction.
To facilitate cloning, a silent mutation was introduced into the MLV gag-pol construct pLGPS by site-directed mutagenesis to create a unique ClaI site. The resulting gag-pol expression construct was named pWZH30. The mutations in NC were introduced into pWZH30 by PCR-based site-directed mutagenesis (30) or by the QuickChange mutagenesis kit (Stratagene). Most of the mutagenic oligonucleotide primers were designed to introduce additional silent mutations and generate new restriction sites. Restriction digestion analysis was performed to identify plasmids containing mutations. Detailed descriptions of the mutagenic oligonucleotides and the strategies used to generate individual mutants are available upon request. The PflMI-ClaI fragment containing mutations in NC was subcloned back into pWZH30. The inserted 577-bp fragment was analyzed by DNA sequencing to verify the presence of the desired mutations and the absence of any undesired mutations.
Cell culture, DNA transfection, and virus infection.
D17 is a dog osteosarcoma cell line permissive to infection by MLV. All D17 cells and D17-derived cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 6% calf serum and maintained in a 37°C incubator with 5% CO2. The cells were transfected and selected for resistance to ouabain, hygromycin, or G418 as previously described (37, 38). In addition, 3′-azido-3′-deoxythymidine (AZT) (1 μM final concentration) was added to all transfected cells to reduce the probability of reinfection of the virus-producing cells (61).
A3 is a single-cell clone that was generated by stably transfecting D17 cells with pSV-A-MLV-env, which expresses the MLV envelope (61). A3 cells were maintained, transfected, and selected for drug resistance in a manner similar to that described above for D17 cells. PG13 is a murine helper cell line expressing MLV gag-pol and the gibbon ape leukemia virus envelope (45). The absence of a gibbon ape leukemia virus receptor on murine cells prevents reinfection of the PG13 helper cells. PG13 cells were grown in DMEM supplemented with 10% calf serum and maintained in a 37°C incubator with 5% CO2. PG13 cells were transfected and selected for resistance to G418 as previously described (2).
Hydroxyurea (HU) treatment was performed as previously described (61). Briefly, D17 cells were treated with DMEM containing 1 mM HU 4 h prior to infection, 4 h during infection, and 24 h postinfection. The cells were then placed under selection with G418 and analyzed by flow cytometry.
Construction of cell lines cGF100Fp and cGFSLFP.
Cell lines cGF100FP and cGFSLFP were constructed in a manner similar to cGF250FP. First, pGF100FP or pGFSLFP was transfected into PG13 cells. The transfected cells were subjected to G418 selection, and the drug-resistant colonies were pooled and expanded. Virus was harvested from the transfected PG13 cells and used to infect A3 cells, which express the MLV amphotropic envelope. After G418 selection was complete, individual drug-resistant colonies that did not exhibit detectable fluorescence were isolated and expanded. One of the GF100FP cell clones, named cGF100FP and used in subsequent studies, was able to produce high viral titers upon transfection with wild-type MLV gag-pol construct. Similarly, a cell clone expressing vGFSLFP, named cGFSLFP, was identified and used in subsequent studies. Genomic DNA was isolated from cell lines cGF100FP and cGFSLFP, and the proviral structures were analyzed by PCR and Southern blot hybridization using standard procedures (58). The Southern analysis verified that only one provirus was integrated in each of the cell lines (data not shown).
Protocol for determination of in vivo direct-repeat deletion frequency.
cGF250FP, cGF100FP, or cGFSLFP cells were plated at a density of 2 × 105 per 60-mm-diameter dish and 24 h later cotransfected with wild-type or mutant MLV gag-pol constructs and pSVhygro. The transfected cells were maintained in the presence of 1 μM AZT and selected for resistance to hygromycin; the resistant colonies were pooled and expanded. Before virus was collected, the culture medium containing AZT was removed and the cells were plated at a density of 5 × 106 per 100-mm-diameter dish. Twenty-four hours later, 7 ml of fresh medium was added to each dish; the culture medium containing vGF250FP, vGF100FP, or vGFSLFP virus was harvested the next day and used to infect D17 target cells. Flow cytometry analysis was performed to ensure that GFP expression was not detected in the virus-producing cells. The infected D17 target cells were subjected to G418 selection; the numbers of G418-resistant colonies were used to determine viral titers. Subsequently, approximately 500 to 5,000 G418-resistant colonies were pooled and analyzed by flow cytometry to obtain the percentage of cells that expressed GFP.
In vivo trans-complementation assay.
A gag-pol expression plasmid containing the replication-defective D150E RT mutation was introduced into cGF250FP, cGF100FP, or cGFSLFP cells by cotransfection with pSVα3.6. Pools of ouabain-resistant colonies (>2,000 colonies) were used to generate cGF250FP(D150E), cGF100FP(D150E), and cGFSLFP(D150E) cells, respectively. The replication-defective NC mutants were then cotransfected into cGF250FP(D150E), cGF100FP(D150E), and cGFSLFP(D150E) cells with pSVhygro and selected for resistance to hygromycin. The assays for determination of in vivo direct-repeat deletion frequency described above were subsequently performed.
Flow cytometry analysis.
To quantify the numbers of cells expressing GFP, approximately 500 to 5,000 drug-resistant colonies were pooled and subjected to flow cytometry analyses using a FACScan flow cytometer and CellQuest software (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). Some of the flow cytometry analyses were performed by the Clinical Support Laboratory, Clinical Services Program, Science Applications International Corporation Frederick.
Statistical analysis.
The statistical analysis consisted of pairwise comparisons of means among the wild-type group, which served as a control, and the various NC mutants (41). Two different strategies were used for the statistical-significance analysis. First, two-tailed two-sample independent t tests were used. Furthermore, Bonferroni adjustment was used to set the criterion for the statistical significance for each comparison at 0.05/N. Thus, the probability of any type I error is maintained at 0.05 across all comparisons. In this study, for each comparison between the mutant and the wild type, the P value for statistical significance was set at 0.05/5 (0.01) for Table 1 and 0.05/10 (0.005) for Table 2. Therefore, a P value of <0.01 for Table 1 and a P value of <0.005 for Table 2 were considered statistically significant.
TABLE 1.
Effects of lysine residue substitutions in zinc finger domain of MLV NC on frequency of direct-repeat deletion
| MLV NC phenotype | No. of expts | Titer (103 CFU/ml) (mean ± SEM)a | Frequency (%) of direct-repeat deletions (mean ± SEM)b | Relative change in direct-repeat deletion frequencyc |
|---|---|---|---|---|
| Wild type | 8 | 49.4 ± 8.5 | 10.8 ± 0.3 | 1.0 |
| Single-substitution mutants | ||||
| K30I | 2 | 0.7 ± 0.4 | 21.5 ± 0.4 | 2.0 |
| K30A | 2 | 8.4 ± 2.8 | 11.5 ± 0.3 | No changed |
| K32A | 2 | 51.5 ± 9.5 | 11.6 ± 1.9 | No change |
| K37Q | 2 | 5.9 ± 0.2 | 17.1 ± 0.3 | 1.6 |
| K37L | 2 | 29.1 ± 13.7 | 11.2 ± 0.7 | No change |
| Multiple-substitution mutants | ||||
| WZH43 | 2 | <0.1e | NDf | ND |
| WZH46 | 2 | <0.1 | ND | ND |
The viral titer for each experimental group was determined by serial dilutions and infections. The standard error of the mean (SEM) was determined by using Sigma Plot 5.0.
Frequency of direct-repeat deletion was determined as the percentage of infected D17 target cells that exhibited fluorescence after G418 selection compared with the negative control.
Calculated as follows: frequency of direct-repeat deletion observed with mutant MLV NC/frequency of direct-repeat deletion observed with wild-type NC. Statistically significant changes in the frequency of direct-repeat deletions, relative to the wild-type NC (set to 1.0), are shown (t test; P < 0.01).
The relative change in frequency of template switching was not statistically different from that of wild-type NC (t test; P ≥ 0.01).
Viral titers are 4 ± 3 and 22 ± 3 CFU/ml for WZH43 and WZH46, respectively.
ND, not determined. The frequency of direct-repeat deletions could not be determined due to insufficient numbers (<100) of colonies available after G418 selection for these mutants.
TABLE 2.
trans-complementation of zinc finger mutants in MLV NC and effects on frequency of direct-repeat deletion
| MLV NC | No. of expts | Titer (103 CFU/ml) (mean ± SE)a | Frequency (%) of direct-repeat deletions (mean ± SE)b | Relative change in direct-repeat deletion frequencyc |
|---|---|---|---|---|
| Wild typed | 8 | 49.4 ± 8.5 | 10.8 ± 0.3 | No change |
| Wild type + D150E | 4 | 31.1 ± 16.0 | 11.0 ± 1.0 | 1.0 |
| WZH43 + D150E | 2 | 2.7 ± 0.4 | 22.0 ± 0.1 | 2.1 |
| WZH46 + D150E | 2 | 1.4 ± 0.1 | 22.1 ± 1.1 | 2.0 |
| W35S + D150E | 2 | 1.7 ± 0.6 | 19.1 ± 1.5 | 1.7 |
| H34C + D150E | 2 | 1.5 ± 1.1 | 21.4 ± 3.4 | 1.9 |
| C39H + D150E | 2 | 1.2 ± 0.2 | 19.5 ± 2.1 | 1.8 |
| C26S + D150E | 2 | 1.4 ± 0.3 | 17.8 ± 0.8 | 1.6 |
| C29S + D150E | 2 | 5.0 ± 0.1 | 16.8 ± 2.8 | 1.5 |
| C26S-C29S + D150E | 2 | 1.3 ± 0.1 | 19.9 ± 1.4 | 1.8 |
| C39S + D150E | 2 | 1.5 ± 0.5 | 20.1 ± 3.1 | 1.8 |
| SSHS + D150E | 2 | 1.9 ± 0.7 | 19.8 ± 0.5 | 1.8 |
See Table 1, footnote a.
See Table 1, footnote b.
Calculated as follows: frequency of direct-repeat deletion observed with mutant MLV NC complemented with D150E in cGF250FP(D150E)/frequency of direct-repeat deletion observed with wild-type NC in the presence of D150E in cGF250FP(D150E). Statistically significant changes in the frequency of direct-repeat deletions, relative to wild-type NC with D150E (set to 1.0), are shown (t test; P < 0.005).
Experiments were performed by using the standard in vivo direct-repeat deletion assay as a control.
Second, the Dunnett test with a built-in control for the accumulation of type I error was used to confirm the t test results. This test permits comparisons among any number of experimental groups and a single control group (wild type) while maintaining the overall rate of type I error at a level of <0.05.
RESULTS
Experimental protocol.
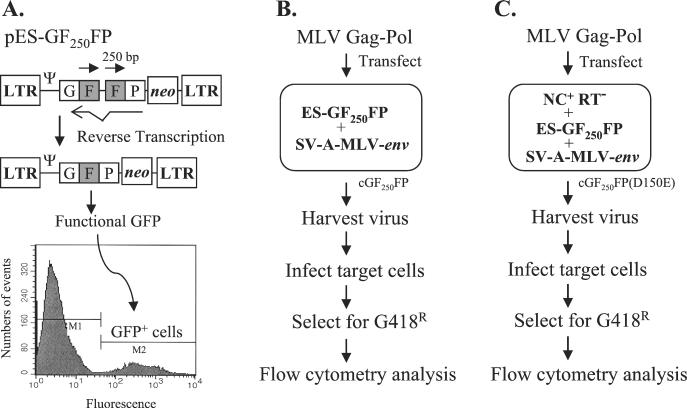
An in vivo RT template-switching assay was previously developed to identify structural determinants of RT that play a role in template switching (61). The MLV-based retroviral vector pES-GF250FP used in this study is shown in Fig. 2A. In this vector, the GFP gene was divided into GF and FP fragments with an overlapping 250-bp F portion. The pES-GF250FP vector did not express a functional GFP gene product (data not shown). During reverse transcription, one of the F portions is deleted upon a template-switching event, resulting in functional reconstitution of the GFP gene. The infected cells containing the reconstituted GFP gene are fluorescent and can be detected by flow cytometry analysis. The stable cell line cGF250FP was constructed in a manner similar to the construction of the previously used cell line B2-1 GFFP (61). Briefly, cGF250FP cells express the amphotropic MLV envelope gene from plasmid pSV-A-MLV-env and contain an integrated copy of the ES-GF250FP provirus (data not shown). Flow cytometry analysis indicated that <0.1% of the cGF250FP cells were fluorescent (data not shown). The experimental protocol used to determine the effects of mutations in NC on template switching during reverse transcription is outlined in Fig. 2B. First, the pLGPS-derived MLV gag-pol constructs containing mutations in MLV NC were transfected into cGF250FP cells. Virus produced from the transfected cells was then used to infect D17 target cells. After G418 selection, drug-resistant colonies were pooled (500 to 5,000 colonies) from six 60-mm-diameter dishes and analyzed by flow cytometry. The proportion of fluorescent cells represents the frequency of direct-repeat deletion resulting from template switching. Viral titers were determined by quantitation of the drug-resistant colonies.
FIG. 2.
Protocol for analyzing the effects of NC mutations on RT template switching in vivo. (A) Structure of MLV-based vector pES-GF250FP, structure of provirus after direct-repeat deletion, and flow cytometry analysis. pES-GF250FP contains LTRs and all of the cis-acting elements of MLV. The GFFP and neo genes are transcribed from the LTR promoter. The IRES of encephalomyocarditis virus is used to express neo. The directly repeated F portions of GFP are shaded and indicated by right-facing arrows. During reverse transcription, one of the repeated F portions may be deleted to reconstitute a functional GFP. The cells containing a functional GFP gene are fluorescent and can be detected by flow cytometry. A typical graph obtained from flow cytometry is shown after one round of viral replication using wild-type MLV gag-pol. The y axis is the number of events scored, which is interpreted as the number of cells, and the x axis is the intensity of the fluorescence. The cell population that does not express GFP is gated as M1, whereas the cell population that expresses GFP is gated as M2 (GFP+ cells). In this plot, M1 is 89.2% and the M2 is 10.8%. Ψ, MLV packaging signal. (B) Protocol for in vivo direct-repeat deletion assay. cGF250FP is a D17-based cell line expressing pES-GF250FP and pSV-A-MLV-env. Wild-type or mutated MLV gag-pol constructs with NC mutations were separately cotransfected with pSVhygro into the cGF250FP cells, and the virus produced was harvested and used to infect D17 cells. After G418 selection, the infected cell clones resistant to G418 were analyzed by flow cytometry, and the frequencies of direct-repeat deletion were determined. (C) Protocol for in vivo trans-complementation assay. The RT mutant D150E was transfected into cGF250FP cells to construct the cell line cGF250FP(D150E). The D150E mutant expresses wild-type NC and defective RT. The wild-type or mutated MLV gag-pol constructs with NC mutations were subsequently transfected into the cGF250FP(D150E) cells, and the virus produced was harvested and used to infect D17 cells. After G418 selection, the infected cell clones resistant to G418 were analyzed by flow cytometry, and the frequencies of direct-repeat deletion were determined.
For NC mutants that were severely defective in viral replication (>10,000-fold reduction in titer), an in vivo trans-complementation assay was developed (Fig. 2C). The general protocol was similar to that described above, except that the cell line used [cGF250FP(D150E)] was generated by stably transfecting the RT mutant D150E, which expresses MLV gag-pol, in the cGF250FP cells. The gag gene from the D150E mutant expressed wild-type NC. However, the substitution mutation D150E in the RT rendered the virus replication defective (28). The replication-defective NC mutants were transfected into cGF250FP(D150E) cells, and the effects of these mutations on RT template switching were determined by infecting target cells, selecting the infected cells for G418 resistance, and performing flow cytometry analysis of the G418-resistant cells.
In this system, viral titers and frequencies of template switching were examined during a single round of the viral replication cycle. The multiplicity of infection for the experiments analyzed by flow cytometry was <0.001; therefore, the probability of double infection was very low. The frequency of direct-repeat deletion provided a measure of the template-switching events.
Substitutions of basic residues and CCHC motif residues in the MLV NC zinc finger domain.
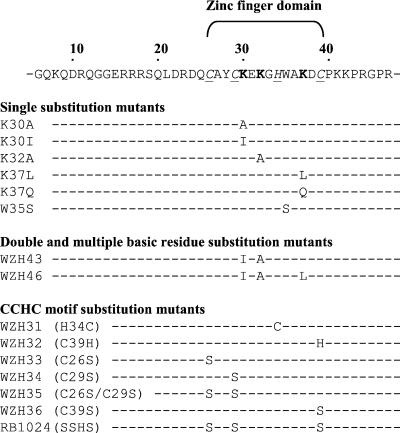
To determine the role of the MLV NC zinc finger domain in RT template switching, we introduced single amino acid substitutions in the zinc finger domain (Fig. 3). The first set of substitution mutations was designed to change positively charged residues in the zinc finger domain to neutral amino acids, because several studies have indicated that the basic residues in NC proteins play an important role in RNA binding (8, 10, 12). The second set of mutations was designed to disrupt the structure of the zinc finger motif, because multiple lines of evidence indicate that this structure is important for various functions of NC (22, 23, 27). The zinc finger domain of MLV NC contains three lysine residues. First, each of the three lysine residues was replaced with a neutral amino acid to generate mutants K30A, K30I, K32A, K37L, and K37Q. In addition, we also constructed one double mutant named WZH43 (K30I-K32A) and one triple mutant named WZH46 (K30I-K32A-K37L) in which two or all three of the lysine residues were simultaneously replaced with neutral residues. Finally, the conserved aromatic amino acid in the zinc finger domain, W35, was replaced with a serine residue to generate the W35S mutant (Fig. 3).
FIG. 3.
Mutations in zinc finger domain of MLV NC. The MLV NC primary sequence (amino acids 6 to 51) containing the zinc finger domain is shown. The numbers above the primary sequence indicate the amino acid positions. The zinc finger domain is bracketed, CCHC residues are in italics and underlined, and the basic residues in the zinc finger domain are in boldface. The substitution mutations analyzed at each amino acid position are indicated below the primary sequence. The names of the corresponding plasmid constructs are shown on the left.
A series of mutants containing substitutions for the retroviral NC CCHC motif residues were used to investigate the role of this zinc-binding motif in RT template switching. These mutants were previously generated and described (21, 24) and were subcloned into the MLV gag-pol expression construct for this study (Fig. 3). Mutants WZH31 (H34C) and WZH32 (C39H) contained the altered zinc finger motifs CCCC and CCHH, respectively, and retained the ability to coordinate the zinc ion. Mutants WZH33 (C26S), WZH34 (C29S), WZH35 (C26S-C29S), and WZH36 (C39S) contained replacements of cysteines with serines, which presumably disrupted their ability to tightly bind the zinc ion. In addition, mutant RB1024 (SSHS) was generated. The RB1024 mutant contained replacements of all three cysteines with serines, which were expected to prevent inter- and intramolecular cross-linking of the NC proteins by disulfide bond formation.
Effects of basic residue substitutions in the MLV NC zinc finger motif on template switching.
The viral titers and frequencies of template switching were determined for MLV NC mutants in which each of the lysine residues in the zinc finger motif was replaced with a neutral residue. In this set of experiments, the pES-GF250FP vector was used, and the frequency of direct-repeat deletion and GFP reconstitution for the wild-type NC was 10.8% (Table 1). Each NC mutant was analyzed in two independent experiments so that the statistical significance of the results could be determined. The single-residue substitution mutations had various effects on the viral titers. The K30I mutant had the most significant reduction in viral titer (70-fold). The K30A and K37Q mutations resulted in approximately 5- to 10-fold reductions in viral titers. The K32A and K37L mutations had no significant effect on viral titers (t test; P > 0.05). Only the K30I and K37Q mutants displayed statistically significant increases in the RT template-switching frequencies of 2.0- and 1.6-fold, respectively. Neutral amino acid substitutions at the same lysine residues had different effects on the RT template-switching frequency, since the K30I mutation increased the template-switching frequency but the K30A mutation had no effect on template switching. This result suggested that the structure of the amino acid side chain at this position had a more significant effect on the viral titer and RT template switching than the change in charge of the side chain.
To further investigate the roles of the positively charged residues of the NC zinc finger domain in template switching, we constructed a double and a triple mutant in which two or all three of the basic residues in the zinc finger domain were replaced by neutral amino acids (Fig. 3). These two mutants were severely defective in viral replication and exhibited viral titers that were 1,000- to 10,000-fold lower than the wild-type titer (Table 1). Therefore, these mutants were not able to produce sufficient numbers of colonies for analysis of the frequency of template switching.
To determine the effects of NC mutants that were severely defective in viral replication on template switching, we used the in vivo trans-complementation assay, which was similar to an assay previously used to analyze complementation between polymerase and RNase H domain mutants of RT (34). In this assay (Fig. 2C), the D150E mutant expressed a wild-type NC but was defective in RT activity, whereas the NC mutants expressed a defective NC and a functional RT. Assuming that the Gag-Pol proteins from the D150E mutant and the NC mutants could coassemble, we expected that the virion produced would contain a functional RT. In addition, the virion would contain a mixture of wild-type and mutant NC proteins. Therefore, complementation of these two gag-pol mutants (NC− RT+ and NC+ RT−) resulted in virions that could complete one cycle of viral replication. In this manner, we could rescue the viral titers of replication-defective NC mutants and determine whether the presence of the defective NC proteins affected the frequency of direct-repeat deletion. It was conceivable that the conditions of the trans-complementation assay affected the frequency of RT template switching. However, the rates of template switching for wild-type NC were not different in the trans-complementation assay and the standard assay (Table 2), suggesting that the conditions of the trans-complementation assay did not cause an increase in RT template switching in this system.
The mutants that exhibited 1,000- to 10,000-fold reduction in viral titer were rescued in the trans-complementation assay (Table 2). After trans-complementation, the titers were 1,000 to 2,000 CFU/ml. The frequencies of direct-repeat deletions of vGF250FP after trans-complementation were 22.0% for WZH43 (K30I-K32A) and 22.1% for WZH46 (K30I-K32A-K37L), which was similar to the frequency for K30I alone (21.5%), suggesting there was no correlation between the degree of positive charge in the zinc finger domain and the frequency of template switching.
trans-complementation of zinc finger CCHC motif mutants and frequency of template switching.
Because most previously characterized zinc finger domain mutants have been shown to be noninfectious, we used the trans-complementation assay to determine the effects of zinc finger domain CCHC motif mutations on RT template switching (Table 2). The zinc finger domain mutants with substitutions in the CCHC motif could be rescued in trans; the titers after trans-complementation were 1,000 to 5,000 CFU/ml. Mutants H34C and C39H showed 1.9- and 1.8-fold increases in the frequency of template switching, respectively. The other five mutants contained substitutions in which one (C26S, C29S, or C39S), two (C26S-C29S), or all three (SSHS) cysteine residues were replaced with serines; this set of mutations severely disrupted the zinc finger motif conformation. All of these mutants were rescued in trans and displayed increased frequencies of template switching, ranging from 1.5- to 1.8-fold higher than that of the wild type. Thus, all NC zinc finger domain mutants exhibited statistically significant increases in the frequency of RT template switching.
Effects of mutations in the zinc finger domain on RT template switching in regions of template containing RNA secondary structure.
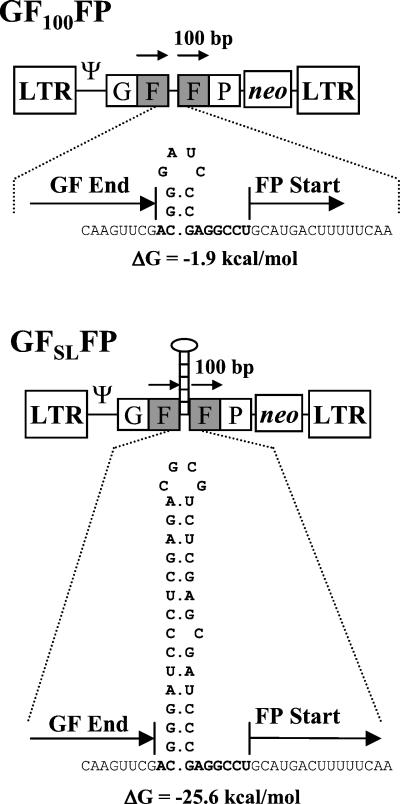
Mutations in the zinc finger domain of MLV NC resulted in modest but statistically significant increases in RT template switching for vGF250FP. We hypothesized that the effects of the NC mutations on RT template switching might be dependent on the extent of template RNA secondary structure. To study the role of NC during reverse transcription of template regions containing RNA secondary structure, we used a pair of retroviral vectors, named pGF100FP and pGFSLFP, with 100-bp direct repeats of the GFP gene (Fig. 4). The vector pGFSLFP was very similar to pGF100FP; however, a predicted stem-loop structure (41 bp; ΔG = −25.6 kcal/mol) was present in pGFSLFP in the noncoding sequence between the two F portions. Cell lines cGF100FP and cGFSLFP, containing one provirus from each vector, were constructed to study the effects of mutations in NC on the frequency of template switching.
FIG. 4.
Structure of MLV-based vectors pGF100FP and pGFSLFP, which contain LTRs and all of the cis-acting elements of MLV. The GF and FP fragments, as well as neo, are transcribed from the LTR promoter. The IRES of encephalomyocarditis virus is used to express neo. The directly repeated F portions of GFP are shaded and indicated by overhead arrows, which are 100 bp in length. GF End and FP Start mark the exact sequence where the first F portion ends and the second F portion starts. The intervening sequence between the two F portions is shown below each vector. The stem-loop structure is predicted by RNAstructure (version 2.5) software. The total RNA secondary-structure energy value of the sequence shown in boldface for vGF100FP is −1.9 kcal/mol, whereas the secondary-structure energy value of the RNA sequence shown in boldface for vGFSLFP is −25.6 kcal/mol (DNASIS version 2.6 software).
First, to determine the effects of the predicted RNA secondary structure on RT template switching, we performed the experiment with wild-type RT and NC. We observed a fivefold increase in the frequency of template switching for vGFSLFP (8.2%) compared to vGF100FP (1.7%) (Fig. 5). As expected, the frequency of RT template switching for the vGF100FP (1.7%) was much lower than for vGF250FP (10.8%). The higher frequency of template switching observed for vGF250FP could have resulted from the larger size of the repeat (250 versus 100 bp), the larger distance between homologous sequences in the repeats, or the presence of small secondary structures in the GF250FP vector that are absent in the GF100FP vector. It was previously shown that increasing the distance between repeats stimulated the frequency of template switching between direct repeats (13). A comparison of the viral titers indicated that the presence of the stem-loop in pGFSLFP did not have a significant effect on the overall efficiency of replication, since the viral titers produced by PG13 cells transfected with pGFSLFP (36,650 ± 2,450 CFU/ml) and pGF100FP (35,850 ± 550 CFU/ml) were not significantly different from each other (two-tailed two-sample t test; P = 0.78). Therefore, the increase in the frequency of GFP reconstitution could not be the result of more efficient expression of the deleted provirus. Because the predicted stem-loop structure was between the two F portions, it was deleted upon reconstitution of GFP and could not have affected the expression of GFP through more efficient transcription or translation. The result indicated that the predicted stem-loop structure enhanced RT template switching.
FIG. 5.
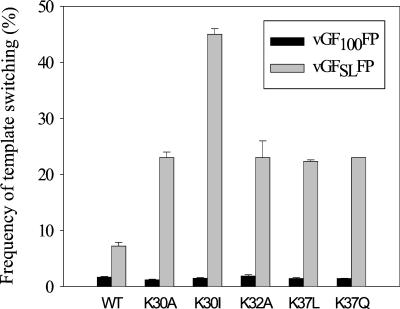
Comparison of the effects of MLV NC basic substitution mutations in the zinc finger domain on frequency of template switching in the presence or absence of a predicted stem-loop structure in the template. The mean is averaged from two to five experiments, and the error bars represent standard errors of the mean.
We then examined the effects of NC zinc finger domain mutations in which the basic residues were replaced with neutral amino acids. The substitutions K30A, K32A, and K37L did not have statistically significant effects on the frequency of template switching when the cGF250FP cells were used (Table 1). When cGF100FP cells were used, the frequencies of template switching ranged from 1.2 to 2.3%, which were not significantly different from the frequency of template switching for wild-type NC (Fig. 5). However, the frequencies of template switching obtained with the mutants exhibited significant increases when cGFSLFP cells were used. The frequencies of template switching were increased to 23 (K30A), 23 (K32A), and 20% (K37L); thus, in comparison to vGF100FP, the frequencies of template switching were 19- (K30A), 14- (K32A), and 26-fold (K37L) higher, respectively. Most importantly, these template-switching frequencies were significantly higher than the 8.2% frequency obtained with wild-type NC and vGFSLFP (t test; P < 0.01).
Both the K30I and K37Q mutants exhibited statistically significant increases in template switching (2.0- and 1.6-fold, respectively) when vGF250FP was used (Table 1). We therefore expected K30I and K37Q mutations to have greater effects on RT template switching for vGFSLFP than wild-type NC. The K30I mutant exhibited only a 1.5% frequency of template switching for vGF100FP, which was similar to the 1.7% frequency observed with wild-type NC; in sharp contrast, the frequency of template switching increased to 45% for vGFSLFP (Fig. 5). Therefore, the predicted RNA secondary structure in vGFSLFP resulted in a 30-fold overall increase in the template switching frequency for the K30I mutant. Similarly, the K37Q mutant displayed a frequency of template switching of 23% for vGFSLFP, which was 16-fold higher than the 1.5% frequency of template switching for vGF100FP.
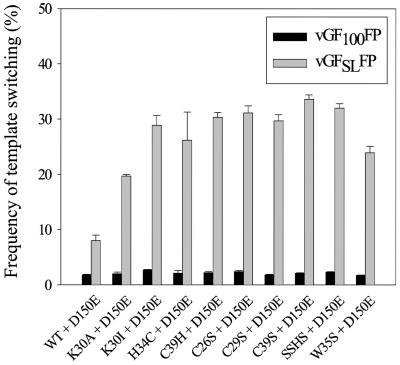
To study the role of the CCHC motif of MLV NC, we used the trans-complementation assays with cGF100FP(D150E) and cGFSLFP(D150E) cells to rescue replication-defective CCHC mutants. As shown in Fig. 5 and 6, the frequencies of template switching for wild-type NC with and without complementation with D150E were not different. As additional controls, the K30I and K30A mutants were also subjected to the trans-complementation assay (Fig. 6). Upon trans-complementation, the template-switching frequency for the K30I mutant was reduced from 45 to 29%, and the frequency for the K30A mutant was reduced from 24 to 19%. These results suggested that, because of the presence of wild-type NC in the infectious virion, the effects of the NC mutations on template switching were reduced. All of the CCHC mutants exhibited frequencies of template switching similar to that of wild-type NC for cGF100FP(D150E) (1.7 to 2.4%). In contrast, when the CCHC motif mutants were introduced into cGFSLFP(D150E) cells, the frequencies of template switching increased significantly, ranging from 24 to 34%. Thus, the substitution mutations in the CCHC motif of MLV NC increased the frequency of template switching approximately 10- to 20-fold when a predicted secondary structure was present in the template. Similar to the lysine substitution mutants, the effect of the predicted secondary structure was substantially greater for the CCHC motif mutants than the 8.2% template-switching frequency obtained with the wild-type NC.
FIG. 6.
trans-complementation of MLV NC zinc finger mutants and effects of substitutions in CCHC motif of MLV NC on frequency of template switching in the presence or absence of a predicted stem-loop structure in the template. The wild type (WT), K30I, and K30A are controls of the experimental system. The mean is averaged from two to four experiments, and the error bars represent standard errors of the mean.
The effects of HU treatment on template switching associated with RNA secondary structure.
Previous studies have shown that HU treatment depletes dNTP pools in cells (39), reduces the rate of DNA polymerization (51), and increases the frequency of RT template switching for wild-type RT and some RT mutants (61). We hypothesized that the RNA secondary structure forms a pause site for RT and increases the time available for annealing between the nascent DNA and acceptor RNA, which in turn leads to a higher frequency of RT template switching. To explore the mechanism by which the predicted RNA secondary structure increased the frequency of template switching, we sought to determine whether the increase in RT template switching by the RNA structure is sensitive to the effects of HU treatment.
The effects of HU treatment on RT template switching are summarized in Table 3. First, the frequencies of RT template switching for wild-type NC were compared in the absence and presence of HU treatment for vGF100FP and vGFSLFP. The frequency of template switching increased 3.7-fold (from 1.7 to 6.3%) with HU treatment for wild-type vGF100FP. In contrast, the frequency of template switching increased only slightly (1.5-fold) with HU treatment for wild-type vGFSLFP. This result suggested that the enhancing effect of the RNA structure on RT template switching was insensitive to HU treatment.
TABLE 3.
Comparison of the effect of HU treatment on frequency of template switching in the presence of predicted secondary structure
| Cell line | MLVNC | Frequency of direct-repeat deletions (mean % ± SE)a
|
Relative change in frequency of template switching (+HU/−HU)d | |
|---|---|---|---|---|
| −HUb | +HUc | |||
| cGF100FP | ||||
| WTe | 1.7 ± 0.1 | 6.3 ± 0.1 | 3.7 | |
| K30A | 1.2 ± 0.1 | 3.0 ± 0.5 | 2.5 | |
| K37L | 1.4 ± 0.2 | 4.2 ± 0.2 | 3.0 | |
| K37Q | 1.5 ± 0.1 | 4.5 ± 0.4 | 3.0 | |
| cGFSLFP | ||||
| WT | 8.2 ± 0.5 | 12.3 ± 1.1 | 1.5 | |
| K30A | 23.0 ± 1.1 | 27.6 ± 2.1 | No change | |
| K37L | 19.7 ± 0.3 | 22.3 ± 0.3 | No change | |
| K37Q | 23.0 ± 0 | 27.6 ± 0.3 | 1.2 | |
Frequency of direct-repeat deletion was determined as the percentage of infected D17 target cells that exhibited fluorescence after G418 selection compared with the negative control. The standard error (SE) was determined by using Sigma Plot version 5.0 software.
Frequency of direct-repeat deletion was determined in the absence of HU treatment during infection.
Frequency of direct-repeat deletion was determined in the presence of HU treatment during infection.
Calculated as follows: frequency of direct-repeat deletion observed with MLV NC in the presence (+) of HU treatment/frequency of direct-repeat deletion observed with MLV NC in the absence (−) of HU treatment. Statistically significant changes in the frequency of direct-repeat deletions with HU treatment, relative to the frequency in the absence of HU treatment, are shown (t test; P < 0.05). No change, P ≥ 0.05 (nt test).
WT, wild type.
Next, we compared the effects of HU treatment for NC mutants K30A, K37L, and K37Q. The template-switching frequencies for the three NC mutants increased 2.5- to 3.0-fold when the target cells were infected with vGF100FP in the presence of HU. Thus, HU treatment during infection with mutant as well as wild-type vGF100FP resulted in significant increases in the RT template-switching frequency. In contrast, when mutant vGFSLFP was used to infect D17 cells in the presence of HU, the template-switching frequencies for the K30A and K37L mutants were not statistically different (t test; P > 0.05) and for the K37Q mutant increased only 1.2-fold relative to the frequencies obtained without HU treatment. Thus, for NC mutants K30A, K37L, and K37Q, HU treatment had little effect on RT template switching in the presence of the RNA secondary structure. These results further suggested that the effect of the RNA secondary structure on enhancing template switching was not sensitive to HU treatment.
It was also interesting that the RT template-switching frequencies for the NC mutants in the presence of HU during vGF100FP infection were 3 to 4.5%, which were significantly lower than the 6.3% frequency obtained with wild-type vGF100FP in the presence of HU treatment (t test; P < 0.05/3 = 0.0167). Therefore, under these conditions, mutations in NC decreased the frequency of RT template switching.
DISCUSSION
In this study, we used a direct-repeat deletion assay to study the role of MLV NC during in vivo DNA synthesis by extensive mutational analysis of the zinc finger domain. Our results showed that mutations in the MLV NC zinc finger domain increased the frequency of RT template switching. The effects of NC mutations on template switching were more dramatic when a predicted stable secondary structure was present in the template RNA.
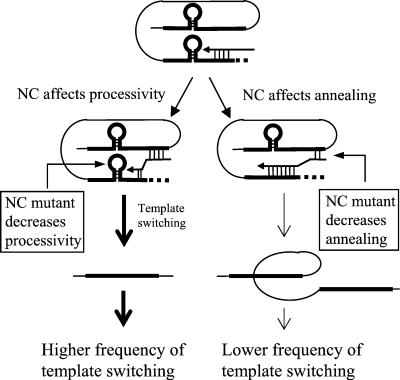
The potential roles of NC in reverse transcription and template switching are illustrated in Fig. 7. NC could increase the overall rate of polymerization by facilitating the unfolding of RNA secondary structures that may act as RT pause sites. Mutations or conditions that slow down DNA polymerization, such as depletion of intracellular dNTP pools with HU treatment, increase the frequency of template switching (34, 61). NC proteins have been shown to facilitate in vitro DNA synthesis by reducing RT pausing caused by secondary structures (26, 27, 36, 65). These studies suggest that NC facilitates the melting of RNA secondary structures and thus increases the processivity of reverse transcription. Based on these in vitro results, mutations in NC would be expected to increase the frequency of template switching. On the other hand, NC proteins have also been found to facilitate the annealing of nascent DNA with acceptor template upon RNase H degradation of the donor template in vitro (16, 50), as well as to facilitate minus-strand transfer (1, 18, 27, 53). These findings suggest that mutations in NC should decrease annealing and thus reduce the frequency of template switching.
FIG. 7.
Roles of NC in RT template switching. For simplicity, only intramolecular template switching is shown. The thick black lines represent direct repeats in an RNA template. The horizontal arrows represent nascent DNA. The thick dotted lines represent RNA template degraded by RNase H. Annealing between the nascent DNA and RNA template is designated by short vertical lines.
The results of our in vivo study indicate that the effect of NC on increasing the rate of DNA synthesis is more pronounced than its effect on annealing the nascent DNA to acceptor RNA. We observed that mutations in the zinc finger domain of MLV NC increased the frequency of template switching of vGF250FP and vGFSLFP, which indicates that the zinc finger domain of MLV NC has a greater effect on increasing the rate of DNA synthesis than on annealing nascent DNA with the acceptor template during viral DNA synthesis, especially when extensive secondary structures are present in the template. The results suggest that the zinc finger domain of MLV NC enhances the rate of DNA polymerization in vivo through its effects on destabilizing RNA secondary structure during viral DNA synthesis.
Retroviral NC protein is generally considered to be an RNA chaperone during reverse transcription. The nucleic acid chaperone activities of NC have been well demonstrated during in vitro tRNA primer placement and minus-strand DNA transfer reactions by catalyzing the annealing of complementary sequences (1, 7, 18, 21, 22, 26, 27, 31, 44, 50, 53). The ability of NC to chaperone processive viral DNA synthesis by destabilizing secondary structures has also been studied in vitro (36, 42, 65). The results of our in vivo study indicate that, during viral DNA synthesis, the chaperone function of NC in destabilizing secondary structure is predominant over the annealing of nascent DNA to acceptor RNA. One possibility is that the annealing reaction between the nascent DNA and acceptor RNA is in competition with elongation of DNA synthesis on the donor template; the annealing of nascent DNA and the acceptor template may be a slow event relative to elongation of DNA synthesis. In this assay, the annealing reaction and the subsequent template switching must occur within the relatively short time required for RT to reverse transcribe through one copy of the direct repeat. On the other hand, NC is bound to the viral RNA as part of the viral nucleoprotein complex and presumably will have ample time to unfold any secondary structures that might act as RT pause sites to facilitate rapid DNA polymerization.
It is conceivable that the chaperone activity of NC might stabilize the DNA-RNA hybrids that form between nascent DNA and donor template RNA; enhancing the stability of these hybrids would suppress annealing between nascent DNA and acceptor RNA and reduce the frequency of template switching. However, the well-defined nucleic acid chaperone activity of NC argues against this possibility. NC, through its nucleic acid chaperone function, acts to maximize base pairing in nucleic acids (reviewed in reference 55 and references therein). Since the template RNA is degraded by RNase H after it is copied, the extent of base pairing between nascent DNA and acceptor nucleic acid should be greater than the base pairing between nascent DNA and donor template RNA fragments. Therefore, the effect of NC on promoting annealing between nascent DNA and acceptor template should be more pronounced than its effect on stabilizing hybridization to donor RNA fragments.
Does NC play a role in annealing nascent DNA and acceptor RNA during DNA synthesis? The results obtained with HU treatment during vGF100FP infection (Table 3) suggest that NC indeed promotes annealing of nascent DNA and acceptor template under certain conditions. HU treatment during infection with wild-type vGF100FP increased the RT template-switching frequency to 6.3%; however, HU treatment during infection with vGF100FP containing mutations in NC increased template switching to only 3 to 4.5%. Thus, under these conditions, mutations in NC actually resulted in a decrease in RT template switching (3 to 4.5% for NC mutants versus 6.3% for the wild type), suggesting that wild-type NC promoted annealing between nascent DNA and the acceptor template. In this case, the direct repeats were rather short (100 bp) and probably did not have extensive secondary structure in the template. In addition, the rate of DNA synthesis was reduced with HU treatment, allowing more time for annealing between the nascent DNA and the acceptor template. This result suggests that NC can facilitate both melting of RNA secondary structure and annealing between complementary sequences. When the repeat length is short and does not contain extensive RNA secondary structures, and the rate of DNA synthesis is low, the effect of NC on annealing is more evident.
Several in vitro studies have indicated that NC enhances minus-strand DNA transfer by promoting annealing between nascent DNA and template RNA (16, 50). During minus-strand DNA transfer, RT reaches the end of the template; therefore, the rate of DNA polymerization is zero. Under this extreme condition, there is no DNA synthesis, so that the effect of NC on annealing nascent DNA and acceptor RNA would predominate and NC would be expected to stimulate minus-strand DNA transfer. However, the minus-strand DNA transfer reaction for HIV-1 is made more complex by the presence of the transactivation response element (TAR) stem-loop structure in the R region. It was recently shown that NC stimulates the HIV-1 minus-strand transfer reaction by unfolding the TAR secondary structure and inhibiting TAR-induced self-priming of DNA synthesis (18, 27). Presumably, the NC-induced unfolding of the TAR secondary structure prevents premature weak-stop DNA transfer caused by RT pausing at the site of TAR. Our proposal that MLV NC denatures RNA secondary structure in vivo is analogous to the NC-induced unfolding of the TAR secondary structure. Other in vitro studies have also shown a stimulation of internal template switching by NC (46, 52, 57). As suggested earlier, the length of homology, the extent of RNA secondary structure, and the rate of DNA synthesis could all play a role in determining whether the net effect of NC is to promote template switching or to increase the rate of DNA synthesis on the donor template.
The results obtained with the basic residues in the zinc finger domain suggest that the structure of the side chain, rather than the loss of a positively charged side chain, has a greater impact on viral titers and RT template switching. For example, the K30A mutant, which contained a small side chain, showed only a fivefold reduction in viral titer, no change in template switching for vGF250FP, and a 15-fold increase in the frequency of template switching for vGFSLFP. In contrast, the K30I mutant containing a bulky side chain had a 70-fold reduction in viral titer, a twofold increase in the frequency of template switching for vGF250FP, and a 30-fold increase in the frequency of template switching for vGFSLFP. We speculate that the bulky isoleucine side chain altered the conformation of the zinc finger motif. The strictly conserved structure of the zinc finger motif has been shown to be critical for viral replication (23, 56).
By using an in vivo trans-complementation assay, we show that replication-defective NC mutants can be rescued by trans-complementation with an RT-inactivating mutant expressing wild-type NC. Infectious viral particles produced after trans-complementation contained a mixture of Gag and Gag-Pol polyproteins expressed from both NC+ RT− and NC− RT+ variants of gag-pol. The functional RT in the infectious virion can only be produced by the Gag-Pol proteins derived from the NC mutants. Because the processing of the Gag-Pol proteins from the NC mutants resulted in production of the mutated NC proteins as well as wild-type RT, the mutated NC proteins must be present in the infectious virion.
The ratio of the wild-type and mutated NC proteins in the infectious virion cannot be determined in this trans-complementation system. If the Gag and Gag-Pol proteins of the NC mutants and the RT mutant are expressed equally and copackaged randomly, then the wild-type and mutant NC proteins are likely to be present in a 1:1 ratio in the infectious virion. However, the efficiency with which the NC mutants bind to RNA and are proteolytically processed could affect the ratio of the wild-type and mutated NC proteins present in the infectious virion. Nevertheless, these infectious viruses produced after trans-complementation of NC zinc finger mutants exhibited an increase in the frequency of template switching.
It was theoretically possible that the inactive RT from the D150E mutant interfered with the efficiency of DNA synthesis by binding to the template and preventing the progression of wild-type RT. However, our results do not support this notion, because the D150E mutation did not affect the frequency of template switching when complemented with wild-type gag-pol (Fig. 6). In addition, trans-complementation of the K30I mutant with the D150E mutant was shown to decrease the frequency of template switching for vGFSLFP from 45 to 29%, representing a 36% decrease. Similarly, the K30A trans-complemented with D150E resulted in a slightly decreased frequency of RT template switching from 23.0 to 19.7%. Because the viral particles contain wild-type NC as well as mutated NC, the enhancing effects of the NC mutations on template switching are most likely to be underestimated in this assay. The decrease in the effect of K30I and K30A mutations on the template-switching frequency in the trans-complementation assay may be due to various effects of these mutations on other functions of NC, such as RNA packaging or tRNA placement, and the extent to which they can be complemented in trans by the replication-defective RT mutant.
The exact nature of the RNA secondary structure present in vGFSLFP that resulted in increasing RT template switching is unknown. Furthermore, the RNA secondary structure may take alternative forms during the process of reverse transcription through interactions with viral proteins as the template RNA is being copied into DNA. Nevertheless, the presence of this 41-bp sequence in vGFSLFP substantially increased the frequency of template switching. Although several mechanisms could be postulated, one attractive hypothesis is that the RNA secondary structure acts as an RT pause site to stall or slow down the rate of DNA synthesis, allowing more time for base pairing between nascent DNA and the acceptor template. The observation in this study that the effect of the RNA secondary structure was not sensitive to HU treatment suggests that the RNA secondary structure causes RT to stall or reduce the rate of DNA synthesis to such a low level that RT binding to the nucleotide substrate is no longer the rate-limiting step in reverse transcription. As a consequence, depletion of intracellular nucleotide pools with HU treatment has little effect on the high frequency of template switching associated with the RNA secondary structure.
In summary, one important in vivo role of MLV NC is to increase the rate of viral DNA synthesis. We hypothesize that MLV NC increases the rate of DNA polymerization through its chaperone activity, which involves breaking base pairs in the viral RNA secondary structure. The effect of NC on DNA synthesis and thus frequency of template switching is highly dependent on the extent of RNA secondary structure present in the template RNA. The zinc finger domain contributes significantly to the chaperone activity of MLV NC. It will be of interest to determine the roles of MLV NC basic regions that flank the zinc finger domain in RT template switching.
Acknowledgments
We especially thank Stephen Hughes and Alan Rein for intellectual input and valuable discussion of the manuscript and Anne Arthur for editorial expertise and revisions. We also thank Sara Cheslock, Que Dang, Krista Delviks, William Fu, Evguenia Svarovskaia, Yegor Voronin, and Yijun Zhang for discussions of the results and critical readings of the manuscript. Finally, we thank Douglas Powell, Computer and Statistical Services, NCI—Frederick, for assistance in statistical analysis of data.
This work was supported by the HIV Drug Resistance Program, National Cancer Institute, and by the National Cancer Institute under contract NO1-CO-56000 with SAIC Frederick.
REFERENCES
- 1.Allain, B., M. Lapadat-Tapolsky, C. Berlioz, and J. L. Darlix. 1994. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 13:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. A., E. H. Bowman, and W. S. Hu. 1998. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J. Virol. 72:1195-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz, R. D., and S. P. Goff. 1994. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 202:233-246. [DOI] [PubMed] [Google Scholar]
- 4.Berthoux, L., C. Pechoux, M. Ottmann, G. Morel, and J. L. Darlix. 1997. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol. 71:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buiser, R. G., R. A. Bambara, and P. J. Fay. 1993. Pausing by retroviral DNA polymerases promotes strand transfer from internal regions of RNA donor templates to homopolymeric acceptor templates. Biochim. Biophys. Acta 1216:20-30. [DOI] [PubMed] [Google Scholar]
- 6.Cen, S., Y. Huang, A. Khorchid, J. L. Darlix, M. A. Wainberg, and L. Kleiman. 1999. The role of Pr55gag in the annealing of tRNA3Lys to human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4485-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, B., K. Weidemaier, W. T. Yip, P. F. Barbara, and K. Musier-Forsyth. 1999. Intra-tRNA distance measurements for nucleocapsid protein-dependent tRNA unwinding during priming of HIV reverse transcription. Proc. Natl. Acad. Sci. USA 96:459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin, J. M., S. H. Hughes, and H. E. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 10.Dannull, J., A. Surovoy, G. Jung, and K. Moelling. 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 13:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darlix, J. L., M. Lapadat-Tapolsky, H. de Rocquigny, and B. P. Roques. 1995. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 254:523-537. [DOI] [PubMed] [Google Scholar]
- 12.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 ψ-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 13.Delviks, K. A., and V. K. Pathak. 1999. Effect of distance between homologous sequences and 3′ homology on the frequency of retroviral reverse transcriptase template switching. J. Virol. 73:7923-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rocquigny, H., D. Ficheux, C. Gabus, B. Allain, M. C. Fournie-Zaluski, J. L. Darlix, and B. P. Roques. 1993. Two short basic sequences surrounding the zinc finger of nucleocapsid protein NCp10 of Moloney murine leukemia virus are critical for RNA annealing activity. Nucleic Acids Res. 21:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeStefano, J. J. 1995. Human immunodeficiency virus nucleocapsid protein stimulates strand transfer from internal regions of heteropolymeric RNA templates. Arch. Virol. 140:1775-1789. [DOI] [PubMed] [Google Scholar]
- 16.DeStefano, J. J. 1996. Interaction of human immunodeficiency virus nucleocapsid protein with a structure mimicking a replication intermediate. Effects on stability, reverse transcriptase binding, and strand transfer. J. Biol. Chem. 271:16350-16356. [PubMed] [Google Scholar]
- 17.DeStefano, J. J., L. M. Mallaber, L. Rodriguez-Rodriguez, P. J. Fay, and R. A. Bambara. 1992. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J. Virol. 66:6370-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driscoll, M. D., and S. H. Hughes. 2000. Human immunodeficiency virus type 1 nucleocapsid protein can prevent self-priming of minus-strand strong stop DNA by promoting the annealing of short oligonucleotides to hairpin sequences. J. Virol. 74:8785-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, Y. X., S. Campbell, D. Harvin, B. Ehresmann, C. Ehresmann, and A. Rein. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 73:4251-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher, R. J., A. Rein, M. Fivash, M. A. Urbaneja, J. R. Casas-Finet, M. Medaglia, and L. E. Henderson. 1998. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 72:1902-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorelick, R. J., D. J. Chabot, D. E. Ott, T. D. Gagliardi, A. Rein, L. E. Henderson, and L. O. Arthur. 1996. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 70:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorelick, R. J., W. Fu, T. D. Gagliardi, W. J. Bosche, A. Rein, L. E. Henderson, and L. O. Arthur. 1999. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J. Virol. 73:8185-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorelick, R. J., T. D. Gagliardi, W. J. Bosche, T. A. Wiltrout, L. V. Coren, D. J. Chabot, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92-104. [DOI] [PubMed] [Google Scholar]
- 24.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like”protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gritz, L., and J. Davies. 1983. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25:179-188. [DOI] [PubMed] [Google Scholar]
- 26.Guo, J., L. E. Henderson, J. Bess, B. Kane, and J. G. Levin. 1997. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J. Virol. 71:5178-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo, J., T. Wu, J. Anderson, B. F. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. G. Levin. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 74:8980-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halvas, E. K., E. S. Svarovskaia, and V. K. Pathak. 2000. Role of murine leukemia virus reverse transcriptase deoxyribonucleoside triphosphate-binding site in retroviral replication and in vivo fidelity. J. Virol. 74:10349-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herschlag, D. 1995. RNA chaperones and the RNA folding problem. J. Biol. Chem. 270:20871-20874. [DOI] [PubMed] [Google Scholar]
- 30.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 31.Housset, V., H. De Rocquigny, B. P. Roques, and J. L. Darlix. 1993. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 67:2537-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 33.Huang, Y., A. Khorchid, J. Gabor, J. Wang, X. Li, J. L. Darlix, M. A. Wainberg, and L. Kleiman. 1998. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA3Lys genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J. Virol. 72:3907-3915. [DOI] [PMC free article] [PubMed]
- 34.Hwang, C. K., E. S. Svarovskaia, and V. K. Pathak. 2001. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc. Natl. Acad. Sci. USA 98:12209-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji, X., G. J. Klarmann, and B. D. Preston. 1996. Effect of human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein on HIV-1 reverse transcriptase activity in vitro. Biochemistry 35:132-143. [DOI] [PubMed] [Google Scholar]
- 37.Julias, J. G., D. Hash, and V. K. Pathak. 1995. E− vectors: development of novel self-inactivating and self-activating retroviral vectors for safer gene therapy. J. Virol. 69:6839-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Julias, J. G., T. Kim, G. Arnold, and V. K. Pathak. 1997. The antiretrovirus drug 3′-azido-3′-deoxythymidine increases the retrovirus mutation rate. J. Virol. 71:4254-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Julias, J. G., and V. K. Pathak. 1998. Deoxyribonucleoside triphosphate pool imbalances in vivo are associated with an increased retroviral mutation rate. J. Virol. 72:7941-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent, R. B., J. R. Emanuel, Y. Ben Neriah, R. Levenson, and D. E. Housman. 1987. Ouabain resistance conferred by expression of the cDNA for a murine Na+,K+-ATPase alpha subunit. Science 237:901-903. [DOI] [PubMed] [Google Scholar]
- 41.Kirk, R. E. 1982. Experimental design, 2nd ed. Brooks-Cole Publishing Company, Pacific Grove, Calif.
- 42.Klasens, B. I., H. T. Huthoff, A. T. Das, R. E. Jeeninga, and B. Berkhout. 1999. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim. Biophys. Acta 1444:355-370. [DOI] [PubMed] [Google Scholar]
- 43.Landau, N. R., K. A. Page, and D. R. Littman. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 65:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, X., Y. Quan, E. J. Arts, Z. Li, B. D. Preston, H. de Rocquigny, B. P. Roques, J. L. Darlix, L. Kleiman, M. A. Parniak, and M. A. Wainberg. 1996. Human immunodeficiency virus type 1 nucleocapsid protein (NCp7) directs specific initiation of minus-strand DNA synthesis primed by human tRNA3Lys in vitro: studies of viral RNA molecules mutated in regions that flank the primer binding site. J. Virol. 70:4996-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Negroni, M., and H. Buc. 1999. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J. Mol. Biol. 286:15-31. [DOI] [PubMed] [Google Scholar]
- 47.Pathak, V. K., and W. S. Hu. 1997. “Might as well jump!” Template switching by retroviral reverse transcriptase, defective genome formation, and recombination. Semin. Virol. 8:141-150. [Google Scholar]
- 48.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc. Natl. Acad. Sci. USA 87:6024-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pathak, V. K., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl. Acad. Sci. USA 87:6019-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peliska, J. A., S. Balasubramanian, D. P. Giedroc, and S. J. Benkovic. 1994. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry 33:13817-13823. [DOI] [PubMed] [Google Scholar]
- 51.Pfeiffer, J. K., M. M. Georgiadis, and A. Telesnitsky. 2000. Structure-based Moloney murine leukemia virus reverse transcriptase mutants with altered intracellular direct-repeat deletion frequencies. J. Virol. 74:9629-9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raja, A., and J. J. DeStefano. 1999. Kinetic analysis of the effect of HIV nucleocapsid protein (NCp) on internal strand transfer reactions. Biochemistry 38:5178-5184. [DOI] [PubMed] [Google Scholar]
- 53.Rascle, J. B., D. Ficheux, and J. L. Darlix. 1998. Possible roles of nucleocapsid protein of MoMuLV in the specificity of proviral DNA synthesis and in the genetic variability of the virus. J. Mol. Biol. 280:215-225. [DOI] [PubMed] [Google Scholar]
- 54.Rein, A., D. P. Harvin, J. Mirro, S. M. Ernst, and R. J. Gorelick. 1994. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J. Virol. 68:6124-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 56.Remy, E., H. de Rocquigny, P. Petitjean, D. Muriaux, V. Theilleux, J. Paoletti, and B. P. Roques. 1998. The annealing of tRNA3Lys to human immunodeficiency virus type 1 primer binding site is critically dependent on the NCp7 zinc fingers structure. J. Biol. Chem. 273:4819-4822. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez-Rodriguez, L., Z. Tsuchihashi, G. M. Fuentes, R. A. Bambara, and P. J. Fay. 1995. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 270:15005-15011. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 59.Schmalzbauer, E., B. Strack, J. Dannull, S. Guehmann, and K. Moelling. 1996. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J. Virol. 70:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuler, W., C. Dong, K. Wecker, and B. P. Roques. 1999. NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes. Biochemistry 38:12984-12994. [DOI] [PubMed] [Google Scholar]
- 61.Svarovskaia, E. S., K. A. Delviks, C. K. Hwang, and V. K. Pathak. 2000. Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J. Virol. 74:7171-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Temin, H. M. 1993. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc. Natl. Acad. Sci. USA 90:6900-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsuchihashi, Z., and P. O. Brown. 1994. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 68:5863-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, W., B. M. Blumberg, P. J. Fay, and R. A. Bambara. 1995. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J. Biol. Chem. 270:325-332. [DOI] [PubMed] [Google Scholar]
- 65.Wu, W., L. E. Henderson, T. D. Copeland, R. J. Gorelick, W. J. Bosche, A. Rein, and J. G. Levin. 1996. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 70:7132-7142. [DOI] [PMC free article] [PubMed] [Google Scholar]