Abstract
We describe the further properties of a protein, designated SOS gp140, wherein the association of the gp120 and gp41 subunits of the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein is stabilized by an intersubunit disulfide bond. HIV-1JR-FL SOS gp140, proteolytically uncleaved gp140 (gp140UNC), and gp120 were expressed in stably transfected Chinese hamster ovary cells and analyzed for antigenic and structural properties before and after purification. Compared with gp140UNC, SOS gp140 reacted more strongly in surface plasmon resonance and radioimmunoprecipitation assays with the neutralizing monoclonal antibodies (MAbs) 2G12 (anti-gp120), 2F5 (anti-gp41), and 17b (to a CD4-induced epitope that overlaps the CCR5-binding site). In contrast, gp140UNC displayed the greater reactivity with nonneutralizing anti-gp120 and anti-gp41 MAbs. Immunoelectron microscopy studies suggested a model for SOS gp140 wherein the gp41 ectodomain (gp41ECTO) occludes the “nonneutralizing” face of gp120, consistent with the antigenic properties of this protein. We also report the application of Blue Native polyacrylamide gel electrophoresis (BN-PAGE), a high-resolution molecular sizing method, to the study of viral envelope proteins. BN-PAGE and other biophysical studies demonstrated that SOS gp140 was monomeric, whereas gp140UNC comprised a mixture of noncovalently associated and disulfide-linked dimers, trimers, and tetramers. The oligomeric and conformational properties of SOS gp140 and gp140UNC were largely unaffected by purification. An uncleaved gp140 protein containing the SOS cysteine mutations (SOS gp140UNC) was also oligomeric. Surprisingly, variable-loop-deleted SOS gp140 proteins were expressed (although not yet purified) as cleaved, noncovalently associated oligomers that were significantly more stable than the full-length protein. Overall, our findings have relevance for rational vaccine design.
The native, fusion-competent form of the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein complex is a trimeric structure composed of three gp120 subunits and three gp41 subunits; the receptor-binding (CD4 and coreceptor) sites are located in the gp120 moieties, and the fusion peptides are located in the gp41 components (10, 33, 34, 52, 69, 78, 83). In the generally accepted model of HIV-1 fusion, the sequential binding of gp120 to CD4 and a coreceptor induces a series of conformational changes in the gp41 subunits, leading to the insertion of the fusion peptides into the host cell membrane in a highly dynamic process (14, 31, 39, 59, 68, 72, 81, 84, 91). The associations between the six components of the fusion-competent complex are maintained via noncovalent interactions between gp120 and gp41 and between the gp41 subunits (52, 84). These interactions are relatively weak, making the fusion-competent complex unstable. This instability perhaps facilitates the conformational changes in the various components that are necessary for the fusion reaction to proceed efficiently, but it greatly complicates the task of isolating the native complex in purified form. Put simply, the native complex falls apart before it can be purified, leaving only the dissociated subunits.
One reason it would be desirable to produce the native HIV-1 envelope complex is to explore its potential as an immunogen, perhaps after modification to improve its exposure of critical neutralization epitopes. The limited neutralizing-antibody response to HIV-1 in infected people is directed at the native complex and is probably raised against it (6, 40, 49, 51); P. W. H. I. Parren, D. R. Burton, and Q. J. Sattentau, Letter, Nat. Med. 3:366, 1997). In contrast, the isolated subunits have not proven efficient at inducing relevant neutralizing antibodies (reviewed in references 6, 49, and 51). We and others are therefore attempting to make more-stable forms of the envelope glycoprotein complex that better mimic the native structure. Usually, these efforts have focused on making various forms of soluble gp140 glycoproteins which contain gp120 but only the ectodomain of gp41 (4, 11, 13, 17, 19-21, 57, 66, 76, 85-87, 90).
An approach to resolving the instability of the native complex is to remove the cleavage site that naturally exists between the gp120 and gp41 subunits. Doing so means that proteolysis of this site does not occur, leading to the expression of gp140 glycoproteins in which the gp120 subunit is covalently linked to the gp41 ectodomain (gp41ECTO) by means of a peptide bond (2, 3, 16-18). Such proteins can be oligomeric, sometimes trimeric (11, 16-21, 54, 66, 85-87, 90). However, it is not clear that they truly represent the structure of the native, fusion-competent complex in which the gp120-gp41 cleavage site is fully utilized. Hence the receptor-binding properties of uncleaved gp140 (gp140UNC) proteins tend to be impaired, and nonneutralizing antibody epitopes are exposed on them that probably are not accessible on the native structure (4, 6, 28, 60, 90).
We have taken an alternative approach to the problem of gp120-gp41 instability, which is to retain the cleavage site but to introduce a disulfide bond between the gp120 and gp41ECTO subunits (4, 57). Properly positioned, this intermolecular disulfide bond forms efficiently during envelope glycoprotein (Env) synthesis, allowing the secretion of gp140 proteins that are proteolytically processed but in which the association between the gp120 and gp41ECTO subunits is maintained by the disulfide bond.
Here we show that the gp41-gp41 interactions are unstable in the SOS gp140 protein, which is expressed and purified primarily as a monomer. In contrast, gp140UNC proteins—with or without the SOS cysteine substitutions—are multimeric, implying that cleavage of the peptide bond between gp120 and gp41 destabilizes the native complex. Despite being monomeric, the purified and unpurified forms of SOS gp140 are better antigenic structural mimics of the native, fusion-competent Env structure than are the corresponding gp120 or gp140UNC proteins. This may be because the presence and orientation of gp41ECTO occludes certain nonneutralization epitopes on SOS gp140 while preserving the presentation of important neutralization sites. This explanation is consistent with immunoelectron microscopy studies of the protein. Unexpectedly, proteolytically mature, but variable-loop-deleted, SOS gp140 glycoproteins have enhanced oligomeric stability, so these molecules warrant further study for their structural and immunogenic properties.
MATERIALS AND METHODS
Plasmids.
The pPPI4 eukaryotic expression vectors encoding SOS and uncleaved forms of HIV-1JR-FL gp140 have been described previously (4, 72). The SOS gp140 protein contains cysteine substitutions at residues A501 in the C5 region of gp120 and T605 in gp41 (4, 57). In gp140UNC, the sequence KRRVVQREKRAV at the junction between gp120 and gp41ECTO has been replaced with a hexameric Leu-Arg motif to prevent scission of gp140 into gp120 and gp41ECTO (4). Plasmids encoding variable-loop-deleted forms of HIV-1JR-FL SOS gp140 have been described (57). In these constructs, the tripeptide GAG is used to replace V1 loop sequences (D133-K155) and V2 loop sequences (F159-I194), alone or in combination. The SOS gp140UNC protein contains the same cysteine substitutions that are present in SOS gp140, but the residues REKR at the gp120-gp41ECTO cleavage site have been replaced by the sequence IEGR, to prevent gp140 cleavage. Relative to the GenBank HIV-1JR-FL env sequence (accession no. AAB05604), the gp140 proteins contained a K668N substitution near the carboxy terminus of the molecules, consistent with the consensus subtype B sequence. The furin gene (71) was expressed from plasmid pcDNA3.1furin (4).
MAbs and CD4-based proteins.
The following anti-gp120 monoclonal antibodies (MAbs) were used: IgG1b12 (against the CD4 binding site [7]), 2G12 (against a unique C3-V4 glycan-dependent epitope [75]), 17b (against a CD4-inducible epitope [70]), 19b (against the V3 loop [42]), and 23A (against the C5 region [41]). The anti-gp41 MAbs were 2F5 (against an epitope centered on the sequence ELDKWA [44, 48, 93]) and 2.2B (against epitope cluster II). MAbs IgG1b12, 2G12, and 2F5 are broadly neutralizing (74). MAb 17b weakly neutralizes diverse strains of HIV-1, more so in the presence of soluble CD4 (70), whereas the neutralizing activity of MAb 19b against primary isolates is limited (73). MAbs 23A and 2.2B are nonneutralizing. Soluble CD4 (sCD4) and the CD4-based molecule CD4-IgG2 have been described elsewhere (1).
HIV-1 gp140 and gp120 glycoproteins.
To create stable cell lines that secrete full-length HIV-1JR-FL SOS gp140 or ΔV1V2 SOS gp140, we cotransfected DXB-11 dihydrofolate reductase-negative Chinese hamster ovary (CHO) cells with pcDNA3.1furin and either pPPI4-SOS gp140 (4) or pPPI4-ΔV1V2∗ SOS gp140 (57), respectively, using the calcium phosphate precipitation method. Doubly transformed cells were selected by passaging the cells in nucleoside-free α-minimum essential medium containing 10% fetal bovine serum, Geneticin (Life Technologies, Rockville, Md.), and methotrexate (Sigma, St. Louis, Mo.). The cells were amplified for gp140 expression by stepwise increases in methotrexate concentration, as described elsewhere (1). Clones were selected for SOS gp140 expression, assembly, and endoproteolytic processing based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses of culture supernatants. CHO cells expressing SOS gp140UNC were created using similar methods, except that pcDNA3.1furin and Geneticin were not used. Full-length SOS gp140 was purified from CHO cell culture supernatants by Galanthus nivalis lectin affinity chromatography (Sigma) and Superdex 200 gel filtration chromatography (Amersham-Pharmacia, Piscataway, N.J.), as described elsewhere (72). The gp140UNC glycoprotein was purified by lectin chromatography only. The concentration of purified Envs was measured by UV spectroscopy as described previously (61) and was corroborated by enzyme-linked immunosorbent assay and densitometric analysis of SDS-PAGE gels. Recombinant HIV-1JR-FL, HIV-1LAI, and HIV-1YU2 gp120 glycoproteins were produced using methods that have been previously described (72, 81).
Where indicated, HIV-1 envelope glycoproteins were transiently expressed in adherent 293T cells by transfection with Env- and furin-expressing plasmids, as described previously (4). For radioimmunoprecipitation assays, the proteins were metabolically labeled with [35S]cysteine and [35S]methionine for 24 h prior to analysis.
SDS-PAGE, radioimmunoprecipitation, Blue Native PAGE, and Western blot analyses.
SDS-PAGE analyses were performed as described elsewhere (4). Reduced and nonreduced samples were prepared by boiling for 2 min in Laemmli sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 25% glycerol, 0.01% bromophenol blue) in the presence or absence, respectively, of 50 mM dithiothreitol (DTT). Protein purity was determined by densitometric analysis of the stained gels followed by the use of ImageQuant software (Molecular Devices, Sunnyvale, Calif.). Radioimmunoprecipitation assays (RIPAs) were performed on Env-containing cell culture supernatants, as previously described (4, 57).
Blue Native (BN)-PAGE was carried out with minor modifications to the published method (62, 63). Thus, purified protein samples or cell culture supernatants were diluted with an equal volume of a buffer containing 100 mM 4-(N-morpholino)propane sulfonic acid (MOPS), 100 mM Tris-HCl, pH 7.7, 40% glycerol, 0.1% Coomassie blue, just prior to loading onto a 4 to 12% Bis-Tris NuPAGE gel (Invitrogen). Typically, gel electrophoresis was performed for 2 h at 150V (∼0.07A) using 50 mM MOPS, 50 mM Tris, pH 7.7, 0.002% Coomassie blue as cathode buffer, and 50 mM MOPS, 50 mM Tris, pH 7.7 as anode buffer. When purified proteins were analyzed, the gel was destained with several changes of 50 mM MOPS, 50 mM Tris, pH 7.7 subsequent to the electrophoresis step. Typically, 5 μg of purified protein were loaded per lane.
For Western blot analyses, gels and polyvinylidene difluoride (PVDF) membranes were soaked for 10 min in transfer buffer (192 mM glycine, 25 mM Tris, 0.05% SDS [pH 8.8] containing 20% methanol). Following transfer, PVDF membranes were destained of Coomassie blue dye using 25% methanol and 10% acetic acid and air dried. Destained membranes were probed using the anti-V3 loop MAb PA1 (Progenics) followed by horseradish peroxidase-labeled anti-mouse immunoglobulin G (IgG) (Kirkegaard & Perry), each used at a final concentration of 0.2 μg/ml. Luminometric detection of the envelope glycoproteins was obtained with the Renaissance Western blot Chemiluminescence Reagent Plus system (Perkin-Elmer Life Sciences, Boston, Mass.). Bovine serum albumin (BSA), apo-ferritin, and thyroglobulin were obtained from Amersham Biosciences (Piscataway, N.J.) and used as molecular mass standards.
Matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry.
Proteins were dialyzed overnight against water prior to analysis. Where indicated, SOS gp140 (1 mg/ml) was reduced with 10 mM DTT (Sigma), after which iodoacetamide (Sigma) was added to a final concentration of 100 mM, before dialysis. The samples were mixed with an equal volume of sinapinic acid matrix solution, dried at room temperature, and analyzed by MALDI-TOF mass spectrometry (37). MALDI-TOF mass spectra were acquired on a PerSeptive Biosystems Voyager-STR mass spectrometer with delayed extraction. Samples were irradiated with a nitrogen laser (Laser Science Inc.) operated at 337 nm. Ions produced in the sample target were accelerated with a deflection voltage of 30,000 V.
Sedimentation equilibrium analysis.
Sedimentation equilibrium measurements were performed on a Beckman XL-A Optima analytical ultracentrifuge with an An-60 Ti rotor at 20°C. Protein samples were dialyzed overnight into run buffer (50 mM sodium phosphate, 150 mM NaCl [pH 7.0]) diluted to 0.15, 0.30, and 0.60 mg/ml and centrifuged in a six-sector cell at rotor speeds of 6,000 and 9,000 rpm. Data were acquired at two wavelengths per rotor speed and were fit using the program NONLIN to a single species model of the natural logarithm of the absorbance versus radial distance squared (29). Solvent density and protein partial specific volume were calculated according to solvent and protein composition, respectively (36).
Size exclusion chromatography.
Purified, CHO cell-expressed SOS gp140, gp140UNC, and gp120 proteins were analyzed by size exclusion chromatography on a TSK G3000SWXL high-performance liquid chromatography column (TosoHaas, Montgomeryville, Pa.) using phosphate-buffered saline as the running buffer. The protein retention time was determined by monitoring the UV absorbance of the column effluent at a wavelength of 280 nm. The column was calibrated using ferritin as a model protein that exists in oligomeric states of 220, 440, and 880 kDa (24).
Surface plasmon resonance measurements.
A Biacore X optical biosensor was used. Each MAb was immobilized at 8,000 to 10,000 resonance units by the amine coupling method to a CM5 sensor chip, according to the manufacturer's instructions (Biacore, Inc., Piscataway, N.J.). A reference surface (lacking antibody) was used as a background control. Binding experiments were performed at 25°C in HSB-EP buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% [vol/vol] Surfactant P20). Purified Envs (25 nM) were run over the test and control chips at a flow rate of 30 μl/min, whereas CHO cell culture supernatants (∼5 nM Env) were analyzed at 10 μl/min. To study the exposure of CD4-induced epitopes, sCD4 was added to the envelope glycoproteins at an 8-molar excess concentration for at least 1 h prior to analysis. The sensor surface was regenerated with a short pulse of 3.5 M MgCl2.
Immunoelectron microscopy.
Immunoelectron-microscopic analyses of SOS gp140 and gp120 alone and in complex with MAb, MAb fragments, and sCD4 were performed by negative staining with uranyl formate as previously described (55, 56). The samples were examined on a JEOL JEM CX-100 electron microscope and photographed at a magnification of 100,000 diameters.
Immune complex image digitalizing and averaging.
The electron micrographs of immune complex images were digitalized on an AGFA (Ridgefield Park, N.J.) DUOSCAN T2500 negative scanner at a scanning resolution of 2,500 pixels per inch. Potentially informative complexes were selected and windowed as 256- by 256-pixel images. Approximately 500 randomly oriented examples of each complex combination were windowed, brought into alignment, and then averaged using the SPIDER software package (23). Windowed images were first normalized by scaling to adjust the image pixel values to a mean of 1. Complexes were then centered and masked for an alignment through classification (multiple references [multireference] alignment [23]). For this alignment procedure, the particles were first assigned to six classes using k-means clustering (23). The images in each class were then averaged, and the six class averages were taken as the multireference for the realignment of all of the original windowed complexes. Complexes with mirrored (inverted) orientation were automatically righted for averaging. The newly aligned complexes were then reclassified, and class averages were calculated as the new references. All of the complexes were then realigned based on the new multiple references. The classification and multireference alignment processes were repeated until no further improvement was evident.
For image subtraction comparisons, a threshold value was first applied to the averaged images. They were then manually aligned for best fit, and one image was subtracted from the other.
Molecular modeling.
The SwissPDBviewer program (25) and Viewerlite software (Accelrys, Inc., San Diego, Calif.) were used to enhance the electron microscopy-based interpretations and to investigate the likely location of the gp41 domain in SOS gp140.
RESULTS
Assembly and cleavage of purified SOS gp140.
We have previously described the antigenic properties of unpurified HIV-1JR-FL SOS gp140 proteins produced via transient transfection of 293T cells (4). To facilitate preparation of larger amounts of this protein for evaluation in purified form, we constructed a stable CHO cell line that expresses both SOS gp140 and human furin. Heterologous furin was expressed to facilitate efficient proteolytic processing of SOS gp140 (4).
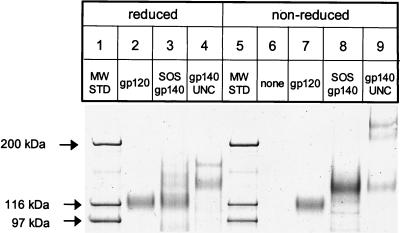
The SOS gp140 protein was purified from CHO cell supernatants to >91% homogeneity as determined by SDS-PAGE and densitometric analysis of the nonreduced protein (Fig. 1, lane 8). Only minor amounts of free gp120 were present in the SOS gp140 preparation, indicating that the intersubunit disulfide bond remained substantially intact during purification. No high-molecular-mass SOS gp140 oligomers or aggregates were observed (Fig. 1, lane 8). Under nonreducing conditions, SOS gp140 migrated as a predominant 140-kDa band. The major contaminant was bovine alpha 2-macroglobulin, which migrates as a ∼170-kDa band on a reducing SDS-PAGE gel (Fig. 1, lane 3) and can be eliminated by adaptation of the CHO cell line to serum-free culture (unpublished results). Upon reduction with DTT, the purified SOS gp140 protein migrated as a predominant 120-kDa band, with a minor (∼14%) fraction of the 140-kDa band present (Fig. 1, lane 3). These data indicated that approximately 86% of the SOS gp140 protein was proteolytically processed.
FIG. 1.
SDS-PAGE analysis of purified HIV-1JR-FL SOS gp140, gp140UNC, and gp120 proteins. CHO cell-expressed proteins (0.5 μg) in Laemmli sample buffer with (reduced) or without (non-reduced) 50 mM DTT were resolved on a 3 to 8% polyacrylamide gradient gel.
The HIV-1JR-FL gp140UNC protein was expressed in CHO cells using similar methods, although without cotransfected furin, and was also obtained at ∼90% purity. It too contained alpha 2-macroglobulin as the major contaminant, but no free gp120 was detectable (Fig. 1, lanes 4 and 9). In the absence of DTT, alpha 2-macroglobulin migrates as a ∼350-kDa dimer and is not clearly resolved from gp140UNC oligomers (Fig. 1, lane 9). Under nonreducing conditions, bands consistent with gp140UNC monomers (140 kDa), dimers (280 kDa), and trimers (420 kDa) were observed in roughly equal amounts (Fig. 1, lane 9). These proteins were reactive with anti-gp120 MAbs in Western blot analysis (data not shown). When treated with DTT, gp140UNC gave rise to an intensified monomer band at 140 kDa and an alpha 2-macroglobulin monomer band at ∼170 kDa; but gp140 oligomers were absent (Fig. 1, compare lanes 4 and 9). Thus, disulfide-linked, reducible oligomers comprise half or more of the gp140UNC preparation. Comparable amounts of reducible oligomers have been observed in gp140UNC protein preparations derived from subtype A, B, and E viruses, with minor strain-to-strain differences (46, 67). Reducible gp160 oligomers of this type have been proposed to contain aberrant intermolecular disulfide bonds (46). If so, at least some of the oligomers present in gp140UNC preparations represent misfolded protein aggregates.
Biophysical properties of purified SOS gp140. (i) Matrix-assisted laser desorption ionization mass spectrometry.
This technique was used to determine the absolute molecular masses of HIV-1JR-FL gp120 and SOS gp140. The measured molecular masses were 121.9 kDa for SOS gp140 and 91.3 kDa for gp120. Reduced SOS gp140 gave rise to a small peak of uncleaved gp140 at 118.5 kDa, a gp120 peak at 91.8 kDa, and a gp41ECTO peak at 27 kDa. Differences in glycosylation between cleaved and uncleaved SOS gp140 proteins could account for the 3.4-kDa difference in their measured masses. A difference of ∼0.5 kDa was observed in the mass of gp120 when expressed alone and in the context of SOS gp140. This difference is not significant but rather is within the range of assay variation for a heavily glycosylated protein. The measured mass of HIV-1JR-FL gp120 is comparable to previously reported molecular masses of CHO cell-expressed HIV-1GB8 gp120 (91.8 kDa) and Drosophila cell-expressed HIV-1WD61 gp120 (99.6 kDa) (30, 45). The anomalously high molecular masses (∼120 and ∼140 kDa, respectively [Fig. 1]) observed for gp120 and SOS gp140 by SDS-PAGE reflect the high carbohydrate content of these proteins. The extended structure of the glycans and their poor reactivity with the dodecyl sulfate anion retard the electrophoretic migration of the glycoproteins through SDS-PAGE gel matrices (30).
(ii) Ultracentrifugation.
Sedimentation equilibrium measurements were used to examine the oligomeric state of purified SOS gp140. Over protein concentrations ranging from 0.15 to 0.60 mg/ml, the apparent molecular mass of SOS gp140 was consistently found to be 155 kDa (Fig. 2A). Hence, the purified SOS gp140 protein is monomeric in solution. There was no systematic dependence of molecular mass on protein concentration over the range studied, since the molecular masses were all within 15% of those calculated for an ideal monomer. However, the residuals (the difference between the data and the theoretical curve for a monomer) deviated from zero in a systematic fashion (Fig. 2A), suggesting the presence of small amounts of oligomeric material. The difference in molecular masses observed using mass spectrometry (122 kDa) and sedimentation equilibrium (155 kDa) may reflect imprecisions in the latter technique. In particular, the uncertain composition of the SOS gp140 glycans introduces an error of this magnitude into calculations of the molecule's partial specific volume. However, this issue does not obscure our chief conclusion that the SOS gp140 protein is a monomer.
FIG. 2.
Biophysical analyses of purified, CHO cell-expressed HIV-1JR-FL envelope glycoproteins. (A) Sedimentation equilibrium studies of SOS gp140 indicate that it is monomeric in solution. Representative data (open circles) obtained at a rotor speed of 9,000 rpm and a protein concentration of 0.6 mg/ml are plotted as ln(absorbance) versus the square of the axis of rotation. The slope is proportional to the molecular weight of the protein oligomer. Dashed lines with increasing slopes indicate, respectively, the predicted data for monomeric (1), dimeric (2), and trimeric (3) states of the protein. Deviations from the calculated values are plotted as residuals in the upper portion of the figure. (B) Analytical size exclusion chromatography. Purified SOS gp140, gp140UNC, and gp120 proteins were resolved on a TSK G3000SWXL column in phosphate-buffered saline, and their retention times were compared with those of known molecular mass (MW) standard proteins of 220, 440, and 880 kDa (arrowed). The main peak retention time of SOS gp140 (5.95 min) is consistent with it being a monomer that is slightly larger than monomeric gp120 (retention time, 6.24 min), whereas gp140UNC (retention time, 4.91 min) migrates as an oligomeric species. (C) The oligomeric status of pure standard proteins, thyroglobulin, ferritin, and albumin, were compared with gp120 and gp120 in complex with soluble CD4 using BN-PAGE. The proteins were visualized on the gel using Coomassie blue. (D) BN-PAGE analysis of CHO cell-derived, purified HIV-1JR-FL gp120, SOS gp140, and gp140UNC glycoproteins.
(iii) Analytical gel filtration chromatography.
Purified HIV-1JR-FL SOS gp140, gp140UNC, and gp120 proteins were also examined using size exclusion chromatography. Monomeric gp120 eluted with a retention time of 6.24 min and an apparent molecular mass of ∼200 kDa (Fig. 2B). The apparently large size of this protein reflects the extended structures of its carbohydrate moieties. The retention time (5.95 min) and apparent molecular mass (∼220 kDa) of the SOS gp140 protein are consistent with it being a monomer that is slightly larger than gp120. In contrast, the gp140UNC protein eluted at 4.91 min as a broad peak with an average molecular mass of >500 kDa, which is consistent with it comprising a mixture of oligomeric species. Although the chromatogram suggests the existence of multiple species in the gp140UNC preparation, this gel filtration technique cannot resolve mixtures of gp140 dimers, trimers, and tetramers.
(iv) Blue Native polyacrylamide gel electrophoresis.
BN-PAGE was used to examine the oligomeric state of the purified SOS gp140 and gp140UNC proteins. In BN-PAGE, most proteins are fractionated according to their Stokes radius (62, 63). We first applied this technique to a model set of soluble proteins, including gp120 alone and in complex with sCD4 (Fig. 2C). The model proteins included thyroglobulin and ferritin, which naturally comprise a distribution of noncovalent oligomers of various sizes. The oligomeric states of these multisubunit proteins, as determined by BN-PAGE, are similar to those observed using other nondenaturing techniques (24, 77). BSA exists as monomers, dimers, and higher-order species in solution (35); the same ladder of oligomers was observed in BN-PAGE. Not surprisingly, the gp120/sCD4 complex, which has an association constant in the nanomolar range (1), remained intact during BN-PAGE analysis.
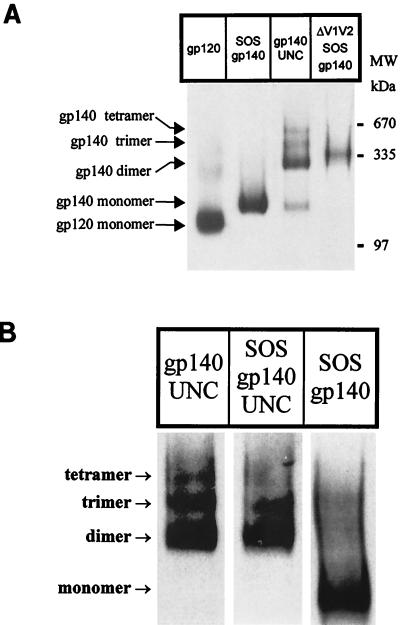
The purified SOS gp140 protein was largely monomeric by BN-PAGE (Fig. 2D), although a minor amount (<10%) of dimeric species was also observed. The purified gp140UNC protein migrated as well-resolved dimers, trimers, and tetramers, with trace amounts of monomer present (Fig. 2D). The gp140UNC dimer represented the major oligomeric form of the protein present under nondenaturing conditions. Although tetrameric gp140UNC is a distinct minor species on BN-PAGE gels (Fig. 2D), it is absent from nonreduced SDS-PAGE gels (Fig. 1). Upon treatment with SDS and heat, the gp140UNC tetramers probably revert to lower-molecular-weight species, such as monomers and/or disulfide-linked dimers. As expected, HIV-1JR-FL gp120 migrated as a predominant 120-kDa monomeric protein. BN-PAGE analyses of unpurified gp140 proteins are described below (see Fig. 7).
FIG. 7.
BN-PAGE analyses of unfractionated cell culture supernatants. (A) Comparison of HIV-1JR-FL gp120, SOS gp140, gp140UNC, and ΔV1V2 SOS gp140 glycoproteins present in culture supernatants from stable CHO cell lines. (B) Proteolytic cleavage destabilizes gp140 oligomers. 293T cells were transfected with furin and plasmids encoding SOS gp140, gp140UNC, or SOS gp140UNC. Cell culture supernatants were combined with MOPS buffer containing 0.1% Coomassie blue and resolved by BN-PAGE. Proteins were then transferred to PVDF membranes and visualized by Western blotting. Thyroglobulin and the BSA dimer were used as molecular mass (MW) markers (see Fig. 2C).
Overall, ultracentrifugation, gel filtration, and BN-PAGE analyses were in excellent agreement as to the oligomeric states of these purified Env proteins. BN-PAGE, however, was the only method capable of clearly resolving the mixture of oligomeric species contained in the gp140UNC preparation.
Immunoelectron microscopy of SOS gp140. (i) SOS gp140 and SOS gp140-MAb complexes.
In the absence of antibodies, the electron micrographs revealed SOS gp140 to be mostly monomeric, randomly oriented, and multilobed (Fig 3A). Qualitatively similar images were obtained with HIV-1JR-FL gp120 (data not shown), and the two proteins could not be clearly distinguished in the absence of MAbs or other means of orienting the images.
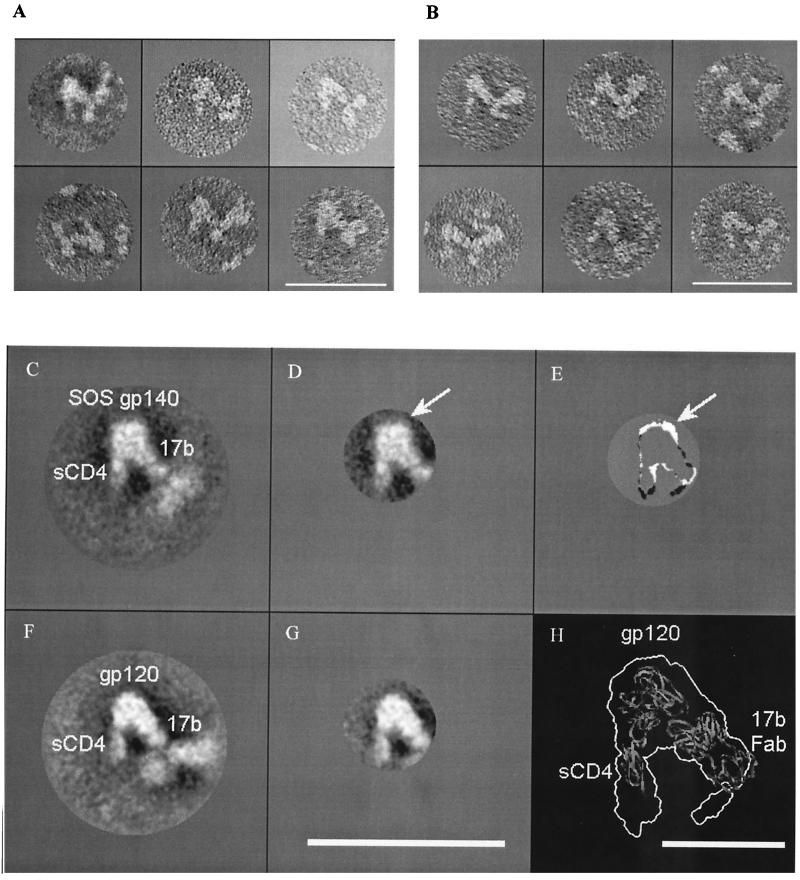
FIG. 3.
Negative-stain electron micrographs of SOS gp140 alone (A) and in complex with MAbs (B to F). Bar, 40 nm. In panels B to F, the panels were masked and rotated so that the presumptive Fc of the MAb is oriented downward. When multiple MAbs were used, the presumptive Fc of MAb 2F5 is oriented downward. In panels B to F, interpretative diagrams are also provided to illustrate the basic geometry and stoichiometry of the immune complexes. SOS gp140, intact MAb, and F(ab)′2 are illustrated by ovals, Y-shaped structures, and V-shaped structures, respectively, in the schematic diagrams, which are not drawn to scale. The MAbs used are as follows: 2F5 (B), IgG1b12 (C), 2G12 (D), MAb 2F5 plus F(ab′)2 IgG1b12 (E), and MAb 2F5 plus MAb 2G12 (F).
Electron micrographs were also obtained of SOS gp140 in complex with MAbs 2F5 (Fig. 3B), IgG1b12 (Fig. 3C), and 2G12 (Fig. 3D). To aid in interpretation, the complexes were masked and rotated such that the presumptive Fc of the MAb points downward. Schematic diagrams are also provided for each complex in order to illustrate the basic geometry and stoichiometry observed. In each case, the complexes shown represent the majority or plurality species present. However, other species, such as free MAb and monovalent MAb-SOS gp140 complexes, were also present in each sample (data not shown).
When combined with IgG1b12 or 2F5, SOS gp140 formed rather typical immune complexes composed of a single MAb and up to two SOS gp140s (Fig. 3B and C). The complexes adopted the characteristic Y-shaped antibody structure, with a variable angle between the Fab arms of the MAb. In contrast, the 2G12-SOS gp140 complexes produced strikingly different images (Fig. 3D). Y-shaped complexes comprising two distinct Fab arms with bound SOS gp140s were rare. Instead, the 2G12-SOS gp140 images were highly linear and appeared to represent one MAb bound to two SOS gp140 proteins aligned in parallel. Similar structures were observed less frequently for 2G12 in a complex with HIV-1JR-FL gp120, but not with HIV-1YU2 gp120 (data not shown). We suspect that the parallel alignment of the SOS gp140s forces the two Fab arms into similar alignment, resulting in an overall linear structure. These complexes are unprecedented in our immunoelectron microscopy studies of Env-MAb complexes (55, 56, 92; K. H. Roux, unpublished observations). One hypothesis is that 2G12 binds to HIV-1JR-FL gp120 and SOS gp140 in an orientation that promotes residual weak gp120-gp120 and/or gp41-gp41 interactions, which then stabilize the complex in the parallel configuration observed. Additional studies are ongoing to further explore this finding.
Combinations of these well-characterized MAbs were used to examine the relative placement of their epitopes on SOS gp140. In the first combination, SOS gp140-2F5-IgG1b12, multiple ring structures were observed which appeared to be composed of two SOS gp140 proteins bridged by two antibody molecules (data not shown). To distinguish between the 2F5 and IgG1b12 MAbs, we examined complexes formed between IgG1b12 F(ab′)2, SOS gp140, and the intact 2F5 MAb. Characteristic ring structures were again observed (Fig. 3E). The ring complexes were then subjected to computational analysis using the SPIDER program package to yield several categories of averaged images (data not shown). The MAb 2F5 and IgG1b12 F(ab′)2 components can be clearly delineated in the images, as can the SOS gp140 molecule. When bound to a given SOS gp140 molecule, the Fab arms of 2F5 and IgG1b12 lie at approximately right angles, as indicated in the schematic diagram (Fig. 3E).
In marked contrast to the IgG1b12-containing ternary complexes, those composed of SOS, 2F5, and 2G12 formed extended chains rather than closed rings (Fig. 3F). These observations place the 2F5 and 2G12 epitopes at opposite ends of the SOS gp140 molecule. There was significant heterogeneity in the stoichiometry of the 2F5/2G12/SOS gp140 complexes, just one example of which is indicated in the schematic diagram.
(ii) Immunoelectron microscopy of SOS gp140 and gp120 in complex with sCD4 and MAb 17b.
In an effort to further characterize the topology of SOS gp140, we reacted it with MAb 17b and/or sCD4. We generated the corresponding YU2 gp120 complexes for comparison. As expected, the combination of MAb 17b plus SOS gp140 or gp120 alone did not form complexes, consistent with the need for sCD4 to induce the 17b epitope. Similarly, unremarkable complexes were obtained when sCD4 was mixed with SOS gp140 or gp120 in the absence of MAb 17b (data not shown). However, complexes with clearly defined geometry were obtained for sCD4/Env/17b (Fig. 4A and B).
FIG. 4.
Individual, averaged and subtracted electron micrographs of SOS gp140 and gp120 in complex with sCD4 and MAb 17b. Panels A and B are individual electron micrographs of ternary complexes of SOS gp140 (A) and YU2 gp120 (B). The Fc region of MAb 17b is aligned downward. Panels C and F are averaged electron micrographs of ternary complexes of SOS gp140 (C) and gp120 (F). Panels D and G are masked and averaged electron micrographs of the SOS gp140 complex (D) and the gp120 complex (G). Panel E represents the density remaining upon subtraction of the gp120 complex (G) from the gp140 complex (D). In panels D and E, the arrow indicates the area of greatest residual density, which represents the presumptive gp41ECTO moiety that is present in SOS gp140 but not in gp120. Panel H indicates the outline of the gp120 complex (G) overlaid upon a ribbon diagram of the X-ray crystal structure of the gp120 core in complex with sCD4 and the 17b Fab fragment (PDB code 1GC1) (33). The gp120 complex was enlarged to facilitate viewing. Bar, 40 nm (A to G) or 10 nm (H).
These complexes were composed of 17b with one or two attached SOS gp140s or gp120s, together with tangentially protruding sCD4 molecules. These complexes were then subjected to computer-assisted averaging (Fig. 4C and F). The free arm and the Fc region of MAb 17b were disordered in these images due to the flexibility of the MAb, so the averaged images, based on multireference alignment (23), were masked to highlight the better-resolved sCD4, Env, and 17b Fab structures (Fig. 4D and G). The gp120 and SOS gp140 images were qualitatively similar, but an image subtraction of one from the other revealed the presence of additional mass on the SOS gp140 protein (arrowed in Fig. 4D and E). For this process, a threshold cutoff value was first applied to the averaged SOS gp140/sCD4/17b (Fig. 4D) and gp120/sCD4/17b (Fig. 4G) images. The two profiles were then manually aligned for best fit, and the image of the gp120 complex was subtracted from that of the SOS gp140 complex. This additional mass may represent gp41ECTO, although we cannot strictly exclude other explanations, such as differences in the primary sequence and/or glycosylation of the gp120 and SOS gp140 proteins used. The resolution of the technique does not enable us to estimate the molecular weight of the additional mass. Moreover, a significant portion of the gp41ECTO moiety of SOS gp140 could reside “under” gp120 and thus be hidden in the image.
In order to orient the putative gp41ECTO moiety in relation to the remaining structures seen in the electron micrographs, the X-ray structure of the gp120 core in complex with the D1D2 domain of sCD4 and Fab 17b (33) was docked, using Program O, into the profile map obtained for the sCD4/gp120/MAb 17b complex (Fig. 4H). Given that there are differences in the gp120 (whole versus core) and CD4 (four-domain versus two-domain) molecules used for the electron microscopy and crystallization studies, there is reasonable agreement in the overall topology of the structures generated.
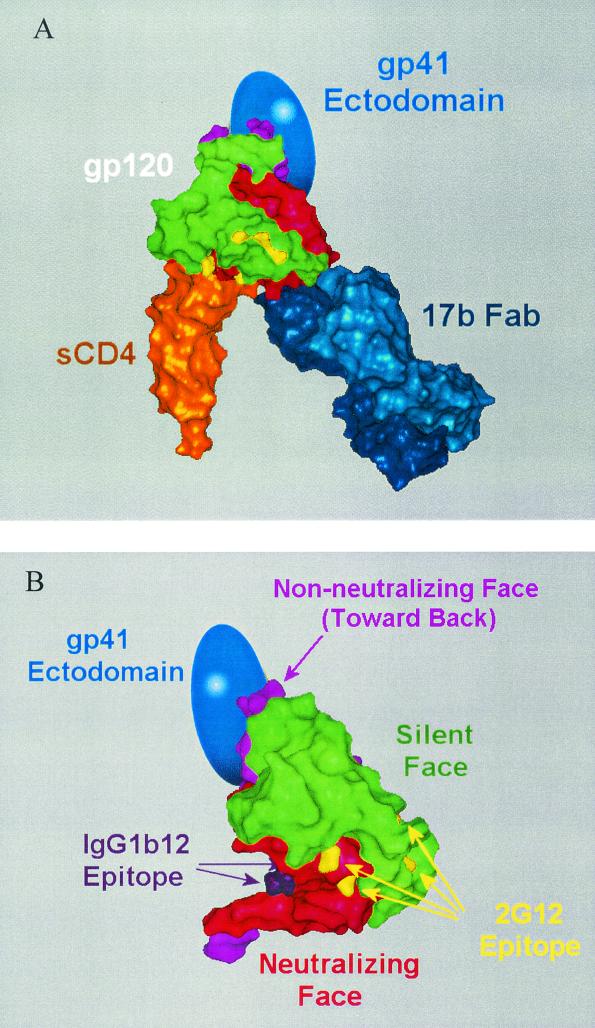
This agreement in structures (Fig. 4H) enabled us to position the putative gp41ECTO moiety in relation to the core gp120 structure (Fig. 5). The previously defined neutralizing, nonneutralizing, and silent faces of gp120 (41, 83) are illustrated, as are the IgG1b12 (58) and 2G12 (83) epitopes. According to this model, the gp41ECTO moiety recognized by MAb 2F5 is located at ∼90° relative to the IgG1b12 epitope and ∼180° from the 2G12 epitope (Fig. 5B). This model is in broad agreement with the independently derived electron microscopy images of the complexes formed between SOS gp140 and combinations of these MAbs (Fig. 3E and F). This putative placement of gp41ECTO would cause it to largely occlude the nonneutralizing face of gp120, a result that is consistent with the MAb reactivity patterns observed for SOS gp140 both here and elsewhere (4).
FIG. 5.
Models indicating the approximate location of gp41ECTO in relation to gp120 as derived from electron microscopy data of SOS gp140. (A) Presumptive location of gp41ECTO (represented by the blue oval) in relation to the X-ray crystal structure of the gp120 core (green, silent face; red, neutralizing face; lavender, nonneutralizing face [41, 83]) in complex with sCD4 (orange) and Fab 17b (dark blue) (PDB code 1GC1) (33). (B) Surface rendering of the gp120 core with faces colored as above. Also indicated are the key residues of the IgG1b12 (58) (purple) and 2G12 (83) (yellow) epitopes. The image of the gp120 core in panel B is rotated 60° to the right about the vertical axis and 15° upward about the horizontal axis with respect to panel A.
Antigenic properties of unpurified SOS gp140 and gp140UNC proteins. (i) RIPAs.
RIPA was used to determine whether the antigenicity of HIV-1JR-FL SOS gp140 differed when the protein was expressed in stably transfected CHO cells from what was observed previously when the same protein was expressed in transiently transfected 293T cells (4). The SOS gp140 proteins in unpurified supernatants expressed from CHO cells were efficiently recognized by neutralizing agents to gp120 epitopes located in the C3/V4 region (MAb 2G12), the CD4 binding site (the CD4-IgG2 molecule), and the V3 loop (MAb 19b) (data not shown). In addition, the conserved CD4-induced neutralization epitope defined by MAb 17b was strongly induced on SOS gp140 by sCD4. SOS gp140 was also efficiently immunoprecipitated by the broadly neutralizing gp41 MAb 2F5. In contrast, SOS gp140 was largely unreactive with the nonneutralizing MAbs 23A and 2.2B to gp120 and gp41, respectively (data not shown). These findings are consistent with our previous observations (4) and indicate that CHO and 293T cell-derived HIV-1JR-FL SOS gp140 proteins possess similar antigenic properties.
Relatively minor amounts of free gp120 were observed in the unpurified SOS gp140 CHO cell supernatants (data not shown), as reported previously for 293T-derived material (4). This free gp120 was preferentially recognized by MAb 23A, suggesting that its C5 epitope is largely obscured in SOS gp140 (data not shown). This is consistent with the electron microscopy-derived topology model described above (Fig. 5B) and with what is known about the gp120-gp41 interface (27, 41, 82). As with 293T-derived material (4), processing of SOS gp140 at the gp120-gp41 cleavage site was efficient in CHO cells, as determined by RIPAs performed under reducing and nonreducing conditions (data not shown). Similar levels of assembly and proteolytic processing were observed when unpurified SOS gp140 was analyzed by Western blotting rather than RIPA (data not shown). Thus, the folding, assembly, and processing of this protein appear to be largely independent of the cell line used for its production.
(ii) SPR assays.
Surface plasmon resonance (SPR) was used to further characterize the antibody- and receptor-binding properties of unpurified, CHO cell-expressed SOS gp140 and gp140UNC proteins. A comparison of results obtained using SPR and RIPA with the same MAbs allows us to determine if the antigenicity of these proteins is method dependent. Whereas SPR is a kinetically limited procedure that is completed in one or more minutes, RIPA is an equilibrium method in which Env-MAb binding occurs over several hours. SPR analysis was also performed on purified and unpurified forms of the SOS gp140 and gp140UNC proteins, to assess whether protein antigenicity was significantly altered during purification. Purified HIV-1JR-FL gp120 was also studied. Although the purified SOS gp140 protein is a monomer, it does contain the gp120 subunit linked to the ectodomain of gp41. Since there is evidence that the presence of gp41 can affect the antigenic structure of gp120 (32, 53, 79), we thought it worth determining whether monomeric SOS gp140 behaved differently than monomeric gp120 in its interactions with neutralizing and nonneutralizing MAbs.
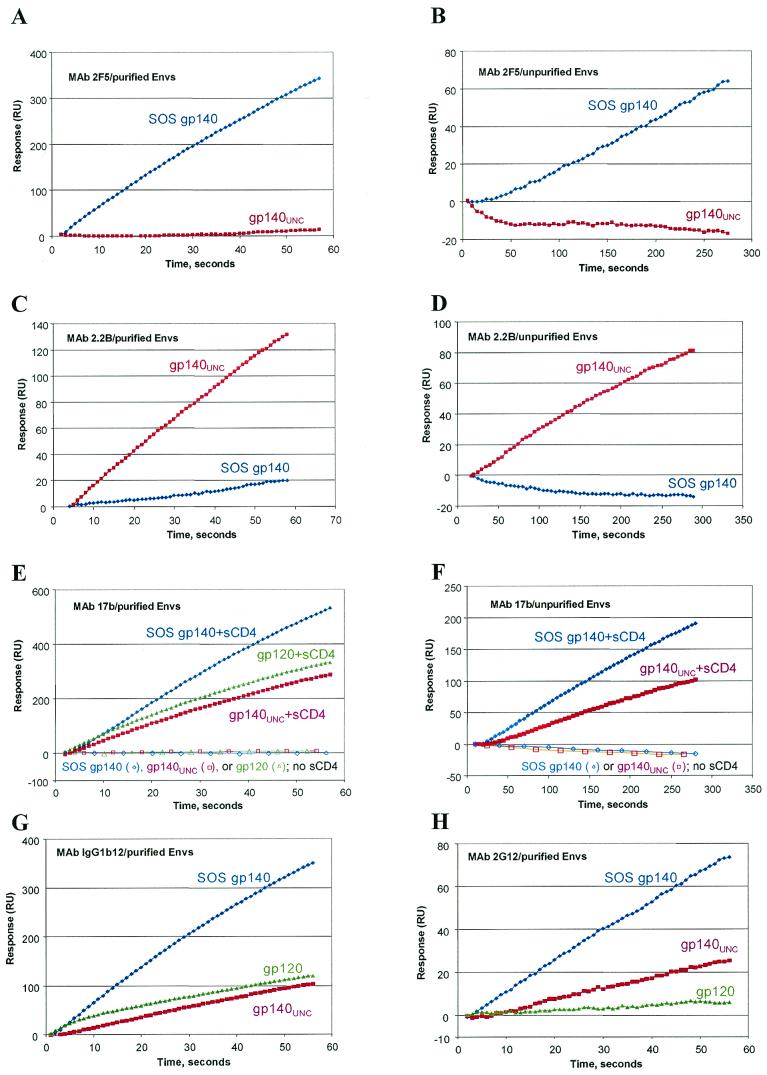
There was good concordance of results between RIPA- and SPR-based (Fig. 6) antigenicity analyses of unpurified SOS gp140 in CHO cell supernatants. For example, SOS gp140 bound the broadly neutralizing anti-gp41 MAb 2F5 (Fig. 6B) but not the nonneutralizing anti-gp41 MAb 2.2B (Fig. 6D). Similarly, binding of MAb 17b was strongly potentiated by sCD4 (Fig. 6F). Unpurified SOS gp140 bound the neutralizing anti-gp120 MAbs 2G12 and 19b but not the nonneutralizing anti-gp120 MAb 23A in both SPR and RIPA experiments (data not shown). Taken together, the RIPA and SPR data indicate that unpurified, CHO cell-derived SOS gp140 rapidly and avidly binds neutralizing anti-gp120 and anti-gp41 MAbs, whereas binding to the present set of nonneutralizing MAbs is not measurable by either technique.
FIG.6.
SPR analysis of CHO cell-expressed HIV-1JR-FL SOS gp140, gp140UNC, and gp120 proteins. Anti-gp120 and anti-gp41 MAbs were immobilized onto sensor chips and exposed to buffers containing the indicated gp120 or gp140 glycoproteins in either purified or unpurified form, as indicated. Where noted, Env proteins were mixed with an eightfold molar excess of sCD4 for 1 h prior to analysis. Culture supernatants from stably transfected CHO cells were used as the source of unpurified SOS gp140 and gp140UNC proteins. The concentrations of these proteins were measured by Western blotting and adjusted so that approximately equal amounts of each protein were loaded. Only the binding phases of the sensorgrams are shown; in general, the dissociation rates were too slow to provide meaningful information.
SPR revealed some significant differences in the reactivities of SOS gp140 and gp140UNC proteins with anti-gp41 MAbs. Thus, SOS gp140 but not gp140UNC bound MAb 2F5 but not MAb 2.2B, whereas the converse was true for gp140UNC. Notable, albeit less dramatic, differences were observed in the reactivity of SOS gp140 and gp140UNC with some anti-gp120 MAbs. Of the two proteins, SOS gp140 had the greater kinetics and magnitude of binding to the neutralizing MAbs IgG1b12 (Fig. 6G), 2G12 (Fig. 6H), and 17b in the presence of sCD4 (Fig. 6E and F). The binding of gp140UNC to 17b was clearly potentiated by sCD4, as has been reported elsewhere (90). Neither SOS gp140 nor gp140UNC bound the anti-gp120 MAb 23A (data not shown). This was expected for gp140UNC, since the C5 amino acid substitutions that eliminate the cleavage site directly affect the epitope for MAb 23A (43).
Qualitatively, the antigenicities of SOS gp140 and gp140UNC were little changed upon purification (Fig. 6, compare panels A, C, and E with panels B, D, and F). Hence the lectin affinity and gel filtration columns used for purification do not appear to significantly affect, or select for, a particular conformational state of these proteins. However, these studies do not allow for direct, quantitative comparisons of SPR data derived using purified and unpurified materials.
Compared with monomeric gp120, the purified gp140UNC protein reacted more strongly with MAb 2G12 but less strongly with MAb IgG1b12. Prior SPR studies have demonstrated that 2G12 avidly binds to oligomeric forms of Env (89), and it is possible that MAb 2G12 is capable of undergoing bivalent binding to oligomeric Envs. It will be informative to perform electron microscopy analyses of 2G12 in complex with gp140UNC or other oligomeric Envs in future studies, given the unusual nature of the 2G12-SOS gp140 complex (Fig. 3D).
Oligomeric properties of unpurified SOS gp140 and gp140UNC proteins.
BN-PAGE was used to examine the oligomeric state of the SOS gp140 and gp140UNC proteins present in freshly prepared, CHO cell culture supernatants. The SOS gp140 protein was largely monomeric by BN-PAGE, with only a minor proportion of higher-order proteins present (Fig. 7A). In some, but not all, 293T cell preparations, greater but highly variable amounts of dimers and higher-order oligomers were observed using BN-PAGE (data not shown, but see Fig. 7B). This probably accounts for our previous report that oligomers can be observed in unpurified SOS gp140 preparations using other techniques (4).
The unpurified gp140UNC protein typically migrated as well-resolved dimers, trimers, and tetramers, with trace amounts of monomer sometimes present (Fig. 7A). Qualitatively similar banding patterns were observed for purified (Fig. 2D) and unpurified (Fig. 7A) gp140UNC proteins. In each case, dimers of gp140UNC were the most abundant oligomeric species. HIV-1JR-FL gp120 ran as a predominant 120-kDa monomeric band, although small amounts of gp120 dimers were observed in some unpurified supernatants. In general, the BN-PAGE analyses indicate that the oligomeric properties of the various Env proteins did not change appreciably upon purification (compare Fig. 7A and 2D).
The same CHO cell supernatants were also analyzed by analytical gel filtration, the column fractions being collected in 0.2-ml increments and analyzed for Env content by Western blotting. The retention times of unpurified gp120, SOS gp140, and gp140UNC proteins were determined to be ∼6.1, ∼5.9, and ∼5.2 min, respectively (data not shown). These values agree with those observed for the purified proteins (Fig. 2B) to within the precision of the method. The gel filtration studies thus corroborate the BN-PAGE data in that unpurified gp120 and SOS gp140 were mostly monomeric, while gp140UNC was mostly oligomeric (data not shown). However, unlike BN-PAGE, this analytical gel filtration procedure does not have sufficient resolving power to characterize the distribution of the oligomeric species present in the gp140UNC preparation.
SDS-PAGE followed by Western blot analyses of supernatants containing unpurified SOS gp140 and gp140UNC proteins yielded banding patterns similar to those shown in Fig. 1 for the purified proteins (data not shown). The gp120 preparation contained ∼10% dimer, which was observed only when SDS-PAGE analyses were carried out under nonreducing conditions. Thus, the gp120 dimer represents disulfide-linked and presumably misfolded material (46).
SOS gp140 glycoproteins with deletions of variable loops form more-stable oligomers.
We previously described HIV-1JR-FL SOS gp140 glycoproteins from which one or more of the gp120 variable loops were deleted to better expose underlying, conserved regions around the CD4- and coreceptor-binding sites. It was possible to remove the V1, V2, and V3 loop structures individually or in pairs without adversely affecting the formation of the intersubunit disulfide bond, proper proteolytic cleavage, or protein folding. However, the triple loop deletant was not efficiently cleaved (57). In order to explore the oligomeric properties of these modified SOS gp140 glycoproteins, the supernatants of 293T cells transiently cotransfected with these gp140 constructs and furin were analyzed by BN-PAGE. Unexpectedly, deletion of the variable loops, both alone and in combination, significantly enhanced the stability of the SOS gp140 oligomers. The ΔV1V2 SOS gp140 preparation contained almost exclusively trimeric and tetrameric species, whereas ΔV1 SOS gp140 formed a mixture of dimers, trimers, and tetramers similar to that seen with gp140UNC (data not shown). The ΔV2 SOS gp140 protein was predominantly oligomeric, but it also contained significant quantities of monomer. Thus, in terms of oligomeric stability, the SOS proteins can be ranked as follows: first ΔV1V2 SOS gp140, then ΔV1 SOS gp140, then ΔV2 SOS gp140, and then full-length SOS gp140. The reasons for this rank order are not yet clear but are under investigation.
Based on the above observations, we chose to generate a CHO cell line that stably expresses the ΔV1V2 SOS gp140 protein. Supernatants from the optimized CHO cell line were first analyzed by SDS-PAGE under reducing and nonreducing conditions, followed by Western blot detection. The major Env band was seen at 120 kDa (ΔV1V2 gp140 protein) in the nonreduced gel and at 100 kDa (ΔV1V2 gp120 protein) in the reduced gel (data not shown). These results are consistent with our prior findings that deletion of the V1V2 loops decreases the apparent molecular mass of the protein by ∼20 kDa (57). Notably, the ΔV1V2 SOS gp140 protein was largely free both of disulfide-linked aggregates and of the ∼100-kDa loop-deleted, free gp120 protein. Thus, proteolytic cleavage and SOS disulfide bond formation occur efficiently in the ΔV1V2 SOS gp140 protein (data not shown).
CHO cell supernatants containing ΔV1V2 SOS gp140, full-length SOS gp140, and gp140UNC were also analyzed by BN-PAGE and Western blotting (Fig. 7A). As was observed with the transiently transfected 293T cells, unpurified CHO cell-derived material was oligomeric. The CHO cell-derived ΔV1V2 SOS gp140 migrated as a distinct single band with a molecular mass consistent with that of a trimer (360 kDa); the ΔV1V2 SOS gp140 band lies between those of the gp140UNC dimer (280 kDa) and the gp140UNC trimer (420 kDa) (Fig. 7A). Hence the ΔV1V2 SOS gp140 protein represents a proteolytically mature form of HIV-1 Env that oligomerizes into presumptive trimers via noncovalent interactions. Purification and additional biophysical studies of this protein are now in progress, and immunogenicity studies are planned.
The uncleaved SOS gp140 and gp140UNC proteins possess similar oligomeric properties.
Overall, the above analyses reveal a clear difference in the oligomeric properties of the SOS gp140 and gp140UNC proteins. One structural difference between these proteins is their proteolytic cleavage status; another is the presence or absence of the intersubunit disulfide bond that defines SOS gp140 proteins. To address the question of whether it is gp120-gp41 cleavage or the introduced cysteine residues that destabilize the SOS gp140 oligomers, we made the SOS gp140UNC protein. Here, the cysteines capable of intersubunit disulfide bond formation are present, but the cleavage site between gp120 and gp41ECTO has also been modified to prevent cleavage. The SOS gp140UNC, SOS gp140, and gp140UNC proteins were all expressed transiently in 293T cells and analyzed by BN-PAGE (Fig. 7B). In this and multiple repeat experiments, SOS gp140UNC and gp140UNC had similar migration patterns on the native gel, with the dimer band predominating and some monomers, trimers, and tetramers also present. In contrast, SOS gp140 was primarily monomeric, although small amounts of dimeric and trimeric species were also observed in this particular analysis (Fig. 7B).
The above results suggest that the SOS gp140UNC protein behaves more like the gp140UNC protein than the SOS gp140 protein. This, in turn, implies that the cleavage of gp140 into gp120 and gp41ECTO has a substantial effect on how gp140 is oligomerized via interactions between the gp41ECTO moieties, whereas the presence of the cysteine substitutions in gp120 and gp41 has little effect on these interactions. We believe that this observation is central to understanding the relative instability of SOS gp140 oligomers, compared to those of the gp140UNC protein. We note, however, that we have not determined whether or not the intermolecular disulfide bond actually forms in SOS gp140UNC; the simple method of DTT treatment to reduce this bond is inadequate, because the uncleaved peptide bond between the gp120 and gp41ECTO moieties still holds the two subunits together. Addressing this issue will require characterizing purified SOS gp140UNC by methods such as peptide mapping. Such studies are now in progress, to further explore the effect of gp140 cleavage on the structure of the gp120-gp41ECTO complex.
DISCUSSION
We have previously described the antigenic properties of SOS gp140, an HIV-1 envelope glycoprotein variant in which an intermolecular disulfide bond has been introduced to covalently link the gp120 and gp41ECTO subunits (4, 57). In the original report, we demonstrated that the SOS gp140 protein, as contained in supernatants of transiently transfected 293T cells, was an antigenic mimic of virion-associated Env (4). In that report, the methods employed were not sufficiently robust to conclusively determine the oligomeric state of unpurified 293T-derived SOS gp140 (4). Here we show with a smaller panel of MAbs that purified and unpurified CHO cell-derived SOS gp140 proteins also mimic native Env in terms of their patterns of antibody reactivity. However, unlike virus-associated Env, SOS gp140 is a monomeric protein.
Antigenicity and immunoelectron microscopy studies support a model for SOS gp140 in which the neutralizing face of gp120 is presented in a native conformation but the nonneutralizing face is occluded by gp41ECTO (Fig. 5). In one set of immunoelectron microscopy studies, SOS gp140 was examined in complex with combinations of anti-gp120 and anti-gp41 MAbs to defined epitopes (Fig. 3). The gp41ECTO subunit, as defined by the position of the anti-gp41 MAb 2F5, was located ∼180° from the MAb 2G12 epitope and ∼90° from the MAb IgG1b12 epitope, as is the nonneutralizing face. A second set of studies compared SOS gp140 and gp120 in complex with sCD4 and MAb 17b (Fig. 4). Here, a region of additional mass in the gp140 complex defined the presumptive gp41ECTO; its location was similarly adjacent to the nonneutralizing face of gp120. This model of the geometry of the gp120-gp41 interaction is consistent with previous models based on mutagenesis techniques and the mapping of MAb epitopes (27, 41, 82). It also provides a basis for interpreting the patterns of MAb reactivity, as discussed below.
In the present report, the antigenicity of CHO-derived SOS gp140 was explored from a number of perspectives: (i) in comparison with gp140UNC and gp120, (ii) before and after purification, and (iii) in an equilibrium-based assay (RIPA) versus a kinetics-based assay (SPR). SOS gp140 proteins expressed in stably transfected CHO cells or transiently transfected 293T cells possessed qualitatively similar antigenic properties that were largely unaffected by purification. These analyses utilized a seven-member panel of MAbs and CD4-based proteins; it is possible that subtle antigenic differences could be discerned with additional MAbs or with peptides that target the amino- or carboxy-terminal heptad regions of gp41. We observed that most neutralizing anti-gp120 MAbs bound more strongly and more rapidly to SOS gp140 than to the gp120 or gp140UNC proteins, whereas the converse was true of nonneutralizing MAbs (Fig. 6). These results were largely independent of the analytical methodology used (RIPA or SPR) or the purification state of the glycoproteins and thus extend our earlier RIPA analyses of unpurified Env glycoproteins (4). We have addressed these issues on a largely qualitative basis in the present study; quantitative comparisons of MAb reactivities are now being explored.
It is not obvious why neutralizing MAbs recognize monomeric SOS gp140 better than monomeric gp120. One possibility relates to differences in the conformational freedom of the two glycoproteins. Monomeric gp120 has considerable conformational flexibility, such that “freezing” of the conformation by CD4 binding results in an unexpectedly large loss in entropy (45). Indeed, it has been suggested that reducing the conformational freedom of a gp120 immunogen may provide a means of generating broadly neutralizing antibodies, which generally recognize conformational epitopes (45). The presence of gp41ECTO may serve to minimize the conformational flexibility of the gp120 subunit of SOS gp140, stabilizing the protein in conformations recognized by neutralizing antibodies. However, the induction of 17b binding by sCD4 demonstrates that SOS gp140 is still capable of sampling multiple, relevant conformations. Studies are in progress to address these issues.
Variations in conformational flexibility may also underlie the antigenic differences observed between the SOS gp140 and gp140UNC proteins. Other possible explanations include differences in oligomerization and cleavage. Further studies using additional Env protein variants (e.g., SOS gp140UNC), a broader range of anti-Env MAbs, and purified or size-fractionated proteins of a homogenous subunit composition will be required to explore these issues more thoroughly.
Standard biophysical techniques were used to demonstrate that the purified HIV-1JR-FL SOS gp140 glycoprotein is a monomer comprising one gp120 subunit that was disulfide linked to gp41ECTO. Since it is generally accepted that the gp41 subunits are responsible for Env trimerization (8, 10, 38, 69, 78), we assume that the gp41-gp41 interactions within the cleaved SOS gp140 glycoprotein are weak, and that this instability precludes the purification of cleaved trimers. Others have reported that gp41-gp41 interactions are unstable in the context of gp140 (21, 85). Moreover, ultracentrifugation and nuclear magnetic resonance spectroscopy studies of the gp41 monomer-trimer equilibrium indicate that monomers are favored at concentrations of <3 μM (9, 80), which are comparable to the highest concentration of SOS gp140 attained in this report. The 1-mg/ml stock solution of purified SOS gp140 is approximately 7 μM, and the protein underwent further dilution during gel filtration, ultracentrifugation, and other biophysical analyses.
We also report the application of a rapid, simple, and high-resolution electrophoretic technique, BN-PAGE, for exploring the oligomeric state of HIV-1 envelope glycoproteins in unpurified as well as purified forms. In this technique, the proteins of interest are combined with the dye Coomassie blue, which binds to the exposed hydrophobic surfaces of proteins and usually enhances their solubility. In the presence of the dye, most proteins adopt a negative charge, migrate towards the anode in an electric field, and so can be sieved according to their Stokes radius in a polyacrylamide gradient gel. Whereas traditional native PAGE methods are typically performed under alkaline conditions (pH 9.5), BN-PAGE uses a physiological pH (pH 7.5), which is more compatible with protein stability. We demonstrate that a gp120/sCD4 complex and a variety of purified, oligomeric model proteins all remain associated during BN-PAGE analysis. When combined with Western blot detection, BN-PAGE can be used to determine the oligomeric state of HIV-1 envelope glycoproteins at all stages of purification. This high-resolution technique can resolve monomeric, dimeric, trimeric, and tetrameric forms of gp140.
As determined by BN-PAGE and other methods, the SOS gp140 protein was secreted in mostly monomeric form. In contrast, gp140UNC formed oligomers that are significantly more stable. Thus, we show that HIV-1JR-FL gp140UNC comprises a mixture of dimers, trimers, and tetramers, with dimers representing the major oligomeric form present under nondenaturing conditions. Although noncovalently associated oligomers constitute a significant percentage of the gp140UNC preparation, half or more of the material consists of disulfide-linked and presumably misfolded material (46). Others have made similar observations with uncleaved gp140 proteins from other HIV-1 strains and from simian immunodeficiency virus (11, 16-21, 28, 46, 54, 66, 67, 85-87). The question then arises as to why the SOS gp140 protein is a monomer but the uncleaved proteins are oligomeric. We believe that the cleavage of the gp120-gp41 peptide bond alters the overall conformation of the envelope glycoprotein complex, rendering it fusion competent but also destabilizing the association between the gp41 subunits. Support for this argument is provided by the evidence that the SOS gp140UNC protein resembles gp140UNC rather than SOS gp140; cleavage is clearly more important than the introduced cysteines in determining the oligomeric stability of gp140 proteins. We hypothesize that destabilization of gp41-gp41 interactions might be necessary for gp41-mediated fusion to occur efficiently upon activation of the Env complex by gp120-receptor interactions. Moreover, having cleavage and activation take place late in the synthetic process minimizes the risk of fusion events occurring prematurely, i.e., during intracellular transport of the envelope glycoprotein complex. Additional studies are in progress to explore the effect of cleavage on Env structure.
We can only speculate on the structure of gp41ECTO in SOS gp140. The strong reactivity of MAb 2F5 suggests that the C-terminal portion mimics the native, pre-fusion conformation, but there are few probes available for other portions of the native molecule. Clearly, gp41ECTO does not adopt a postfusion, coiled-coil conformation in the SOS gp140 monomer. It's not obvious how coiled-coil interactions are abrogated by a single cysteine substitution in an area outside the heptad repeat regions of gp41, but one possibility is that covalent attachment of gp120 at this “hinge” region of gp41 sterically prevents formation of intermediate or final intra- or intermolecular interactions that are required to achieve the coiled-coil state.
Taken together, the antigenic and biophysical data of SOS gp140, gp120, and gp140UNC suggest that SOS gp140 represents an improved yet clearly imperfect mimic of native Env. It is perhaps surprising that an SOS gp140 monomer mimics virus-associated Env in its reactivity with the present panel of MAbs. Immunochemical studies and the X-ray crystal structure of the gp120 core in complex with CD4 and MAb 17b have together defined the surface of gp120 in terms of neutralizing, nonneutralizing, and silent faces (33, 83). The data presented here and elsewhere (4) demonstrate the neutralizing face is readily accessible on SOS gp140, whereas the nonneutralizing face is not. There are still no immunologic ways to probe the exposure of the silent face of gp120 (41). A source of purified SOS gp140 glycoprotein, as described herein, will facilitate further studies of the antigenic structure of SOS gp140 in comparison with that of native Env.
Do gp140UNC proteins mimic the structure of the native, fusion-competent envelope glycoprotein complex on virions? We believe not, based on their exposure of nonneutralization epitopes in both gp120 and gp41 that are not accessible on the surface of native envelope glycoprotein complexes (4, 50, 61, 88). Similarly, the Env proteins of several other viruses undergo dramatic refolding upon cleavage as determined by electron microscopy and patterns of antibody reactivity (5, 15, 22, 26, 47, 64).
However, cleavage does not induce global conformational changes in all viral Envs. High-resolution crystal structures have been obtained for both cleaved and uncleaved forms of the influenza hemagglutinin protein, which is perhaps the best-characterized viral coat protein. Hemagglutinin cleavage induced mostly localized refolding and had little impact on the overall conformation of the molecule (12, 65).
Given that SOS gp140 is monomeric, what can be done to further stabilize the structure of fully cleaved, envelope glycoprotein complexes? The immunoelectron microscopy data of the 2G12/SOS gp140 complex suggest that appropriately directed antibodies could strengthen weak oligomeric interactions. The immunogenicity of such complexes may be worth testing, although a bivalent MAb might be expected to promote formation of Env dimers rather than trimers. We have already attempted to combine the SOS gp140 disulfide bond stabilization strategy with one in which the gp41 subunits were also stabilized by an intermolecular disulfide bond—this was unsuccessful, in that the mutated protein was poorly expressed and could not be cleaved into gp120 and gp41 subunits, even in the presence of cotransfected furin (R. W. Sanders et al., unpublished results). Similarly, adding GCN-4 domains onto the C terminus of gp41 hindered the proper cleavage of gp140 into gp120 and gp41 furin (Sanders et al., unpublished). Other approaches, based on site-directed mutagenesis of selected gp41 residues, are currently being evaluated.
Fortuitously, we have found that variable-loop-deleted forms of HIV-1JR-FL SOS gp140 form more-stable oligomers than their full-length counterparts. Thus, the SOS gp140 proteins lacking either the V1 or V2 variable loops contain a greater proportion of oligomers than the full-length protein, and the V1V2 double loop deletant is expressed primarily as noncovalently associated trimers. One hypothesis is that the extended and extensively glycosylated variable loops sterically impede the formation of stable gp41-gp41 interactions in the context of the full-length SOS gp140 protein. Indeed, using the crystal structure of the gp120/CD4/17b complex, Kwong et al. have developed a model of oligomeric gp120 that places V1V2 sequences at the trimer interface (34). The variable-loop-deleted SOS gp140 proteins may therefore represent proteolytically mature HIV-1 envelope glycoproteins that can perhaps eventually be produced and purified as oligomers. We previously demonstrated that unpurified forms of variable-loop-deleted SOS gp140 proteins possess favorable antigenic properties (57). These proteins are therefore worth further evaluation in structural and immunogenicity studies.
Acknowledgments
We thank Dennis Burton, Hermann Katinger, and James Robinson for the gifts of several MAbs. Additional MAbs were obtained through the AIDS Research and Reference Reagents Program. We thank Joseph Sodroski and Richard Wyatt for generously providing MAbs and recombinant HIV-1YU2 gp120 protein.
This work was funded by NIH grants and contracts R01 AI39420, R01 AI42382, R01 AI45463, R21 AI44291, R21 AI49566, and U01 AI49764. J.P.M. is a Stavros S. Niarchos Scholar. The Department of Microbiology and Immunology at Weill Medical College gratefully acknowledges the support of the William Randolph Hearst Foundation.
REFERENCES
- 1.Allaway, G. P., K. L. Davis-Bruno, G. A. Beaudry, E. B. Garcia, E. L. Wong, A. M. Ryder, K. W. Hasel, M.-C. Gauduin, R. A. Koup, J. S. McDougal, and P. J. Maddon. 1995. Expression and characterization of CD4-IgG2, a novel heterotetramer which neutralizes primary HIV-1 isolates. AIDS Res. Hum. Retrovir. 11:533-539. [DOI] [PubMed] [Google Scholar]
- 2.Berman, P. W., T. J. Gregory, L. Riddle, G. R. Nakamura, M. A. Champe, J. P. Porter, F. M. Wurm, R. D. Hershberg, E. K. Cobb, and J. W. Eichberg. 1990. Protection of chimpanzees from infection by HIV-1 after vaccination with recombinant glycoprotein gp120 but not gp160. Nature 345:622-625. [DOI] [PubMed] [Google Scholar]
- 3.Berman, P. W., W. M. Nunes, and O. K. Haffar. 1988. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J. Virol. 62:3135-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, A. Master, C. S. Cayanan, C. L. Wiley, L. Schiffner, B. Travis, S. Kuhmann, D. R. Burton, S. L. Hu, W. C. Olson, and J. P. Moore. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):S87-S98. [PubMed] [Google Scholar]
- 7.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. H. I. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 8.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caffrey, M., J. Kaufman, S. Stahl, P. Wingfield, A. M. Gronenborn, and G. M. Clore. 1999. Monomer-trimer equilibrium of the ectodomain of SIV gp41: insight into the mechanism of peptide inhibition of HIV infection. Protein Sci. 8:1904-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 11.Chen, B., G. Zhou, M. Kim, Y. Chishti, R. E. Hussey, B. Ely, J. J. Skehel, E. L. Reinherz, S. C. Harrison, and D. C. Wiley. 2000. Expression, purification, and characterization of gp160e, the soluble, trimeric ectodomain of the simian immunodeficiency virus envelope glycoprotein, gp160. J. Biol. Chem. 275:34946-34953. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 13.Cherpelis, S., I. Shrivastava, A. Gettie, X. Jin, D. D. Ho, S. W. Barnett, and L. Stamatatos. 2001. DNA vaccination with the human immunodeficiency virus type 1 SF162ÄV2 envelope elicits immune responses that offer partial protection from simian/human immunodeficiency virus infection to CD8+ T-cell-depleted rhesus macaques. J. Virol. 75:1547-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutch, R. E., R. N. Hagglund, M. A. Nagel, R. G. Paterson, and R. A. Lamb. 2001. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 281:138-150. [DOI] [PubMed] [Google Scholar]
- 16.Earl, P. L., C. C. Broder, R. W. Doms, and B. Moss. 1997. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J. Virol. 71:2674-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edinger, A. L., M. Ahuja, T. Sung, K. C. Baxter, B. Haggarty, R. W. Doms, and J. A. Hoxie. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farzan, M., H. Choe, E. Desjardins, Y. Sun, J. Kuhn, J. Cao, D. Archambault, P. Kolchinsky, M. Koch, R. Wyatt, and J. Sodroski. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferlenghi, I., B. Gowen, F. de Haas, E. J. Mancini, H. Garoff, M. Sjoberg, and S. D. Fuller. 1998. The first step: activation of the Semliki Forest virus spike protein precursor causes a localized conformational change in the trimeric spike. J. Mol. Biol. 283:71-81. [DOI] [PubMed] [Google Scholar]
- 23.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 24.Gerl, M., R. Jaenicke, J. M. Smith, and P. M. Harrison. 1988. Self-assembly of apoferritin from horse spleen after reversible chemical modification with 2,3-dimethylmaleic anhydride. Biochemistry 27:4089-4096. [DOI] [PubMed] [Google Scholar]
- 25.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 26.Heinz, F. X., S. L. Allison, K. Stiasny, J. Schalich, H. Holzmann, C. W. Mandl, and C. Kunz. 1995. Recombinant and virion-derived soluble and particulate immunogens for vaccination against tick-borne encephalitis. Vaccine 13:1636-1642. [DOI] [PubMed] [Google Scholar]
- 27.Helseth, E., U. Olshevsky, C. Furman, and J. Sodroski. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65:2119-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffman, T. L., G. Canziani, L. Jia, J. Rucker, and R. W. Doms. 2000. A biosensor assay for studying ligand-membrane receptor interactions: binding of antibodies and HIV-1 Env to chemokine receptors. Proc. Natl. Acad. Sci. USA 97:11215-11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, M. L., J. J. Correia, D. A. Yphantis, and H. R. Halvorson. 1981. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys. J. 36:575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, D. H., B. W. McBride, M. A. Roff, and G. H. Farrar. 1995. Efficient purification and rigorous characterisation of a recombinant gp120 for HIV vaccine studies. Vaccine 13:991-999. [DOI] [PubMed] [Google Scholar]
- 31.Jones, P. L., T. Korte, and R. Blumenthal. 1998. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J. Biol. Chem. 273:404-409. [DOI] [PubMed] [Google Scholar]
- 32.Klasse, P. J., J. A. McKeating, M. Schutten, M. S. Reitz, Jr., and M. Robert-Guroff. 1993. An immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 (HXB2-Env:Ala 582(→Thr)) decreases viral neutralization by monoclonal antibodies to the CD4-binding site. Virology 196:332-337. [DOI] [PubMed] [Google Scholar]
- 33.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambin, P., D. Rochu, N. Herance, and J. M. Fine. 1982. High molecular-weight polymers in human albumin solutions. Quantitative determination by polyacrylamide gradient electrophoresis. Rev. Fr. Transfus. Immunohematol. 25:487-498. [DOI] [PubMed] [Google Scholar]
- 36.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, United Kingdom.
- 37.Lewis, J. K., M. Bendahmane, T. J. Smith, R. N. Beachy, and G. Siuzdak. 1998. Identification of viral mutants by mass spectrometry. Proc. Natl. Acad. Sci. USA 95:8596-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 39.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 41.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore, J. P., A. Trkola, B. Korber, L. J. Boots, J. A. Kessler, F. E. McCutchan, J. Mascola, D. D. Ho, J. Robinson, and A. J. Conley. 1995. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J. Virol. 69:122-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, J. P., R. L. Willey, G. K. Lewis, J. Robinson, and J. Sodroski. 1994. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J. Virol. 68:6836-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owens, R. J., and R. W. Compans. 1999. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology 179:827-833. [DOI] [PubMed] [Google Scholar]
- 47.Paredes, A. M., H. Heidner, P. Thuman-Commike, B. V. Prasad, R. E. Johnston, and W. Chiu. 1998. Structural localization of the E3 glycoprotein in attenuated Sindbis virus mutants. J. Virol. 72:1534-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker, C. E., L. J. Deterding, C. Hager-Braun, J. M. Binley, N. Schülke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing antibody, 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parren, P. W. H. I., and D. R. Burton. 2001. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 77:195-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parren, P. W. H. I., P. Fisicaro, A. F. Labrijn, J. M. Binley, W. P. Yang, H. J. Ditzel, C. F. Barbas, III, and D. R. Burton. 1996. In vitro antigen challenge of human antibody libraries for vaccine evaluation: the human immunodeficiency virus type 1 envelope. J. Virol. 70:9046-9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 52.Poignard, P., E. O. Saphire, P. W. H. I. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 53.Reitz, M. S. J., C. Wilson, C. Naugle, R. C. Gallo, and M. Robert-Guroff. 1988. Generation of a neutralization-resistant variant of HIV-1 is due to selection for a point mutation in the envelope gene. Cell 54:57-63. [DOI] [PubMed] [Google Scholar]
- 54.Richardson, T. M. J., B. L. Stryjewski, C. C. Broder, J. A. Hoxie, J. R. Mascola, P. L. Earl, and R. W. Doms. 1996. Humoral response to oligomeric human immunodeficiency virus type 1 envelope protein. J. Virol. 70:753-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roux, K. H. 1989. Immunoelectron microscopy of idiotype-anti-idiotype complexes. Methods Enzymol. 178:130-144. [DOI] [PubMed] [Google Scholar]
- 56.Roux, K. H. 1996. Negative-stain immunoelectron-microscopic analysis of small macromolecules of immunologic significance. Methods 10:247-256. [DOI] [PubMed] [Google Scholar]
- 57.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saphire, E. O., P. W. H. I. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 59.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 61.Scandella, C. J., J. Kilpatrick, W. Lidster, C. Parker, J. P. Moore, G. K. Moore, K. A. Mann, P. Brown, S. Coates, and B. Chapman. 1993. Nonaffinity purification of recombinant gp120 for use in AIDS vaccine development. AIDS Res. Hum. Retrovir. 9:1233-1244. [DOI] [PubMed] [Google Scholar]
- 62.Schägger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 63.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 64.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 66.Stamatatos, L., M. Lim, and C. Cheng-Mayer. 2000. Generation and structural analysis of soluble oligomeric gp140 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 16:981-994. [DOI] [PubMed] [Google Scholar]
- 67.Staropoli, I., C. Chanel, M. Girard, and R. Altmeyer. 2000. Processing, stability, and receptor binding properties of oligomeric envelope glycoprotein from a primary HIV-1 isolate. J. Biol. Chem. 275:35137-35145. [DOI] [PubMed] [Google Scholar]
- 68.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan, K., J. Liu, J. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas, G., B. A. Thorne, L. Thomas, R. G. Allen, D. E. Hruby, R. Fuller, and J. Thorner. 1988. Yeast KEX2 endopeptidase correctly cleaves a neuroendocrine prohormone in mammalian cells. Science 241:226-230. [DOI] [PubMed] [Google Scholar]
- 72.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 73.Trkola, A., T. Ketas, V. N. KewalRamani, F. Endorf, J. M. Binley, H. Katinger, J. Robinson, D. R. Littman, and J. P. Moore. 1998. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J. Virol. 72:1876-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trkola, A., A. P. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas, D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG2. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.VanCott, T. C., J. R. Mascola, R. W. Kaminski, V. Kalyanaraman, P. L. Hallberg, P. R. Burnett, J. T. Ulrich, D. J. Rechtman, and D. L. Birx. 1997. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J. Virol. 71:4319-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Venkatesh, S. G., and V. Deshpande. 1999. A comparative review of the structure and biosynthesis of thyroglobulin. Comp. Biochem. Physiol. C. Pharmacol. Toxicol. Endocrinol. 122:13-20. [DOI] [PubMed] [Google Scholar]
- 78.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 79.Wilson, C., M. S. J. Reitz, K. Aldrich, P. J. Klasse, J. Blomberg, R. C. Gallo, and M. Robert-Guroff. 1990. The site of an immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 does not constitute the neutralization epitope. J. Virol. 64:3240-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wingfield, P. T., S. J. Stahl, J. Kaufman, A. Zlotnick, C. C. Hyde, A. M. Gronenborn, and G. M. Clore. 1997. The extracellular domain of immunodeficiency virus gp41 protein: expression in Escherichia coli, purification, and crystallization. Protein Sci. 6:1653-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 82.Wyatt, R., E. Desjardin, U. Olshevsky, C. Nixon, J. Binley, V. Olshevsky, and J. Sodroski. 1997. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J. Virol. 71:9722-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 84.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 85.Yang, X., M. Farzan, R. Wyatt, and J. Sodroski. 2000. Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 74:5716-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang, X., L. Florin, M. Farzan, P. Kolchinsky, P. D. Kwong, J. Sodroski, and R. Wyatt. 2000. Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J. Virol. 74:4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. H. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J. Virol. 75:2741-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zeder-Lutz, G., J. Hoebeke, and M. H. Van Regenmortel. 2001. Differential recognition of epitopes present on monomeric and oligomeric forms of gp160 glycoprotein of human immunodeficiency virus type 1 by human monoclonal antibodies. Eur. J. Biochem. 268:2856-2866. [DOI] [PubMed] [Google Scholar]
- 90.Zhang, C. W., Y. Chishti, R. E. Hussey, and E. L. Reinherz. 2001. Expression, purification, and characterization of recombinant HIV gp140. The gp41 ectodomain of HIV or simian immunodeficiency virus is sufficient to maintain the retroviral envelope glycoprotein as a trimer. J. Biol. Chem. 276:39577-39585. [DOI] [PubMed] [Google Scholar]
- 91.Zhang, W., G. Canziani, C. Plugariu, R. Wyatt, J. Sodroski, R. Sweet, P. Kwong, W. Hendrickson, and I. Chaiken. 1999. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry 38:9405-9416. [DOI] [PubMed] [Google Scholar]
- 92.Zhu, P., W. C. Olson, and K. H. Roux. 2001. Structural flexibility and functional valency of CD4-IgG2 (PRO 542): potential for crosslinking HIV-1 envelope spikes. J. Virol. 75:6682-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]