Abstract
Human papillomavirus type 11 (HPV-11) infection causes genital warts and recurrent respiratory papillomatosis. While there is compelling evidence that CD4+ T cells play an important role in immune surveillance of HPV-associated diseases, little is known about human CD4+ T-cell recognition of HPV-11. We have investigated the CD4+ T-cell responses of 25 unrelated healthy donors to HPV-11 L1 virus-like particles (VLP). CD4+ T-cell lines from 21 of 25 donors were established. Cell sorting experiments carried out on cells from six donors demonstrated that the response was located in the CD45RAlow CD45ROhigh memory T-cell population. To determine the peptide specificity of these responses, epitope selection was analyzed by using 95 15-mer peptides spanning the entire HPV-11 L1 protein. No single region of L1 was immunodominant; responders recognized between 1 and 10 peptides, located throughout the protein, and peptide responses fell into clear HLA class II restricted patterns. Panels of L1 peptides specific for skin and genital HPV were used to show that the L1 CD4+ T-cell responses were cross-reactive. The degree of cross-reactivity was inversely related to the degree of L1 sequence diversity between these viruses. Finally, responses to HPV-11 L1 peptides were elicited from ex vivo CD45RO+ peripheral blood mononuclear cells, demonstrating that recognition of HPV-11 was a specific memory response and not due to in vitro selection during tissue culture. This is the first study of CD4+ T-cell responses to HPV-11 in healthy subjects and demonstrates marked cross-reactivity with other skin and genital HPV types. This cross-reactivity may be of significance for vaccine strategies against HPV-associated clinical diseases.
Human papillomaviruses (HPV) are small double-stranded DNA viruses and are responsible for pathological conditions ranging from benign skin warts to invasive cervical carcinomas (24). More than 95 types of HPV, with considerable sequence variability, have been described (6). The closely related HPV type 6 (HPV-6) and HPV-11 are associated with benign genital warts, the most common sexually transmitted disease (22), and recurrent respiratory papillomatosis (RRP), a rare but serious condition characterized by recurrent wart formation within the aerodigestive tract (12). Although a variety of medical treatments for RRP have been tried, none has been universally successful, and repeated surgery remains the mainstay of therapy. Thus new treatment options are urgently required (reviewed in references 12 and 16). It has been suggested that the juvenile form of RRP is transmitted from mother to child in the birth canal and that the adult form may be transmitted sexually (21). However, since HPV has been detected in the mouths of healthy school children (5, 31) and yet RRP remains a rare disease (12), there is a strong suggestion that a host factor(s) plays a central role in disease pathogenesis. Delineation of this factor(s) could substantially facilitate the rational design of new therapies for RRP.
CD4+ T cells are prominent in resolving cutaneous (20) and genital (9) warts, and patients with acquired CD4+ T-cell deficiency (e.g., immunosuppression) (28, 30, 33) or advanced human immunodeficiency virus infection (8, 23) often develop florid cutaneous and genital warts. Thus, a compelling theory for RRP susceptibility is that some patients develop the disease because they make an abnormal CD4+ T-cell response to HPV-6 and/or HPV-11. This hypothesis cannot be directly tested because little is known about normal human CD4+ T-cell responses to HPV-6 and HPV-11. Information about HPV-6 recognition from a disease population is available: Hong and coworkers have examined CD4+ T-cell recognition of the L1 protein of HPV-6 in patients with anogenital warts (19). In their study, responses of at least twice background to L1 in peripheral blood mononuclear cells (PBMC) from 7 of 11 patients and in wart-infiltrating lymphocytes from 5 of 13 patients were seen. Two regions of L1 appeared to be immunodominant (Table 1). More information on CD4+ T-cell responses to HPV-16, which is very strongly implicated in the pathogenesis of cervical carcinoma, is available (37). Of particular note are two studies demonstrating CD4+ T-cell proliferative responses to a partial-length HPV-16 L1 fusion protein in healthy donors and patients with cervical intraepithelial neoplasia III (11, 35) (see Discussion) and a detailed investigation of CD4+ T-cell responses to a panel of peptides partially spanning HPV-16 L1 in two healthy donors (36). Data indicating that the pattern of antibody and T-cell responses in patients clearing cervical HPV infection are different from those in patients progressing to cervical cancer are also available (25).
TABLE 1.
Previously defined CD4+ T-cell epitopes
| HPV type | L1 Epitope location | Epitope sequence | HLA restriction | Reference |
|---|---|---|---|---|
| 6 | 311-330 | KAQGHNNGICWGNQLFVTV | Not identified | 19 |
| 6 | 411-430 | DTYRYVQSQAITCQKPTPEK | Not identified | 19 |
| 16 | 40-63 | PPVPVSKVVSTDEYVARTNIYYHA | DRB1*04 | 36 |
| 16 | 219-244 | TVIQDGDMVHTGFGAMDFTTLQANKS | DRB1*04 | 36 |
| 16 | 279-294 | EQMFVRHLFNRAGTVG | DRB1*07 | 36 |
| 16 | 91-106 | VSGLQYRVFRIHLPDP | DRB1*03 | 36 |
| 16 | 311-335 | NLASSNYFPTPSGSMVTSDAQIFNK | Not identified | 35 |
| 16 | 311-335 | NLASSNYFPTPSGSMVTSDAQIFNK | DRB1*11 | 11 |
As a first step in testing the hypothesis of an abnormal CD4+ T-cell response in RRP pathogenesis, we examined CD4+ T-cell responses to HPV-11 L1, the principal virus coat protein, in 20 HLA class I- and II-typed healthy donors. Responses to HPV-11, rather than HPV-6, were examined since the former is associated with a more aggressive phenotype (29, 32). Unlike previous studies of human CD4+ T-cell responses to HPV antigens, this study used full-length HPV-11 L1 virus-like particles (VLP) to generate short-term CD4+ T-cell lines and a panel of 95 15-mer HPV-11 L1-specific peptides spanning the entire L1 molecule to analyze epitope selection. To our knowledge, this is the first study reporting CD4+ T-cell responses to HPV-11 in healthy individuals and demonstrates cross-reactive memory responses to a range of skin and genital HPV types in the majority of subjects tested.
MATERIALS AND METHODS
Donors.
Blood was obtained from 25 healthy, unrelated, human immunodeficiency virus- and hepatitis B virus-negative adult donors. All donors were HLA class I and II typed by PCR with sequence-specific primers (4). Only one donor had a history of previous genital warts (donor 11). The use of volunteers in this research complied with the policies of the University of Wales College of Medicine and Bro Taf Research Ethics Committee.
HPV-11 antigens.
HPV-11 L1 VLP were a generous gift from Merck Sharp & Dohme (West Point, Pa.). The HPV-11 L1 major coat protein was produced by using a Saccharomyces cerevisiae expression system as previously described (10, 27). A panel of 95 synthetic 15-mer peptides spanning the entire sequence of the HPV-11 L1 VLP was synthesized by Eurogentec (Seraing, Belguim). The peptides were designed by using the sequence of the HPV-11 L1 VLP (accession no. AAC53712) and are shown in Fig. 1. Peptides overlapped by a minimum of nine residues. To facilitate peptide synthesis, glycine and proline residues in the first two C-terminal positions were avoided, and thus some peptides overlapped by more than nine residues. Peptide design was not influenced by any prior consideration of published HPV epitopes or known HLA peptide binding motifs. Peptides were initially employed in 19 pools of five peptides (5 μg of each peptide/ml). Once responses to peptide pools had been examined, responses to individual peptides within recognized pools were examined. Panels of 15-mer peptides representing regions of the L1 antigens of HPV-1 to -4, -6, -16, and -18 were synthesized as detailed above. A T-cell line specific for the hemagglutinin (HA) of influenza virus A/Beijing/32/92 was used to test the peptide pools for nonspecific mitogenic activity. This line was established and assayed as previously described (15) and was found to mount a strong proliferative response to influenza virus HA but not to proliferate in response to the HPV-11 L1 VLP or any of the HPV-11 L1 peptide pools (data not shown).
FIG. 1.
Sequences of HPV-11 L1, HPV-6 L1, and HPV-11 L1 peptides. The sequences of HPV-11 L1 (accession no. AAC53712), HPV-6 L1 (accession no. CAA25026), and the HPV-11 L1-specific peptides are shown. Open boxes, sequence differences between HPV-11 and HPV-6 L1. Peptides were employed in 19 pools, each containing 5 peptides and labeled by the N-terminal residue of the first peptide in the pool and the C-terminal residue of the last peptide in the pool. Peptides were designated by their N-terminal amino acids.
Establishment of CD4+ T-cell lines.
PBMC were separated from whole blood by Ficoll-Hypaque centrifugation. HPV-11 L1 VLP-specific lines were established by culturing PBMC at a density of 106 cells/2-ml well in flat-bottom tissue culture plates (Greiner) with HPV-11 L1 (10 μg/ml) in complete medium (minimal essential medium [MEM; Sigma, Pool, Dorset, England], 5% heat-inactivated pooled AB serum, 100 IU of penicillin-streptomycin [Gibco, Life Technologies, Paisley, Scotland]/ml, 2 mM l-glutamine [Gibco]) at 37°C and 5.8% CO2 for 2 h. Cells were washed twice, and 4 × 106 autologous PBMC were added to each well. A single batch of MEM and AB serum was used for all experiments. On day 7, the emerging line was recovered, washed twice, and restimulated with autologous, irradiated (3,000 rads) PBMC prepulsed for 2 h with HPV-11 L1 (1 μg/ml in complete MEM), and then washed. The restimulated lines were supplemented with 10% (vol/vol) Lymphocult-LT (Biotest Folex, Frankfurt, Germany) at days +1 and +3. Every 7 days, the line was recovered, washed twice, and restimulated as described above.
Cell sorting.
PBMC were isolated as described above, resuspended in 1 ml of complete medium, and stained at 4°C for 30 min with anti-CD45RA-fluorescein isothiocyanate and anti-CD45RO-phycoerythrin (both from Becton Dickinson, San Diego, Calif.) at final concentrations of 10 and 1.25 μg/ml, respectively. Immunostained PBMC were washed twice with complete medium and resuspended at ∼2 × 107 cells/ml for cell sorting. Sorting was carried out on a MoFlo cell sorter (Cytomation, Freiburg, Germany) using standard forward versus side scatter lymphocyte gates and either CD45RAhigh CD45ROlow (RA+) or CD45RAlow CD45ROhigh (RO+) populations. Purities were confirmed on a FACScan flow cytometer (Becton Dickinson) and were >94%.
Definition of L1 epitopes.
At days 14, 21, 28, and 35 the proliferative responses of the emerging lines to HPV-11 L1 and the HPV-11 L1 peptide pools were examined. T cells from the lines were washed and cultured (4 × 104 cells/150 μl) in complete medium in 96-well, U-bottom tissue culture plates with 104 autologous, irradiated (3,000 rads) PBMC prepulsed for 2 h with media (control), HPV 11 L1 VLP (1, 5, and 10 μg/ml), or L1 peptide pools (19 peptide pools each containing 5 peptides at individual concentrations of 5 μg/ml). At 48 h, the cells were pulsed with 0.5 μCi of tritiated thymidine (Amersham Pharmacia Biotech UK Limited, Buckinghamshire, United Kingdom)/well, and harvested 16 h later with a Tomtec cell harvester (Hamden). Proliferation was measured by liquid scintillation spectroscopy. Experiments were conducted in triplicate, and a response three times the geometric mean of controls (T cells, antigen-presenting cells [APC], and media) was taken to be significant. Once responses to pools had been defined, responses to individual peptides (5 μg/ml) within the dominant pools were examined as detailed above. The cross-reactivities of the short-term T-cell lines to panels of 15-mer peptides representing HPV-1 to -4, -6, -16, and -18 were examined as detailed above.
RESULTS
Development of T-cell lines.
HPV-11 L1 VLP-specific T-cell lines were established in 21 of 25, unrelated, healthy adult donors examined. The CD4+ T-cell content of the developing T-cell lines increased from 50 to 60% at the end of the first week of in vitro culture, to 70% at the end of the second week, to 80% at the end of the third week, to 90% at the end of the fourth week, and to 95% at the end of the fifth week (data not shown). All four nonresponders were DR7 positive (one was HLA DRB1*0701 DRB4*01 DQB1*02 positive and three were HLA DRB1*0701 DRB4*01 null DQB1*0303 positive; all were heterozygotes).
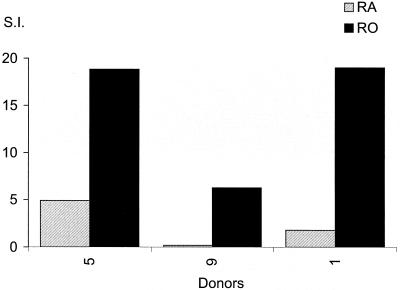
As >80% of our healthy donors had responded to the HPV-11 L1 VLP, we were interested to discover if the responses resided in the memory or naive T-cell populations. These subsets, at least those within the CD4+ T-cell population, can be broadly defined by the expression of CD45RO and CD45RA, respectively, with additional markers identifying effector memory or central memory populations (17, 34). Fresh PBMC from six healthy, responding donors were sorted into RO+ and RA+ populations and stimulated with HPV-11 L1 in short-term, 8-day proliferation assays. Results for all six subjects showed that the majority of their HPV-11 L1 response (range, 88 to 99%) was derived from the RO+ memory T-cell population (Fig. 2). Only three of six had significant (more than three times background) L1 proliferative responses from the naive RA+ lymphocyte fraction (donors 5, 9, and 16), and all of these responses were <10% of the response recorded from the RO+ subset. Furthermore, the magnitude of the response seen in the RA+ lymphocyte fraction was inversely related to the purity of the cell sort.
FIG. 2.
Analysis of the nature of the L1 proliferative response by study of RO+ and RA+ populations. Fresh PBMC from six responding donors were sorted into RO+ and RA+ fractions and used in proliferation assays as described in Materials and Methods. Experiments were conducted on days 6, 7, and 8 in triplicate. Responses three times the geometric mean of controls (T cells, APC, and media) were taken to be significant. The maximal geometric mean of the PBMC responses to HPV-11 L1 VLP at day 6, 7, or 8 is shown. This was determined independently for the RA+ and RO+ fractions. Data are presented as SIs (mean counts per minute for APC prepulsed with antigen/mean counts per minute for APC prepulsed with media).
To confirm this observation, attempts to establish HPV-11 L1-specific CD4+ T-cell lines from RA+ and RO+ cell fractions obtained from three of the above donors were made. Figure 3 shows the proliferative responses to L1 by these lines after 2 weeks of culture (third in vitro restimulation). Donors 9 and 16 showed significant proliferative responses to HPV-11 L1 only in the lines selected from the RO+ PBMC fraction. Donor 5's line derived from the RA+ fraction responded to L1 (stimulation index [SI], 4.9); however, this response was weaker than the response seen in the T-cell line derived from the same donor's RO+ cells (SI, 18.8). The lines selected from the RA+ fractions could not be maintained in culture beyond week 2.
FIG. 3.
Responses of HPV-11 VLP-selected short-term lines derived from RO+ and RA+ PBMC populations to L1. Fresh PBMC from six responding donors were sorted into RO+ and RA+ fractions as described in Materials and Methods and stimulated with 10 μg of HPV-11 VLP/ml. After 7 days, the developing line was restimulated with irradiated, autologous PBMC prepulsed with HPV-11 L1 and supplemented at days 8 and 11 with 10% Lymphocult-LT. The line was maintained for a further week in culture as detailed above. To assess proliferative responses to HPV-11 L1, the line was recovered and cultured in complete MEM at a density of 4 × 104 cells/150 μl in 96-well U-bottom tissue culture plates with 104 autologous, irradiated PBMC prepulsed with media (control) or HPV 11 L1 VLP (5 μg/ml). After 48 h, the cells were pulsed with tritiated thymidine and harvested 16 h later. Proliferation was measured by liquid scintillation spectroscopy. Experiments were conducted in triplicate, and data are presented as SIs (geometric mean of responses to APC prepulsed with antigen/geometric mean of response to APC prepulsed with media).
Response of short-term CD4+ T-cell lines to L1 peptides.
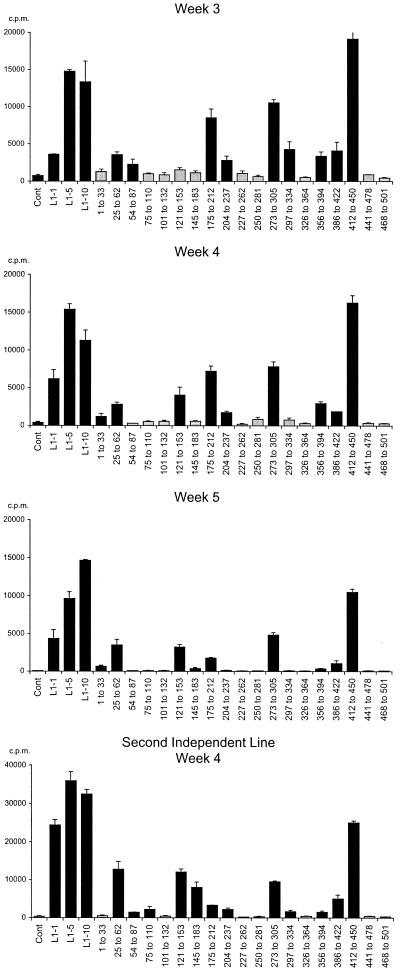
We were interested to determine which regions of L1 were recognized and therefore analyzed HPV-11 L1 epitope selection using a panel of 95 15-mer peptides overlapping by 9 residues and spanning the entire sequence of L1. Figure 4 shows the responses of a representative T-cell line at weeks 3, 4, and 5 of in vitro culture and the results from an independent line derived from the same donor. With time, the background proliferative responses of the T-cell lines (to autologous irradiated APC and media) decline and a stable pattern of HPV-11 L1 peptide pool recognition emerges. Once the pattern of L1 peptide pool response was established, the recognition of individual peptides within the dominant L1 peptide pools was examined, and responses could usually be localized to either individual L1 peptides or pairs of overlapping peptides (Fig. 5).
FIG. 4.
Development of an HPV-11 L1-specific T-cell line in vitro. The proliferative responses of a representative T-cell line to HPV-11 L1 (5 μg/ml) and the L1 peptides (5 μg of each individual peptide/ml) at 3, 4, and 5 weeks of in vitro culture are shown. Results obtained from a second independent line raised from the same donor at week 4 of in vitro culture are also shown. A short-term T-cell line was established and restimulated as described for Fig. 3. The line was maintained for a further 3 weeks of culture as detailed above. To assess proliferative responses to HPV-11 L1 and L1 peptides, a fraction of the line was recovered at days 21, 28, and 35 and cultured with 104 autologous, irradiated PBMC prepulsed with media (control), HPV 11 L1 VLP (1, 5, and 10 μg/ml), or L1 peptide pools (five peptides, at an individual concentration of 5 μg/ml). All other experimental details are as described for Fig. 3. Data are presented as the geometric means of three individual data points. Error bars, standard errors. Black bars, responses more than three times the geometric mean of the control; grey bars, responses less than three times the geometric mean of the control.
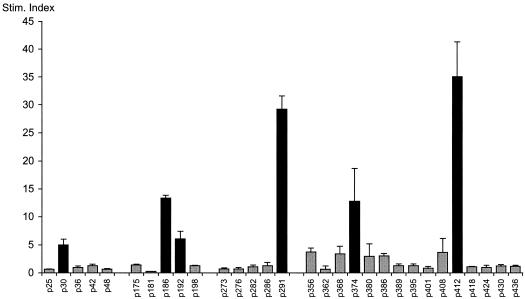
FIG. 5.
Fine specificity of an HPV-11 L1 specific T-cell line. Proliferative responses of a representative T-cell line to individual HPV-11 L1 peptides (5 μg/ml) within dominant peptide pools are shown (experimental details are as for Fig. 3). Geometric means of three individual data points are shown. Data are presented as SIs (mean response to APC prepulsed with antigen/mean response to APC prepulsed with media, with both responses in counts per minute). Error bars, standard errors. Black and grey bars are as defined for Fig. 4.
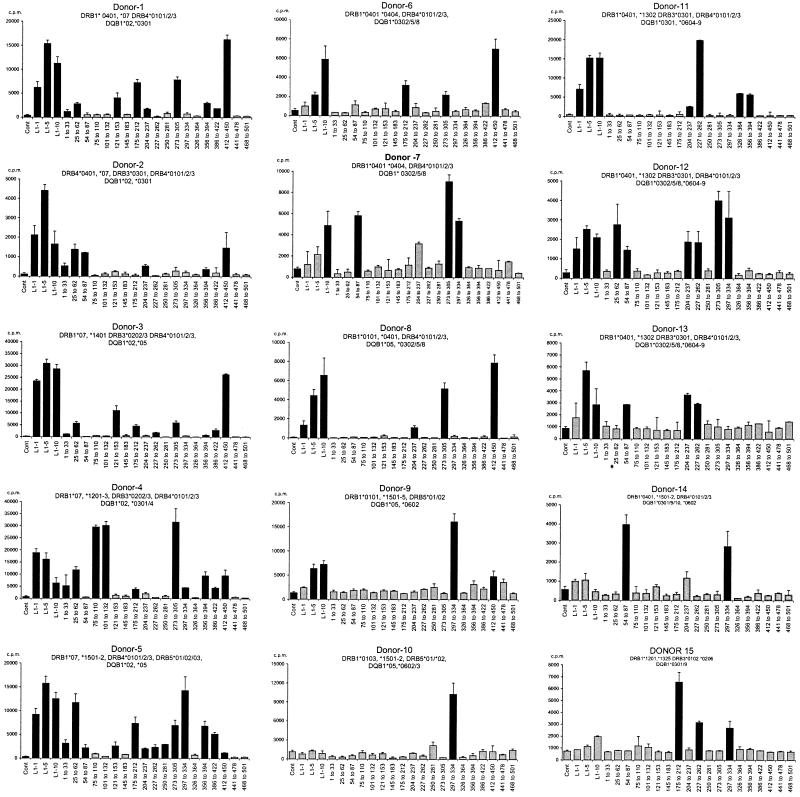
Figure 6 shows the results from 15 of 21 donors whose cells responded to HPV-11 L1 VLP and the HPV-11 L1 peptide pools. The responses of six DRB1*0301- DQB1*02-positive controls have been reported elsewhere (C. M. Gelder, O. M. Williams, K. W. Hart, S. Wall, G. Williams, D. Ingrams, P. Bull, M. Bunce, K. Welsh, S. E. F. Marshall, and L. Borysiewicz, submitted for publication). Eighteen of the 19 L1 peptide pools were recognized by cells from at least one donor, the exception being peptide pool 468-501. This pool represents the C terminus of the L1 VLP. Cells from every donor responded to at least 1 HPV-11 L1 peptide pool, and cells from donor 8 recognized 10 L1 peptide pools. The observed patterns of L1 peptide pool recognition differed between donor cells with different HLA class II haplotypes, and no single peptide pool was immunodominant. In contrast, with the exception of DRB1*04-positive subjects (see below), the observed patterns of peptide pool recognition were found to be similar when cells from HLA class II-matched donors were compared. Where responses were localized to individual peptides within the dominant pools, the donor cells recognized identical peptides or pairs of overlapping peptides. Furthermore, the majority of recognized peptides contained HLA class II peptide binding motifs listed in The HLA Facts Book (26) (Table 2). It was also striking that there was considerable variation in the number of responses associated with individual HLA class II haplotypes. For example, cells from subjects 1 to 5, who share the DRB1*07 DQB1*02 haplotype, all recognized pools 25 to 62, and cells from four out of five recognized pools 175-212, 273-305, 386-422, and 412-450. When the responses to individual peptides were analyzed, cells from the majority of these donors focused on peptides p25, p186, p380/p386, and p412, all of which contain a recognized DRB1*0701 peptide binding motif (Table 2) (26). In contrast, cells from donors 9 and 10, who share the DRB1*15 DQB1*06 haplotype (along with donors 5 and 14), focused on a single peptide pool (297-334). Within the pool, they recognized the p304 peptide, which contains an identifiable DRB1*0602 binding motif (26).
FIG. 6.
Response of HPV-11 L1-specific T-cell lines from 15 unrelated healthy adult donors to HPV-11 L1 VLPs and L1 peptides. T-cell lines were established and proliferative responses to HPV-11 L1 and L1 peptide pools were examined as detailed for Fig. 3. Experiments were conducted in triplicate, geometric means of three individual data points are shown, and responses three times the geometric mean of controls (T cells, APC, and media) were taken to be significant. Error bars, standard errors. Grey and black bars are as defined for Fig. 4. Donor HLA class I types were as follows: donor 1, A*0301/2, *2301, B*0702/4/5, *4402-5, Bw4,6, Cw*0401-3; donor 2, A*2402-5/7, *2901/2, B*2701-9, *4402-6 Bw4, Cw*0202, *1601; donor 3, HLA-A*0301/2, *6801/2 B*0702/5, *4701, Bw4,6 Cw*0602, *0702/3; donor 4, A*0201-17, B*4006-8, *2701-9, Bw4, Cw*0202, *0501; donor 5, A*0101, *0201-17, B*0702-5, *4402-5 Bw4,6, Cw*0602 *0702/3; donor 6, A*2402-5/7, B*3906, *2701-9, Bw4,6, Cw*0202, *0702-3; donor 7, A*3101/2, B*1501/5-7/12/14/19/20, *4001/7 Bw6, Cw*0303,*0701; donor 8, A*0101/2, *0201-17, B*1801-2, Bw6; donor 9, A*03, B*0702/4/7, *3701, Bw4,6, Cw*0602, *0702; donor 10, A*0201-17, *6801/2, B*3501-9, *5101-5, Bw4,6, Cw*0103, *1501-5; donor 11, A*0301-2, B*40011, *4402-5, Bw4,6, Cw*0304, *0501; donor 12, A*0301/2, B*4001, *5701-3, Bw4,6 Cw*0701,*0304; donor 13, A*0201-17, B*5701, *5801, Bw4, Cw*0602, *0701; donor 14, A*0101/2, *0201-17, B*0702/4, *4402-4/6, Bw6, Cw*0702/3, *1502-5.
TABLE 2.
CD4+ T-cell epitopes defined in this studya
| HLA | Pool recognized | Responding donors | Individual peptide specificity | Sequence | Motif |
|---|---|---|---|---|---|
| DRB1*0101 | 412-450 | 8, 9 | p412 | D T Y R Y V Q S Q A I T C Q K | DRB1*0101 |
| DRB1*0401 | 54-87 | 2, 6, 12, 13, 14 | p68 | R V F K V V L P D P N K F A L | DRB1*0401/DRB4 |
| DRB1*0401 | 204-237 | 1, 2, 6, 8, 11, 12, 13, 14 | p204 | A M N F A D L Q T N K S D V P | DRB1*0401/DRB4 |
| DRB1*0401 | 273-305 | 5, 6, 11, 13 | p286 | S I Y V H T P S G S L V S S E | DRB1*0401/DRB4 |
| DRB1*0701 | 25-62 | 1, 2, 3, 4, 5 | p30 | T N I F Y H A S S S R L L A V | DRB1*0701 |
| DRB1*0701 | 175-212 | 1, 3, 4, 5 | p186 | E L I T S V I Q D G D M V D T | No motif |
| DRB1*0701 | 273-305 | 1, 3, 4, 5 | p286 | S I Y V H T P S G S L V S S E | DRB1*0701 |
| DRB1*0701 | 356-394 | 1, 2, 4, 5 | p380 | S A E V M A Y I H T M N P S V | DRB1*0701 |
| DRB1*0701 | 412-450 | 1, 2, 3, 4, 5 | p412 | D T Y R Y V Q S Q A I T C Q K | DRB1*0701 |
| DRB1*1302 | 227-262 | 11, 12, 13 | p237, p243 | D P Y G D RL F F Y L R K E QM F A R HF | DRB1*1302 |
| DQB1*02 | 356-394 | 1, 2, 4, 5 | p356 | Y K E Y M R H V E E F D L Q F | DQB1*02 |
| DQB1*0301 | 227-262 | 11, 12, 13, 15 | p237 | D P Y G D R L F F Y L R K E Q | DQB1*0301 |
| DQB1*0602 | 297-334 | 9, 10, 14 | p303 | F N K P Y W L Q K A Q G H N N | DQB1*0602 |
Binding motifs are listed in reference (26). Proposed anchor residues within a peptide sequence are in boldface, and the section of each peptide thought to occupy the HLA class II binding groove is underlined. P30 potentially fits into the proposed DRB1*0701 binding motif in three orientations (residues 3 to 5 of this peptide all match the P1 specificity, and residues 7 and 8 match the p6 binding specificity). For overlapping peptides p237 and p243, which were both recognized the nonoverlapping residues are double underlined.
Even though the repertoire of VLP peptide pool responses associated with HLA DRB1*0401 was less rigid than that observed for HLA DRB1*0701, cells from five of nine DRB1*0401-positive donors recognized peptide pool 204-237 and those from five of nine recognized pool 273-305. Within these pools, responses focused on peptides p204 and p286, which contain identifiable DRB1*0401 binding motifs (Table 2). Identifiable patterns of L1 peptide recognition were also observed for DRB1*1302, DQB1*02, DQB1*0301, and DQB1*0602. Once again, appropriate peptide motifs could be identified within selected peptides (Table 2). No corresponding patterns of L1 peptide recognition were seen for donors sharing HLA class I alleles unless they also shared HLA class II alleles.
Cross-reactivity of HPV-11 L1-selected CD4+ T cells.
To discover whether these HPV-11 L1 CD4+ T-cell responses were cross-reactive with common skin and genital HPV types, we aligned the amino acid sequences of six frequently recognized HPV-11 L1 peptides (p30, p139, p186, p243, p304, and p412) with the corresponding regions of four skin HPVs (HPV-1 to -4) and three genital HPVs (HPV-6, -16, and -18) (Table 3). The responses of six HPV-11 L1-specific lines were then tested against panels of peptides representing these regions in the individual HPV types listed above (Fig. 7). As the sequences of HPV-11 and HPV-6 differ in only one of six regions (residues 243 to 258), only a single HPV-6 peptide was synthesized. All the lines recognized at least one peptide representing a skin or genital HPV, indicating that the lines were cross-reactive. Interestingly, the strength of the cross-reactive, proliferative response was inversely related to the number of amino acid differences between the variant and HPV-11 peptides contained within the defined putative HLA binding motifs (Table 3). Thus, cells from donors 1, 3, and 5, who are DRB1*0701 positive, all proliferated in response to HPV-11 L1 p412 and cells from donors 1 and 3 also recognized the HPV-2 equivalent of this peptide. Comparison between HPV-11 and HPV-2 sequences showed only one conservative amino acid difference within the region of this peptide believed to interact with HLA/T-cell receptor (V to L at residue 418), while there are between two and six amino acid differences between HPV-11 and other HPV types. Similarly, cells from all three donors recognized HPV-11 p30 and the HPV-1 equivalent of this peptide, though the magnitudes of their individual responses varied considerably. There are two conservative substitutions between HPV-11 and HPV-1 in this region (I to L at residue 32 and S to T at residue 37), while all the other HPV types contain nonconservative substitutions (S to G at residue 37 and F to Y at residue 33 in all except HPV-18). Cells from donors 5 and 9, who are DQB1*0602 positive, responded to HPV-11 L1 p304, and this response was again cross-reactive, the strongest response being that to HPV-16. Only a single conservative residue substitution was found in this region when HPV-11 and HPV-16 were compared (K to R at residue 312). Cells from donor 11, who is DRB1*1302 positive, recognized HPV-11 p243, and this response was cross-reactive with the strongest response being that to HPV-16. Once more, there is only a single conservative residue substitution between HPV-11 and HPV-16 in this region (K to R at residue 249). Finally, cells from donor 17, who is DRB1*0301 positive, responded to p139, and this response was cross-reactive with HPV-2, HPV-16, and HPV-18. HPV-11 has two conservative amino acid substitutions compared to the other HPV types in this region (HPV-2 and HPV-16: V to I at residue 144 and G to S at residue 145; HPV-18: G to S at residue 145 and M to V at residue 146).
TABLE 3.
Sequence differences between HPV-11 and representative skin (HPV-1 to -4) and genital (HPV-6, -16, and -18) HPV typesa
| Peptide | HPV type | Sequence |
|---|---|---|
| 30-45 | HPV-11 | T N I F Y H A S S S R L L A V |
| HPV-6b | ||
| HPV-1a | LTEL | |
| HPV-2a | V YG GT | |
| HPV-3 | YYGT | |
| HPV-4 | S L Y FG T ET | |
| HPV-16 | YG T | |
| HPV-18 | SGT | |
| 139-144 | HPV-11 | D N R V N V G M D Y K Q T Q L |
| HPV-6b | ||
| HPV-1a | SQT A FAM | |
| HPV-2a | GEI S | |
| HPV-3 | SDI S VN | |
| HPV-4 | Q DS LPM | |
| HPV-16 | E C I S | |
| HPV-18 | VDS V | |
| 186-201 | HPV-11 | E L I T S V I Q D G D M V D T |
| HPV-6b | ||
| HPV-1a | Q MEEMI | |
| HPV-2a | Q F T N T TEE | |
| HPV-3 | A P | |
| HPV-4 | V NYCI | |
| HPV-16 | N T | |
| HPV-18 | K N TL E | |
| 243-258 | HPV-11 | L F F Y L R K E Q M F A R H F |
| HPV-6b | F | |
| HPV-1a | MF ARY T | |
| HPV-2a | MSRT | |
| HPV-3 | ML | |
| HPV-4 | F GRL Y | |
| HPV-16 | RVL | |
| HPV-18 | MCRL | |
| 304-319 | HPV-11 | F N K P Y W L Q K A Q G H N N |
| HPV-6b | ||
| HPV-1a | R SR CQ | |
| HPV-2a | R R | |
| HPV-3 | R R | |
| HPV-4 | RN RT | |
| HPV-16 | R | |
| HPV-18 | H | |
| 412-417 | HPV-11 | E D T Y R Y V Q S Q A I T C Q |
| HPV-6b | ||
| HPV-1a | QF L GS L A A K C | |
| HPV-2a | QL | |
| HPV-3 | F L TS | |
| HPV-4 | QF LRT RP | |
| HPV-16 | FTA | |
| HPV-18 | VFV |
The region of each peptide believed to lie within the HLA class II peptide binding groove is in boldface. GenBank accession numbers for the sequences of individual HPV types are as follows: HPV-11, NC_001525; HPV-6b, NC_001355; HPV-1, NC_001356; HPV-2, NC_001352; HPV-3, NC_001588; HPV-4, X70827; HPV-16, AF125673; HPV-18, NC_001357.
FIG. 7.
Cross-reactivity of short-term HPV-11 L1-selected T-cell lines to a panel of peptides representing immunodominant regions of common skin and genital HPV types. Shown are the responses of T-cell lines selected from six donors to a panel of peptides representing the equivalent regions of the immunodominant HPV-11 L1 peptides from common skin HPV (HPV-1 to -4) and genital HPV (HPV-6, -16, and -18). Experimental details are as for Fig. 3. Experiments were conducted in triplicate, geometric means of three individual data points are shown, and responses three times the geometric mean of controls (Cont) (T cells, APC, and media) were taken to be significant. Error bars, standard errors. Grey and black bars are as defined for Fig. 4.
To confirm that cross-reactivity was not simply due to in vitro selection during tissue culture, the ex vivo responses of RA+ and RO+ populations of PBMC from six donors to the variant HPV peptides were examined in 8-day proliferation assays (Fig. 8). Responses were seen in the RO+ fractions from five donors, and cells from all five recognized the appropriate HPV-11 L1 peptide(s) and their skin and genital HPV equivalents. This demonstrates that both the HPV-11 peptide response and the reactivity to skin and genital HPV were present in the peripheral blood memory T-cell pool.
FIG. 8.
Ex vivo responses to HPV peptides by RO+ and RA+ PBMC. Fresh PBMC from six responding donors were sorted into RO+ and RA+ fractions. The proliferative responses of these cells to immunodominant HPV-11 L1 peptides and the equivalent regions of common skin and genital HPV at days 6, 7, and 8 were examined in triplicate. All peptides were used at a final concentration of 5 μg/ml. The HPV-11 peptides (and HPV-6 variant of p243) were employed individually, and the skin and genital HPV were pooled (the skin pool [SKIN] contains HPV-1 to -4; the genital pool [GEN] contains HPV-16 and -18). Responses three times the geometric mean of controls (CONT) (T cells, APC, and media) were taken to be significant. The maximal geometric means of the PBMC responses to HPV L1 peptides at day 6, 7, or 8 are shown. These was determined independently for the RA+ and RO+ fractions. Error bars, standard errors.
Interestingly donors 6 and 15 had different patterns of reactivity to L1 and the L1 peptide variants in their CD45RA+ and CD45RO+ PBMC fractions.
DISCUSSION
In this study, we examined the repertoire of CD4+ T-cell responses to HPV-11 L1 antigen using a panel of 25 healthy, adult donors. CD4+ T-cell lines that recognized between 1 and 10 HPV-11 peptides, representing epitopes that were distributed throughout the HPV-11 L1 molecule, from 21 donors could be established. The exception was the pool covering the extreme C-terminal tail of L1, previously reported to be prone to degradation (7, 38). Cells from donors sharing HLA class II haplotypes displayed remarkably similar patterns of L1 peptide selection. Indeed this similarity was comparable to that previously observed with HA from influenza A virus, a considerably more immunogenic virus (13, 15).
Although HPV infection rates of up to 50% have been reported in some studies of young sexually active adults (2, 3), it was particularly striking that we were able to establish HPV-11 L1-specific CD4+ T-cell lines in >80% of donors. This figure is similar to that obtained by Hong and coworkers in a disease population (19) and considerably in excess of the 12% rate of positive immunoglobulin G responsiveness to HPV-11 VLP previously reported in a healthy control population (18). Cell sorting experiments revealed that most (88 to 99%) of the responses were made by RO+ memory T cells (17, 34). This population may, in fact, represent all of the responding cells as cell sorting resulted in 94 to 99% purity (data not shown). Therefore, any responses from the RA+ population may have been derived from contaminating RO+ cells.
Because one possible explanation for this finding was that the observed CD4+ T-cell responses in our study were due to cross-reactivity with other HPV types (particularly skin HPV), we aligned the amino acid sequences of HPV types commonly infecting the skin (HPV-1 to -4) and genital tract (HPV-6, -16, and -18) and compared them to the sequences of the HPV-11 L1 peptides recognized in this study. The great majority of recognized peptides represented regions of HPV-11 L1 that were identical to those of HPV-6 L1. However, there were between 1 and 10 amino acid differences between HPV-11 and the other HPV types in these regions. Using panels of peptides representing the corresponding regions of these other HPV types, we examined the cross-reactivities of six L1 T-cell lines. Every line proliferated in response to at least one peptide representing a skin or genital HPV type. The strength of the cross-reactive, proliferative response was inversely related to the number of amino acid differences between the variant peptides and HPV-11. By examining the proliferative responses of RA+ and RO+ PBMC fractions, we demonstrated that the response to HPV-11 L1 peptides are present in the ex vivo cell population and not simply due to in vitro selection by the highly immunogenic HPV-11 VLP during tissue culture. Thus, the most likely explanation for the high degree of CD4+ reactivity to HPV-11 L1 (>80% donors) seen in this study is that there is cross-reactivity between the CD4+ T-cell responses to common skin and genital HPV and the response to HPV-11. Though this is the first demonstration of cross-reactivity in the human CD4+ T-cell response to HPV, cross-reactive T-cell responses in the presence of strain-specific antibody responses are a feature of other human viral infections, most notably influenza virus infection (15). As CD4+ T cells are prominent in resolving cutaneous (20) and genital warts (9), the question of whether T-cell responses selected by one HPV type might modulate the immune response to a second HPV type arises. Even a weak response early in the course of infection might alter the resulting viral load and the subsequent disease process. This is a complex issue to test experimentally due to the paucity of suitable animal models of HPV infection. In addition more information about the cytokine response by HPV-11 T-cell lines in response to variant HPV peptides is required, and this is the subject of ongoing research. However the data presented here raise the intriguing possibility that one might be able to boost CD4+ T-cell responses to a broad range of HPV types by using a univalent HPV vaccine, considerably simplifying vaccination strategies.
Of 25 subjects, cells from only 4 did not respond to HPV-11 L1, and these were all DRB1*0701 positive. Responses to HPV-11 L1 and the L1 peptides in five other DRB1*0701 positive donors were seen, and peptides recognized by cells from these donors contained a DRB1*0701 peptide binding motif. HLA-DRB1*0701 has not been reported to increase the likelihood of developing genital warts or RRP. Clearly the fact that all four nonresponders were DRB1*0701 positive might be a chance observation and might relate to differences in history of exposure to HPV. However, DRB1*0701 is associated with nonresponsiveness to two other highly purified protein vaccines: hepatitis B and influenza vaccines (1, 14). It is conceivable that the nonresponders and responders may have differences in antigen processing due to polymorphism in a gene linked to the HLA DRB1*0701. Alternatively, nonresponders may have a deficit in their T-cell receptor repertoire. This is currently the subject of further investigation.
We were interested in comparing the peptides selected in this study to previously published HPV epitopes. Table 1 summarizes previously published CD4+ T-cell epitopes from HPV-6 and -16 and B-cell epitopes from HPV-11. Hong and coworkers, while investigating T-cell proliferative responses of wart-infiltrating lymphocytes in patients with genital warts to a panel of peptides spanning HPV-6 L1, reported that two peptides representing residues 311 to 330 and 411 to 430 of HPV-6 induced proliferation in 4 of 13 donors (19). The L1 proteins of HPV-6 and -11 have similar amino acid sequences (Fig. 1). In the present study, responses to L1 localized to residues 304 to 318 in DRB1*1501-positive donors and to residues 412 to 426 in donors positive for DRB1*0101 and DRB1*0701. Both regions are conserved between HPV-6 and HPV-11. Interestingly, two of the donors in the study by Hong and coworkers whose cells recognized residues 311 to 330 were also DRB1*15 positive and another two donors whose cells recognized residues 411 to 430 were DRB1*01 positive. Shepherd and coworkers used an incomplete panel of peptides spanning a partial-length HPV 16 L1 fusion protein in healthy donors and patients with cervical intraepithelial neoplasia III to define an immunodominant region between residues 311 and 335 (equivalent to residues 282 to 306 in HPV-11) (35). This region was recognized by cells from 73% of patients and 18% of controls. In our study cells from 8 of 20 (40%) donors responded to the equivalent region in HPV-11 L1, which has nine amino acid substitutions between HPV-11 and HPV-16. Finally, Strang and coworkers examined CD4+ T-cell responses of two healthy donors to eight synthetic peptides partially spanning HPV-16 L1 (36). They identified two DR4-restricted epitopes at residues 40 to 63 and 219 to 244 (equivalent to HPV-11 residues 12 to 36 and 190 to 215), one DR7-restricted epitope at residues 279 to 294 (equivalent to HPV-11 residues 250 to 65), and one epitope at residues 91 to 106 (equivalent to HPV-11 residues 62 to 77) that may have been DR3 restricted. Only the response to residues 219 to 244 (equivalent to residues 190 to 215 in HPV-11) was reflected in the pattern of peptide recognition seen in this study.
In conclusion, we have examined the repertoire of CD4+ T-cell responses to HPV-11 L1 antigen using a panel of 25 healthy adult donors. Short-term CD4+ T-cell lines could be established in >80% of the donors. A remarkably similar pattern of L1 epitope selection was seen in HLA class II-matched donors. The response resided in the RO+ memory fraction and appears to be principally due to cross-reactivity with skin and/or other genital HPV types. The effectiveness of these responses in controlling HPV infection and their potential bearing on vaccination strategies are important areas for further study.
Acknowledgments
This study was funded by The Wellcome Trust. C.M.G. is a Wellcome Senior Fellow in Clinical Research, and O.M.W. is a Wellcome Training Fellow.
We are grateful to Lezsek Borysiewicz, Martin Rowe, and Ita Askonas for encouragement and helpful discussions. We thank James Cook and Merck Research Laboratories, West Point, Pa., for the kind provision of the HPV-11 VLP. Thanks also to Terry Hoy, Janet Fisher, and the staff of the Flow Cytometry Facility of the Central Biotechnology Service, University of Wales College of Medicine, for cell sorting.
REFERENCES
- 1.Alper, C. A., M. S. Kruskall, D. Marcus-Bagley, D. E. Craven, A. J. Katz, S. J. Brink, J. L. Dienstag, Z. Awdeh, and E. J. Yunis. 1989. Genetic prediction of nonresponse to hepatitis B vaccine. N. Engl. J. Med. 321:708-712. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, H. M., A. Hildesheim, M. H. Schiffman, A. G. Glass, B. B. Rush, D. R. Scott, D. M. Cadell, R. J. Kurman, and M. M. Manos. 1993. Determinants of genital human papillomavirus infection in low-risk women in Portland, Oregon. Sex. Transm. Dis. 20:274-278. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, H. M., Y. Ting, C. E. Greer, J. C. Chambers, C. J. Tashiro, J. Chimera, A. Reingold, and M. M. Manos. 1991. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA 265:472-477. [PubMed] [Google Scholar]
- 4.Bunce, M., C. M. O'Neill, M. C. Barnardo, P. Krausa, M. J. Browning, P. J. Morris, and K. I. Welsh. 1995. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP). Tissue Antigens 46:355-367. [DOI] [PubMed] [Google Scholar]
- 5.Cason, J., J. N. Kaye, R. J. Jewers, P. K. Kambo, J. M. Bible, B. Kell, B. Shergill, F. Pakarian, K. S. Raju, and J. M. Best. 1995. Perinatal infection and persistence of human papillomavirus types 16 and 18 in infants. J. Med. Virol. 47:209-218. [DOI] [PubMed] [Google Scholar]
- 6.Chan, S. Y., H. Delius, A. L. Halpern, and H. U. Bernard. 1995. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J. Virol. 69:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 8.Chirgwin, K. D., J. Feldman, M. Augenbraun, S. Landesman, and H. Minkoff. 1995. Incidence of venereal warts in human immunodeficiency virus-infected and uninfected women. J. Infect. Dis. 172:235-238. [DOI] [PubMed] [Google Scholar]
- 9.Coleman, N., H. D. Birley, A. M. Renton, N. F. Hanna, B. K. Ryait, M. Byrne, D. Taylor-Robinson, and M. A. Stanley. 1994. Immunological events in regressing genital warts. Am. J. Clin. Pathol. 102:768-774. [DOI] [PubMed] [Google Scholar]
- 10.Cook, J. C., J. G. Joyce, H. A. George, L. D. Schultz, W. M. Hurni, K. U. Jansen, R. W. Hepler, C. Ip, R. S. Lowe, P. M. Keller, and E. D. Lehman. 1999. Purification of virus-like particles of recombinant human papillomavirus type 11 major capsid protein L1 from Saccharomyces cerevisiae. Protein Expr. Purif. 17:477-484. [DOI] [PubMed] [Google Scholar]
- 11.de Gruijl, T. D., H. J. Bontkes, J. M. Walboomers, P. Coursaget, M. J. Stukart, C. Dupuy, E. Kueter, R. H. Verheijen, T. J. Helmerhorst, M. F. Duggan-Keen, P. L. Stern, C. J. Meijer, and R. J. Scheper. 1999. Immune responses against human papillomavirus (HPV) type 16 virus-like particles in a cohort study of women with cervical intraepithelial neoplasia. I. Differential T-helper and IgG responses in relation to HPV infection and disease outcome. J. Gen. Virol. 80:399-408. [DOI] [PubMed] [Google Scholar]
- 12.Derkay, C. S. 2001. Recurrent respiratory papillomatosis. Laryngoscope 111:57-69. [DOI] [PubMed] [Google Scholar]
- 13.Gelder, C., M. Davenport, M. Barnardo, T. Bourne, J. Lamb, B. Askonas, A. Hill, and K. Welsh. 1998. Six unrelated HLA-DR-matched adults recognize identical CD4+ T cell epitopes from influenza A haemagglutinin that are not simply peptides with high HLA-DR binding affinities. Int. Immunol. 10:211-222. [DOI] [PubMed] [Google Scholar]
- 14.Gelder, C. M., R. Lambkin, K. W. Hart, D. Fleming, O. M. Williams, M. Bunce, K. I. Welsh, S. E. Marshall, and J. Oxford. 2002. Associations between human leukocyte antigen and nonresponsiveness to influenza vaccine. J. Infect. Dis. 185:114-117. [DOI] [PubMed] [Google Scholar]
- 15.Gelder, C. M., K. I. Welsh, A. Faith, J. R. Lamb, and B. A. Askonas. 1995. Human CD4+ T-cell repertoire of responses to influenza A virus hemagglutinin after recent natural infection. J. Virol. 69:7497-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, G. E., N. M. Bauman, and R. J. Smith. 2000. Pathogenesis and treatment of juvenile onset recurrent respiratory papillomatosis. Otolaryngol. Clin. N. Am. 33:187-207. [DOI] [PubMed] [Google Scholar]
- 17.Hamann, D., P. A. Baars, M. H. Rep, B. Hooibrink, S. R. Kerkhof-Garde, M. R. Klein, and R. A. van Lier. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186:1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heim, K., N. D. Christensen, R. Hoepfl, B. Wartusch, G. Pinzger, A Zeimet, P. Baumgartner, J. W. Kreider, and O. Dapunt. 1995. Serum IgG, IgM, and IgA reactivity to human papillomavirus types 11 and 6 virus-like particles in different gynaecologic patient groups. J. Infect. Dis. 172:395-402. [DOI] [PubMed] [Google Scholar]
- 19.Hong, K., C. E. Greer, N. Ketter, G. Van Nest, and X. Paliard. 1997. Isolation and characterization of human papillomavirus type 6-specific T cells infiltrating genital warts. J. Virol. 71:6427-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwatsuki, K., H. Tagami, M. Takigawa, and M. Yamada. 1986. Plane warts under spontaneous regression. Immunopathologic study on cellular constituents leading to the inflammatory reaction. Arch. Dermatol. 122:655-659. [DOI] [PubMed] [Google Scholar]
- 21.Kashima, H. K., and K. Shah. 1987. Recurrent respiratory papillomatosis. Clinical overview and management principles. Obstet. Gynecol. Clin. N. Am. 14:581-588. [PubMed] [Google Scholar]
- 22.Koutsky, L. 1997. Epidemiology of genital human papillomavirus infection. Am. J. Med. 102:3-8. [DOI] [PubMed] [Google Scholar]
- 23.Laga, M., J. P. Icenogle, R. Marsella, A. T. Manoka, N. Nzila, R. W. Ryder, S. H. Vermund, W. L. Heyward, A. Nelson, and W. C. Reeves. 1992. Genital papillomavirus infection and cervical dysplasia—opportunistic complications of HIV infection. Int. J. Cancer 50:45-48. [DOI] [PubMed] [Google Scholar]
- 24.Lowy, D. R., and P. M. Howley. 2001. Papillomaviruses, p. 2231-2264. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Field's virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 25.Luxton, J. C., A. J. Rowe, J. C. Cridland, T. Coletart, P. Wilson, and P. S. Shepherd. 1996. Proliferative T cell responses to the human papillomavirus type 16 E7 protein in women with cervical dysplasia and cervical carcinoma and in healthy individuals. J. Gen. Virol. 77:1585-1593. [DOI] [PubMed] [Google Scholar]
- 26.Marsh, S. G. E., P. Parham, and L. D. Barber. 2000. The HLA facts book. Academic Press, London, United Kingdom.P. P.B. L. D.
- 27.Neeper, M. P., K. J. Hofmann, and K. U. Jansen. 1996. Expression of the major capsid protein of human papillomavirus type 11 in Saccharomyces cerevisiae. Gene 180:1-6. [DOI] [PubMed] [Google Scholar]
- 28.Petry, K. U., D. Scheffel, U. Bode, T. Gabrysiak, H. Kochel, E. Kupsch, M. Glaubitz, S. Niesert, H. Kuhnle, and I. Schedel. 1994. Cellular immunodeficiency enhances the progression of human papillomavirus-associated cervical lesions. Int. J. Cancer 57:836-840. [DOI] [PubMed] [Google Scholar]
- 29.Pou, A. M., F. L. Rimell, J. A. Jordan, D. L. Shoemaker, J. T. Johnson, P. Barua, J. C. Post, and G. D. Ehrlich. 1995. Adult respiratory papillomatosis: human papillomavirus type and viral coinfections as predictors of prognosis. Ann. Otol. Rhinol. Laryngol. 104:758-762. [DOI] [PubMed] [Google Scholar]
- 30.Reid, T. M., N. G. Fraser, and I. R. Kernohan. 1976. Generalized warts and immune deficiency. Br. J. Dermatol. 95:559-564. [DOI] [PubMed] [Google Scholar]
- 31.Rice, P. S., C. Mant, J. Cason, J. M. Bible, P. Muir, B. Kell, and J. M. Best. 2000. High prevalence of human papillomavirus type 16 infection among children. J. Med. Virol. 61:70-75. [DOI] [PubMed] [Google Scholar]
- 32.Rimell, F. L., D. L. Shoemaker, A. M. Pou, J. A. Jordan, J. C. Post, and G. D. Ehrlich. 1997. Pediatric respiratory papillomatosis: prognostic role of viral typing and cofactors. Laryngoscope 107:915-918. [DOI] [PubMed] [Google Scholar]
- 33.Rudlinger, R., I. W. Smith, M. H. Bunney, and J. A. Hunter. 1986. Human papillomavirus infections in a group of renal transplant recipients. Br. J. Dermatol. 115:681-692. [DOI] [PubMed] [Google Scholar]
- 34.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708-712. [DOI] [PubMed] [Google Scholar]
- 35.Shepherd, P. S., A. J. Rowe, J. C. Cridland, T. Coletart, P. Wilson, and J. C. Luxton. 1996. Proliferative T cell responses to human papillomavirus type 16 L1 peptides in patients with cervical dysplasia. J. Gen. Virol. 77:593-602. [DOI] [PubMed] [Google Scholar]
- 36.Strang, G., J. K. Hickling, G. A. McIndoe, K. Howland, D. Wilkinson, H. Ikeda, and J. B. Rothbard. 1990. Human T cell responses to human papillomavirus type 16 L1 and E6 synthetic peptides: identification of T cell determinants, HLA-DR restriction and virus type specificity. J. Gen. Virol. 71:423-431. [DOI] [PubMed] [Google Scholar]
- 37.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, J., J. Doorbar, X. Y. Sun, L. V. Crawford, C. S. McLean, and I. H. Frazer. 1991. Identification of the nuclear localization signal of human papillomavirus type 16 L1 protein. Virology 185:625-632. [DOI] [PubMed] [Google Scholar]