Abstract
Proteolytic cleavage of the hemagglutinin (HA) of human influenza viruses A/Aichi/2/68 (H3N2) and A/WSN/34 (H1N1) from HA0 to HA1/HA2 was studied in primary human adenoid epithelial cells (HAEC). HAEC contain a mixture of ciliated and nonciliated secretory cells and mimic the epithelium membrane of the human respiratory tract. Pulse-chase labeling with [35S]methionine and Western blot analysis with anti-HA antibodies of cellular and virion polypeptides showed that HAEC cleaved newly synthesized HA0 to HA1/HA2 (“cleavage from within”) and significant amounts of cleaved HA accumulated within cells. It was also shown that HAEC was able to cleave HA0 of incoming virions (“cleavage from without”), whereas the HA0 of nonabsorbed virions free in extracellular fluid were not cleaved, supporting the conclusion that HA0 cleavage in HAEC is cell associated. Low-molecular-weight inhibitors of serine proteases, aprotinin and leupeptin, when added to influenza virus-infected HAEC suppressed HA0 cleavage and reduced the amount of cleaved HA1/HA2 both in cells and in progeny virions and thus diminished the infectivity of the virus. In contrast, the addition of fetal bovine serum, containing a number of high-molecular-weight antiproteases that compete for proteases in the extracellular environment, did not inhibit influenza virus growth in HAEC. These data suggest that in human respiratory epithelium the cleavage of influenza virus HA containing a single arginine in the proteolytic site (i) is a cell-associated process accomplished by serine-type protease(s) and (ii) is sensitive to low-molecular-weight exogenous inhibitors of serine proteases.
Influenza A viruses possess two envelope glycoproteins—hemagglutinin (HA) and neuraminidase (NA)—which protrude from virions as spikes. HA mediates influenza attachment to sialylic acid cell receptors and virus entry into target cells (39). NA facilitates elution of virus progeny from infected cells (28). HA is composed of two subunits, HA1 (molecular mass of 55 kDa) and HA2 (molecular mass of 25 kDa), that are cleaved by host proteases from their precursor HA0 (molecular mass of 75 kDa) (30). The cleavage of HA0 into HA1/HA2 activates virus infectivity (17, 21) and is important for influenza virus pathogenicity in human and avian hosts (18, 32). There are three sets of available data on regulation of influenza A virus HA cleavage.
The first data set is on the structure of HA molecules. The major characteristic of the HA that determines sensitivity to host proteases is the composition of the proteolytic site in the external loop in the HA0 molecule which links HA1 and HA2 (6). This loop may contain either a single Arg or Lys residue (monobasic cleavage site) or several Lys and/or Arg residues, with an R-X-K/R-R motif, which form a multibasic cleavage site. The multibasic cleavage site of HA exists in influenza A virus subtypes H5 and H7. All other influenza A viruses and influenza B and C viruses contain HAs with a monobasic cleavage site (18). Influenza A viruses having multibasic cleavage sites are more virulent and induce systemic infection in hosts, whereas viruses with a monobasic HA site initiate infection only in the respiratory tract in mammals or in the respiratory and enteric tracts in avian species (18, 32). Carbohydrate side chains in the vicinity of the HA cleavage site may interfere with proteases reaching the cleavage site. When the masking effect of oligosaccharide chains is abolished an increase may occur in sensitivity of HA to proteolytic cleavage (13, 24, 25).
A second set of data has defined the proteases involved in influenza virus activation in different hosts. The subtilizin-like serine type endoproteases are responsible for the cleavage of influenza virus HA subtypes 5 and 7 which possess multibasic sites (22, 31). These subtilizin-like endoproteases cleave influenza virus HAs intracellularly in the constitutive exocytic pathway (31, 37). These proteases cleave multibasic HA0s in many cell types, thus, allowing the virulent, systemic infection seen with such viruses (for reviews, see references 18 and 32).
Influenza virus HAs with monobasic cleavage sites are activated by secreted trypsin-like proteases (17, 21). Such secreted enzymes include plasmin (20), kallikrein, urokinase, thrombin (29), blood clotting factor Xa (12), acrosin (9), tryptase Clara (16), mini-plasmin (23), proteases from human respiratory lavage (2), and bacterial proteases from Staphylococcus aureus (33) and Pseudomonas aeruginosa (5). Kawaoka's group has demonstrated that the neuraminidase of A/WSN/34 has the ability to bind plasminogen allowing plasmin activation at the cell membrane with resultant HA cleavage (11). Therefore, cleavage activation of influenza virus monobasic HAs by host proteases is generally thought to occur extracellularly either at the plasma membrane or after virus release from the cell (18, 32). Alternatively, Boycott et al. have proposed that A/WSN/34 HA may be cleaved intracellularly in bovine MDBK cells during the initial steps of virus internalization (4).
The third data set examines the effects of protease inhibitors on influenza virus HA cleavage. Peptidil chloromethylketones mimicking a proteolytic site sequence, such as decREKR-CMK, have been shown to suppress HA cleavage of the multibasic HA of FPV/34 (H7N1) virus (10, 31). In influenza viruses with monobasic HAs, inhibitors of serine proteases, including e-aminocaproic acid, an inhibitor of plasminogen activation (11, 41), aprotinin (42), leupeptin (34), pulmonary surfactant (15), human mucus protease inhibitor (3), have been shown to reduce HA cleavage and virus activation in cultured cells, in chicken embryos, and in lungs of infected mice. Aerosol inhalations of aprotinin have a therapeutic effect against influenza and parainfluenza in mice (27) and humans (47).
In the references above, there are relatively little data on influenza virus HA cleavage in the human respiratory tract. Specifically, the influenza virus activating protease (IAP) in the human respiratory tract remains unidentified. To better define the site of protease activation of influenza virus in humans, we have prepared primary epithelial cell cultures from human adenoids (HAEC). HAEC contain a mixture of highly differentiated ciliated and nonciliated epithelial cells that model the human respiratory epithelial membrane and are permissive for influenza virus replication (7).
In this paper, we have used HAEC to dissect the timing and location of influenza virus HA cleavage and tested the suppression of cleavage by exogenous protease inhibitors. We will show that influenza virus HA cleavage in HAEC is (i) cell associated and (ii) sensitive to low-molecular-weight exogenous inhibitors of serine proteases.
MATERIALS AND METHODS
Viruses and cells.
Influenza A viruses A/Aichi/2/68 (H3N2), A/WSN/34 (H1N1), and an A/Beijing/89 (H3N2)-like virus isolated in our laboratory in Nashville, Tenn., were grown in 10-day-old embryonated chicken eggs as previously described (43). Madin-Darby canine kidney (MDCK) cells were passaged in Dulbecco's minimal essential medium (DMEM) containing 10% bovine fetal calf serum (FCS) and used to prepare influenza with an HA0 phenotype as previously described (40). As previously described (7), adenoids weighing an average of 1.4 g were obtained at the time of adenoidectomy and were incubated in minimal essential medium with 10 ml of 0.1% pronase type 14 overnight at 4°C. The resultant HAEC and lymphocyte suspension was treated with 10% FCS (Atlanta Biologicals, Atlanta, Ga.) to inactivate pronase. Cells were centrifuged and resuspended in 50% Ham's F-12 medium-50% Eagle's minimal essential medium with the following supplements: insulin (5 μg/ml), transferrin (5 μg/ml), epidermal growth factor (10 ng/ml), cholera toxin (10 ng/ml), hydrocortisone (1 μM), bovine hypothalamic extract (15 μg/ml), HEPES buffer (0.015 M), retinol (0.1 μM), gentamicin (50 μg/ml), penicillin G (15 U/ml), streptomycin (15 U/ml), and FCS (0.5%). Cells were placed on an underlying collagen matrix of Vitrogen 100 (Cohesion Technologies, Inc., Palo Alto, Calif.) that had been prepared with 5× minimal essential medium, 75 mM HEPES, and NaOH to bring the pH to 7.0 (7). Media were changed every second day, resulting in removal of the nonadherent lymphocyte population. The HAEC reached confluency after 10 to 14 days. Typically, 24 35-mm wells can be prepared from a single adenoid. The HAEC have subpopulations of cells that include those with active cilia, those with immunohistochemical markers for Clara cells, and those with mucin production (7). When adenoid cells were grown in medium containing 10% FCS, human adenoid fibroblasts (HAF) overgrew the epithelial cells. Influenza virus grown in HAF has an uncleaved HA0. All cells were maintained at 37°C in a 5% CO2 atmosphere.
Protease inhibition.
In protease inhibitor experiments, stock solutions of 5-mg/ml aprotinin (a natural 6-kDa polypeptide protease inhibitor from bovine lungs [8]) (AWD, Dresden, Germany), 1-mg/ml leupeptin (a 0.45-kDa oligopeptide protease inhibitor from bacteria; Sigma) (1); and 1-mg/ml Cucurbita maxima trypsin inhibitor (CMTI) (a 3-kDa polypeptide protease inhibitor from squash seeds [26]) (a kind gift of J. Otlewsky from Warsaw University, Warsaw, Poland) were sterilely prepared in water. They were immediately added to HAEC culture medium in a range of final concentrations.
Polyacrylamide gel electrophoresis (PAGE).
Polypeptides were electrophoresed in 12% polyacrylamide gels using Tris-glycine-sodium dodecyl sulfate (SDS) buffer followed by autoradiography as previously described (41). Gels were stained with Coomassie blue R-350 by following a protocol recommended by Pharmacia. For electrophoresis, cellular polypeptides were treated in a dissociation buffer (2% SDS, 0.01 dithiotreitol, 0.02 M Tris-HCl, pH 6.8) for 2 min at 96°C. A molecular weight marker kit (Amersham Pharmacia Biotech) was used containing myosin (200 kDa), phosphorylase b (97.4 kDa), bovine serum albumin (69 kDa), ovalbumin (46 kDa), carbonic anhydrase (30 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.3 kDa).
Western blot (WB) analysis.
After SDS-PAGE, the polypeptides were transferred from the gel onto polyvinylidene difluoride 0.45-μm-pore-size membranes (Bio-Rad) by semidry electroblotting with a Tris-HCl-aminocaproic acid buffer system (pH ∼9.8) containing 0.004% of SDS (19). Membranes were washed with 150 mM phosphate-buffered saline (PBS) and incubated overnight at 4°C in 10% dried milk in PBS. After washing with PBS, membranes were incubated for 2 h at room temperature in PBS containing 1% bovine serum albumin and anti-influenza virus antibodies anti-A/Shangdong/9/93 (H3N2) sheep serum (Centers for Disease Control and Prevention, Atlanta, Ga.) and anti-A/USSR/77 (H1N1) goat immunoglobulin G (ViroStat, Portland, Maine). Membranes were then exposed to anti-sheep and anti-goat horseradish peroxidase-conjugated secondary anti-species antibodies (Dako, Copenhagen, Denmark), followed by visualization of positive bands with the Pierce (Rockford, Ill.) enhanced chemiluminescence (ECL) procedure by using Kodak BioMax film.
Double plaque assay titration.
MDCK cells grown in a 24-well plate were incubated with 0.4 ml of 10-fold virus sample dilutions in PBS/well. After a 60-min incubation at room temperature, the virus inoculum was removed and the cells were covered with 2.5 ml of agarose (culture quality; Sigma) at a final concentration of 0.9% in DMEM containing 0.25% bovine serum albumin-0.075% sodium bicarbonate. In the case of the standard plaque assay, 0.3 μg of acetylated trypsin (Sigma)/ml was added to the agarose. In the modified assay method, no trypsin was added initially, but at 24 h of incubation, the cells were overlaid with an additional 0.7% agarose layer (0.5 ml per well) containing 6 μg of acetylated trypsin/ml. Three days after infection, cells were fixed with a 10% formaldehyde solution, agarose layers were removed, fixed cells were stained with hematoxylin-eosin solution, and plaques were counted. This double titration distinguishes between viruses containing cleaved or uncleaved HA (40). The modified assay, with addition of a trypsin overlay only after 24 h, allows determination of the real infectivity of the virus population, i.e., virions with cleaved HA1/2; whereas the potential infectivity, i.e., virions with cleaved and uncleaved HAs, is measured by the immediate addition of trypsin to the initial agarose overlay.
Proteolytic activity of HAEC homogenates.
A/Aichi/2/68 virus was grown in MDCK cells without trypsin, the culture fluid was clarified at 4,000 rpm for 20 min, and the virus containing supernatant was pelleted through 25% glycerol in PBS at 22,000 rpm for 2 h at 4°C (SW-41 rotor; Spinko L7-55). The virus pellet was resuspended in DMEM (final virus concentration, 29 to 210 HAU/ml) and either added to growing HAEC or mixed with freshly prepared HAEC homogenates and incubated for 3.5 h at 37°C. In some experiments, the samples of clarified culture fluids were used as reference virus and mixed with HAEC homogenate. HAEC homogenates were prepared from mock-infected and influenza WSN/34-infected cells. For this, HAEC were scraped and centrifuged at 1,500 rpm for 15 min at +4°C. The HAEC pellet containing approximately 106 cells was pipetted into 100 to 150 μl of DMEM and subdivided into two equal parts. One part was left in ice and the other was additionally treated by two cycles of freeze-thawing. After incubation, infectivity titers of the virus-HAEC homogenate mixtures were estimated by the double plaque assay and viral HA polypeptide patterns were analyzed by PAGE-WB and identified by ECL with anti-H3N2 virus antibodies.
Pulse-chase labeling and immune precipitation of cellular polypeptides.
HAEC were infected with influenza A/Aichi/2/68 or A/WSN/34 virus at a multiplicity of infection (MOI) of 1 PFU per cell. At 6.5 h postinfection (hpi), the culture was incubated for 45 min in MEM lacking cold methionine and containing 35S-labeled methionine (Amersham Pharmacia Biotech) at a concentration of 100 μCi/ml (pulse). Thereafter, the cells were washed two times with MEM containing a 10-fold concentration of cold methionine and incubated in MEM for additional 1.5 and 3.0 h (chase). After labeling, the cells were mechanically removed from the plate and pelleted. The cell pellets were dissolved in 200 μl of radioimmunoprecipitation assay (RIPA) buffer containing 250 mM NaCl, 50 mM Tris-HCl (pH 7.8), 0.4% of nonionic detergent NP-40, 200 μM of phenylmethylsulfonyl fluoride, 50 TIU of aprotinin per ml, and 100 μM of benzamidine (Sigma) and were repeatedly centrifuged at 12,000 × g to remove undissolved debris. Aliquots (10 to 20 μl) of prepared supernatant were diluted in 150 μl of PBS and mixed with anti-influenza virus antibodies prepared against A/Shangdong/9/89 (H3N2) virus in sheep (Centers for Disease Control and Prevention), incubated 1 h at room temperature, and then mixed with 25 μl of a 40% suspension of protein G-Sepharose (Amersham Pharmacia Biotech) and additionally incubated for 45 min. Immune complexes were washed five times with PBS and dissolved in SDS-containing dissociation buffer. Samples were fractionated by PAGE followed by autoradiography of the gels on Kodak BioMax film.
Virus growth in presence of serum.
MDCK cells were cultured in 12-well plates and HAEC were grown in 6-well plates. Cells at 90% confluency were washed twice with PBS and infected with A/Beijing-like with an MOI of 0.01 for 90 min at room temperature and washed twice with MEM. HAEC were incubated with F12 media with 0, 1, 5, and 10% heat-inactivated FBS. MDCK cells were incubated with MEM Eagle media containing 0.5 μg of trypsin/ml and an amount of serum as listed above. All cells were maintained at 37°C in a 5% CO2 atmosphere. Supernatant was harvested at 1, 24, 48, 72 hpi in a 1:3 dilution with Hanks balanced salt solution (GIBCO/BRL) transport media containing 10% gelatin and antibiotics and quick-frozen in an ethanol dry ice bath. Viral titrations were performed in MDCK cells as described above.
RESULTS
It has been shown that HAEC are permissive for influenza virus reproduction and produced virions containing both cleaved (HA1/HA2) and noncleaved (HA0) (7). However, we wanted to determine (i) whether HA0 cleavage takes place in cells or in the extracellular environment, (ii) whether HA0 cleavage observed in HAEC increases virus infectivity, and (iii) whether HA0 cleavage in HAEC is sensitive to exogenous inhibitors of serine proteases added to the infected cells. To answer these questions the following experiments were done.
HA cleavage of virus grown in HAEC.
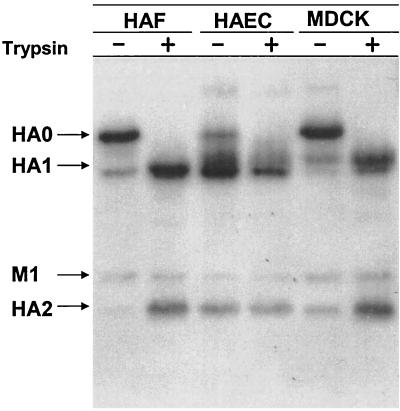
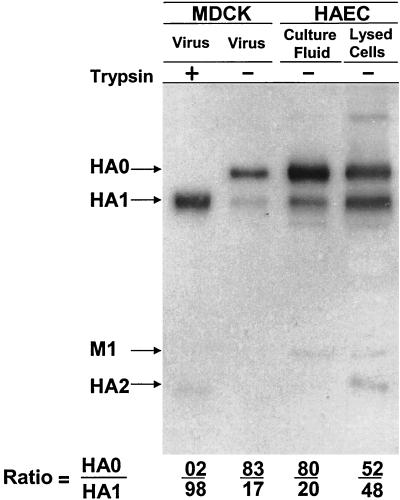
Cleavage of HA was compared in HAEC, which have several human epithelial cell types, with cleavage in HAF and MDCK. HAEC, HAF, and MDCK cultures without exogenous trypsin were infected with A/Aichi/2/68 virus at 2 to 4 PFU per cell and incubated for between 24 and 32 hpi. The progeny viruses were analyzed by PAGE and plaque assays. Viruses grown in HAF and MDCK cells contained predominantly noncleaved HA0, whereas virus produced in HAEC contained predominantly cleaved HA1/2 (Fig. 1). Consistent with the gel patterns, MDCK- and HAF-grown viruses were found to have low real infectivity while HAEC-grown viruses possessed high titers of cleaved virus that could initiate infection without exogenous trypsin (Table 1). These observations show that HAEC infected with an influenza A virus containing a single arginine in the HA proteolytic site are capable of cleaving of viral HA0 and producing infectious virions containing cleaved HA1/2.
FIG. 1.
Polypeptide patterns of A/Aichi/2/68 virus grown in HAF, MDCK, and HAEC. Cultures of HAF, MDCK, and HAEC were infected with influenza A/Aichi/2/68 virus at an MOI of about 0.5 PFU per cell and then were incubated in F12 medium alone for 24 to 32 h. After incubation, culture fluids were divided into two equal parts, and one of them was treated with 2.5 μg of trypsin/ml for 1 h. Virus particles were pelleted through 20% sucrose and analyzed by double plaque assay (Table 1) and by PAGE-WB. Viral proteins were identified by ECL with anti-H3N2 sheep antibodies and anti-sheep horseradish peroxidase conjugate.
TABLE 1.
Infectivity of influenza A/Aichi/68 virus grown in HAF, MDCK, and HAEC
| Virus samplesa | Trypsin treatment | Infectivity titers (PFU/ml)
|
||
|---|---|---|---|---|
| Real | Potential | Ratiob | ||
| HAF | − | 3.0 × 103 | 1.0 × 106 | 33.3 |
| + | 1.2 × 106 | 1.1 × 106 | 0.9 | |
| MDCK | − | 2.0 × 104 | 5.0 × 106 | 25.0 |
| + | 6.0 × 106 | 1.1 × 107 | 1.8 | |
| HAEC | − | 8.0 × 105 | 1.2 × 106 | 1.5 |
| + | 8.0 × 105 | 6.0 × 105 | 0.75 | |
Samples of A/Aichi/68 virus grown in cultures of HAF, MDCK, and HAEC and treated with trypsin in vitro as described for Fig. 1 were evaluated by the double plaque assay as described in Materials and Methods.
The ratio of potential infectivity to real infectivity reflects the overall number of potentially active virions per one activated virion in the virus population analyzed.
Inhibition of HA cleavage in HAEC cell pellets by low-molecular-weight protease inhibitors.
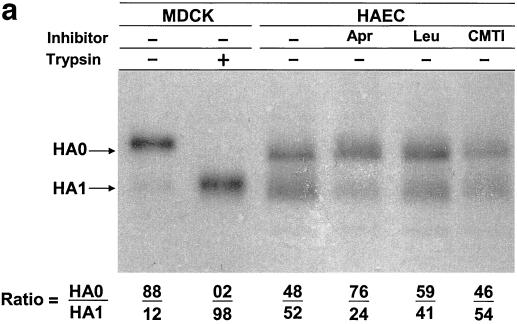
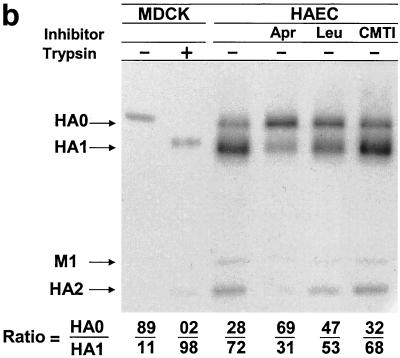
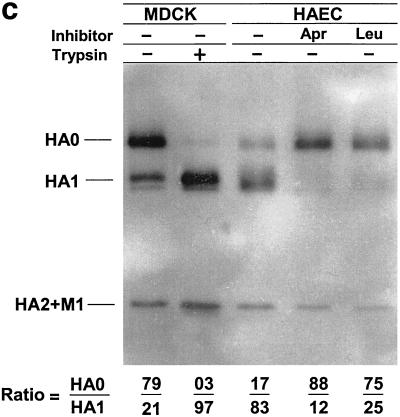
HAEC infected with influenza A/Aichi/2/68 (H3N2) and A/WSN/34 (H1N1) viruses were incubated in medium alone or in medium containing the low-molecular-weight protease inhibitors aprotinin, leupeptin, and CMTI. All three inhibitors studied possessed similar antiproteolytic activity as measured by trypsin inhibition. Aprotinin, leupeptin, and CMTI caused 50% inhibition of 1 μM trypsin at 1.1, 1.4, and 1.3 μM, respectively (data not shown). At between 20 and 24 hpi, cell pellets and culture fluids were harvested and assayed by PAGE and by plaque assays to distinguish between viruses containing cleaved or uncleaved HA (40). It was found that a significant amount of the cleaved HA accumulated in infected cells and about 52% of A/Aichi/2/68 HA was in the cleaved HA1 configuration in the HAE cell pellet (Fig. 2a, lane 3). The addition of the protease inhibitors aprotinin and leupeptin (Fig. 2a, lanes 4 and 5) to culture medium reduced the amount of cleaved HA1 accumulating in cells. In contrast, CMTI (Fig. 2a, lane 6) had no effect. Aprotinin caused the greatest inhibition of HA cleavage, with reduction of cleaved HA1 to 24%.
FIG. 2.
Influence of protease inhibitors on viral polypeptides synthesized in HAEC infected with influenza A virus. Cultures of HAEC were infected with influenza A/Aichi/2/68 (a and b) and A/WSN/34 (c) viruses at an MOI of about 1 PFU per cell. (a) Cell pellets; (b) cleavage patterns of released A/Aichi/2/68 virus. Cells were incubated in either F12 medium alone or in F12 medium containing 50 μM aprotinin, 140 μM leupeptin, or CMTI. As controls, the patterns of A/Aichi/2/68 (a and b) and A/WSN/34 (c) virus polypeptides grown in MDCK cells with (+) and without (−) trypsin are shown. Cellular polypeptides were analyzed by PAGE-WB and identified by ECL with anti-H3N2 virus (a and b) and anti-H1N1 virus (c) antibodies and anti-species conjugates. Western blots were scanned, and HA0/HA1 ratios were calculated as indicated at the bottom of the gels.
Effect of low-molecular-weight protease inhibitors on HA cleavage of released virus particles.
In the set of experiments on HA cleavage of released virus particles, we investigated the influence of protease inhibitors on the HA0/HA1 ratio in released virions after assembly in HAEC. It was found that the percent of cleaved HA1 had increased in the released virions to 72% (Fig. 2b, lane 3) when compared with 52% HA1 within cells (Fig. 2a, lane 3). Aprotinin and leupeptin decreased the percent of HA1 to 31 and 53%, respectively (Fig. 2b, lanes 4 and 5). As with cell-associated protein, CMTI did not change the HA0/HA1 ratio in virions (Fig. 2b, lane 6). Even greater inhibition of viral HA0 cleavage by aprotinin and leupeptin was seen in HAEC infected with WSN/34 virus (Fig. 2c, lanes 4 and 5), indicating that suppression of the HA cleavage is not only an inhibitor-dependent event but also a virus-dependent event. It follows from the above results that (i) substantial virus HA cleavage takes place in intracellular or plasma membranes of HAEC and (ii) both cleaved and uncleaved HA molecules are assembled into virions.
Effect of antiproteases on infectivity of released influenza virus.
The effect of the protease inhibitors of virus yield in HAEC was determined. The infectivity of released virus, as determined by the double plaque assay, was found to correlate with the percent HA1 cleavage (Fig. 2b and c; Table 2). Both A/Aichi/2/68 and A/WSN/34 viruses grown in HAEC without protease inhibitors had similar titers by standard and modified methods showing high real infectious activity. Leupeptin and CMTI did not inhibit the real infectivity of A/Aichi/2/68 virus in the modified assay, but leupeptin inhibited the more susceptible A/WSN/34 virus (Table 2). There was about a 10-fold decrease in the real infectivity of progeny of both viruses grown in the presence of aprotinin, the most effective inhibitor of cleavage (Table 2).
TABLE 2.
Infectivity of influenza A virus grown in HAEC and the effect of protease inhibitors
| Virus samplesa | HA titer (HAU/ml)b | Infectivity titer (PFU/ml)c
|
||
|---|---|---|---|---|
| Apparent | Potential | Ratiod | ||
| A/Aichi/2/68 | ||||
| Control | 32 | 2.3 × 107 | 2.5 × 107 | 1.1 |
| Aprotinin | 8 | 1.2 × 106 | 1.0 × 107 | 8.3 |
| Leupeptin | 8 | 1.1 × 107 | 1.3 × 107 | 1.2 |
| CMTI | 32 | 2.2 × 107 | 2.3 × 107 | 1.0 |
| A/WSN/34 | ||||
| Control | 16 | 4.4 × 106 | 4.0 × 106 | 0.9 |
| Aprotinin | 4 | 0.5 × 105 | 3.4 × 105 | 6.8 |
| Leupeptin | 4 | 1.3 × 105 | 7.4 × 105 | 5.7 |
One-week-old cultures of HAEC were infected with indicated virus at an MOI of 1 PFU per cell and then were incubated in either F12 medium alone (control) or in F12 medium containing aprotinin (50 μM), leupeptin (140 μM), or CMTI (50 μM).
The HA titer of the culture medium was evaluated at 28 hpi using a 1% suspension of chicken erythrocytes.
Real and potential infectivity titers of the culture media were measured at 28 hpi by double plaque assay (40).
The ratio of potential infectivity to real infectivity reflects the overall number of potentially active virions per one activated virion in the virus population analyzed.
Effect of serum on influenza virus growth in HAEC.
Further support for the concept of cell-associated HA0 cleavage was obtained from examining the effect of serum on the growth of influenza A/Beijing-like virus in HAEC. Influenza virus-infected MDCK cells supplemented with trypsin in their culture fluid was used as positive control. Serum has a number of high-molecular-weight antiproteases, such as alpha2-macroglobulin, alpha1-antitrypsin, alpha2-antiplasmin, and antithrombin III (36), that may compete for proteases on the cell surface or in the extracellular fluid of influenza virus-infected HAEC. HAEC and MDCK were infected with influenza A/Beijing-like virus at an MOI of 0.01 PFU per cell, and after adsorption, cells were incubated in medium containing increasing amounts of heat-inactivated FCS. The yield of influenza A/Beijing-like virus at 48 h in HAEC was unaffected by serum concentrations of up to 10% (Table 3). In contrast, in MDCK cells supplemented with trypsin influenza, virus was undetected when serum concentrations were greater than 1%. Thus, virus activation by exogenous trypsin, which is necessary for virus growth in MDCK, was effectively inhibited in serum-containing medium.
TABLE 3.
Effect of serum on growth of influenza A/Beijing virus in MDCK and HAECa
| Cell line | Viral yield (PFU/ml) at FBS concn of:
|
||||
|---|---|---|---|---|---|
| 0% | 1% | 2.5% | 5% | 10% | |
| MDCK | 3.2 × 106 | 9.0 × 103 | 0 | 0 | 0 |
| HAEC | 3.8 × 106 | 6.6 × 106 | 8.5 × 106 | 3.7 × 106 | 3.0 × 106 |
Cultures of MDCK and HAEC were infected with A/Beijing/89/H3N2 virus at a multiplicity of input of 0.01 PFU per cell. After adsorption, the infected cells were washed and incubated with F12 medium containing the indicated concentrations of heat-inactivated FCS. Trypsin was added to the culture fluid of MDCK cells at a final concentration of 5 μg/ml. The overall virus yield was evaluated by the standard plaque assay after 48 h of incubation.
Timing of HA cleavage during virus entry into HAEC.
The above observations have shown cleavage of de novo-synthesized HA0 (“cleavage from within”); we next examined the ability of HAEC to cleave the HA0 of parental incoming virus (“cleavage from without”). Influenza Aichi/2/68 virus grown in MDCK cells without trypsin and containing uncleaved HA0 and low real infectivity (Fig. 1 and Table 1) was used as the inoculum in these experiments. This uncleaved virus was inoculated onto HAEC for various periods of time. After these incubations, the culture fluids and pelleted cell samples were analyzed by PAGE and modified plaque assay to study the site and rapidity of HA cleavage and infectivity of incubated virus.
It was found that virus remaining in the culture fluid retained its HA0 configuration over at least 4 h of incubation (Fig. 3, lane 3). In contrast, electrophoretic analysis showed that HA0 of parental virus was cleaved into HA1/2 shortly after adsorption to cells. After 90 min, about 50% of cell-associated HA0 was cleaved into HA1/2, well before de novo synthesis of HA could occur (Fig. 3, lane 4). In these experiments, approximately one-third of virions were absorbed by cells based on the relative amounts of HA0 plus HA1 in the cell pellet and culture fluid by electrophoresis (Fig. 3, lanes 3 and 4). However, the infectivity of the virus in cell pellets was about 100 times lower than in culture fluid, suggesting that HA0 virus had entered the cell, been activated, undergone fusion with the endosomal membrane, and entered the noninfectious eclipse phase. Further incubation of such cells for about 20 h led to the expected intracellular synthesis and accumulation of typical virus polypeptide pattern and virus production (data not shown). These observations indicate that an additional pathway for intracellular cleavage of influenza virus HA0 by cell-associated proteases(s) exists at the cell membrane on entry and/or in the endosomal compartment, a process we are calling cleavage from without.
FIG. 3.
Polypeptides of influenza A virus incubated with growing HAEC. A/Aichi/2/68 virus grown in MDCK cells was incubated with growing HAEC for 3.5 h at 37°C. After incubation, culture fluid (lane 3) was withdrawn and cells (lane 4) were removed and homogenized. Viral polypeptides were analyzed by PAGE-WB and identified by ECL with anti-H3N2 virus sheep antibodies and anti-sheep conjugate. The controls include A/Aichi virus grown in MDCK cells (lane 1) and after treatment in vitro with trypsin (lane 2). Western blots were scanned and HA0/HA1 ratios were calculated (bottom).
Lack of viral HA cleavage by HAEC homogenates.
In the next set of experiments, we asked whether IAP accumulates in HAEC as an active enzyme. For this, a cell homogenate was prepared from influenza virus-infected and -noninfected HAEC and the abilities of this homogenate to cleave virus HA0 and activate virus infectivity were investigated. As in the experiments with growing cells, a nonactivated A/Aichi/68 virus grown in MDCK cells was used as reference virus. Cell homogenates were prepared from mock-infected HAEC and HAEC infected with A/WSN/34 (H1N1) virus at 7.5 hpi—a time at which there was not yet infectious A/WSN/34 progeny virus (A/WSN/34 virus-infected cells were included to see if there was an increase of IAP activity in cells in the course of influenza virus infection). A/Aichi/2/68-MDCK virus was incubated with A/WSN/34 virus-infected and -uninfected cell homogenates for 4 h at 37°C and then was analyzed by PAGE and infectivity. In contrast to virus exposed to growing HAEC, after incubation with cell homogenate, the virus retained noncleaved HA0 and had low infectivity without trypsin (data not shown). These data suggested either that IAP of HAEC is a short-lived enzyme rapidly suppressed by antiproteases in the cell homogenate or that its function depends on cell metabolism.
Cleavage of newly formed influenza virus HA.
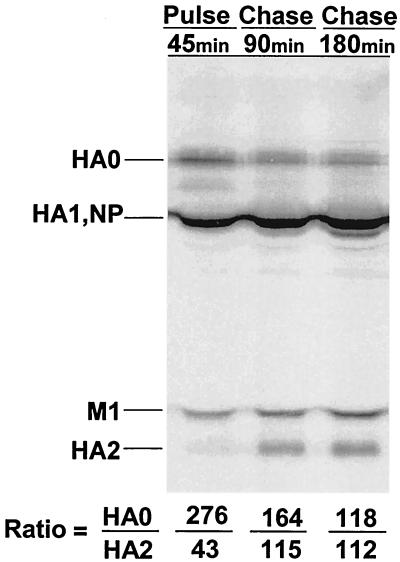
In the final set of experiments, we estimated the rate of cleavage of de novo-formed influenza virus HA in HAEC. For this, a pulse-chase experiment with 35S-labeled methionine was used. At 6.5 hpi, A/Aichi/2/68 or A/WSN/34 HAEC was labeled with [35S]methionine for 45 min (pulse) and then was incubated in cold methionine-containing medium for 1.5 and 3.0 h (chase). Polypeptides of pulse-chase-labeled cells were precipitated by antibodies specific for influenza proteins, and these immune precipitates were analyzed by PAGE followed by autoradiography of the gels (Fig. 4). By the end of the 45-min pulse, some of the HA molecules were already cleaved and a small HA2 band had appeared. The HA1 subunit is not visible in this gel because the major viral nucleocapsid protein NP comigrated with HA1 and masked it on the gel. During the next 1.5 h, HA2 significantly increased while the HA0 decreased. An additional 1.5-h incubation (3-h chase) only slightly increased the amount of HA2 (Fig. 4, bottom lines). Similar rates of intracellular cleavage of A/WSN/34 HA were obtained in HAEC cells infected with influenza A/WSN/34 virus (not shown). These data show that (i) cleavage of newly synthesized influenza virus HA is cell associated in HAEC and (ii) this cleavage is a rapid process in which the majority of HA0 molecules are cleaved preferentially within 1 to 2 h after synthesis.
FIG. 4.
Pulse-chase-labeled pattern of viral polypeptides synthesized in HAEC infected with influenza A virus. HAEC were infected with A/Aichi/2/68 (H3N2) virus and labeled at 6.5 hpi with [35S]methionine for 45 min (pulse) followed by 90 and 180 min chase in cold-methionine medium. Labeled HAEC were dissolved in RIPA buffer, and viral polypeptides were precipitated by anti-H3N2 virus-specific antibodies and analyzed by PAGE followed by autoradiography of gel. Gel autoradiographs were scanned and HA0/M1 and HA1/M1 ratios, where the M1 is 100%, were calculated (bottom).
DISCUSSION
In the experiments reported here, we examined the fate of influenza virus HA in cultures of human cells prepared from adenoids. The primary HAEC are similar to a normal epithelium of the upper respiratory tract of humans with differentiated cells that maintain ciliary motion, secrete mucin, and have markers for the polymeric Ig receptor and Clara cells (7, 38). Thus, the cleavage of influenza virus HA protein and virus activation obtained in HAEC should reflect the conditions that exist in the human respiratory tract.
We have shown that cleavage of influenza virus HA in HAEC is a cell-associated process mediated by the cells in which the virus is replicating. We have immunohistochemical evidence that the primary target for influenza virus replication in HAEC is the ciliated cell (data not published). It appears that such cleavage may occur in HAEC both for newly synthesized HA molecules (cleavage from within) and for the HA of incoming (input) virions (cleavage from without). A persistent and important question in influenza virus pathogenesis has been the origin of IAP in respiratory epithelium. The question has revolved around whether it is synthesized directly in virus-infected ciliated epithelial cells or produced by specialized cells, such as Clara cells (which are found in HAEC [7]), from which it may act in a paracrine fashion to cleave HA (14, 16, 35).
Currently, it is not possible to resolve the role of Clara cell protease or other secreted proteases in influenza virus pathogenesis. However, the absence of influenza virus HA cleavage in HAEC culture fluid, the rapid rate and high efficiency of HA cleavage in HAEC both on entry and after de novo HA synthesis, and the accumulation of cleaved HA1/2 associated with HAEC suggest the existence of an intracellular IAP in human respiratory epithelium cells acting directly on the cells in which influenza virus is replicating. It is tempting to speculate that such a hypothetical enzyme may be similar in terms of intracellular transport and turnover to those in the subtilizin-like family that provide the intracellular cleavage of viral glycoproteins containing multibasic proteolytic sites in trans-Golgi area (18, 32). However, in distinction from subtilizin-like proteases, this IAP enzyme would have to possess a specificity for single-arginine proteolytic sites. A final possibility is that the more recently described mini-plasmin (23) may be a key enzyme and that IAP is a membrane-associated enzyme that can be recycled into the endosome to cleave HA0 on virus entry. The cleavage of A/WSN/34 has been attributed to plasmin activation on the cell surface (11), but the HAEC system had no exogenous plasmin and the two prototype viruses (A/WSN/34 and A/Aichi/2/68) used in the experiments behaved similarly in HAEC. It is possible that several mechanisms may contribute to HA0 cleavage or that mechanisms differ at different levels of the human respiratory tract.
An important feature of this cleavage process was our inability to detect IAP activity after homogenization of the cell pellet, i.e., its apparent functionality only in viable cells. IAP activity did not accumulate in HAEC and disappeared after cell disruption. It follows from this observation that host IAP has a short period of activity that may be regulated either by unstable cofactors and natural antagonists, such as protease inhibitors, or by high functional lability of IAP enzyme itself. It is evident that, in any case, synthesis of newly active enzyme and/or its regulatory cofactors are necessary and require host cell metabolism.
The data presented here that exogenous protease inhibitors suppressed influenza virus activation in human respiratory epithelium suggest that they may have therapeutic value. An important aspect in this context is the specific sensitivity of influenza virus HA cleavage to inhibitors of proteases. The inhibition by aprotinin and leupeptin, inhibitors of serine-type proteases, indicate that human respiratory IAP or its cofactor belong to this protease class. Leupeptin is composed of only three amino acids (molecular mass, 0.5 kDa) (1), and aprotinin is a 58-amino-acid polypeptide (molecular mass, 6 kDa) isolated from bovine lungs (8). The ability of these low-molecular-weight inhibitors to suppress viral HA cleavage in cells implies either that they are effectively internalized into infected cells or that HA cleavage is accomplished in sites at the plasma membrane easily accessible to low-molecular-weight inhibitors but inaccessible to the high-molecular-weight serum exogenous protease inhibitors.
There is therapeutic experience with aprotinin. It has been shown to effectively suppress influenza virus HA cleavage and virus activation in cultured cells (42), embryonated chicken eggs (44), and mouse lungs (43) and provide protection against lethal influenza virus infection of mice when administered intraperitoneally (45) and by aerosol inhalation (29). The aerosol inhalations in mice did not have side effects during application for 1 month (46, 47). Aprotinin aerosol inhalations in humans with influenza and parainfluenza have shown therapeutic efficacy, including reduction in duration of fever, headache, sore throat, hoarseness, cough, and earlier resolution of illness and discharge from the hospital (48). The present study strengthens the idea that the antiviral target of aprotinin in humans is inhibition of viral HA cleavage and that it or other proteases inhibitors remain potential anti-influenza drugs (49).
Acknowledgments
We thank H. D. Klenk for helpful discussions.
This work was supported by NATO linkage grant HT-974619, NIAID RPRU Contract AI-65298, Howard Hughes Medical Institute grant 75195546302, Russian Basic Research Foundation grants 48442 and 04000, German-Russian (DFG-RFBR) grant 436 RUS 113/587, and ASGL-Research Laboratories grant 11-D/1/CHT-99.
REFERENCES
- 1.Aoyagi, T., T. Takeuchi, A. Matsuzaki, K. Kawamura, and S. Kondo. 1969. Leupeptins, new protease inhibitors from Actinomycetes. J. Antibiot. 22:283-289. [DOI] [PubMed] [Google Scholar]
- 2.Barbey-Morel, C. L., T. N. Oeltmann, K. M. Edwards, and P. F. Wright. 1987. Role of respiratory tract proteases in infectivity of influenza A virus. J. Infect. Dis. 155:667-672 . [DOI] [PubMed] [Google Scholar]
- 3.Beppu, Y., Y. Imamura, M. Tashiro, T. Towatari, H. Ariga, and H. Kido. 1997. Human mucus protease inhibitor in airway fluids is a potential defensive compound against infection with influenza A and Sendai virus. J. Biochem. 121:309-316. [DOI] [PubMed] [Google Scholar]
- 4.Boycott, R., H. D. Klenk, and M. Ohuchi. 1994. Cell tropism of influenza A virus mediated by hemagglutinin activation at the stage of virus entry. Virology 203:313-319. [DOI] [PubMed] [Google Scholar]
- 5.Callan, R. J., F. A. Hartmann, S. E. West, and V. S. Hinshaw. 1997. Cleavage of influenza A virus H1 hemagglutinin by swine respiratory bacterial proteases. J. Virol. 71:7579-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 7.Endo, Y., K. N. Carroll, M. R. Ikizler, and P. F. Wright. 1996. Growth of influenza A virus in primary, differentiated epithelial cells derived from adenoids. J. Virol. 70:2055-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritz, H., and G. Wunderer. 1983. Biochemistry and applications of aprotinin, the kallikrein inhibitor from bovine organs. Arzneimittel-Forschung 33:479-494. [PubMed] [Google Scholar]
- 9.Garten, W., F. X. Bosch, D. Linder, R. Rott, and H. D. Klenk. 1981. Proteolytic activation of the influenza virus hemagglutinin: the structure of the cleavage site and the enzymes involved in cleavage. Virology 115:361-374. [DOI] [PubMed] [Google Scholar]
- 10.Garten, W., A. Steineke, E. Shaw, P. Wikstrom, and H. D. Klenk. 1989. Inhibition of proteolytic activation of influenza virus hemagglutinin by specific peptidyl chloroalkyl ketones. Virology 172:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goto, H., and Y. Kawaoka. 1998. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc. Natl. Acad. Sci. USA 95:10224-10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotoh, B., T. Ogasawara, T. Toyoda, N. Inocencio, M. Hamaguchi, and Y. Nagai. 1990. An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo. EMBO J. 9:4189-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaoka, Y., C. W. Naeve, and R. G. Webster. 1984. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology 139:303-316. [DOI] [PubMed] [Google Scholar]
- 14.Kido, H., Y. Yokogoshi, K. Sakai, N. Tashiro, Y. Kishino, A. Fukutomi, and N. Katanuma. 1992. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J. Biol. Chem. 267:13573-13579. [PubMed] [Google Scholar]
- 15.Kido, H., K. Sakai, Y. Kishino, and M. Tashiro. 1993. Pulmonary surfactant is a potential endogenous inhibitor of proteolytic activation of Sendai virus and influenza A virus. FEBS Lett. 322:115-119. [DOI] [PubMed] [Google Scholar]
- 16.Kido, H., Y. Chen, and M. Murakami. 1999. Cellular proteinases and viral infection: influenza virus, sendai virus, and HIV-1, p. 205-217. In B. Dunn (ed.), Proteases of infectious agents. Academic Press, New York, N.Y.
- 17.Klenk, H. D., R. Rott, M. Orlich, and J. Blodorn. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426-439. [DOI] [PubMed] [Google Scholar]
- 18.Klenk, H. D., and W. Garten. 1994. Host cell proteases controlling virus pathogenecity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 19.Kyhse-Anderson, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 20.Lazarowitz, S. G., A. R. Goldberg, and P. W. Choppin. 1973. Proteolytic cleavage by plasmin of the HA polypeptide of influenza virus: host cell activation of serum plasminogen. Virology 56:172-180. [DOI] [PubMed] [Google Scholar]
- 21.Lazarowitz, S. G., and P. W. Choppin. 1975. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68:440-454. [DOI] [PubMed] [Google Scholar]
- 22.Morsy, J., W. Garten, and R. Rott. 1994. Activation of an influenza virus A/turkey/Oregon/71 HA insertion variant by the subtilisin-like endoprotease furin. Virology 202:988-991. [DOI] [PubMed] [Google Scholar]
- 23.Murakami, M., T. Towatari, M. Ohuchi, M. Shiota, M. Akao, Y. Okumura, M. A. A. Parry, and H. Kido. 2001. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur. J. Biochem. 268:2847-2855. [DOI] [PubMed] [Google Scholar]
- 24.Ohuchi, M., M. Orlich, R. Ohuchi, B. E. Simpson, W. Garten, H. D. Klenk, and R. Rott. 1989. Mutations at the cleavage site of the hemagglutinin alter the pathogenicity of influenza virus A/chick/Penn/83 (H5N2). Virology 168:274-280. [DOI] [PubMed] [Google Scholar]
- 25.Ohuchi, R., M. Ohuchi, W. Garten, and H. D. Klenk. 1991. Human influenza virus hemagglutinin with high sensitivity to proteolytic activation. J. Virol. 65:3530-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otlewski, J., and D. Krowarsch. 1996. Squash inhibitor family of serine proteinases. Acta Biochem. Polonica. 43:431-444. [PubMed] [Google Scholar]
- 27.Ovcharenko, A. V., and O. P. Zhirnov 1994. Aprotinin aerosol treatment of influenza and paramyxovirus bronchopneumonia of mice. Antivir. Res. 23:107-118. [DOI] [PubMed] [Google Scholar]
- 28.Palese, P., K. Tobita, M. Ueda, and R. W. Compans. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397-410. [DOI] [PubMed] [Google Scholar]
- 29.Scheiblauer, H., M. Reinacher, M. Tashiro, and R. Rott. 1992. Interactions between bacteria and influenza A virus in the development of influenza pnneumonia. J. Infec. Dis. 166:783-791. [DOI] [PubMed] [Google Scholar]
- 30.Skehel, J. J., and M. D. Waterfield. 1975. Studies on the primary structure of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 72:93-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stieneke-Grober, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H. D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with a multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1-20. [DOI] [PubMed] [Google Scholar]
- 33.Tashiro, M., P. Ciborowski, M. Reinacher, G. Pulverer, H. D. Klenk, and R. Rott. 1987. Synergistic role of staphylococcal proteases in the induction of influenza virus pathogenecity. Virology 157:421-430. [DOI] [PubMed] [Google Scholar]
- 34.Tashiro, M., H. D. Klenk, and R. Rott. 1987. Inhibitory effect of a protease inhibitor, leupeptin, on the development of influenza pneumonia, mediated by concomitant bacteria. J. Gen. Virol. 68:2039-2043. [DOI] [PubMed] [Google Scholar]
- 35.Tashiro, M., and R. Rott. 1996. The role of proteolytic cleavage of viral glycoproteins in the pathogenesis of influenza virus infections. Sem. Virol. 7:237-245. [Google Scholar]
- 36.Travis, J., and G. S. Salvesen. 1983. Human plasma proteinase inhibitors. Annu. Rev. Biochem. 52:655-709. [DOI] [PubMed] [Google Scholar]
- 37.Walker, J. A., T. Sakaguchi, Y. Matsuda, T. Yoshida, and Y. Kawaoka. 1992. Location and character of the cellular enzyme that cleaves the hemagglutinin of the virulent avian influenza virus. Virology 190:278-287. [DOI] [PubMed] [Google Scholar]
- 38.Widdicombe, J. G., and R. J. Pack. 1982. The Clara cell. Eur. J. Respir. Dis. 63:202-220. [PubMed] [Google Scholar]
- 39.White, J. M. 1992. Membrane fusion. Science 258:917-922. [DOI] [PubMed] [Google Scholar]
- 40.Zhirnov, O. P., A. G. Ovcharenko, and A. G. Bukrinskaya. 1982. A modified plaque assay method for accurate analysis of infectivity of influenza viruses with uncleaved hemagglutinin. Arch. Virol. 71:177-183. [DOI] [PubMed] [Google Scholar]
- 41.Zhirnov, O. P., A. V. Ovcharenko, and A. G. Bukrinskaya. 1982. Proteolytic activation of influenza WSN virus in cultured cells is performed by homologous plasmam enzymes. J. Gen. Virol. 63:469-474. [DOI] [PubMed] [Google Scholar]
- 42.Zhirnov, O. P., A. V. Ovcharenko, and A. G. Bukrinskaya. 1984. Suppression of influenza virus replication in infected mice by protease inhibitors. J. Gen. Virol. 65:191-196. [DOI] [PubMed] [Google Scholar]
- 43.Zhirnov, O. P., A. V. Ovcharenko, and A. G. Bukrinskaya. 1985. Myxovirus replication in chicken embryos can be suppressed by aprotinin due to the blockage of viral glycoprotein cleavage. J. Gen. Virol. 66:1633-1638. [DOI] [PubMed] [Google Scholar]
- 44.Zhirnov, O. P. 1987. High protection of animals lethally infected with influenza virus by aprotinin-rimantadine combination. J. Med. Virol. 21:161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhirnov, O. P., A. V. Ovcharenko, P. B. Golyando, T. P. Svinigeeva, A. V. Dolgova, and A. V. Nikitin. 1994. Antiviral aerosol of aprotinin. 1. General action on the organism during the inhalation treatment. Antibiot. Themother. 5:25-32. (In Russian.) [Google Scholar]
- 46.Zhirnov, O. P., A. V. Ovcharenko, P. B. Golyando, T. P. Svinigeeva, A. V. Dolgova, and A. V. Nikitin. 1994. Antiviral aerosol of aprotinin. 2. The lack of local irritant and allergic complications. Antibiot. Themother. 9/10:53-57. (In Russian.) [Google Scholar]
- 47.Zhirnov, O. P., L. S. Kirzhner, A. V. Ovcharenko, and N. A. Malyshev. 1996. Aerosolized aprotinin is an effective drug against viral respiratory illness. Antiinfect. Drug Chemother. 14:209-216.
- 48.Zhirnov, O. P. 1983. Proteolytic activation of myxoviruses and a new strategy in the treatment of viral diseases. Problems Virol. 4:9-21. (In Russian.) [PubMed] [Google Scholar]