Abstract
Evolution of fitness values upon replication of viral populations is strongly influenced by the size of the virus population that participates in the infections. While large population passages often result in fitness gains, repeated plaque-to-plaque transfers result in average fitness losses. Here we develop a numerical model that describes fitness evolution of viral clones subjected to serial bottleneck events. The model predicts a biphasic evolution of fitness values in that a period of exponential decrease is followed by a stationary state in which fitness values display large fluctuations around an average constant value. This biphasic evolution is in agreement with experimental results of serial plaque-to-plaque transfers carried out with foot-and-mouth disease virus (FMDV) in cell culture. The existence of a stationary phase of fitness values has been further documented by serial plaque-to-plaque transfers of FMDV clones that had reached very low relative fitness values. The statistical properties of the stationary state depend on several parameters of the model, such as the probability of advantageous versus deleterious mutations, initial fitness, and the number of replication rounds. In particular, the size of the bottleneck is critical for determining the trend of fitness evolution.
The initial steps in virus evolution are generation of diversity (through mutation, recombination, and genome segment reassortment in multipartite genomes), competition among the generated variants, and selection of those mutants showing the largest phenotypic advantage in a given environment (see overviews in references 9, 14, 17, 28, 30, and 40). Because of high mutation rates during viral RNA biosynthesis (2, 18), RNA viruses replicate as complex mutant swarms termed viral quasispecies (14, 21-24). The degree of adaptation of a viral quasispecies to a given environment is often quantitated by a relative fitness value, which measures the replication capacity of a viral population relative to a reference mutant distinguishable either genotypically or phenotypically (15, 26, 33, 37). The long-term survival probability of a virus should consider parameters other than replication capacity (such as particle stability, transmissibility, etc. [11, 12, 15]). Despite their limitations, relative fitness values, determined by growth competition experiments between two viral populations in cell culture and in vivo, are providing insights into basic features of quasispecies evolution as well as viral disease progression (5, 15, 49).
Because of the large variations in viral population size during infections in vivo, a focus of interest has been the study of the influence of virus population size on the evolution of fitness values (reviewed in reference 15). Experimental results with several RNA viruses have documented that large population passages often result in fitness gains (7, 26, 43, 45, 58) while repeated bottleneck events (serial plaque-to-plaque transfers being an extreme example) lead to average fitness losses (6, 19, 25, 27, 59, 60), as predicted by the operation of Muller's ratchet (38, 41). According to this mechanism, asexual populations of organisms will tend to accumulate deleterious mutations in an irreversible fashion unless accumulation is retarded by recombination. Studies with vesicular stomatitis virus (VSV) established that the size of the population bottleneck that led to either an increase, a decrease, or maintenance of a fitness value was determined by the initial fitness of the population (44). Consistent with this observation, exponential fitness gains observed upon large population passages were eventually limited by the population size, resulting in stochastic fitness variations (46).
In a recent study of the fitness evolution of foot-and-mouth disease virus (FMDV) clones subjected to at least 100 successive plaque-to-plaque transfers, a linear accumulation of mutations together with a biphasic fitness decrease was observed (27). The second phase showed fitness fluctuations around a fitness mean value without perceivable fitness loss. This fact is remarkable since it could underlie a mechanism for low-fitness viral populations to elude extinction. For the present report we developed a numerical model that investigates fitness losses of fast-replicating genomes displaying high mutation rates and subjected to plaque-to-plaque transfers. The results with this new numerical model are in agreement with experimental results showing a biphasic fitness evolution of FMDV clones and fitness stability upon plaque transfers of very low fitness clones (27). The model predicts also the effect of transmission size on fitness evolution, as documented previously with experiments using VSV (44). Given its relevance to virus survival, the fitness stability of low-fitness clones has been further supported by 30 additional plaque-to-plaque transfers of highly debilitated FMDV clones. An essential feature of the model is the consideration of advantageous compensatory mutations as a molecular mechanism to cause fitness recovery, a feature adopted on the basis of experimental observations made with FMDV (1, 26) and also with unrelated systems such as human immunodeficiency virus (HIV) type 1 (4, 42) and some antibiotic-resistant bacteria (3, 50).
MATERIALS AND METHODS
Plaque-to-plaque transfers of FMDV clones.
The origin of BHK-21 cells, procedures for cell culture, and plaque assays with FMDV have been previously described (25-27). Relative fitness values of FMDV clones were estimated from the average number of PFU present in a plaque grown on a BHK-21 cell monolayer, determined upon triplicate platings; this procedure is identical to that used in previous studies (27) and differs from the standard measurement based on growth competition experiments (33). FMDV clone  was obtained after subjecting clone
was obtained after subjecting clone  to 130 plaque-to-plaque transfers as previously described (27).
to 130 plaque-to-plaque transfers as previously described (27).  is a clone derived from C-S8c1, the standard FMDV clone used in our laboratory and which was obtained from natural isolate C-Sta. Pau Sp/70 (53); the isolation of
is a clone derived from C-S8c1, the standard FMDV clone used in our laboratory and which was obtained from natural isolate C-Sta. Pau Sp/70 (53); the isolation of  was described in reference 25. In the present experiments four subclones derived from clone
was described in reference 25. In the present experiments four subclones derived from clone  were subjected to 30 additional plaque-to-plaque transfers following the plating procedure described previously (27).
were subjected to 30 additional plaque-to-plaque transfers following the plating procedure described previously (27).
Numerical model.
The model developed in the present study delineates fitness evolution upon plaque-to-plaque transfers, mimicking the experiments carried out elsewhere with RNA viruses and in particular with FMDV (25, 27). To study the effect of population size on fitness variation, transfers involving pools of virus from a number (Np) of plaques to Np plaques in the way described by Novella et al. (44) have also been modeled. The model is divided into two main steps.
Step 1: simulation of the development of a lytic plaque.
The process starts with a “virtual” single virus genome which after several replication cycles (denoted by r) will generate progeny, in a way analogous to the viral population produced in a lytic plaque. Each individual virus is characterized only by a parameter, W, standing for its fitness, which can take discrete values ranging from 1 to ∞. Fitness values are taken as the mean direct progeny per individual, which is proportional to the replication rate. Other factors that may have an influence on fitness (11, 12) are not considered. Individuals in the model are referred to as sequences or genomes instead of viruses. Each sequence has a certain probability of generating a number k (k = 1, 2, 3,…) of direct descendants after replication. The probability P(k) of generating k descendants is given by a Poisson distribution, {P(k) = [(W)k e−W]/k!}, in which W is the viral fitness, given here as the mean number of direct descendants (54).
During progeny formation, mutations take place, giving rise to genomes with the same fitness as, or lower or higher fitness than, the starting one. Each time that a deleterious mutation occurs (probability of occurrence of deleterious mutation = p), the fitness of the new genome decreases by one unit, and when the mutations are advantageous (compensatory and back mutations) the fitness increases by one unit (probability of increase of fitness = q). In the model, q ≪ p < 1 and the highest probability (1 − p − q) corresponds to occurrence of mutations with no discernible effects on fitness. As only phenotypic variations are observed, the model does not distinguish between sequences with neutral mutations and those with no mutations. The first replication cycle (r = 1) corresponds to the replication of the founder sequence to give a number (k) of progeny with probability P(k). Each new sequence can also initiate its own replication, originating a new ensemble of daughter sequences whose number is also described by a Poisson distribution. The process is performed for a given number of replication cycles (r), r being proportional to the time of plaque development (24 to 48 h in the experiments with FMDV [27]).
Step 2: selection of the sequences for the subsequent transfers.
In the previous step, an ensemble of sequences with different fitness values was generated. Fitness always has a positive value, as sequences with W ≤ 0 do not replicate and are lost. The transition to W ≤ 0 defines an extinction threshold which is parallel to the case of the experimental system when viruses which have accumulated many deleterious mutations or a lethal mutation do not originate plaques and do not participate in the next round of plaque formation. One of the plaque-forming genomes is randomly chosen and used as input for repetition of step 1. Steps 1 and 2 are then repeated a number of times (T), analogous to a number of plaque-to-plaque transfers carried out in the laboratory. Each time that a simulated lytic plaque develops, it is possible to determine the distribution of fitness values of the genomes composing the plaque and the number of genomes generated from the parental one.
Numerical computations.
The computer programs were written in FORTRAN language. Short runs were carried out on a Pentium 3, and long runs were performed on an Alpha 64-bit DS10 466 MHz, computer.
RESULTS AND DISCUSSION
Effect of initial fitness.
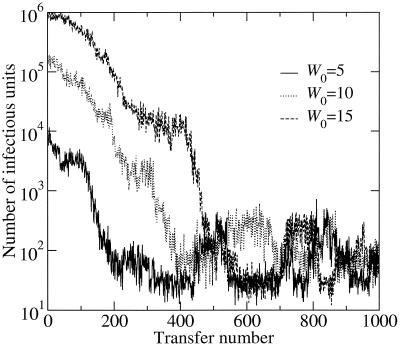
The objective of the present study is to develop a model that simulates the evolution of fitness values of viral populations subjected to serial plaque-to-plaque transfers, by using parameter values derived directly from experimental results. In the numerical model the process is started with genomes of different initial fitness values (W0). In all cases studied, there was a time interval, corresponding to the first transfers, in which the number of infectious individuals generated (which is proportional to the fitness of the population at each transfer) decreased in a roughly exponential manner. After a variable number of transfers (T ∼ 200 for W0 = 5 and T ∼ 500 for W0 = 15 [Fig. 1 ]), the decreasing trend stopped and a statistically stationary state of fitness values was reached. In this second phase, the number of individuals generated at each passage showed large fluctuations around a mean value which was independent of the fitness of the sequence which started the process. The values of the slopes of fitness decrease and the mean values attained at the stationary state were calculated for different W0. When the parameters p and q were kept constant throughout the successive transfers, there were no variations, either in the rate of fitness decrease or in the mean fitness value reached at the stationary state (Fig. 1). To attain a more realistic description of the process of fitness evolution, some modifications were introduced in the numerical simulations. In the real experimental process of serial plaque transfers, the higher the fitness of the initial quasispecies, the higher the probability of obtaining variants with deleterious mutations (44); thus, the effects of population bottlenecks leading to a fitness decrease were more drastic. This was introduced in the simulation by making the probability of fitness decrease (p) proportional to the fitness (p = 0.01 W). In this case, the model shows that the higher the fitness of the starting genome, the higher the absolute value of the slope of the decreasing phase (Table 1). However, the average fitness value reached in the stationary state and the passage number at which the stationary state started were essentially independent of the initial fitness (Table 1).
FIG. 1.
Numerical simulation of the evolution of the number of infectious units produced as a function of the number of plaque-to-plaque transfers. The parameters kept constant in the simulations are p = 0.01, q = 0.001, and r = 5; three different initial fitness values for the founder sequence (W0 = 5, 10, or 15) were assayed. Each curve represents an average of five independent runs. The numerical model and computer programs used are described in Materials and Methods.
TABLE 1.
Effect of the initial fitness on the process of fitness evolution through plaque-to-plaque transfers
| W0 | Slopea (T1-T50) | Meanb (T200-T1,000) | Transfer no. at which stationary state starts |
|---|---|---|---|
| 2 | −0.009 | 1.1 | ∼200 |
| 3 | −0.017 | 1.1 | ∼200 |
| 4 | −0.021 | 1.1 | ∼200 |
| 8 | −0.026 | 1.1 | ∼200 |
| 10 | −0.020 | 1.1 | ∼200 |
Slope of the regression line estimated when the logarithm of the fitness is represented as a function of the transfer number (transfers 1 to 50).
Mean is the value of the average fitness at the stationary state (transfers 200 to 1,000). The fitness values have been averaged over 200 independent trials. The parameter values used in the simulations are r = 5, p = 0.01 W (p is proportional to the fitness of the genomes), and q = 0.001. The standard deviation of fitness values (an indicator of the amplitude of the fluctuations) was the same (0.02) for all the values of W0 assayed.
Effect of the frequency of deleterious mutations.
The reductions in fitness upon successive transfers and the time span before the stationary state was achieved were closely related to the frequency of deleterious mutations (Table 2). The rate of decrease in the number of infectious individuals at each plaque transfer (proportional to the average fitness) increased with p. Both the mean fitness value at the stationary phase and the number of passages required to reach the stationary fitness level decreased as p increased (Table 2).
TABLE 2.
Effect of the probability of occurrence of deleterious mutations (p) on the process of fitness evolution through plaque-to-plaque transfers
| p | Slopea (T1-T50) | Meanb (Tc-T1,000) | Transfer no. at which stationary state starts (Tc) |
|---|---|---|---|
| 0.0025 | −0.0007 | ND | >1,000 |
| 0.005 | −0.002 | ND | >1,000 |
| 0.01 | −0.005 | 1.3 | ∼500 |
| 0.02 | −0.01 | 1.1 | ∼250 |
| 0.04 | −0.03 | 1.0 | ∼150 |
Slope of the regression line estimated when the logarithm of the fitness is represented as a function of the transfer number.
Mean is the value of the average fitness at the stationary state (fitness values from Tc to T1,000). The fitness values have been averaged over 200 independent trials. The parameter values used in the simulations are r = 5 and q = 0.001. ND, not determined.
Basis for a stationary state of fitness equilibrium.
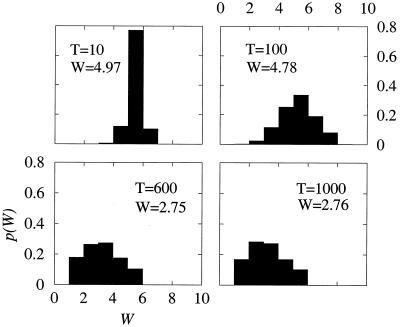
To investigate the possible origin of fitness equilibrium, the statistical distribution of fitness values at different transfers (WT) was analyzed (Fig. 2). The initial sequence was given a fitness of 5, and during a certain number of transfers the average fitness of the distributions decreased (W10 = 4.97, W100 = 4.78, and W600 = 2.75) until a constant average value was obtained (W600 = W1,000 ∼ 2.7). The results show that the shape of the distributions changes with the transfer number. At the stage of fitness decrease (i.e., T = 10 and T = 100) the distributions are quite symmetric, indicating that during the development of a lytic plaque similar amounts of genomes with fitness lower or higher than the average were generated. A distribution of fitness values was previously reported for subclones of VSV (20). However, the distribution of fitness values at the stationary stage (i.e., T = 600 and T = 1,000) is more asymmetric, reflecting the existence of a boundary on the lower values of fitness indicative of an extinction threshold, which causes the distributions to be skewed toward higher fitness values (Fig. 2). The distributions at T = 600 and T = 1,000 were very similar, suggesting that in the stationary state the probability of choosing a genome with a given fitness for the next transfer does not change with time, and consequently, the average number of infectious individuals generated when the process has reached this point is the same at different transfers. Due to the broad distribution of fitness obtained, large fluctuations in fitness values at different transfers are observed.
FIG. 2.
Distribution of fitness values at different plaque transfers. The probability distributions P(W) were calculated at transfers T = 10, 100, 600, and 1,000, by using the following constant values for the parameters: W0 = 5, p = 0.01, q = 0.001, and r = 5. Each distribution has been obtained as an average of 200 independent trials. The W value in each panel is the average fitness of the genome distribution (filled boxes). The numerical model and computer programs used are described in Materials and Methods.
Infective population size and fitness evolution.
The effect of the transmission size (number of infectious units from a virus population used in successive infections) on fitness evolution was studied by choosing different numbers of sequences to initiate the replication in the next transfer. The transmission size was critical to determine the trend of fitness evolution. For W0 = 6, fitness losses occurred when the transmission size Np was 1 or 2, whereas a transmission size of 3 or higher resulted in fitness gains (Fig. 3A). As expected from the results of the plaque transfers involving one genome, an initial phase of fitness variation was followed by a stationary state which was more evident when the transmission size was small (Fig. 3A). The average fitness value at the stationary state was completely determined by the transmission size of the population (keeping constant the values of p, q, and r). This implies that, for a given transmission size, the fitness of the population may increase or decrease, depending on whether the initial fitness is lower or higher than the characteristic fitness determined by the transmission size (Fig. 3B). After a sufficient number of transfers, all the populations converged toward a similar average fitness value which was independent of the initial fitness.
FIG. 3.
Evolution of fitness values during Np plaque-to-Np plaque transfers as a function of the population transmission size (Np) (A) or the initial fitness (B). In panel A the initial fitness was W0 = 6 in all cases. In panel B the transmission size was Np = 3 in all cases, and the lines from bottom to top correspond to W = 1, 3, 5, 7, and 9, respectively. In all cases the values kept constant for the parameters were p = 0.01, q = 0.001, and r = 5. Values are the average of 200 independent trials. The numerical model and computer programs are as described in Materials and Methods.
Exploration of different parameter values and consistency with experimental results.
The influence of p, q, and r on fitness evolution has also been explored. In all cases the qualitative behavior of the model was the same provided q ≪ p < 1. The biphasic evolution of fitness, the presence of a stationary state, and the statistical properties of the fluctuations did not depend on the particular parameters chosen. Some combinations of parameters gave quantitative results quite similar to those observed in the actual process of plaque-to-plaque transfers carried out with FMDV (Fig. 4). With p = 0.06 and q = 0.001, both the rate of decrease of fitness as a function of plaque transfer and the transfer number at which the stationary state starts were very close to the values obtained experimentally (compare Fig. 4A and B). The stationary fitness level with r = 8 was closer to the experimental value than was the level predicted with r = 5. As examples of gross discrepancy with experimental results, two simulations with p = q are included in Fig. 4A. When p = q = 0 (no mutations) (Fig. 4A, lower left panel), population fluctuations (due to the distribution of fitness according to a Poisson distribution) without a transient phase of fitness change were observed. When p = q = 0.02 (Fig. 4A, lower right panel), an increase in the number of infectious units occurred due to the faster replication of sequences that increase their fitness as a consequence of beneficial mutations. Longer simulations (data not shown) indicated that the average fitness grew in an unrestricted fashion and that no stationary state was reached. We have also explored a number of variations of the rules of our model (real values for fitness instead of integer numbers, different values of the extinction threshold, continuous replication of sequences instead of discrete replication cycles, etc. [S. Manrubia, M. Arribas, and E. Lázaro, submitted for publication]). In all cases the qualitative behavior was unchanged, and the conclusions were not altered.
FIG. 4.
Comparison of the infectious units produced as a function of plaque transfer number according to the numerical model and in actual experiments. (A) Time series of the number of infectious units predicted by using several combinations of parameters. (Upper left) p = 0.06, q = 0.001, r = 5, and W0 = 6. (Upper right) p = 0.06, q = 0.001, r = 8, and W0 = 6. (Lower left) p = q = 0, r = 5, and W0 = 6. (Lower right) p = q = 0.02, r = 5, and W0 = 6. (B) Number of infectious units per plaque (plaque titer) of clone  as a function of plaque transfer number. (Left) Plaque titers obtained when
as a function of plaque transfer number. (Left) Plaque titers obtained when  was subjected to 100 successive plaque-to-plaque transfers (27). (Right) Plaque titers obtained with four subclones (a, b, c, and d) were isolated from
was subjected to 100 successive plaque-to-plaque transfers (27). (Right) Plaque titers obtained with four subclones (a, b, c, and d) were isolated from  plaque transfer 100 and subjected to 30 transfers; the time of plaque development was 26 to 30 h. For some plaque transfers of clone
plaque transfer 100 and subjected to 30 transfers; the time of plaque development was 26 to 30 h. For some plaque transfers of clone  plaque transfer 100c (marked with an asterisk), the time of plaque development was increased to 46 h, since plaque transfer 27 contained less than 10 PFU after 27 h of plaque development. Note that the scale of the ordinate in the four right panels is different from that in the left panel. Simulations and experimental procedures are described in Materials and Methods.
plaque transfer 100c (marked with an asterisk), the time of plaque development was increased to 46 h, since plaque transfer 27 contained less than 10 PFU after 27 h of plaque development. Note that the scale of the ordinate in the four right panels is different from that in the left panel. Simulations and experimental procedures are described in Materials and Methods.
The robustness of average fitness stability has been further explored experimentally by subjecting four subclones derived from an FMDV clone which had already reached a stationary state of fitness values to 30 additional plaque-to-plaque transfers (Fig. 4B). It is very remarkable that after 30 additional plaque transfers—a total of 90 plaque-to-plaque transfers since the stationary state was reached (27)—the mean average fitness of the virus remained constant. As also observed in the numerical simulations, the successive fitness values determined experimentally showed large fluctuations. These are expected from the broad fitness distribution of the individuals which compose the population ensemble and which are the origin of the single individual genome that participates in the subsequent transfer. The presence within a plaque of genomes with different fitness values was also documented experimentally by analyzing subclones isolated from the same parental clone (27).
Main features of the model, biological implications, and connections with present views of RNA virus evolution.
The numerical model proposed here includes as a main feature the occurrence, with a low probability, of advantageous mutations that play a key role in reaching a stationary state of fitness, after a transient phase of variable fitness values. The model incorporates also the likely higher frequency of neutral and deleterious mutations than of advantageous mutations in the course of virus plaque formation. However, the frequency of neutral mutations during virus evolution is unknown. Often synonymous mutations are the best candidates to be neutral. Yet the picornavirus coding regions may contain cis-acting elements which confer on open reading frames a phenotypic involvement in addition to their protein coding function (31, 39). Evidence of a selective value of synonymous replacements has been obtained with several RNA viruses (13, 52). Therefore, synonymous mutations within these elements are unlikely to be neutral. Furthermore, recent evidence has shown that a corrected ratio of nonsynonymous to synonymous mutations may be taken as indicative of positive selection in an experimental setting in which positive selection is impossible (56), making quantifications of neutral sites on the basis of the ratio of nonsynonymous to synonymous mutations questionable. Given also the contingent nature of some neutral mutations (48), it is likely that the number of truly neutral sites in RNA genomes with compact genetic information is limited, albeit the number is unknown and difficult to estimate. This uncertainty should not affect the numerical model proposed here, since one of its basic quantitative features is that the combined number of deleterious and neutral mutations largely exceeds the number of advantageous mutations, irrespective of the proportion of deleterious to neutral mutations.
The stationary state of fitness values results from an equilibrium between the trend to eliminate individuals as their fitness falls below the extinction threshold and the probability of selecting for the subsequent transfer individuals with compensatory mutations. This probability is higher in the stationary state than in the transient phase, as a consequence of the increased number of individuals whose fitness becomes zero. The occurrence of compensatory, advantageous mutations was not introduced in most models of Muller's ratchet (32, 35), which considered that back mutations (true reversions) constituted the only mechanism to compensate for the negative effects of deleterious mutations. Wagner and Gabriel (57) considered the effect of compensatory mutations in a model of fitness evolution in finite parthenogenetic populations subjected to random drift. They also observed a biphasic dynamics in which, after an initial transient phase, a stationary state was reached. That compensatory mutations are far more frequent than back mutations has been directly demonstrated experimentally by determining the entire genomic nucleotide sequences of FMDV produced within individual plaques without any biological amplification (27). A similar absence of back mutations was found in the process of fitness gain of FMDV clones upon large population passages (1, 26). The molecular basis for fitness increase by compensatory mutations includes restoration of secondary and higher-order RNA structures in regulatory elements and recovery of catalytic activities in viral enzymes, among others (14). Even in models that did not incorporate the occurrence of beneficial mutations, the predicted speed of the ratchet was higher when the number of deleterious mutations accumulated in a population was low (35). When the number of deleterious mutations reached a certain value, the speed of the ratchet could be dramatically decreased through epistatic effects. In yet another theoretical model, it has been recently shown that the time between successive clicks of the ratchet (corresponding to the total elimination of the fittest type) grew exponentially with any additional mutation (8). In the latter model, despite the fact that the system never reached a truly stationary state, the ratchet was, in practice, arrested.
Kondrashov (35) discussed the case of truncation selection (an extreme form of epistasis), which is directly comparable to our model in the marginal case where beneficial mutations are absent (q = 0). Indeed, if the fitness of a population takes a zero value when the number of mutations exceeds a threshold, the population reaches a “stationary” state where all individuals have the same positive minimum fitness level and have accumulated in their genome exactly the maximum number of mutations compatible with viability. The population sits then precisely at the extinction threshold with no diversity in the system. Consequently, under these conditions there is no possibility of fitness recovery. This threshold would be represented in our model by the minimum allowed fitness, thus mimicking a sort of truncation in the sense of the model proposed by Kondrashov (35). However, as soon as q is set to a positive value, the following effects, which have been indeed observed in experimental settings, are obtained: (i) the stationary state is not at the minimum value of fitness; (ii) occasionally, the whole population can attain states of higher fitness; and (iii) states with low fitness can be reversed through large population passages.
The model predicts that, in the case of HIV infections, low-fitness virus could be endowed with mechanisms to elude extinction. Prolonged survival of HIV could occur despite remarkable fitness decreases associated with incorporation of mutations that confer resistance to antiretroviral inhibitors (4, 42) and the effect of fitness decrease in delaying disease progression (49) and despite reductions in population size following highly active antiretroviral therapy (reviewed in reference 9).
There are ways, other than consecutive plaque transfers, to push a viral population toward accumulation of deleterious mutations. Among them, the use of drugs that increase the mutation frequency is of special relevance (14, 34, 51). The use of mutagenic agents either alone or in combination with antiviral inhibitors can drive viral populations to extinction (10, 36, 47). Our numerical model predicts that an increase in the rate of deleterious mutations (assumed to be directly related to the total mutation frequency) will drive the population to a stationary state with a lower average fitness level. This suggests that a high-enough deleterious mutation rate could push the population toward extinction, since all the individual genomes would be eliminated due to the negative selection. It is very intriguing how some organisms or simple replicons such as viruses have evolutionarily selected for the replication of their genomes an average mutation frequency which allows them to adapt rapidly to environmental changes while evading extinction despite the likely occurrence of bottleneck events in the course of infections within hosts and in the process of viral transmissions between infected and susceptible hosts (29).
Both the biphasic fitness evolution accompanying serial plaque transfers and the effect of transmission size and initial fitness on fitness variations predicted by the numerical model described here are in complete agreement with experimental results (27, 44). Such an agreement constitutes a proof of the validity of our model and of the dynamics predicted at very high passage number, under conditions not yet explored in laboratory experiments. In particular, the model encourages additional experiments to study the molecular basis of long-term fitness stability upon many additional plaque-to-plaque transfers, despite continuing high mutation rates. This equilibrium state offers an interesting counterpart to the population equilibrium (constancy of consensus or average nucleotide sequence despite a complex and dynamic mutant spectrum) attained after many large population passages (16, 55). It would be extremely informative for an understanding of quasispecies dynamics to compare the composition of mutant spectra in low-fitness and in high-fitness equilibrium states.
Acknowledgments
We are indebted to J. Pérez-Mercader for promoting interdisciplinary projects at CAB and F. Morán for valuable suggestions.
Work at CAB was supported by grants from INTA and CAM, and work at CBMSO was supported by grants PM 97-0060-C02-01 and BMC 2001-1823-C02-01 and an institutional grant from the Fundación Ramón Areces.
REFERENCES
- 1.Arias, A., E. Lázaro, C. Escarmís, and E. Domingo. 2001. Molecular intermediates of fitness gain of an RNA virus: characterization of a mutant spectrum by biological and molecular cloning. J. Gen. Virol. 82:1049-1060. [DOI] [PubMed] [Google Scholar]
- 2.Batschelet, E., E. Domingo, and C. Weissmann. 1976. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene 1:27-32. [DOI] [PubMed] [Google Scholar]
- 3.Björkman, J., D. Hughes, and D. I. Andersson. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman, A. M., S. Paulous, and F. Clavel. 1996. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J. Gen. Virol. 77:419-426. [DOI] [PubMed] [Google Scholar]
- 5.Carrillo, C., M. Borca, D. M. Moore, D. O. Morgan, and F. Sobrino. 1998. In vivo analysis of the stability and fitness of variants recovered from foot-and-mouth disease virus quasispecies. J. Gen. Virol. 79:1699-1706. [DOI] [PubMed] [Google Scholar]
- 6.Chao, L. 1990. Fitness of RNA virus decreased by Muller's ratchet. Nature 348:454-455. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, D. K., E. A. Duarte, A. Moya, S. F. Elena, E. Domingo, and J. Holland. 1993. Genetic bottlenecks and population passages cause profound fitness differences in RNA viruses. J. Virol. 67:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colato, A., and J. F. Fontanari. 2001. Soluble model for the accumulation of mutations in asexual populations. Phys. Rev. Lett. 87:238102.. [DOI] [PubMed] [Google Scholar]
- 9.Crandall, K. A. (ed.). 1999. The evolution of HIV. The Johns Hopkins University Press, Baltimore, Md.
- 10.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeFilippis, V. R., and L. P. Villarreal. 2000. An introduction to the evolutionary ecology of viruses, p. 125-208. In C. J. Hurst (ed.), Viral ecology. Academic Press, San Diego, Calif.
- 12.DeFilippis, V. R., and L. P. Villarreal. 2001. Virus evolution, p. 353-370. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 13.de la Torre, J. C., C. Giachetti, B. L. Semler, and J. J. Holland. 1992. High frequency of single-base transitions and extreme frequency of precise multiple-base reversion mutations in poliovirus. Proc. Natl. Acad. Sci. USA 89:2531-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingo, E., C. Biebricher, M. Eigen, and J. J. Holland. 2001. Quasispecies and RNA virus evolution: principles and consequences. Landes Bioscience, Austin, Tex.
- 15.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 16.Domingo, E., D. Sabo, T. Taniguchi, and C. Weissmann. 1978. Nucleotide sequence heterogeneity of an RNA phage population. Cell 13:735-744. [DOI] [PubMed] [Google Scholar]
- 17.Domingo, E., R. G. Webster, and J. J. Holland (ed.). 1999. Origin and evolution of viruses. Academic Press, San Diego, Calif.
- 18.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duarte, E., D. Clarke, A. Moya, E. Domingo, and J. Holland. 1992. Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proc. Natl. Acad. Sci. USA 89:6015-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duarte, E. A., I. S. Novella, S. Ledesma, D. K. Clarke, A. Moya, S. F. Elena, E. Domingo, and J. J. Holland. 1994. Subclonal components of consensus fitness in an RNA virus clone. J. Virol. 68:4295-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eigen, M. 2000. Natural selection: a phase transition? Biophys. Chem. 85:101-123. [DOI] [PubMed] [Google Scholar]
- 22.Eigen, M. 1996. On the nature of virus quasispecies. Trends Microbiol. 4:216-218. [DOI] [PubMed] [Google Scholar]
- 23.Eigen, M., and C. K. Biebricher. 1988. Sequence space and quasispecies distribution, p. 211-245. In E. Domingo, P. Ahlquist, and J. J. Holland (ed.), RNA genetics, vol. 3. CRC Press, Inc., Boca Raton, Fla.
- 24.Eigen, M., and P. Schuster. 1979. The hypercycle. A principle of natural self-organization. Springer, Berlin, Germany. [DOI] [PubMed]
- 25.Escarmís, C., M. Dávila, N. Charpentier, A. Bracho, A. Moya, and E. Domingo. 1996. Genetic lesions associated with Muller's ratchet in an RNA virus. J. Mol. Biol. 264:255-267. [DOI] [PubMed] [Google Scholar]
- 26.Escarmís, C., M. Dávila, and E. Domingo. 1999. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller's ratchet. J. Mol. Biol. 285:495-505. [DOI] [PubMed] [Google Scholar]
- 27.Escarmís, C., G. Gómez-Mariano, M. Dávila, E. Lázaro, and E. Domingo. 2002. Resistance to extinction of low fitness virus subjected to plaque-to-plaque transfers: diversification by mutation clustering. J. Mol. Biol. 315:647-661. [DOI] [PubMed] [Google Scholar]
- 28.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of virology: molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 29.Gerone, P. J., R. B. Couch, G. V. Keefer, R. G. Douglas, E. B. Derrenbacher, and V. Knight. 1966. Assessment of experimental and natural viral aerosols. Bacteriol. Rev. 30:576-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbs, A., C. Calisher, and F. García-Arenal (ed.). 1995. Molecular basis of virus evolution. Cambridge University Press, Cambridge, United Kingdom.
- 31.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordo, I., and B. Charlesworth. 2000. The degeneration of asexual haploid populations and the speed of Muller's ratchet. Genetics 154:1379-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland, J. J., J. C. de la Torre, D. K. Clarke, and E. Duarte. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holland, J. J., E. Domingo, J. C. de la Torre, and D. A. Steinhauer. 1990. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 64:3960-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondrashov, A. S. 1994. Muller's ratchet under epistatic selection. Genetics 136:1469-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loeb, L. A., J. M. Essigmann, F. Kazazi, J. Zhang, K. D. Rose, and J. I. Mullins. 1999. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. USA 96:1492-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez, M. A., C. Carrillo, F. González-Candelas, A. Moya, E. Domingo, and F. Sobrino. 1991. Fitness alteration of foot-and-mouth disease virus mutants: measurement of adaptability of viral quasispecies. J. Virol. 65:3954-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maynard-Smith, J. 1976. The evolution of sex. Cambridge University Press, Cambridge, United Kingdom.
- 39.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morse, S. S. (ed.). 1994. The evolutionary biology of viruses. Raven Press, New York, N.Y.
- 41.Muller, H. J. 1964. The relation of recombination to mutational advance. Mutat. Res. 1:2-9. [DOI] [PubMed] [Google Scholar]
- 42.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 43.Novella, I. S., E. A. Duarte, S. F. Elena, A. Moya, E. Domingo, and J. J. Holland. 1995. Exponential increases of RNA virus fitness during large population transmissions. Proc. Natl. Acad. Sci. USA 92:5841-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novella, I. S., S. F. Elena, A. Moya, E. Domingo, and J. J. Holland. 1995. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J. Virol. 69:2869-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novella, I. S., C. L. Hershey, C. Escarmis, E. Domingo, and J. J. Holland. 1999. Lack of evolutionary stasis during alternating replication of an arbovirus in insect and mammalian cells. J. Mol. Biol. 287:459-465. [DOI] [PubMed] [Google Scholar]
- 46.Novella, I. S., J. Quer, E. Domingo, and J. J. Holland. 1999. Exponential fitness gains of RNA virus populations are limited by bottleneck effects. J. Virol. 73:1668-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pariente, N., S. Sierra, P. R. Lowenstein, and E. Domingo. 2001. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J. Virol. 75:9723-9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quer, J., C. L. Hershey, E. Domingo, J. J. Holland, and I. S. Novella. 2001. Contingent neutrality in competing viral populations. J. Virol. 75:7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quiñones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. van Der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrag, S. J., and V. Perrot. 1996. Reducing antibiotic resistance. Nature 381:120-121. [DOI] [PubMed] [Google Scholar]
- 51.Schuster, P., and P. F. Stadler. 1999. Nature and evolution of early replicons, p. 1-24. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, San Diego, Calif.
- 52.Simmonds, P., and D. B. Smith. 1999. Structural constraints on RNA virus evolution. J. Virol. 73:5787-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobrino, F., M. Dávila, J. Ortín, and E. Domingo. 1983. Multiple genetic variants arise in the course of replication of foot-and-mouth disease virus in cell culture. Virology 128:310-318. [DOI] [PubMed] [Google Scholar]
- 54.Sokal, R. R., and F. J. Rohlf. 1981. Biometry. W. H. Freeman & Co., San Francisco, Calif.
- 55.Steinhauer, D. A., J. C. de la Torre, E. Meier, and J. J. Holland. 1989. Extreme heterogeneity in populations of vesicular stomatitis virus. J. Virol. 63:2072-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vartanian, J. P., M. Henry, and S. Wain-Hobson. 2001. Simulating pseudogene evolution in vitro: determining the true number of mutations in a lineage. Proc. Natl. Acad. Sci. USA 98:13172-13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner, G. P., and W. Gabriel. 1990. Quantitative variation in finite parthenogenetic populations: what stops Muller's ratchet in the absence of recombination? Evolution 44:715-731. [DOI] [PubMed] [Google Scholar]
- 58.Weaver, S. C., A. C. Brault, W. Kang, and J. J. Holland. 1999. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J. Virol. 73:4316-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuste, E., C. López-Galíndez, and E. Domingo. 2000. Unusual distribution of mutations associated with serial bottleneck passages of human immunodeficiency virus type 1. J. Virol. 74:9546-9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuste, E., S. Sánchez-Palomino, C. Casado, E. Domingo, and C. López-Galíndez. 1999. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J. Virol. 73:2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]