Abstract
A membranous fraction that could synthesize viral RNA in vitro in the presence of magnesium salt, ribonucleotides, and an ATP-regenerating system was isolated from feline calicivirus (FCV)-infected cells. The enzymatically active component of this fraction was designated FCV replication complexes (RCs), by analogy to other positive-strand RNA viruses. The newly synthesized RNA was characterized by Northern blot analysis, which demonstrated the production of both full-length (8.0-kb) and subgenomic-length (2.5-kb) RNA molecules similar to those synthesized in FCV-infected cells. The identity of the viral proteins associated with the fraction was investigated. The 60-kDa VP1 major capsid protein was the most abundant viral protein detected. VP2, a minor structural protein encoded by open reading frame 3 (ORF3), was also present. Nonstructural proteins associated with the fraction included the precursor polypeptides Pro-Pol (76 kDa) and p30-VPg (43 kDa), as well as the mature nonstructural proteins p32 (derived from the N-terminal region of the ORF1 polyprotein), p30 (the putative “3A-like” protein), and p39 (the putative nucleoside triphosphatase). The isolation of enzymatically active RCs containing both viral and cellular proteins should facilitate efforts to dissect the contributions of the virus and the host to FCV RNA replication.
Feline calicivirus (FCV), a member of the genus Vesivirus in the family Caliciviridae, is a major agent of respiratory disease in cats. The FCV genome is an approximately 7.7-kb single-strand positive-sense RNA molecule that is covalently linked to a protein designated VPg (for virus protein, genome) at the 5′ end and polyadenylated at the 3′ end (8, 18). The genome is organized into three open reading frames (ORFs). ORF1 encodes an approximately 200-kDa polyprotein that is processed by the virus-encoded 3C-like cysteine proteinase into the mature nonstructural proteins p5.6, p32, p39 (nucleoside triphosphatase [NTPase]), p30, VPg, and Pro-Pol (36a, 38). ORF2 encodes a 73-kDa capsid precursor (preVP1) that is cleaved in trans by the same virus-encoded proteinase to yield the 14-kDa capsid leader (LC) and the approximately 60-kDa mature major capsid protein VP1 (9, 31, 37). ORF3 encodes a 12-kDa basic protein of unknown function, designated VP2, that is associated with mature virions (17, 36).
RNA purified from virus particles and capped RNA transcripts derived from a full-length cDNA clone are infectious when transfected into feline kidney cells (21, 35). Two major polyadenylated positive-sense RNA molecules have been detected in FCV-infected cells (7, 18, 30). The 7.7- to 8-kb genomic RNA serves as a message for translation of the viral nonstructural proteins, and the ∼2.6-kb subgenomic RNA serves as a bicistronic template for translation of structural proteins VP1 and VP2 (18, 31). Several additional species of positive- and negative-sense RNA have been detected in FCV-infected cells, but their significance is not known (7, 30).
All of the positive-strand RNA viruses examined thus far form replication complexes (RCs) associated with intracellular membranes (10, 33, 39, 47). The potential association of FCV replication with cellular membranes was first reported in 1975 by Studdert and O'Shea and by Love and Sabine (23, 40). In both studies, feline cells infected with FCV showed extensive rearrangements of intracellular membranes when cross sections were observed by electron microscopy. The association of RNA replication with membranes in infected cells has been studied extensively for the polioviruses (4, 6, 34, 42), and it has been demonstrated that certain viral proteins, such as 2C, 2BC, and 3A, can independently alter intracellular membranes in transfected cells (2, 11, 14, 41, 43).
The replication strategy of the caliciviruses has not been characterized as extensively as for many positive-strand RNA viruses. The caliciviruses have certain features in common with picornaviruses (relatedness in nonstructural protein motifs and similarities in infected-cell morphology) and alphaviruses (the presence of a subgenomic RNA for translation of the structural proteins) (7, 29, 30). Our study was initiated to examine how caliciviruses might utilize this potential combination of elements from the replication strategies of other positive-strand RNA viruses, as well as to identify features unique to calicivirus replication.
Techniques developed for the isolation and assay of RCs from poliovirus-infected cells were applied to the study of FCV-infected cells, and enzymatically active FCV RCs were obtained. Characterization of the FCV RCs demonstrated both similarities to and differences from the picornaviruses and established a new system for the study of FCV replication in vitro.
MATERIALS AND METHODS
Viruses and cells.
The Urbana strain of FCV (Fe/VV/FCV/Urbana/1968/US) used in this study has been characterized previously (Fig. 1) (35; Sosnovtsev et al., unpublished). Crandell-Rees feline kidney (CRFK) cells were maintained in Eagle's minimum essential medium with chlortetracycline (25 μg/ml), penicillin (250 U/ml), streptomycin (250 μg/ml), amphotericin B (2.5 μg/ml), and 10% fetal bovine serum (FBS). The medium and antibiotics were purchased from Quality Biological, Inc. (Gaithersburg, Md.), and the FBS was from HyClone (Logan, Utah). Virus growth medium was identical to the maintenance medium, except for the use of FBS at a concentration of 2%.
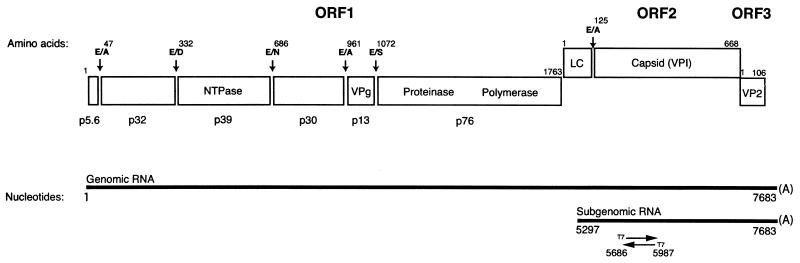
FIG. 1.
Genome organization and cleavage map of FCV. The RNA genome of the Urbana strain of FCV is 7,683 nt in length, excluding the poly(A) tail, and is organized into ORF1 (nt 20 to 5311), ORF2 (nt 5314 to 7320), and ORF3 (nt 7317 to 7637). The proteolytic cleavage sites recognized by the virus-encoded proteinase that define the borders of the nonstructural proteins (encoded in ORF1) and the mature capsid protein (encoded in ORF2) are indicated (36a, 38). Cleavage site abbreviations are as follows: E, glutamic acid; A, alanine; D, aspartic acid; N, asparagine; and S, serine. Two major species of polyadenylated RNAs have been observed in infected cells that correspond to the full-length genome and the 3′-terminal end of the genome (7, 17, 30). The location of the region in the genome (nt 5686 to 5987) from which the sense and antisense RNA transcript probes were designed for Northern blot assays is indicated.
Electron microscopy.
FCV at a multiplicity of infection (MOI) of 10 was adsorbed to a CRFK monolayer for 1 h at 37°C. A noninfected (mock-infected) CRFK control was prepared concurrently in which the same procedure was followed in the absence of virus. After 1 h, the inoculum was removed and 2 ml of virus growth medium was added. Five hours later, the cells were prepared for conventional transmission microscopy. Cells were fixed in 2% glutaraldehyde-0.1 M sodium cacodylate buffer at room temperature for 1 h and then washed in 0.1 M sodium cacodylate buffer at room temperature for 15 min. They were then postfixed with reduced osmium tetroxide and then washed in buffer. Cells were dehydrated in a series of ethanol treatments at 50, 70, and 100% and then soaked in propylene oxide. The cells were embedded in EMbed 812, and sections were cut on a Leica Ultracut FCS ultramicrotome. Thin sections were stained with 7% uranyl acetate in 50% ethanol, followed by staining with 0.01% lead citrate. Sections were reviewed and photographed on an FEI (Philips) CM100 transmission electron microscope. The RC material was adsorbed directly to a carbon-coated grid and stained with 2% aqueous uranyl acetate for 30 s. The stain was removed by wicking with filter paper and observed with the electron microscope. Chemicals for electron microscopy were purchased from Electron Microscopy Sciences, Fort Washington, Pa.
Isolation of RCs from cells.
A monolayer of CRFK cells was prepared in a 150-cm2 flask. FCV at an MOI of 10 was adsorbed to the monolayer for 1 h at 37°C. A mock-infected CRFK control monolayer in a 150-cm2 flask was prepared concurrently. The medium was removed, and cells were rinsed with plain Eagle's minimum essential medium. Cells were then maintained in virus growth medium until the desired time point, when infected or mock-infected cells were removed from the surface of the flask with a cell scraper and pelleted in a 15-ml conical tube by centrifugation at 300 × g for 5 min at 4°C. The supernatant was removed, and the cell pellet was resuspended in 1 ml of cold TN buffer (10 mM Tris [pH 7.8], 10 mM NaCl). After 15 min on ice, the cells were lysed with 60 strokes in a cold 2-ml glass Dounce homogenizer (Bellco Glass, Inc., Vineland, N.J.). The lysate was transferred to a 1.5-ml microcentrifuge tube and subjected to centrifugation for 5 min at 900 × g at 4°C to remove nuclei and unlysed cells. The resulting pellet (designated P1) was resuspended in 120 μl of TN buffer with 15% glycerol and stored at −70°C. The supernatant from P1 was transferred to a fresh microcentrifuge tube and subjected to centrifugation for 20 min at 20,800 × g at 4°C. The resulting pellet (designated P2) was resuspended in 120 μl of TN buffer with 15% glycerol and stored at −70°C. P2 was subsequently designated as either FCV RCs or mock RCs. The supernatant fluid (designated S) remaining from the centrifugation of P2 was stored at −70°C. In order to compare fractions obtained during the isolation of RCs with a total-cell lysate (designated TL), the cell pellet (obtained by centrifugation at 300 × g for 5 min) from one infected or mock-infected 150-cm2 flask was resuspended in 1 ml of cold TN buffer with 15% glycerol and placed immediately at −70°C. The amount of protein in each fraction was determined with the Coomassie Plus Protein Assay Reagent (Pierce, Rockford, Ill.).
RNA replication assay.
The RNA synthesis assay used was similar to that described by Bienz et al. (5). Briefly, an aliquot (18 μl) of the FCV- or mock-infected RC pellet was added to the following reagents in a final volume of 50 μl: 50 mM HEPES (pH 8), 10 mM dithiothreitol, 3 mM MgCl2, 0.25 mM GTP, 0.25 mM UTP, 1 mM ATP, CTP to 0.04 mM with [α-32P]CTP (Amersham Pharmacia Biotech, Piscataway, N.J.) at 400 Ci/mmol and unlabeled CTP, 10 μg of actinomycin D per ml, 25 μg of creatine phosphokinase per ml (Roche, Indianapolis, Ind.), 5 mM creatine phosphate (Roche), 800 U of RNasin per ml (Promega Corporation, Madison, Wis.), and 50 mM potassium acetate. In some experiments, [α-32P]UTP (Amersham) was used as the radioisotope. The mixture was incubated at 30°C for 1 h. Extraction of RNA from the total mixture was performed with the RNeasy Mini Kit (Qiagen, Valencia, Calif.). The RNA was eluted in 50 μl of diethyl pyrocarbonate (DEPC)-treated water and concentrated by precipitation in ethanol at −20°C. The pelleted RNA was resuspended in DEPC-treated water and stored at −70°C.
Gel electrophoresis of RNA and autoradiography.
Gel electrophoresis of RNA was performed with reagents in the NorthernMax-Gly kit (Ambion, Inc., Austin, Tex.). Briefly, RNA was denatured at 65°C in glyoxal-dimethyl sulfoxide sample buffer for 40 min before being loaded onto a 1% low electroendosmosis (LE)-agarose (Ambion) gel. Ethidium bromide staining was used to visualize RNA, and a 0.24- to 9.5-kb RNA ladder (Invitrogen) was included on each agarose gel for sizing. Following electrophoresis, the gels were photographed and dried and the gels were exposed to X-Omat AR film (Kodak, Rochester N.Y.) or the signal was quantified with a 445 SI PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Preparation of RNA transcripts and biotinylated RNA probes.
Primers were designed into which a T7 RNA polymerase promoter sequence (underlined below) was incorporated to allow in vitro transcription of RNA probes from a DNA fragment generated by PCR. For synthesis of a capsid gene-specific RNA probe (corresponding to nucleotides [nt] 5686 to 5987 of the genome, as indicated in Fig. 1) that would detect positive-strand RNA, primers 5′GCTGATGACGGATCCATAACTGCCCCAGAGC (designated FCVcapF) and 5′AATTCGTCTCACTGGTAATACGACTCACTATAGGCTTGTGGTCAATTCTTAAACAAATTCCTATATGATAGCC (designated FCVcapR/T7) were used. For detection of negative-strand RNA, primers AATTCGTCTCACTGGTAATACGACTCACTATAGGGCTGATGACGGATCCATAACTGCCCCAGAGC (designated FCVcapF/T7) and CCAGACCCAGAGATAGAAAACCTTACCTCAACAGATCC (designated FCVcapR) were used.
Plasmid pQ14 (35) was used as the template to generate DNA fragments by PCR that were agarose gel purified and extracted from the agarose with the Qiagen gel extraction kit. One microgram of the DNA fragment was used as a template for in vitro transcription with the Ribomax T7 transcription kit (Promega). Following digestion of the DNA template, the desired RNA band was isolated in a 6% Tris-borate-EDTA-urea polyacrylamide gel (Invitrogen, Carlsbad, Calif.) and excised and the RNA was eluted in DEPC-treated water at 4°C overnight. The eluted RNA was then extracted with phenol-chloroform and then subjected to chloroform extraction and precipitation in ethanol.
RNA probes were produced with the BrightStar psoralen-biotin nonisotopic labeling kit (Ambion). Briefly, approximately 0.5 μg of the purified transcript RNA was cross-linked to the psoralen-biotin reagent on ice with a 365-nm UV light for 45 min. Unincorporated label was removed by n-butanol extraction. The biotinylated RNA probes were stored at −20°C.
Plasmid pQ14 was also used as a template for the synthesis of full-length transcript RNA as previously described (35) for use as a control in Northern blot analysis.
Northern blot analysis.
Northern blot analysis was conducted with the reagents and conditions specified in the NorthernMax-Gly kit (Ambion). The RNA samples used in the Northern blot assays were obtained by extraction of the total RNA from a single replication assay mixture (at various time points). The ethanol-precipitated RNA was resuspended in DEPC-treated water and divided into two aliquots. Both aliquots were denatured in glyoxal sample buffer with heating as described above and loaded onto different halves of the same 1% LE-agarose gel. The RNA was transferred to an Ambion BrightStar-Plus membrane by capillary blotting with the transfer buffer in the NorthernMax-Gly kit (Ambion). Following transfer, the RNA was cross-linked to the membrane with UV light (Stratalinker X linker; Stratagene, La Jolla, Calif.) and the membrane was subjected to a prehybridization step by incubation in Ultrahyb buffer (Ambion) at 68°C for 30 min. The membrane was cut, and each half was incubated with either the sense or the antisense biotinylated RNA transcript probe in UltraHyb buffer at 68°C overnight. Following washing in accordance with the NorthernMax-Gly kit protocol, detection of the bound biotinylated probe was accomplished with the Bright Star BioDetect kit (Ambion). The membranes were exposed to BioMax MS film (Kodak).
RNase treatment.
Radiolabeled RNA synthesized by the FCV RCs and purified as described above was analyzed in RNase digestion experiments. Treatment with RNase A (100 μg/ml; Ambion), which cleaves 3′ of C and U residues in single-stranded RNA, was performed for 1 h at 37°C under high-salt conditions (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]), which are less likely to expose single-stranded regions of double-stranded RNA for cleavage, and low-salt conditions (0.2× SSC), which are more likely to expose single-stranded regions for cleavage. The SSC buffers were prepared from a stock of 20× SSC buffer (3 M sodium chloride, 0.3 M sodium citrate, pH 7.0; Research Genetics, Huntsville, Ala.). Treatment with RNase One (5 U; Promega), which cleaves between any two ribonucleotides in single-stranded RNA, was performed with the buffer provided by the manufacturer (final concentrations, 10 mM Tris [pH 7.5], 5 mM EDTA, and 200 mM sodium acetate). Following treatment, the RNA in the digestion mixture (and a buffer-only control) was purified with the RNeasy Mini Kit (Qiagen), precipitated, and analyzed in an agarose gel as described above.
An aliquot (18 μl) of the FCV RC pellet was incubated with RNase A (100 μg/ml) in 0.2× SSC buffer for 1 h at 37°C. The RNA was purified with the RNeasy Mini Kit, precipitated, and examined by Northern blot analysis as described above.
Radiolabeling of viral RNA synthesized in infected cells.
Five 150-cm2 flasks containing confluent monolayers of CRFK cells were maintained in phosphate-free Dulbecco's modified Eagle's medium (DMEM; Invitrogen) for 16 h. The medium was removed, and FCV at an MOI of 10 was adsorbed to the cells for 1 h at 37°C. The inoculum was removed, and the monolayers were rinsed with phosphate-free DMEM. Phosphate-free DMEM containing 5 mCi of 32Pi and 10 μg of actinomycin D per ml was added to each flask. The infection was allowed to proceed for a total time of approximately 4.5 h. The cells were then collected from the five flasks, pooled, and pelleted at 300 × g for 5 min. The cell pellet was washed with cold phosphate-buffered saline, after which the cells were lysed and the polyadenylated RNA was purified with PolyAT tract system 1000 (Promega).
PAGE of proteins and Western blot analysis.
Proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in Novex 10 to 20% gradient gels (Invitrogen) with Tris-glycine running buffer (Invitrogen). Samples were heated at 95°C for 10 min in 1× Tris-glycine-SDS sample buffer (Invitrogen) containing 2% mercaptoethanol prior to being loaded onto the gel. Coomassie blue staining of proteins directly in gels was accomplished with the GelCode Reagent (Pierce) as recommended by the manufacturer.
Western blot assays were performed as described previously (37). Briefly, proteins were resolved by SDS-PAGE as described above and transferred electrophoretically to nitrocellulose. A TL was prepared from FCV-infected cells as a positive control for the Western blot assays. Briefly, one 150-cm2 flask of CRFK cells was infected with FCV at an MOI of 10 as described above and incubated for 8 h. The cells were collected and pelleted in a 15-ml conical tube by centrifugation at 300 × g for 5 min at 4°C. The pellet was resuspended in 0.5 ml of phosphate-buffered saline and stored at −70°C.
Antisera prepared in guinea pigs against purified FCV virions (35), recombinant VP2 (36), the N terminus of the proteinase (Pro; corresponding to amino acids 1072 to 1192 of the FCV ORF1 polyprotein) (36a), and regions of recombinant nonstructural proteins p30 and p39 were characterized previously (36a). New antisera were prepared in guinea pigs against recombinant FCV Pro-Pol (designated p76 in this study) (46) and recombinant VPg (rVPg) (C. E. Cameron et al., unpublished data) proteins. The specificity of the new antisera was determined by Western blot analysis and immunoprecipitation (data not shown) and is further illustrated by the inclusion of positive and negative antigen controls in the Western blot assays in this study.
Guinea pig antibodies were detected with anti-guinea pig immunoglobulins conjugated to alkaline phosphatase, and color development was accomplished with 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt (BCIP) and nitroblue tetrazolium chloride (Alkaline Phosphatase Substrate Package; Invitrogen).
FCV infection of CRFK cells in the presence of brefeldin A.
Brefeldin A (Sigma, St. Louis, Mo.) was resuspended to 10 mg/ml in methanol. CRFK cells were pretreated with brefeldin A diluted to various concentrations (1 to 100 μg/ml) in virus growth medium for 1 h prior to virus infection. FCV was then added directly to the medium at an MOI of 10 and allowed to adsorb to the cells for 1 h for 37°C. The virus inoculum was then removed, and virus growth medium containing the same concentration of brefeldin A used in the pretreatment was added. After 5 h, when rounding of cells and a cytopathic effect were apparent, the medium was removed and cells were fixed in ice-cold methanol. Immunofluorescence was carried out with hyperimmune serum prepared against virus particles as previously described (35).
RESULTS
Rearrangement of membranes in FCV-infected cells and isolation of a membranous pellet.
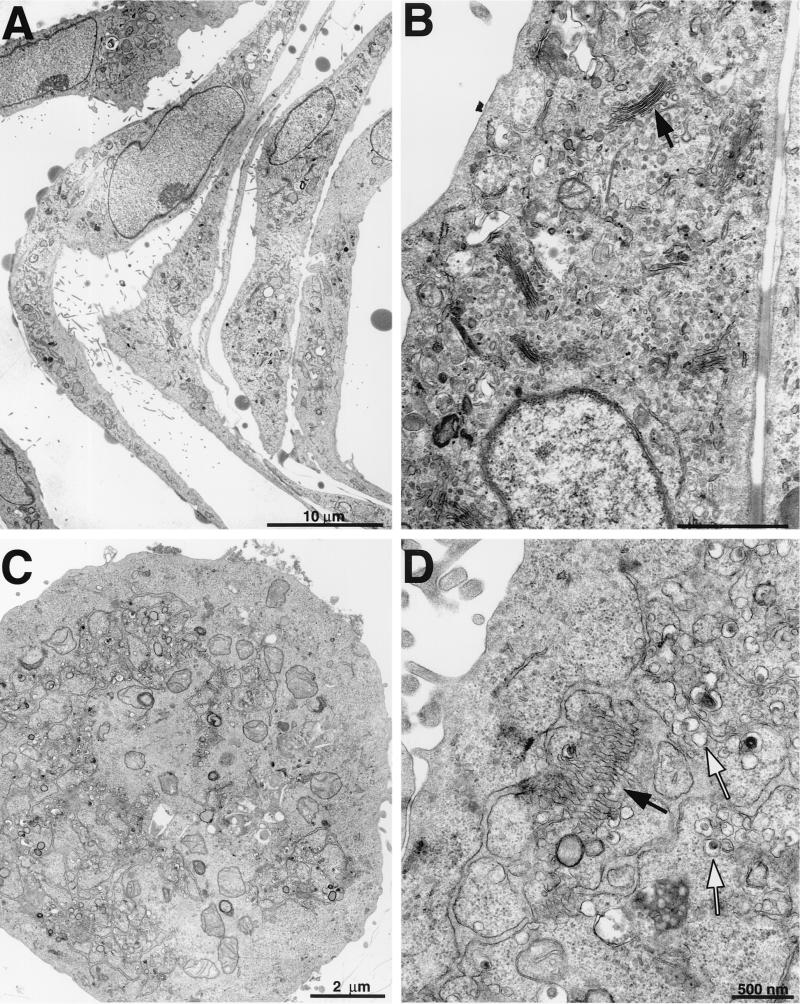
Previous studies by Studdert and O'Shea examined the morphology of FCV-infected feline embryonic cells (40), and those of Love and Sabine examined kitten primary kidney cells (23). We have used CRFK cells extensively in our studies, so we compared the morphology of these cells with that of those examined previously. Mock-infected CRFK cells were elongated in appearance when observed at a low magnification (Fig. 2A). At a higher magnification, the intact Golgi apparatus (indicated by an arrow in Fig. 2B) and endoplasmic reticulum membranes were apparent in these cells. In contrast, CRFK cells infected with FCV at an MOI of 10 showed evidence of rounding at 6 h postinfection (Fig. 2C) and of extensive internal membrane rearrangements with the accumulation of vesicles (indicated by white arrows in Fig. 2D). Structures reminiscent of the Golgi apparatus (indicated by a black arrow in Fig. 2D) were observed in virus-infected cells at this time point, but the membranes were less compact and the organelle appeared distended.
FIG. 2.
Membrane rearrangements in FCV-infected cells. Transmission electron microscopy of mock- or FCV-infected CRFK cells obtained 6 h postinfection. (A) Mock-infected cells at low magnification showing elongated appearance. (B) Mock-infected cells at higher magnification showing normal intracellular arrangement of membranes (arrow, Golgi apparatus). (C) FCV-infected cells at low magnification showing a rounded appearance. (D) FCV-infected cells at higher magnification showing membrane rearrangements (black arrow, putative Golgi apparatus) and vesicles (white arrows).
The conditions for isolation of a membranous pellet from FCV-infected cells were adapted from those used for poliovirus-infected cells (5). The pellets from FCV and mock-infected cells were examined by negative-stain electron microscopy (data not shown). The aggregated membranous materials recovered in both pellets appeared similar, although FCV particles were readily observed in the pellet obtained from FCV-infected cells but not in the pellet from mock-infected cells.
The FCV membranous pellet is enzymatically active.
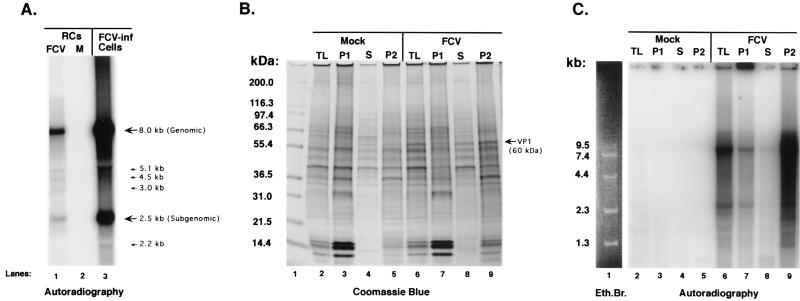
The FCV pellet was analyzed for the ability to synthesize RNA from endogenous templates (Fig. 3A). The pellet was added to a replication assay mixture containing [α-32P]CTP and incubated at 37°C for 1 h. The RNA was purified from the reaction mixture, denatured, and analyzed in an agarose gel. Two major radiolabeled RNA species were detected—one of approximately 8.0 kb and the other of approximately 2.5 kb, with the 8.0-kb band stronger in intensity, presumably because of larger amounts of incorporated radiolabeled nucleotide (Fig. 3A, lane 1). Additional faint bands, corresponding to sizes of 5.1, 4.5, 3.0, and 2.2 kb, were detected. Radiolabeled RNA was not detected when pellets derived from mock-infected CRFK cells were analyzed (Fig. 3A, lane 2). The radiolabeled RNA synthesized from the pellets was compared to the polyadenylated RNA species purified from FCV-infected cells maintained in the presence of 32Pi (Fig. 3A, lane 3). The two major polyadenylated RNA species from FCV-infected cells were similar in size to the RNA synthesized by the FCV pellets, with the 8.0-kb genomic RNA band again stronger in intensity. Additional minor bands similar to those produced by the FCV pellets were also detected. The ability of these membranous pellets from FCV-infected cells to synthesize viral RNA in vitro suggests that they contain the viral RNA RC, analogous to those found in cells infected with poliovirus and other positive-strand viruses.
FIG. 3.
Characteristics of RNA synthesized from endogenous templates in FCV RCs. The RC pellet was isolated from mock- and FCV-infected cells and analyzed in a replication assay that included [α-32P]CTP. Total RNA was extracted from the mixture and analyzed on a 1% agarose gel that was dried and exposed to film. (A) Lanes: 1, RNA from FCV-infected (FCV-inf) RC pellet assay; 2, RNA from mock-infected RC pellet assay; 3, total 32P-labeled RNA isolated from FCV-infected CRFK cells by oligo(dT) selection. (B) Proteins associated with TLs and fractions obtained during purification of RCs (as described in Materials and Methods) were subjected to SDS-PAGE in a 10 to 20% Tris-glycine gel, followed by staining with Coomassie blue. Lanes: 1, Mark 12 protein standards (Invitrogen); 2, mock-infected CRFK TL; 3, mock-infected P1 nuclear pellet; 4, mock-infected supernatant (S) from P2 pellet; 5, mock-infected P2 RC pellet; 6, FCV-infected TL; 7, FCV-infected P1; 8, FCV-infected S; 9, FCV P2 RC pellet. (C) The mock- and FCV-infected TLs and fractions shown in panel B were each analyzed in a replication assay that contained [α-32P]UTP. The radiolabeled RNA was purified and subjected to electrophoresis in a 1% agarose gel, and the signal was measured on a PhosphorImager. The RNA was derived from replication assays with the following samples: lane 2, mock-infected CRFK TL; lane 3, mock-infected P1 nuclear pellet; lane 4, mock-infected supernatant (S) from P2 pellet; lane 5, mock-infected P2 RC pellet; lane 6, FCV-infected TL; lane 7, FCV-infected P1; lane 8, FCV-infected S; lane 9, FCV P2 RCs. The amounts of total protein in each replication assay (which used 18 μl of sample) in lanes 2 to 9 were 6.8, 10.1, 3.7, 11.1, 9.9, 7.0, 5.2, and 17.7 μg, respectively, and reflected the nonadjusted yield of protein in each sample from the isolation procedures described in Materials and Methods. Lane 1 depicts the RNA Ladder (Invitrogen) from the same gel stained with ethidium bromide (Eth.Br.).
The protein content and enzymatic activity of each fraction obtained during the RC isolation procedure were compared to those of a total lysate (TL) in order to confirm the enrichment of an enzymatically active fraction from infected cells. Proteins present in a TL from one 150-cm2 flask (mock or FCV infected for 6 h) were compared by SDS-PAGE with those in three different fractions obtained during the purification of RCs from one mock- or FCV-infected 150-cm2 flask prepared concurrently (Fig. 3B). These fractions were designated P1 (the 900 × g nuclear pellet), S (the supernatant remaining after pelleting of the RCs), and P2 (the final 20,800 × g pellet containing the RCs). Although differences in protein quantity and composition could be detected when these fractions were compared within their respective mock- and FCV-infected groups, the observed protein profile of the TL and the P2 RC pellet was similar, except for the enrichment of an approximately 38-kDa protein, in both the mock- and FCV-infected RCs (Fig. 3B, lane 2 compared with lane 5 and lane 6 compared with lane 9). A major difference between the samples from mock- and FCV-infected cells was the appearance of the 60-kDa VP1 capsid protein in the TL, S, and P2 FCV samples (Fig. 3B, lanes 6, 8, and 9, respectively). Each TL and isolated fraction was tested in the RNA replication assay (Fig. 3C). Again, the samples obtained from mock-infected cells did not show evidence of de novo RNA synthesis (Fig. 3C, lanes 2 to 5). In contrast, all of the samples from FCV-infected cells yielded radiolabeled RNA but in various amounts (Fig. 3C, lanes 6 to 9). These results indicate that the TL from FCV-infected cells is enzymatically active (Fig. 3C, lane 6) and that an enzymatically active fraction (P2) can be isolated from the TL (Fig. 3C, lane 9).
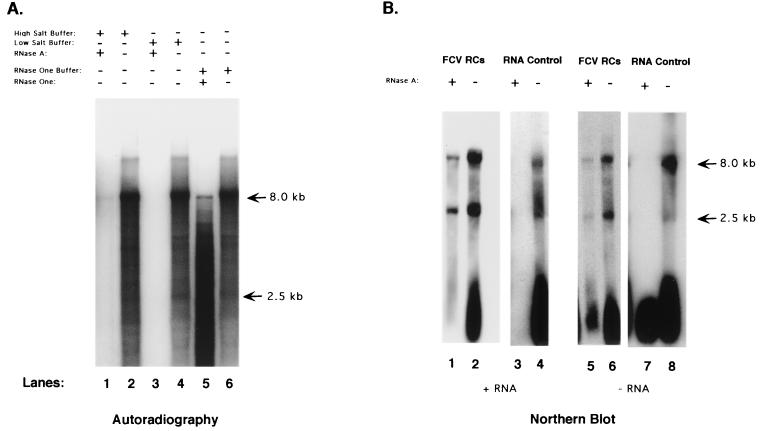
The RNA synthesized by the FCV RCs was analyzed in RNase digestion experiments in order to investigate the presence of double-stranded replicative intermediate forms of viral RNA. Radiolabeled RNA synthesized by the RCs was purified from the reaction mixture with the RNeasy system, precipitated, and then treated with RNase A under high-salt conditions and with RNase One, each of which preferentially cleaves single-stranded rather than double-stranded RNA. Analysis of the treated RNA in a denaturing gel showed evidence of the presence of some digestion-resistant RNA molecules, consistent with the presence of double-stranded RNA forms (Fig. 4A, lanes 1 and 5). Compared with control RNA incubated with buffer only (Fig. 4A, lanes 2, 4, and 6), the RNase-resistant material represented a smaller fraction of the total product RNA. Treatment of the purified RNA with RNase A under low-salt buffer conditions (which relaxes double-stranded RNA interactions) showed total digestion of the RNA (Fig. 4A, lane 3).
FIG. 4.
(A) 32P-labeled RNA was purified from an FCV-infected RC pellet replication assay and subjected to the following treatments: lane 1, high-salt buffer (2× SSC) with RNase A; lane 2, high-salt buffer only; lane 3, low-salt buffer (0.2× SSC) with RNase A; lane 4, low-salt buffer only; lane 5, RNase One buffer plus RNase One enzyme; lane 6, RNase One buffer only. (B) Northern blot analysis of RNA purified directly from FCV RCs collected at 7 h after infection. Lanes: 1 to 4, RNA probed with an antisense RNA probe that detects positive-sense RNA; 5 to 8, RNA probe with a sense RNA probe that detects negative-sense RNA. RNA in lanes 1 and 5 was purified from RCs that had been treated with RNase A (0.2× SSC) for 1 h. The RNA in lanes 3 and 7 was purified from RCs prior to RNase A treatment.
The RNA associated with the isolated RCs was characterized further by Northern blot analysis. RNA purified directly from the RCs (collected at 7 h postinfection and without additional RNA synthesis in a replication assay mixture) was resolved by electrophoresis under denaturing conditions and transferred to a nylon membrane. A biotinylated RNA transcript probe complementary to a region of the subgenomic RNA (nt 5686 to 5987 located within the capsid-encoding gene of the FCV genome) designed for detection of positive-strand RNA synthesis showed the presence of full-length genomic and subgenomic RNAs in the RCs (Fig. 4B, lane 2). The corresponding probe (nt 5686 to 5987) designed to detect the negative strand also showed the presence of full-length genomic and subgenomic RNAs in the RCs (Fig. 4B, lane 6). It should be noted that, in this and other Northern blot experiments, longer exposure times were required for visualization of the negative-strand probe.
The accessibility of the RNA within the RCs to RNase A digestion was examined. An aliquot of the RC pellet was treated with RNase A in low-salt buffer for 1 h at 37°C. The control for this experiment was a concurrent RNase A digestion of RNA that had been previously purified from FCV RCs. Following digestion, the RNA from each experiment was purified by spin column chromatography (RNeasy), resolved in an agarose gel under denaturing conditions, and analyzed in a Northern blot with the biotinylated probes. Direct treatment of the RCs with RNase A resulted in an apparent decrease in the amount of RNA associated with the RCs, although both full-length genomic and subgenomic RNA (positive and negative sense) could still be detected (Fig. 4B, lanes 1 and 5). The full-length purified RNA in the control was completely digested under the conditions used (Fig. 4B, lanes 3 and 7). These data were consistent with the presence of protected endogenous RNA in the FCV RCs.
Time course of RNA accumulation in vitro.
A time course experiment was performed in which the accumulation of RNA in a single FCV RC replication reaction mixture was analyzed. The starting time was designated as time zero, and samples were collected at the 1-, 10-, 30-, and 60-min time points. One-half of the reaction mixture was incubated in the presence of radiolabeled NTP, and one-half was not. Radiolabeled RNA could be detected as early as 10 min after initiation of the reaction, and the signal increased in intensity to the final 60-min time point (Fig. 5A, lanes 1 to 5).
FIG. 5.
Time course analysis of RNA synthesis in an individual replication assay. A replication assay was initiated in the presence or absence of [α-32P]CTP, and aliquots were taken at the indicated time points. (A) Analysis of radiolabeled RNA collected at 0, 1, 10, 30, and 60 min (lanes 1, 2, 3, 4, and 5, respectively). (B) Northern blot analysis of nonradiolabeled RNA samples collected at 0, 1, 10, 30, and 60 min (lanes 1, 2, 3, 4, and 5, respectively) and probed with an antisense RNA probe that detects positive-sense RNA synthesis. Lane 6 contains full-length positive-sense RNA transcripts (designated + RNA) derived from linearized pQ14. (C) Northern blot analysis of nonradiolabeled RNA samples collected at 0, 1, 10, 30, and 60 min (lanes 1, 2, 3, 4, and 5, respectively) and probed with a sense RNA probe that detects negative-sense RNA synthesis. Lane 6 contains full-length RNA transcripts (positive sense) (designated + RNA) derived from linearized pQ14.
A Northern blot analysis was performed on the corresponding RNA samples that were generated with nonradiolabeled NTPs. The biotinylated RNA transcript probe designed for detection of positive-strand RNA synthesis showed the presence of the full-length genomic and subgenomic RNAs in the RCs (collected at 6 h postinfection) at the beginning of the reaction (Fig. 5B, lane 1). The amount of positive-strand genomic and subgenomic RNA increased over time in the individual replication assay, consistent with de novo synthesis of full-length molecules (Fig. 5B, lanes 2 to 5). However, in contrast to the radiolabeling experiments, the subgenomic RNA signal was stronger than that of the genomic RNA in the Northern blot. Features related to technical differences between autoradiography and Northern blot analysis that could likely account for this observed variation were not addressed in this study. A full-length positive-strand RNA transcript derived by in vitro transcription of a linearized full-length cDNA clone of FCV, pQ14 (35), was included as a control in the Northern blot assay. The probe recognized the full-length transcript RNA and an approximately 4-kb truncated species of this transcript (Fig. 5B, lane 6).
A biotinylated RNA transcript probe designed for detection of negative-strand RNA synthesis showed the presence of a subgenomic negative-sense RNA molecule in the RCs at the beginning of the reaction (Fig. 5C, lane 1). An increase in the amount of negative-sense subgenomic RNA synthesis was observed over time (Fig. 5C, lanes 2 to 5), and the probe did not hybridize with the full-length positive-sense RNA transcript derived from pQ14 (Fig. 5C, lane 6). The presence of genomic-size negative-strand RNA could be detected only after prolonged exposure of the film (data not shown), indicating relatively small quantities of this RNA species. The genomic-size negative-strand RNA did not appear to increase in quantity over time as did the genomic-size positive-strand RNA.
Time course analysis of RNA synthesis associated with RCs purified from FCV-infected cells.
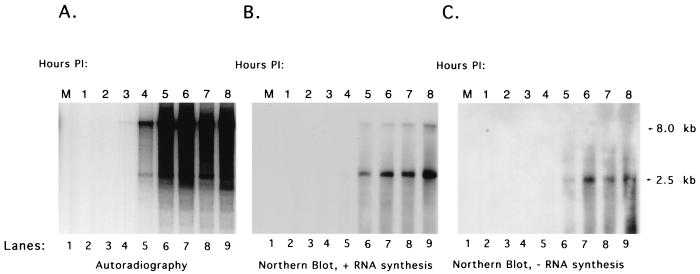
The time at which enzymatically active RCs could be isolated from FCV-infected cells after infection was examined in a time course experiment. Cells were infected with FCV at an MOI of 10, and monolayers were collected hourly through 8 h. The RC pellet isolated at each time point was then analyzed for the ability to synthesize viral RNA in a replication assay. As described above, the replication assay was performed in the presence or absence of radiolabeled NTP. In the replication assays to which [α-32P]CTP was added, RNA synthesis could be detected in isolated RCs collected at the third hour postinfection (Fig. 6A, lane 4). A longer exposure of this gel to film could detect evidence of the synthesis of radiolabeled RNA in RCs collected as early as 2 h postinfection (data not shown). A marked increase in the amount of radiolabeled RNA was detected between RCs isolated at hours 4 and 5 (Fig. 6A, lanes 5 and 6). The isolated RCs continued to support active RNA synthesis through the last time point tested (8 h) (Fig. 6A, lane 9), when nearly all cells were detached from the surface of the flask.
FIG. 6.
Analysis of RNA synthesis from RC pellets collected over time from FCV-infected cells. Cells were infected with FCV at an MOI of 10 and, beginning at 1 h postinfection (PI), collected hourly for isolation of RCs. The RCs isolated at each time point were analyzed for the ability to synthesize RNA in an in vitro replication assay. (A) Analysis of radiolabeled RNA synthesized from RC pellets collected hourly for up to 8 h after infection (lanes 2 to 9). (B) Northern blot analysis of nonradiolabeled RNA synthesized from RC pellets collected at 1 to 8 h postinfection (lanes 2 to 9) probed with an antisense RNA probe that detects positive-strand RNA synthesis. (C) Northern blot analysis of nonradiolabeled RNA synthesized from RC pellets collected at 1 to 8 h postinfection (lanes 2 to 9) probed with a sense RNA probe that detects negative-strand RNA synthesis. Lane 1 contains RNA purified from a replication assay performed under the same conditions with a pellet isolated from mock-infected cells (M).
The RNA associated with the RCs collected over the 8-h time course was analyzed in Northern blot assays with the positive- and negative-sense subgenomic RNA probes. The RNA probe designed to hybridize to positive-sense subgenomic RNA detected both subgenomic- and genomic-length RNA molecules in RCs collected at 5 h postinfection (Fig. 6B, lane 6). However, a longer exposure of the Northern blot showed evidence of the presence of positive-strand subgenomic RNA in RCs collected as early as 2 h after infection (data not shown). The probe for the negative-sense RNA showed an increase in the amount of the negative-strand subgenomic RNA present in RCs collected over time (Fig. 6C, lanes 6 to 9). Again, the genomic-size negative-strand RNA could be detected only after prolonged exposure (data not shown).
Identification of viral structural proteins associated with the FCV RCs.
Analysis of the total proteins associated with the RC pellet by SDS-PAGE had shown the presence of an approximately 60-kDa protein consistent in size with the mature capsid protein (VP1) of FCV (Fig. 3B, lane 9). The identity of this protein was confirmed by a Western blot analysis in which the proteins associated with the RCs collected over the 8-h time course were analyzed (Fig. 7A, lanes 2 to 9). Capsid protein was detected in RCs collected as early as 1 h after infection (Fig. 7A, lane 2) and may have represented the large amount of input virus used for infection of the cells. To examine the earliest time point at which new capsid protein synthesis could be detected in isolated RC pellets, an early time course analysis was conducted. Virus (MOI = 10) was added to a cell monolayer at 4°C, and the cells were immediately rinsed and processed to obtain the RC pellet for the first sample (designated 0). Additional monolayers were placed at 37°C and then collected at 15, 30, 45, 60, and 120 min after infection for the isolation of RC pellets. The capsid protein could be detected readily in RCs collected at 15 min postinfection (Fig. 7B, lane 3), again likely representing capsid protein derived from input virus that adsorbed to the cells at 37°C. The amount of capsid protein detected in the Western blot clearly began to increase in the 45-min RC sample (Fig. 7B, lane 5) and reached levels at 1 to 2 h (Fig. 7B, lanes 6 and 7) similar to the 1- and 2-h levels in the 8-h time course (Fig. 7A, lanes 2 and 3). The amount of capsid protein remained relatively constant until a marked increase was observed in the RCs collected between 4 and 5 h (Fig. 7A, lanes 5 and 6). This increase in capsid protein synthesis correlated with a marked increase in the 2.5-kb subgenomic positive-strand RNA synthesis that was generated by RCs collected between 4 and 5 h (Fig. 6B, lanes 5 and 6).
FIG. 7.
Analysis of structural proteins present in RC pellets. (A) Proteins from RCs collected over an 8-h time course were transferred to nitrocellulose and incubated with hyperimmune serum prepared against purified FCV virions. Lanes: 1, mock-infected (M) RCs; 2 to 9, FCV RCs collected at 1 to 8 h postinfection, respectively. (B) An early time course study was conducted, and the following RCs were collected as indicated and analyzed in a Western blot assay with FCV virion-specific hyperimmune serum: mock-infected (M) RCs (lane 1) and FCV RCs collected at 0, 15, 30, 25, 60, and 120 min (lanes 2 to 7, respectively). The VP1 present in RCs (collected at 8 h) (lane 8) was compared directly in size with that in purified FCV virions (lane 9). The 60-kDa VP1 protein is indicated by an arrow. (C) Antiserum prepared against recombinant VP2 protein (encoded in ORF3) was incubated with a nitrocellulose membrane containing mock-infected (M) RCs (lane 2) and FCV-infected RCs (lane 3). Lane 1 contains See Blue Plus protein molecular size markers (Invitrogen). The VP2 protein is indicated by an arrow. The values on the left are molecular sizes in kilodaltons.
The presence of major structural capsid protein VP1 in the RCs prompted us to examine whether minor structural protein VP2 was also present (Fig. 7C). VP2, encoded by ORF3 of the FCV genome and translated from the 2.5-kb subgenomic RNA (17), is present at one or two copies per virion (36), but its function is unknown. Western blot analysis (Fig. 7C, lane 3) of FCV RCs with antisera prepared against recombinant VP2 showed the presence of an approximately 8-kDa protein consistent with the migration of VP2 in SDS-PAGE (36).
Identification of viral nonstructural proteins associated with the FCV RCs.
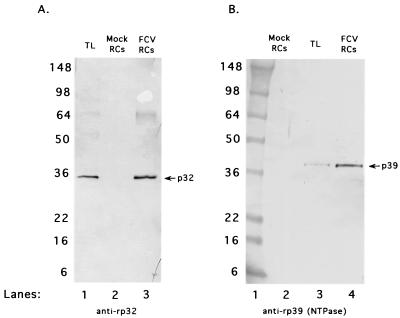
Proteolytic mapping studies of the nonstructural polyprotein encoded by FCV ORF1 (36a) have defined the gene order as p5.6-p32-p39 (NTPase)-p30-p13 (VPg)-p76 (Pro-Pol) (Fig. 1). Analysis of protein synthesis in FCV-infected cells has identified the presence of both fully processed and precursor forms of certain nonstructural proteins. Two nonstructural proteins encoded near the N terminus of the ORF1 polyprotein that appear predominantly as fully processed forms in infected cells are p32 (a protein of unknown function but presumably 2B like) and p39, the putative FCV NTPase. A Western blot assay with antisera specific for these two proteins showed their presence in FCV RCs collected at 6 h postinfection (Fig. 8).
FIG. 8.
Analysis of nonstructural viral proteins encoded near the N-terminal region of ORF1 that are associated with FCV RCs. (A) Antiserum prepared against recombinant p32 protein (anti-rp32) was incubated with FCV 8-h postinfection TL (lane 1), mock-infected RCs (lane 2), and FCV RCs (lane 3). (B) Antiserum prepared against recombinant p39 (anti-rp39) was incubated with mock-infected RCs (lane 2), FCV 8-h postinfection TL (lane 3), and FCV RCs (lane 4). Lane 1 contains See Blue Plus protein molecular size markers (Invitrogen). The values on the left are molecular sizes in kilodaltons.
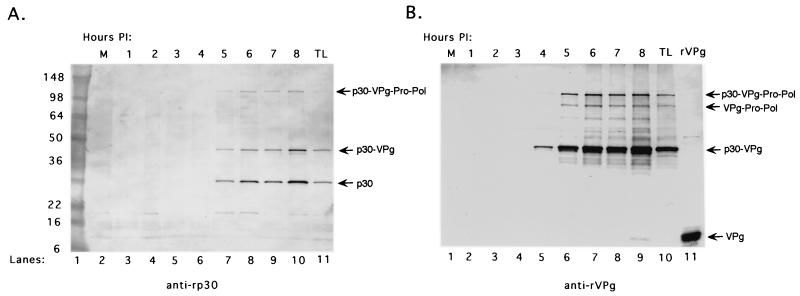
We had previously identified a 43-kDa precursor complex of p30-VPg in FCV-infected cells and proposed that this protein could be analogous to the poliovirus 3AB precursor (36, 36a). We examined whether the 43-kDa precursor was present in the FCV RCs. Antiserum specific for either the FCV recombinant p30 or rVPg protein was used to analyze the RCs collected over the 8-h time course in a Western blot assay in order to examine precursor and product relationships. The antiserum specific for the FCV p30 protein (anti-rp30) detected both the p30-VPg precursor and the cleaved p30 protein that accumulated in the RCs over time (Fig. 9A, lanes 3 to 10). This serum reacted with these two proteins in an FCV-infected TL prepared at 8 h postinfection (Fig. 9A, lane 11) and did not detect these proteins in the mock-infected RCs (Fig. 9A, lane 2). In addition, an approximately 120-kDa protein, previously identified as a precursor from the C-terminal part of ORF1 (p30-VPg-Pro-Pol) (36a), was detected in the RCs (Fig. 9A, lanes 7 to 10). The anti-rp30 serum also reacted with a minor band of approximately 20 kDa that was observed in several lanes, including the mock-infected RC control. An antiserum raised against FCV rVPg (anti-rVPg) also showed the accumulation of the 43-kDa p30-VPg precursor in the RCs over time (Fig. 9B, lanes 2 to 9) and recognized this protein in an FCV-infected TL (Fig. 9B, lane 10). This antiserum recognized the 120-kDa precursor described above and an approximately 88-kDa protein consistent in size with a VPg-Pro-Pol precursor we had identified previously (36a). Additional minor bands of unknown identity and larger than 43 kDa were also detected. The rVPg antiserum did not react with the mock-infected RCs (Fig. 9B, lane 1).
FIG. 9.
Analysis of the p30 and VPg proteins in RCs collected over time from FCV-infected cells. (A) The presence of the p30 protein in RCs collected over the 8-h time course was analyzed in a Western blot assay with antiserum specific for recombinant p30 (anti-rp30). Lanes: 1, See Blue Plus protein molecular size markers (Invitrogen); 2, mock-infected (M) RCs; 3 to 10, FCV RCs collected over an 8-h time course; 11, FCV-infected TL prepared from CRFK cells at 8 h postinfection (PI). (B) The presence of the VPg protein in RCs collected over the 8-h time course was analyzed in a Western blot assay with antiserum specific for rVPg (anti-rVPg). Lanes: 1, mock-infected (M) RCs; 2 to 9, FCV RCs collected over an 8-h time course. Controls in the Western blot assay include FCV-infected TL prepared from CRFK cells at 8 h postinfection (lane 10) and rVPg (lane 11). The values on the left are molecular sizes in kilodaltons.
We had previously reported evidence of two forms of the FCV VPg—a free 12.7-kDa cleaved form found in infected cells, and a modified 15-kDa form found in virions and infected cells that is presumably linked to RNA (36). The release of p30 from the p30-VPg precursor suggested that processed VPg must also be generated from this cleavage event. The rVPg antiserum recognized the approximately 13-kDa purified rVPg protein (Fig. 9B, lane 11). A faint band representing an approximately 13-kDa protein in the FCV RCs was detected on the original membrane beginning at 6 h postinfection (but it is not visible in Fig. 9B) and increased in intensity to the 8-h time point (Fig. 9B, lane 9). The 15-kDa modified form of VPg was not apparent in this Western blot but was detected in immunoprecipitation experiments with the FCV RCs, indicating that linkage of VPg to RNA does occur in the RCs (S. V. Sosnovtsev and K. Y. Green, unpublished data).
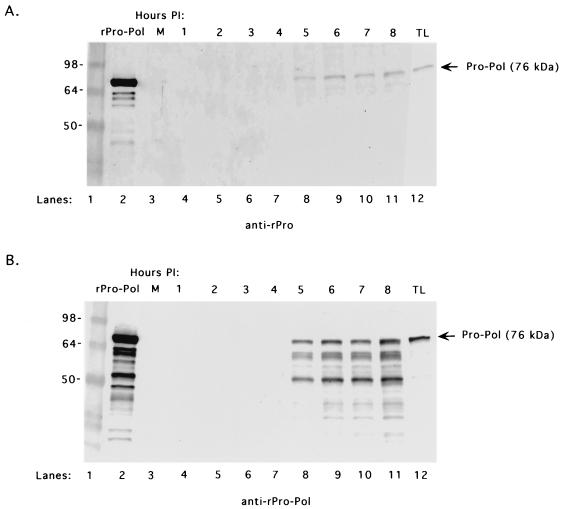
Manifestation of de novo RNA synthesis by the FCV RCs was consistent with the presence of an active RNA-dependent RNA polymerase. Previous studies reported the presence of an approximately 76-kDa stable proteinase-polymerase precursor complex (designated Pro-Pol) in FCV-infected cells (38). Purified, recombinant FCV Pro-Pol (with a genetically inactivated proteinase domain and designated rPro-Pol) expressed in bacteria was shown to be a highly active polymerase (46). The proteins from the RC pellets collected over the 8-h time course were examined in Western blot assays with region-specific antisera. An antiserum prepared against the N terminus of Pro reacted with the approximately 76-kDa rPro-Pol control (Fig. 10A, lane 2) and with the Pro-Pol precursor present in an FCV-infected TL (Fig. 10A, lane 12). This antiserum also recognized a 76-kDa protein that was observed in the FCV RCs beginning at 5 h postinfection and throughout the time course of the experiment (Fig. 10A, lanes 4 to 11). Antiserum prepared against rPro-Pol recognized the rPro-Pol control (Fig. 10B, lane 2) and the Pro-Pol precursor in FCV-infected cells (Fig. 10B, lane 12). The Pro-Pol antiserum also detected a 76-kDa protein in RCs beginning at 5 h postinfection and extending through the time course of the experiment (Fig. 10B, lanes 4 to 11). In addition, the rPro-Pol antiserum detected an approximately 50-kDa protein and intermediate minor bands that accumulated in the RCs collected over time. The rPro-Pol antiserum did not react with the mock-infected RCs (Fig. 10B, lane 3).
FIG. 10.
Analysis of the 76-kDa proteinase-polymerase precursor (Pro-Pol) in RC pellets collected over time from FCV-infected cells. The presence of Pro-Pol in RCs collected over the 8-h time course was analyzed with two different antisera. (A) Mock-infected (M) RCs (lane 3) or FCV-infected RCs collected over an 8-h time course (lanes 4 to 11) were probed with guinea pig hyperimmune sera prepared against the N terminus (amino acids 1072 to 1192) of the recombinant proteinase (anti-rPro). The rPro-Pol protein (lane 2) and FCV-infected TL collected 8 h postinfection (lane 12) were included as controls. (B) Mock-infected (M) RCs (lane 3) or FCV-infected RCs collected over an 8-h time course (lanes 4 to 11) were probed with guinea pig hyperimmune sera prepared against the rPro-Pol protein (anti-rPro-Pol). The rPro-Pol protein (lane 2) and FCV-infected TL collected at 8 h postinfection (lane 12) were included as controls. Lane 1 in each panel contains See Blue Plus protein molecular size markers (Invitrogen). The values on the left are molecular sizes in kilodaltons.
Effect of brefeldin A on FCV growth in infected cells.
The ability to isolate enzymatically active RCs from FCV-infected cells under the same conditions used for poliovirus, as well as the appearance of membrane rearrangements in FCV-infected cells, suggested that additional similarities between these viruses might be found. Poliovirus replication is severely inhibited in the presence of brefeldin A at concentrations as low as 1 μg/ml in HeLa cells (13, 24), and we examined whether FCV might also be inhibited. Brefeldin A is a fungal metabolite that disrupts the secretory pathway in brefeldin A-susceptible cells, presumably by interfering with vesicle-mediated intracellular transport (32). In order to examine whether FCV growth is inhibited in the presence of brefeldin A, FCV infection was analyzed in the presence or absence of brefeldin A. Even at a brefeldin A concentration as high as 100 μg/ml, there was no detectable inhibitory effect on FCV growth, as determined by a visible cytopathic effect, expression of the capsid protein in an immunofluorescence assay (Fig. 11), or the virus titer (data not shown).
FIG. 11.
Effect of brefeldin A on the growth of FCV in CRFK cells. An immunofluorescence assay with hyperimmune serum prepared against purified FCV virions was performed on CRFK cells subjected to the following conditions: (A) Infection with FCV in the absence of brefeldin A. (B) Pretreatment of cells with 100 μg of brefeldin A per ml, followed by infection with FCV in the presence of 100 μg of brefeldin A per ml. (C) Mock infection of CRFK cells without brefeldin A.
CRFK cells were treated with brefeldin A alone in order to determine whether the failure of brefeldin A to inhibit FCV replication might be explained by an inherent resistance of CRFK cells to the drug. Brefeldin A was diluted to concentrations of 1, 10, and 100 μg/ml in virus growth medium (without virus) and added to CRKF monolayers. After 24 h, rounded cells were present in wells containing brefeldin A, with greater numbers of rounded cells as the concentration of brefeldin A increased. After 72 h, only a few viable cells remained compared to the intact control monolayers that received either virus growth medium alone or virus growth medium with the same amount of methanol solvent that would be present in the corresponding brefeldin A dilution (data not shown).
DISCUSSION
FCV was initially classified as a member of the Picornaviridae family because of its positive-sense RNA genome and similar structural morphology (25). It was subsequently placed into the family Caliciviridae, which was created when it became evident that caliciviruses have important differences from picornaviruses (reviewed in reference 16). These differences included the presence of a single major calicivirus structural protein, the synthesis of a subgenomic-size RNA in calicivirus-infected cells, and genome organization (structural proteins are encoded beginning at the 5′ end of picornavirus genomes and nearer the 3′ end in calicivirus genomes). However, certain similarities between picornavirus- and calicivirus-infected cells, such as the presence of extensive membrane rearrangements (23, 40), as well as sequence homology in nonstructural protein motifs (29), were strongly suggestive of common themes in their replication strategies. Furthermore, caliciviruses and picornaviruses, along with poty-like, sobemo-like, and arteri-like viruses, were grouped together in supergroup I of positive-strand RNA viruses on the basis of similarities in their RNA-dependent RNA polymerase proteins (20). In this study, we were able to isolate RCs from FCV-infected cells and show that they are enzymatically active under the same conditions described for the isolation of such complexes from poliovirus-infected cells. The isolation of these active FCV complexes provides an opportunity to compare the characteristics of this in vitro replication system of the caliciviruses with those of other positive-strand RNA viruses.
As in poliovirus, evidence of the synthesis, in isolated RCs, of full-length FCV RNA molecules was found. However, in contrast to poliovirus, a 2.5-kb positive- and negative-sense subgenomic RNA was synthesized. The synthesis of a subgenomic RNA for translation of the structural proteins is unusual for supergroup I picorna-like viruses because many of these viruses produce a large polyprotein from which the capsid proteins are proteolytically cleaved. Exceptions include the plant luteoviruses, which produce a subgenomic RNA, the animal arteriviruses, which produce a nested set of capped, subgenomic-size RNA transcripts that are coterminal with the 3′ end of the genome, and possibly the astroviruses, which may be distant members of supergroup I (20). Several species of RNA molecules were observed in the FCV RC complexes and in the polyadenylated RNAs isolated from FCV-infected cells that were similar to those described previously (7, 30). However, RNA species other than the approximately 8.0-kb genomic-size and 2.5-kb subgenomic-size molecules did not appear to accumulate to high levels over time in our Northern blot analyses. Herbert et al. (17) had reported the presence of only two major polyadenylated RNAs in FCV-infected cells, and our data support a role for two major positive-strand RNA species (8.0 and 2.5 kb) in replication. Furthermore, our data demonstrate the accumulation of an approximately 2.5-kb negative-sense subgenomic RNA molecule during replication, consistent with previous studies of FCV-infected cells by Carter (7). Two major models have been proposed for subgenomic RNA synthesis in positive-strand RNA viruses (reviewed in reference 26). Both models involve initiation of negative-strand synthesis from the 3′ end of the genomic RNA, a feature common to all positive-strand RNA viruses. In the internal-initiation model, a subgenomic promoter is recognized by the polymerase on the newly synthesized negative strand and this is followed by transcription of a positive-sense subgenomic RNA to the 3′ end of the genome. In the premature-termination model, the polymerase dissociates from the genomic template at a terminator as it is synthesizing the negative strand. The truncated negative strand then functions as a template for end-to-end synthesis of a new subgenomic positive strand. The accumulation of a negative-sense subgenomic RNA could occur in both models, assuming that dead-end synthesis might result from the accumulating positive strand in the first model. Although our present study did not address the mechanism used for generation of the FCV subgenomic RNA, the presence of conserved sequences at the 5′ ends of the genomic and subgenomic RNA of FCV argues for the presence of a subgenomic promoter on the negative strand for transcription of the positive strand (31). The ability to study FCV subgenomic RNA synthesis in cell extracts, in concert with the availability of active FCV rPro-Pol (46), should facilitate studies to map the promoters and signals involved in FCV RNA replication.
Analysis of the proteins associated with the FCV RCs showed that nearly all of the virus-encoded proteins are present at the presumed site of RNA replication. Certain nonstructural proteins were present predominantly in their cleaved, mature forms, and these were p32 (a protein of unknown function encoded in the N-terminal region of ORF1) and p39 (a putative NTPase). However, the proteinase and polymerase were both detected as part of a stable precursor complex, Pro-Pol, which had previously been shown to be a highly active form of the FCV polymerase (46). Cleavage and release of the poliovirus 3D polymerase from 3CD is thought to be important for the RNA-dependent RNA polymerase activity of 3D (27, 45), but it is not clear whether a similar mechanism exists in the caliciviruses. An interesting observation in this study was the appearance of an approximately 50-kDa band in the Western blot analysis of the RCs with Pro-Pol-specific antiserum. It will be important to characterize this protein further in order to determine whether it is a biologically relevant product derived from Pro-Pol or a nonspecific degradation product. We cannot rule out the possibility that additional, active forms of the FCV polymerase function during replication.
Western blot analysis of the p30 and VPg proteins in the FCV RCs showed evidence of their association as part of a stable 43-kDa protein p30-VPg precursor. FCV p30 is analogous in genomic location to the 3A protein of poliovirus, which maps immediately upstream of VPg (3B). The 3A component of the poliovirus 3AB protein has been proposed to anchor this precursor in membranes so that VPg is positioned for RNA replication (12, 22, 44). Although the presence of the FCV 43-kDa protein in the FCV RCs is consistent with a similarity to poliovirus 3AB in its function, the role of this FCV precursor protein, as well as the cleaved p30 and VPg proteins, in calicivirus replication requires additional study.
An interesting observation in this study was the marked abundance of capsid protein VP1 in the FCV RCs. FCV particles contain VP1, VP2, and the VPg-linked genomic RNA (36), so it is likely that packaging of the newly synthesized RNA into viral capsids would be coordinated at the site of RNA synthesis. FCV, like other members of the genus Vesivirus, translates its capsid protein as a 73-kDa precursor that is cleaved by the viral proteinase. This cleavage event is essential for the production and spread of infectious virus particles (37). The early appearance of the mature capsid protein after infection in this study indicates that translation of the subgenomic RNA is an early event in replication and is consistent with previous studies that showed evidence of new FCV viral progeny within 30 min after infection (19). The coordination of VPg linkage, packaging of the genome, assembly, and incorporation of VP2 into maturing particles is not yet defined for the caliciviruses. FCV capsid protein can self-assemble independently of RNA and VP2, indicating that the structural determinants for assembly are carried by VP1 alone (15). It is possible that the membranes of the RC might form a scaffold for packaging of the viral RNA into assembling capsids. Localization of the viral proteins within the RCs by immunostaining may give insight into this process.
The demonstration that synthesis of both positive- and negative-sense FCV RNAs can be accomplished in an in vitro system is a promising advance in calicivirus research. Although our data suggest that RNA replication occurs in isolated RCs, further work is needed to distinguish between completion of RNA synthesis on preexisting endogenous templates (elongation) and de novo synthesis of RNA (including initiation of new genomic and subgenomic strands). Further characterization of these complexes may give insight into both the viral and host proteins essential for calicivirus replication. This information could facilitate the development of a cell-free calicivirus in vitro recovery system based on an infectious cDNA clone, similar to that available for poliovirus (28), as well as coupled transcription-translation systems similar to those developed for other positive-strand RNA viruses (1, 3). The development of such systems for FCV, which currently is the only cultivatable calicivirus with a reverse genetics system, may also facilitate the development of replication assays for important noncultivable caliciviruses, including the Norwalk-like viruses, which are associated with epidemic gastroenteritis.
Acknowledgments
We thank Albert Z. Kapikian for continuing support of our work and Natalya Teterina and Jerry M. Keith for helpful discussions.
REFERENCES
- 1.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 2.Barco, A., and L. Carrasco. 1995. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 14:3349-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienz, K., D. Egger, and T. Pfister. 1994. Characteristics of the poliovirus replication complex. Arch. Virol. Suppl. 9:147-157. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, M. J. 1990. Transcription of feline calicivirus RNA. Arch. Virol. 114:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter, M. J., I. D. Milton, J. Meanger, M. Bennett, R. M. Gaskell, and P. C. Turner. 1992. The complete nucleotide sequence of a feline calicivirus. Virology 190:443-448. [DOI] [PubMed] [Google Scholar]
- 9.Carter, M. J., I. D. Milton, P. C. Turner, J. Meanger, M. Bennett, and R. M. Gaskell. 1992. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch. Virol. 122:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 12.Datta, U., and A. Dasgupta. 1994. Expression and subcellular localization of poliovirus VPg-precursor protein 3AB in eukaryotic cells: evidence for glycosylation in vitro. J. Virol. 68:4468-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doedens, J., L. A. Maynell, M. W. Klymkowsky, and K. Kirkegaard. 1994. Secretory pathway function, but not cytoskeletal integrity, is required in poliovirus infection. Arch. Virol. Suppl. 9:159-172. [DOI] [PubMed] [Google Scholar]
- 14.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geissler, K., K. Schneider, A. Fleuchaus, C. R. Parrish, G. Sutter, and U. Truyen. 1999. Feline calicivirus capsid protein expression and capsid assembly in cultured feline cells. J. Virol. 73:834-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181:S322-S330. [DOI] [PubMed] [Google Scholar]
- 17.Herbert, T. P., I. Brierley, and T. D. Brown. 1996. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 77:123-127. [DOI] [PubMed] [Google Scholar]
- 18.Herbert, T. P., I. Brierley, and T. D. Brown. 1997. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 78:1033-1040. [DOI] [PubMed] [Google Scholar]
- 19.Komolafe, O. O., and O. Jarrett. 1986. A possible maturation pathway of calicivirus particles. Microbios 46:103-111. [PubMed] [Google Scholar]
- 20.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. (Erratum, 28:546, 1993.) [DOI] [PubMed] [Google Scholar]
- 21.Kreutz, L. C., B. S. Seal, and W. L. Mengeling. 1994. Early interaction of feline calicivirus with cells in culture. Arch. Virol. 136:19-34. [DOI] [PubMed] [Google Scholar]
- 22.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J. Biol. Chem. 269:66-70. [PubMed] [Google Scholar]
- 23.Love, D. N., and M. Sabine. 1975. Electron microscopic observation of feline kidney cells infected with a feline calicivirus. Arch. Virol. 48:213-228. [DOI] [PubMed] [Google Scholar]
- 24.Maynell, L. A., K. Kirkegaard, and M. W. Klymkowsky. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 66:1985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melnick, J. L., V. I. Agol, H. L. Bachrach, F. Brown, P. D. Cooper, W. Fiers, S. Gard, J. H. Gear, Y. Ghendon, L. Kasza, M. LaPlaca, B. Mandel, S. McGregor, S. B. Mohanty, G. Plummer, R. R. Rueckert, F. L. Schaffer, I. Tagaya, D. A. Tyrrell, M. Voroshilova, and H. A. Wenner. 1974. Picornaviridae. Intervirology 4:303-316. [DOI] [PubMed] [Google Scholar]
- 26.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 27.Molla, A., K. S. Harris, A. V. Paul, S. H. Shin, J. Mugavero, and E. Wimmer. 1994. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J. Biol. Chem. 269:27015-27020. [PubMed] [Google Scholar]
- 28.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 29.Neill, J. D. 1990. Nucleotide sequence of a region of the feline calicivirus genome which encodes picornavirus-like RNA-dependent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 17:145-160. [DOI] [PubMed] [Google Scholar]
- 30.Neill, J. D., and W. L. Mengeling. 1988. Further characterization of the virus-specific RNAs in feline calicivirus infected cells. Virus Res. 11:59-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neill, J. D., I. M. Reardon, and R. L. Heinrikson. 1991. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J. Virol. 65:5440-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orci, L., M. Tagaya, M. Amherdt, A. Perrelet, J. G. Donaldson, J. Lippincott-Schwartz, R. D. Klausner, and J. E. Rothman. 1991. Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell 64:1183-1195. [DOI] [PubMed] [Google Scholar]
- 33.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VPg for infectivity. Virology 210:383-390. [DOI] [PubMed] [Google Scholar]
- 36.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193-203. [DOI] [PubMed] [Google Scholar]
- 36a.Sosnovtsev, S. V., M. Garfield, and K. Y. Green. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 76:7060-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sosnovtseva, S. A., S. V. Sosnovtsev, and K. Y. Green. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studdert, M. J., and J. D. O'Shea. 1975. Ultrastructural studies of the development of feline calicivirus in a feline embryo cell line. Arch. Virol. 48:317-325. [DOI] [PubMed] [Google Scholar]
- 41.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 45.Van Dyke, T. A., and J. B. Flanegan. 1980. Identification of poliovirus polypeptide P63 as a soluble RNA-dependent RNA polymerase. J. Virol. 35:732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei, L., J. S. Huhn, A. Mory, H. B. Pathak, S. V. Sosnovtsev, K. Y. Green, and C. E. Cameron. 2001. Proteinase-polymerase precursor as the active form of feline calicivirus RNA-dependent RNA polymerase. J. Virol. 75:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]