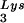Abstract
Human immunodeficiency virus type 1 (HIV-1) uses tRNA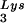 as a primer for reverse transcription and, during viral assembly, this tRNA is selectively packaged into the virus along with the other major tRNALys, tRNA
as a primer for reverse transcription and, during viral assembly, this tRNA is selectively packaged into the virus along with the other major tRNALys, tRNA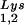 . Increasing the cytoplasmic concentration of tRNA
. Increasing the cytoplasmic concentration of tRNA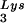 through transfection of cells with a plasmid containing both HIV-1 proviral DNA and a tRNA
through transfection of cells with a plasmid containing both HIV-1 proviral DNA and a tRNA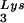 gene results in a greater incorporation of tRNA
gene results in a greater incorporation of tRNA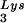 into virions, which is accompanied by increased annealing of tRNA
into virions, which is accompanied by increased annealing of tRNA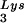 to the viral genome and increased infectivity of the viral population. Increased viral tRNA
to the viral genome and increased infectivity of the viral population. Increased viral tRNA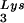 is accompanied by decreased viral tRNA
is accompanied by decreased viral tRNA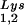 , with the total tRNALys/virion and the GagPol/Gag ratios remaining unchanged. Viral tRNALys can be doubled, with increases in both tRNA
, with the total tRNALys/virion and the GagPol/Gag ratios remaining unchanged. Viral tRNALys can be doubled, with increases in both tRNA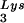 and tRNA
and tRNA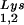 concentrations, by overexpressing lysyl tRNA synthetase. This also results in increased tRNA
concentrations, by overexpressing lysyl tRNA synthetase. This also results in increased tRNA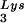 annealing to the viral RNA and increased viral infectivity but, again, no change in the GagPol/Gag ratio was observed. This result indicates that GagPol, whose interaction is required during packaging, is not a limiting factor during tRNALys incorporation into HIV-1, whereas LysRS is.
annealing to the viral RNA and increased viral infectivity but, again, no change in the GagPol/Gag ratio was observed. This result indicates that GagPol, whose interaction is required during packaging, is not a limiting factor during tRNALys incorporation into HIV-1, whereas LysRS is.
During retroviral assembly, particular species of cellular tRNA are selectively packaged into the virus, where they are placed onto the primer binding site (PBS) of the viral genome and are used to initiate the reverse transcriptase (RT)-catalyzed synthesis of minus-strand cDNA. The primer tRNA for members of the avian sarcoma and leukosis virus group is tRNATrp (1, 4, 16, 19, 23, 24), and it is tRNAPro for murine leukemia virus (MuLV) (3, 15, 21). In mammalian cells, there are three major tRNALys isoacceptors (18). tRNA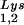 , representing two tRNALys isoacceptors differing by 1 bp in the anticodon stem, is the primer tRNA for several mammalian retroviruses, including Mason-Pfizer monkey virus and human foamy virus, while tRNA
, representing two tRNALys isoacceptors differing by 1 bp in the anticodon stem, is the primer tRNA for several mammalian retroviruses, including Mason-Pfizer monkey virus and human foamy virus, while tRNA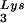 is the primer tRNA for lentiviruses, including human immunodeficiency virus type 1 (HIV-1) (10).
is the primer tRNA for lentiviruses, including human immunodeficiency virus type 1 (HIV-1) (10).
Selective packaging of primer tRNA is defined as an increase in the percentage of the low-molecular-weight RNA population representing primer tRNA in moving from the cytoplasm to the virus. For example, in avian myeloblastosis virus, the relative concentration of tRNATrp changes from 1.4% in the cytoplasm to 32% in the virus (23). In HIV-1 produced from COS7 cells transfected with HIV-1 proviral DNA, both primer tRNA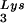 and primer tRNA
and primer tRNA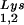 are selectively packaged, and the relative concentration of tRNALys changes from 5 to 6% to 50 to 60% (14). Both tRNA
are selectively packaged, and the relative concentration of tRNALys changes from 5 to 6% to 50 to 60% (14). Both tRNA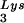 and tRNA
and tRNA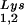 are packaged into HIV-1 with equal efficiency since the tRNA
are packaged into HIV-1 with equal efficiency since the tRNA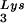 /tRNA
/tRNA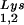 ratio in the virus reflects the cytoplasmic ratio, even when the cytoplasmic ratio is altered (5). In AKR MuLV, selective packaging of primer tRNAPro is less dramatic, going from a relative cytoplasmic concentration of 5 to 6% to 12 to 24% of low-molecular-weight RNA (23). Selective packaging of primer tRNA occurs independently of viral genomic RNA packaging in MuLV, HIV-1, and avian sarcoma virus (13, 14, 16) and has been shown in HIV-1 to occur independently of Gag and GagPol processing as well (8, 14).
ratio in the virus reflects the cytoplasmic ratio, even when the cytoplasmic ratio is altered (5). In AKR MuLV, selective packaging of primer tRNAPro is less dramatic, going from a relative cytoplasmic concentration of 5 to 6% to 12 to 24% of low-molecular-weight RNA (23). Selective packaging of primer tRNA occurs independently of viral genomic RNA packaging in MuLV, HIV-1, and avian sarcoma virus (13, 14, 16) and has been shown in HIV-1 to occur independently of Gag and GagPol processing as well (8, 14).
While it is suggestive that the selective packaging of primer tRNAs into the virion would occur in order to facilitate the annealing of the primer tRNA to the PBS through achieving higher viral concentrations of primer tRNA, experimental proof for this assumption has been absent. In avian retroviruses (2, 16) and HIV-1 (14), virions lacking functional RT [RT(−)] are unable to either selectively package primer tRNA or anneal it to the PBS. However, these observations do not make clear whether reduced genomic placement of primer tRNA is due to the reduction of primer tRNA in the virus or to the absence of functional RT sequences that might be required to place the tRNA on the genome. Also, RT(−) MuLV, although unable to selectively incorporate tRNAPro, is still capable of achieving wild-type levels of annealing of tRNAPro to the PBS (2, 11, 12).
Therefore, we artificially altered viral tRNA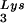 concentrations in HIV-1-transfected COS cells through expression from exogenous plasmids of either tRNALys isoacceptors or lysyl tRNA synthetase (LysRS). We found a direct correlation between viral tRNA
concentrations in HIV-1-transfected COS cells through expression from exogenous plasmids of either tRNALys isoacceptors or lysyl tRNA synthetase (LysRS). We found a direct correlation between viral tRNA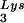 concentrations, annealing to the viral RNA, and infectivity of the viral populations.
concentrations, annealing to the viral RNA, and infectivity of the viral populations.
(This work was performed by J.G. in partial fulfillment of a Ph.D. degree from McGill University, Montreal, Quebec, Canada.)
MATERIALS AND METHODS
Plasmid construction.
SVC21BH10 is a simian virus 40 (SV40)-based vector containing wild-type HIV-1 proviral DNA. SVC21BH10Lys3 and SVC21BH10Lys2 contain both wild-type HIV-1 proviral DNA and a human tRNA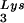 or tRNA2Lys gene, respectively. These vectors were constructed as previously described (5). SVC21BH10.P(−) is a SV40-based vector that contains full-length wild-type HIV-1 proviral DNA containing an inactive viral protease (D25G) and was a gift from E. Cohen, University of Montreal. SVC21BH10.P(−)Lys3 contains both protease-negative HIV-1 proviral DNA and a human tRNA
or tRNA2Lys gene, respectively. These vectors were constructed as previously described (5). SVC21BH10.P(−) is a SV40-based vector that contains full-length wild-type HIV-1 proviral DNA containing an inactive viral protease (D25G) and was a gift from E. Cohen, University of Montreal. SVC21BH10.P(−)Lys3 contains both protease-negative HIV-1 proviral DNA and a human tRNA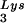 gene, cloned in the same way as SVC21BH10Lys3. Plasmid pM368 contains cDNA encoding full-length (1 to 597 amino acids) human LysRS, as previously described (20). The cDNA was PCR amplified and digested with EcoRI and XhoI, whose sites were placed in each of the PCR primers. For expression in COS7 cells, the PCR DNA fragments were cloned into pcDNA3.1 (Invitrogen) to obtain pLysRS.F, expressing full-length LysRS.
gene, cloned in the same way as SVC21BH10Lys3. Plasmid pM368 contains cDNA encoding full-length (1 to 597 amino acids) human LysRS, as previously described (20). The cDNA was PCR amplified and digested with EcoRI and XhoI, whose sites were placed in each of the PCR primers. For expression in COS7 cells, the PCR DNA fragments were cloned into pcDNA3.1 (Invitrogen) to obtain pLysRS.F, expressing full-length LysRS.
Virus infection, transfection, and purification.
COS7 cells were transfected by using the calcium phosphate method as previously described (14) or, for cotransfections, with Lipofectamine (Invitrogen). Supernatant was collected 63 h posttransfection. Viruses were pelleted from culture medium by centrifugation in a Beckman 45 Ti rotor at 35,000 rpm for 1 h. The viral pellets were then purified by centrifugation in a Beckman SW41 rotor at 26,500 rpm for 1 h through 15% sucrose onto a 65% sucrose cushion. The band of purified virus was removed and pelleted in 1× TNE in a Beckman 45 Ti rotor at 40,000 rpm for 1 h. Viral genomic RNA was extracted by using guanidinium isothiocyanate as previously described (6).
One- and two-dimensional polyacrylamide gel electrophoresis (1D- and 2D-PAGE).
Electrophoresis of [32P]pCp-labeled viral RNA was carried out at 4°C with the Hoeffer SE620 gel electrophoresis apparatus. The gel size was 14 by 32 cm. The first dimension was run in an 11% polyacrylamide-7 M urea gel for 16 h at 800 V. After autoradiography, the piece of gel containing RNA was cut out and run for 30 h (25 W, limiting); this step was followed by autoradiography. All electrophoretic runs were carried out in 0.5× TBE (1× TBE is 50 mM Tris, 5 mM boric acid, plus 1 mM EDTA-Na2). The electrophoretic gel patterns shown in the present study show only the findings with low-molecular-weight RNA, since the high-molecular-weight viral genomic RNA cannot enter into the polyacrylamide gels. Furthermore, these patterns represent only the most abundant tRNA species present, since longer film exposures reveal the presence of the more minor-abundance species.
Packaging of tRNA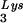 .
.
The relative amount of tRNA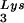 per copy of HIV-1 genomic RNA was determined by dot blot hybridization. Each sample of total viral RNA was blotted onto Hybond N+ nylon membranes (Amersham Pharmacia) and was probed with a 5′ 32P-end-labeled 18-mer DNA probe specific for the 3′ end of tRNA
per copy of HIV-1 genomic RNA was determined by dot blot hybridization. Each sample of total viral RNA was blotted onto Hybond N+ nylon membranes (Amersham Pharmacia) and was probed with a 5′ 32P-end-labeled 18-mer DNA probe specific for the 3′ end of tRNA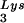 (5′-TGGCGCCCGAACAGGGAC-3′) or tRNA
(5′-TGGCGCCCGAACAGGGAC-3′) or tRNA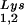 (5′-TGGCGCCCAACGTGGGGC-3′). Experiments were done in tripicate. Determination of tRNALys (i.e., both tRNA
(5′-TGGCGCCCAACGTGGGGC-3′). Experiments were done in tripicate. Determination of tRNALys (i.e., both tRNA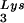 and tRNA
and tRNA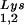 ) used both probes together. The relative amounts of tRNALys isoacceptor per sample were analyzed by using phosphorimaging (Bio-Rad). The blots were then stripped according to the manufacturer's instructions and were reprobed with a 5′ 32P-end-labeled 17-mer DNA probe specific for the 5′ end of HIV-1 genomic RNA, upstream of the PBS (5′-CTGACGCTCTCGCACCC-3′). Phosphorimaging was used to quantitate the relative amount of HIV-1 genomic RNA per sample and the relative amount of tRNA
) used both probes together. The relative amounts of tRNALys isoacceptor per sample were analyzed by using phosphorimaging (Bio-Rad). The blots were then stripped according to the manufacturer's instructions and were reprobed with a 5′ 32P-end-labeled 17-mer DNA probe specific for the 5′ end of HIV-1 genomic RNA, upstream of the PBS (5′-CTGACGCTCTCGCACCC-3′). Phosphorimaging was used to quantitate the relative amount of HIV-1 genomic RNA per sample and the relative amount of tRNA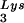 or tRNA
or tRNA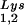 per copy of HIV-1 genomic RNA was determined. The amount of total viral RNA used in these determinations contained 3 × 108 to 10 × 108 copies of genomic RNA, an amount producing signals within the linear range of measurement for hybridization of both tRNALys isoacceptors and genomic RNA, as shown by standard curves generated by using a dilution series of total viral RNA which is hybridized with the DNA probes complementary to either tRNALys or genomic RNA.
per copy of HIV-1 genomic RNA was determined. The amount of total viral RNA used in these determinations contained 3 × 108 to 10 × 108 copies of genomic RNA, an amount producing signals within the linear range of measurement for hybridization of both tRNALys isoacceptors and genomic RNA, as shown by standard curves generated by using a dilution series of total viral RNA which is hybridized with the DNA probes complementary to either tRNALys or genomic RNA.
Primer extension.
tRNA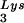 -primed initiation of reverse transcription was measured by the ability of tRNA
-primed initiation of reverse transcription was measured by the ability of tRNA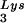 to be extended by six bases in an in vitro HIV-1 reverse transcription reaction. Total viral RNA was used as the source of primer tRNA and template. Each sample contained 5 × 108 copies of genomic RNA, measured as previously described (5), and produced signals falling within the linear range of measurement, as shown by generating a standard curve by using a dilution series of total BH10 viral RNA as the source of primer and template. The sequence of the first six deoxynucleoside triphosphates incorporated is CTGCTA. The reactions were carried out in a volume of 20 μl containing 50 mM Tris-HCl (pH 7.8), 100 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 0.2 mM dCTP, 0.2 mM dTTP, 5 μCi of [α-32P]dGTP, and 0.05 mM ddATP (instead of dATP, thereby terminating the reaction at six bases), 50 ng of HIV-1 RT, and RNase inhibitor (Amersham Pharmacia). After incubation for 15 min at 37°C, the samples were precipitated with isopropanol and were electrophoresed in a 6% polyacrylamide gel at 70 W for 1.5 h. The relative amounts of tRNA
to be extended by six bases in an in vitro HIV-1 reverse transcription reaction. Total viral RNA was used as the source of primer tRNA and template. Each sample contained 5 × 108 copies of genomic RNA, measured as previously described (5), and produced signals falling within the linear range of measurement, as shown by generating a standard curve by using a dilution series of total BH10 viral RNA as the source of primer and template. The sequence of the first six deoxynucleoside triphosphates incorporated is CTGCTA. The reactions were carried out in a volume of 20 μl containing 50 mM Tris-HCl (pH 7.8), 100 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 0.2 mM dCTP, 0.2 mM dTTP, 5 μCi of [α-32P]dGTP, and 0.05 mM ddATP (instead of dATP, thereby terminating the reaction at six bases), 50 ng of HIV-1 RT, and RNase inhibitor (Amersham Pharmacia). After incubation for 15 min at 37°C, the samples were precipitated with isopropanol and were electrophoresed in a 6% polyacrylamide gel at 70 W for 1.5 h. The relative amounts of tRNA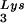 placement were determined by comparing the intensity of bands with phosphorimaging.
placement were determined by comparing the intensity of bands with phosphorimaging.
Statistical analysis of dot blots and primer extension.
All analyses were done in triplicate, with triplicate samples in each experiment. The statistical analyses employed herein include column statistics and one-way analysis of variance (ANOVA). The lowest level of significance was set at P < 0.05.
Viral infectivity.
Viral infectivity was measured by the MAGI assay (9). MAGI cells are CD4+ HeLa cells containing an HIV-1 long terminal repeat fused to a β-galactosidase reporter gene. A total of 4 × 104 cells per well were cultured in 1 ml of medium in 24-well plates. After 24 h, the medium was removed and replaced with 150 μl of culture medium containing various dilutions of virus. DEAE-dextran was added to a final concentration of 20 μg/ml, and viral absorption took place for 2 h, after which 1 ml of fresh culture medium was added. After 48 h, the medium was removed and fixative (1% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline) was added for 5 min. The fixative was removed, and 200 μl of staining solution was added [for 1 ml of solution, we used 950 μl of phosphate-buffered saline, 20 μl of 0.2 M potassium ferrocyanide, 20 μl of 0.2 M potassium ferricyanide, 1.0 μl of 2 M MgCl2, and 10 μl of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) stock (40 mg/ml in dimethyl sulfoxide)]. The cells were washed twice with phosphate-buffered saline, and the numbers of blue cells per well per equal amount of p24 were determined. Only wells containing 20 to 100 blue cells were analyzed, keeping within linear range of analysis (9).
Protein analysis.
Viral particles were purified as described above, and viral proteins were extracted with radioimmunoprecipitation assay buffer (10 mM Tris, pH 7.4; 100 mM NaCl; 1% sodium deoxycholate; 0.1% sodium dodecyl sulfate [SDS]; 1% NP-40; 2 mg of aprotinin/ml; 2 mg of leupeptin/ml; 1 mg of pepstatin A/ml; 100 mg of phenylmethylsulfonyl fluoride/ml). The viral lysates were analyzed by SDS-PAGE (on 10% acrylamide), followed by blotting onto nitrocellulose membranes (Amersham Pharmacia). Detection of protein by Western blotting utilized monoclonal antibodies that are specifically reactive with HIV-1 capsid (Zepto Metrocs, Inc.), RT (NIH AIDS Research and Reference Reagent Program) and a polyclonal antibody for human lysyl tRNA synthetase (a kind gift from Kiyotaka Shiba, Tokyo, Japan). Detection of HIV proteins was performed by enhanced chemiluminescence (NEN Life Sciences Products) with, as secondary antibodies, anti-mouse (for capsid and RT) and anti-rabbit (lysyl tRNA synthetase) antibodies, both of which were obtained from Amersham Life Sciences.
RESULTS
Overexpression of tRNALys isoacceptors from exogenous plasmids.
We have previously shown that the viral tRNA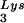 content can be increased by transfecting COS7 cells with an SV40-based plasmid containing both the HIV-1 proviral DNA and a human tRNA
content can be increased by transfecting COS7 cells with an SV40-based plasmid containing both the HIV-1 proviral DNA and a human tRNA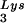 gene (BH10Lys3) and that, as a result, tRNA
gene (BH10Lys3) and that, as a result, tRNA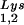 packaging into the virus decreases (5). In a similar manner we have also produced viruses with an excess of tRNA2Lys and a decrease in viral tRNA
packaging into the virus decreases (5). In a similar manner we have also produced viruses with an excess of tRNA2Lys and a decrease in viral tRNA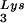 (BH10Lys2) by transfecting COS7 cells with a plasmid containing the HIV-1 proviral DNA and a human gene for tRNA2Lys (obtained from Robert M. Pirtle, University of North Texas). The inverse relationship between viral concentrations of tRNA
(BH10Lys2) by transfecting COS7 cells with a plasmid containing the HIV-1 proviral DNA and a human gene for tRNA2Lys (obtained from Robert M. Pirtle, University of North Texas). The inverse relationship between viral concentrations of tRNA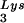 and tRNA
and tRNA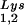 can be seen in Fig. 1. The data in Fig. 1A show the 2D-PAGE patterns of low-molecular-weight viral RNA in wild-type HIV-1 (BH10), BH10Lys3, and BH10Lys2. The identity of the tRNALys isoacceptors found in spots 1 and 2 (tRNA
can be seen in Fig. 1. The data in Fig. 1A show the 2D-PAGE patterns of low-molecular-weight viral RNA in wild-type HIV-1 (BH10), BH10Lys3, and BH10Lys2. The identity of the tRNALys isoacceptors found in spots 1 and 2 (tRNA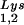 ) and spot 3 (tRNA
) and spot 3 (tRNA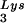 ) have been previously determined (6). BH10Lys3 has an additional small dark spot (3′) which has been identified as an additional tRNA
) have been previously determined (6). BH10Lys3 has an additional small dark spot (3′) which has been identified as an additional tRNA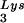 species by a partial T1 digestion pattern (data not shown) that is identical to the partial T1 digestion pattern of the major tRNA
species by a partial T1 digestion pattern (data not shown) that is identical to the partial T1 digestion pattern of the major tRNA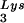 spot (6). This species can sometimes be seen as a very light spot in wild-type virus.
spot (6). This species can sometimes be seen as a very light spot in wild-type virus.
FIG. 1.
Alteration of tRNALys in HIV-1 as a result of overexpression of tRNA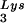 or tRNA
or tRNA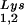 . Wild-type viruses were produced from COS7 cells transfected with BH10, BH10Lys3, or BH10Lys2, and total viral RNA was extracted. (A) 2D-PAGE analysis of low-molecular-weight viral RNA. Total viral RNA was 3′ end labeled with [32P]pCp and then electrophoresed in 11% polyacrylamide in the first dimension and in 20% polyacrylamide in the second dimension. Only low-molecular-weight RNA moves into the gel and is detected by autoradiography. Spots 3 and 3′, tRNA
. Wild-type viruses were produced from COS7 cells transfected with BH10, BH10Lys3, or BH10Lys2, and total viral RNA was extracted. (A) 2D-PAGE analysis of low-molecular-weight viral RNA. Total viral RNA was 3′ end labeled with [32P]pCp and then electrophoresed in 11% polyacrylamide in the first dimension and in 20% polyacrylamide in the second dimension. Only low-molecular-weight RNA moves into the gel and is detected by autoradiography. Spots 3 and 3′, tRNA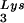 ; spots 1 and 2, tRNA
; spots 1 and 2, tRNA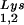 (partial sequencing of these two spots does not distinguish between tRNA1Lys and tRNA2Lys, which differ by only 1 bp in the anticodon stem). BH10, wild-type HIV-1 produced from cells transfected with a plasmid containing wild-type HIV-1 proviral DNA; BH10Lys3, HIV-1 produced from cells transfected with a plasmid containing both HIV-1 proviral DNA and a human gene for tRNA
(partial sequencing of these two spots does not distinguish between tRNA1Lys and tRNA2Lys, which differ by only 1 bp in the anticodon stem). BH10, wild-type HIV-1 produced from cells transfected with a plasmid containing wild-type HIV-1 proviral DNA; BH10Lys3, HIV-1 produced from cells transfected with a plasmid containing both HIV-1 proviral DNA and a human gene for tRNA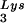 ; BH10Lys2, HIV-1 produced from cells transfected with a plasmid containing both HIV-1 proviral DNA and a human gene for tRNA,Lys. The pattern indicates that tRNA2Lys is found in spot 2 and not spot 1. (B) Analysis of the viral concentrations of tRNALys. Dot blots of viral RNA (containing 3 × 108 to 10 × 108 copies genomic RNA) were hybridized with DNA probes complementary to both tRNA
; BH10Lys2, HIV-1 produced from cells transfected with a plasmid containing both HIV-1 proviral DNA and a human gene for tRNA,Lys. The pattern indicates that tRNA2Lys is found in spot 2 and not spot 1. (B) Analysis of the viral concentrations of tRNALys. Dot blots of viral RNA (containing 3 × 108 to 10 × 108 copies genomic RNA) were hybridized with DNA probes complementary to both tRNA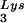 and tRNA
and tRNA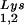 or to viral genomic RNA. Hybridization signals were analyzed by phosphorimaging, and the tRNALys (both tRNA
or to viral genomic RNA. Hybridization signals were analyzed by phosphorimaging, and the tRNALys (both tRNA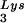 and tRNA
and tRNA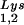 )/genomic RNA ratio was determined for virions produced from cells transfected with BH10, BH10Lys3, or BH10Lys2. The standard curves shown in the left part of the blot in panel B contain a dilution series of BH10 viral RNA, hybridized with the DNA probes complementary to either tRNALys or genomic RNA. The statistical analyses used here include column statistics and one-way ANOVA, where n = 3 and P < 0.05.
)/genomic RNA ratio was determined for virions produced from cells transfected with BH10, BH10Lys3, or BH10Lys2. The standard curves shown in the left part of the blot in panel B contain a dilution series of BH10 viral RNA, hybridized with the DNA probes complementary to either tRNALys or genomic RNA. The statistical analyses used here include column statistics and one-way ANOVA, where n = 3 and P < 0.05.
In Fig. 1B, the relative amount of tRNALys/virion was determined in the three types of virions by using hybridization probes for both tRNA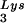 and tRNA
and tRNA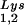 to determine the relative amounts of tRNALys/genomic RNA. The experimental data are shown in Fig. 1B and indicate little change in the total tRNALys/virion.
to determine the relative amounts of tRNALys/genomic RNA. The experimental data are shown in Fig. 1B and indicate little change in the total tRNALys/virion.
This conclusion is supported by the data in Fig. 2A. In these experiments, the relative concentration of tRNA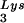 /genomic RNA, normalized to wild-type, was determined by hybridizing dot blots of total viral RNA with DNA probes specific for tRNA
/genomic RNA, normalized to wild-type, was determined by hybridizing dot blots of total viral RNA with DNA probes specific for tRNA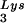 and for genomic RNA, and the values obtained are graphed in Fig. 2A. BH10Lys3 has ca. 1.6 times more tRNA
and for genomic RNA, and the values obtained are graphed in Fig. 2A. BH10Lys3 has ca. 1.6 times more tRNA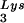 than the wild type, whereas BH10Lys2 has less than one-fifth the amount of tRNA
than the wild type, whereas BH10Lys2 has less than one-fifth the amount of tRNA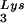 found in wild-type virions. Although the tRNALys species that decreases can often not be seen by 2D-PAGE (Fig. 1), it is quite measurable by hybridization. Thus, the [32P]pCp ligation reaction used to label the tRNALys species detected in 2D-PAGE appears to show a greater insensitivity to lower amounts of viral tRNALys than does the hybridization reaction.
found in wild-type virions. Although the tRNALys species that decreases can often not be seen by 2D-PAGE (Fig. 1), it is quite measurable by hybridization. Thus, the [32P]pCp ligation reaction used to label the tRNALys species detected in 2D-PAGE appears to show a greater insensitivity to lower amounts of viral tRNALys than does the hybridization reaction.
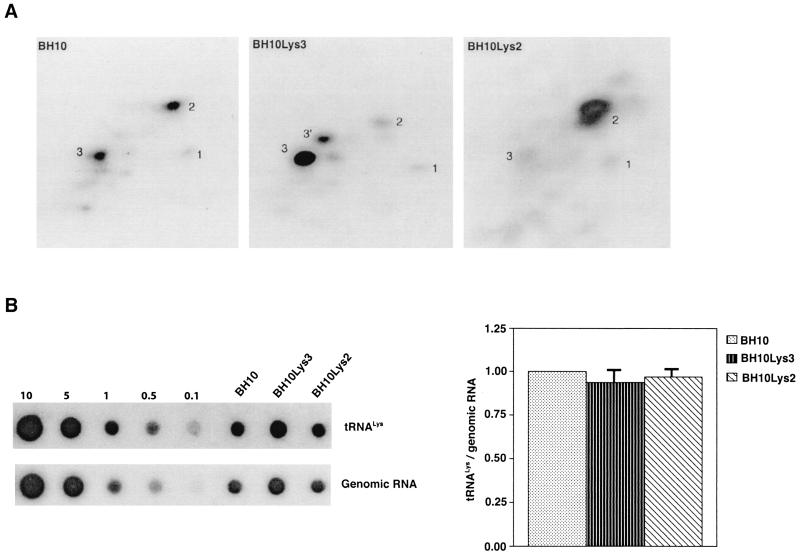
FIG. 2.
Effect of tRNA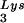 concentrations in wild-type virions upon tRNA
concentrations in wild-type virions upon tRNA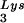 annealing and viral infectivity. Wild-type viruses were produced from COS7 cells transfected with BH10, BH10Lys3, or BH10Lys2. (A) Total viral RNA was extracted, and dot blots of viral RNA were hybridized with DNA probes complementary to either tRNA
annealing and viral infectivity. Wild-type viruses were produced from COS7 cells transfected with BH10, BH10Lys3, or BH10Lys2. (A) Total viral RNA was extracted, and dot blots of viral RNA were hybridized with DNA probes complementary to either tRNA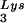 or viral genomic RNA. Hybridization signals were analyzed by phosphorimaging, and the tRNA
or viral genomic RNA. Hybridization signals were analyzed by phosphorimaging, and the tRNA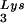 /genomic RNA ratio was determined for virions produced from cells transfected with BH10, BH10Lys3, or BH10Lys2. The standard curves shown in the left part of the blots were generated as described for Fig. 1B. (B) tRNA
/genomic RNA ratio was determined for virions produced from cells transfected with BH10, BH10Lys3, or BH10Lys2. The standard curves shown in the left part of the blots were generated as described for Fig. 1B. (B) tRNA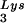 annealing to viral RNA. Total viral RNA was extracted and used as the source of primer tRNA
annealing to viral RNA. Total viral RNA was extracted and used as the source of primer tRNA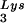 /genomic RNA template in an in vitro reverse transcription reaction, carried out in the presence of α-32P-labeled dGTP; dCTP, dTTP, and ddATP are not labeled. This will result in a six-base extension product since the first six bases incorporated are CTGCTA. Products were analyzed by 1D-PAGE, with samples containing equal amounts of genomic RNA. The standard curve shown in the left part of the blot in panel B contains a dilution series of total BH10 viral RNA, which is used as the source of primer or template. Statistical analyses employed in panels A and B include column statistics and one-way ANOVA, where n = 3 and P < 0.05. ✽, Statistically significant differences. (C) Viral infectivity. Infectivity was determined by using the MAGI assay as described in the text.
/genomic RNA template in an in vitro reverse transcription reaction, carried out in the presence of α-32P-labeled dGTP; dCTP, dTTP, and ddATP are not labeled. This will result in a six-base extension product since the first six bases incorporated are CTGCTA. Products were analyzed by 1D-PAGE, with samples containing equal amounts of genomic RNA. The standard curve shown in the left part of the blot in panel B contains a dilution series of total BH10 viral RNA, which is used as the source of primer or template. Statistical analyses employed in panels A and B include column statistics and one-way ANOVA, where n = 3 and P < 0.05. ✽, Statistically significant differences. (C) Viral infectivity. Infectivity was determined by using the MAGI assay as described in the text.
We next investigated in these three viral preparations whether the amount of tRNA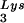 packaged into the virus influences the amount of extendable tRNA
packaged into the virus influences the amount of extendable tRNA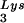 placed onto the PBS. The first six bases incorporated into DNA during the initiation of reverse transcription are CTGCTA. tRNA
placed onto the PBS. The first six bases incorporated into DNA during the initiation of reverse transcription are CTGCTA. tRNA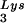 extension was measured in an in vitro reaction by using equal amounts of total viral RNA as the source of primer tRNA
extension was measured in an in vitro reaction by using equal amounts of total viral RNA as the source of primer tRNA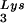 -genomic RNA template, exogenous HIV-1 RT, dCTP, dTTP, [α-32P]dGTP, and ddATP. This will result in a six-base DNA extension of the
-genomic RNA template, exogenous HIV-1 RT, dCTP, dTTP, [α-32P]dGTP, and ddATP. This will result in a six-base DNA extension of the 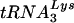 , and the amount of DNA extension-genomic RNA was determined by 1D-PAGE, as shown in Fig. 2B. Relative signal intensities were measured by phosphorimaging, the results of which are presented in Fig. 2B. These data indicate a correlation between tRNA
, and the amount of DNA extension-genomic RNA was determined by 1D-PAGE, as shown in Fig. 2B. Relative signal intensities were measured by phosphorimaging, the results of which are presented in Fig. 2B. These data indicate a correlation between tRNA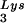 incorporated into the viruses and the amount of extendable tRNA
incorporated into the viruses and the amount of extendable tRNA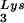 placed onto the PBS.
placed onto the PBS.
The relative infectivity of the three viral preparations was also measured by using the MAGI assay (9), which measures single-round infectivity. CD4+ HeLa cells containing the β-galactosidase gene fused to the HIV-1 long terminal repeat are infected with virus. Cells infected with HIV-1 will have the β-galactosidase gene expressed, and such cells can be detected by using an appropriate substrate for the enzyme, such as X-Gal, whose metabolism turns the cells blue. The number of blue cells is a measure of viral infectivity. The relative infectivity of the different viral populations is graphed in Fig. 2C and indicates that infectivity is directly correlated with tRNA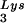 packaging and annealing of extendable tRNA
packaging and annealing of extendable tRNA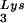 onto the PBS.
onto the PBS.
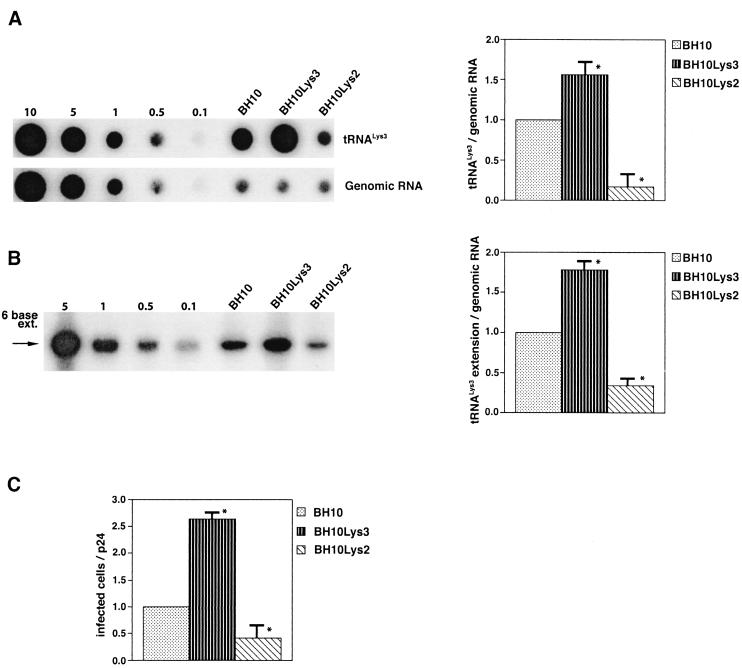
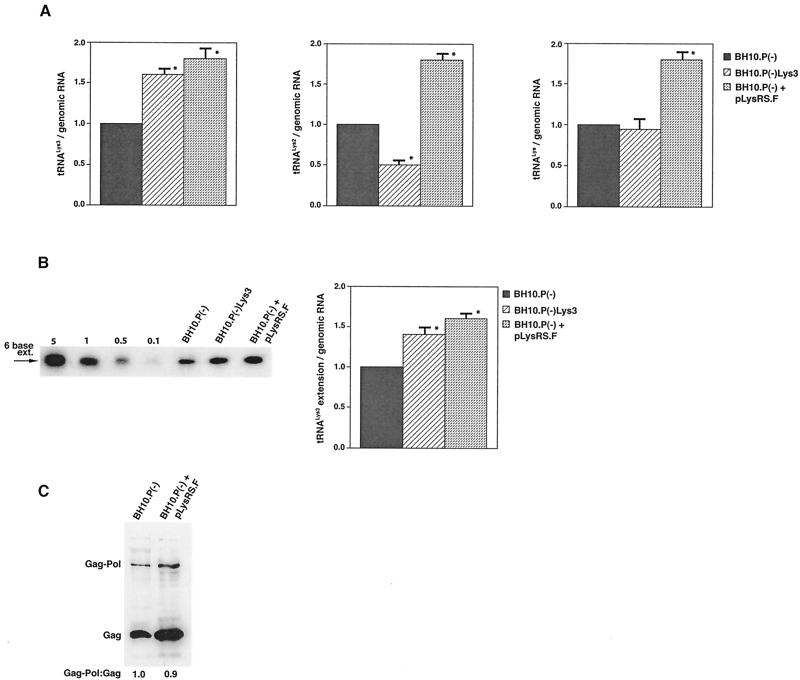
The increase in tRNA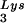 /virus does not result in any increase in GagPol incorporation. This was studied in protease-negative virions to facilitate detection of the GagPol precursor. Figure 3A shows the 2D-PAGE patterns of low-molecular-weight viral RNA in protease-negative BH10.P(−) and BH10.P(−)Lys3 virions. The patterns are nearly identical to their protease-positive counterparts and, as shown in these gels and in Fig. 3C, the increase in tRNA
/virus does not result in any increase in GagPol incorporation. This was studied in protease-negative virions to facilitate detection of the GagPol precursor. Figure 3A shows the 2D-PAGE patterns of low-molecular-weight viral RNA in protease-negative BH10.P(−) and BH10.P(−)Lys3 virions. The patterns are nearly identical to their protease-positive counterparts and, as shown in these gels and in Fig. 3C, the increase in tRNA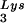 is accompanied by a decrease in tRNA
is accompanied by a decrease in tRNA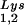 , with the total tRNALys/virion remaining the same. In Fig. 3B, Western blots of viral lysates probed with anti-CA and anti-RT indicate little change in the GagPol/Gag ratios (listed in Fig. 3C).
, with the total tRNALys/virion remaining the same. In Fig. 3B, Western blots of viral lysates probed with anti-CA and anti-RT indicate little change in the GagPol/Gag ratios (listed in Fig. 3C).
FIG. 3.
Alteration of tRNA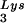 in protease-negative HIV-1 as a result of overexpression of tRNA
in protease-negative HIV-1 as a result of overexpression of tRNA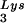 . Protease-negative viruses were produced from COS7 cells transfected with BH10.P(−) or BH10.P(−)Lys3. (A) 2D-PAGE analysis of low-molecular-weight viral RNA. Total viral RNA was 3′ end labeled with [32P]pCp and then electrophoresed. Conditions for 2D-PAGE and labeling of spots is as described in Fig. 2A. (B) Western blots of viral lysates, probed with anti-CA and anti-RT. The results, quantitated by phosphorimaging, are listed in panel C as the GagPol/Gag ratios. (C) Incorporation of tRNALys into HIV-1. Dot blots of viral RNA were hybridized with DNA probes complementary to tRNA
. Protease-negative viruses were produced from COS7 cells transfected with BH10.P(−) or BH10.P(−)Lys3. (A) 2D-PAGE analysis of low-molecular-weight viral RNA. Total viral RNA was 3′ end labeled with [32P]pCp and then electrophoresed. Conditions for 2D-PAGE and labeling of spots is as described in Fig. 2A. (B) Western blots of viral lysates, probed with anti-CA and anti-RT. The results, quantitated by phosphorimaging, are listed in panel C as the GagPol/Gag ratios. (C) Incorporation of tRNALys into HIV-1. Dot blots of viral RNA were hybridized with DNA probes complementary to tRNA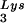 alone, to tRNALys (both tRNA
alone, to tRNALys (both tRNA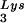 and tRNA
and tRNA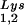 ), and to viral genomic RNA. The results were quantitated by phosphorimaging, and the tRNA
), and to viral genomic RNA. The results were quantitated by phosphorimaging, and the tRNA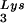 /genomic RNA or tRNALys/genomic RNA ratios are listed in panel C.
/genomic RNA or tRNALys/genomic RNA ratios are listed in panel C.
Overexpression of LysRS from an exogenous plasmid.
The stable viral concentration of tRNALys during overexpression of tRNA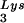 or tRNALys, indicates that there is some factor limiting the incorporation of tRNALys isoacceptors. Although the incorporation of GagPol into viral particles does not change when tRNA
or tRNALys, indicates that there is some factor limiting the incorporation of tRNALys isoacceptors. Although the incorporation of GagPol into viral particles does not change when tRNA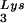 packaging is increased, this could reflect the fact that the total tRNALys packaged does not change. We have, however, recently found that overexpression of LysRS in protease-negative virions can result in up to a twofold increase in both tRNA
packaging is increased, this could reflect the fact that the total tRNALys packaged does not change. We have, however, recently found that overexpression of LysRS in protease-negative virions can result in up to a twofold increase in both tRNA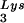 and tRNA
and tRNA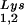 in the virions (S. Cen, M. Niu, K. Musier-Forsyth, and L. Kleiman, unpublished data). Thus, LysRS could be the limiting factor, but its expression might also stimulate GagPol incorporation. We have therefore investigated the effect of overexpression of LysRS upon both tRNA
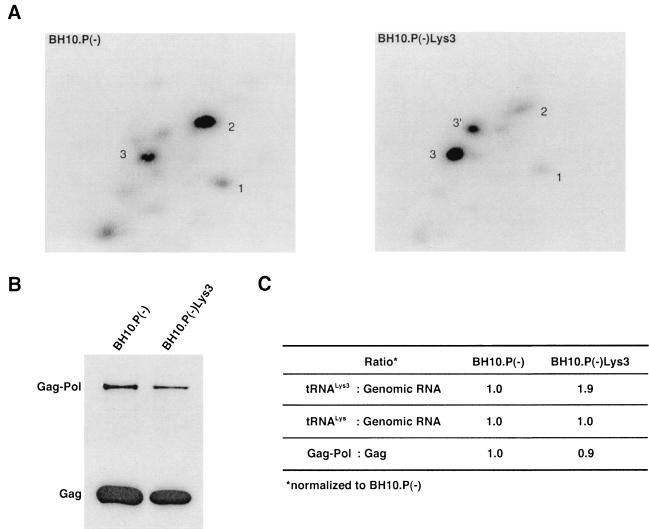
in the virions (S. Cen, M. Niu, K. Musier-Forsyth, and L. Kleiman, unpublished data). Thus, LysRS could be the limiting factor, but its expression might also stimulate GagPol incorporation. We have therefore investigated the effect of overexpression of LysRS upon both tRNA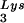 annealing to the PBS and upon GagPol incorporation. COS7 cells were cotransfected with both protease-negative HIV-1 proviral DNA BH10.P(−) and a plasmid encoding human LysRS, pLysRS.F. We have previously shown that this results in greater incorporation of both LysRS and tRNALys into virions (Cen et al., unpublished data), and Fig. 4A shows that, unlike overexpression of tRNA
annealing to the PBS and upon GagPol incorporation. COS7 cells were cotransfected with both protease-negative HIV-1 proviral DNA BH10.P(−) and a plasmid encoding human LysRS, pLysRS.F. We have previously shown that this results in greater incorporation of both LysRS and tRNALys into virions (Cen et al., unpublished data), and Fig. 4A shows that, unlike overexpression of tRNA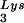 , overexpression of LysRS results in increases in both tRNA
, overexpression of LysRS results in increases in both tRNA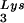 /genomic RNA and tRNA
/genomic RNA and tRNA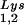 /genomic RNA, resulting in a significant increase in tRNALys in the virion. Figure 4B shows that, like overexpression of tRNA
/genomic RNA, resulting in a significant increase in tRNALys in the virion. Figure 4B shows that, like overexpression of tRNA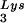 , overexpression of LysRS results in an increase in annealing of tRNA
, overexpression of LysRS results in an increase in annealing of tRNA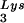 to the viral RNA, as determined by using total viral RNA isolated from the two types of virions as the source of primer or template in the six-base in vitro RT extension reaction. This provides further proof that tRNA
to the viral RNA, as determined by using total viral RNA isolated from the two types of virions as the source of primer or template in the six-base in vitro RT extension reaction. This provides further proof that tRNA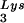 annealing is proportional to the amount of tRNA
annealing is proportional to the amount of tRNA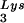 packaged into the virions. The Western blot shown in Fig. 4C shows that the GagPol/Gag ratio in either type of virion remains similar, even though the amount of tRNALys/genomic RNA has significantly increased. This indicates that, during the packaging of tRNALys, GagPol is in excess and LysRS is limiting.
packaged into the virions. The Western blot shown in Fig. 4C shows that the GagPol/Gag ratio in either type of virion remains similar, even though the amount of tRNALys/genomic RNA has significantly increased. This indicates that, during the packaging of tRNALys, GagPol is in excess and LysRS is limiting.
FIG. 4.
The effect of overexpression of LysRS upon tRNALys incorporation and annealing, GagPol incorporation, and viral infectivity. Protease-negative viruses were produced from COS7 cells transfected with BH10.P(−) or BH10.P(−)Lys3 or cotransfected with BH10.P(−) and pLysRS.F. (A) tRNALys incorporation. Dot blots of viral RNA were hybridized with DNA probes complementary to either tRNA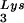 or tRNA
or tRNA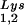 alone, to tRNALys (both tRNA
alone, to tRNALys (both tRNA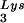 and tRNA
and tRNA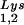 ), and to viral genomic RNA. The results were quantitated by phosphorimaging, and the ratios of tRNA
), and to viral genomic RNA. The results were quantitated by phosphorimaging, and the ratios of tRNA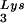 , tRNA
, tRNA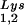 , or tRNALys to genomic RNA are plotted for the three viral types. Statistical analyses of the results are as described in the legend to Fig. 1. (B) tRNA
, or tRNALys to genomic RNA are plotted for the three viral types. Statistical analyses of the results are as described in the legend to Fig. 1. (B) tRNA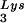 annealing to viral RNA. Total viral RNA was extracted and used as the source of primer tRNA
annealing to viral RNA. Total viral RNA was extracted and used as the source of primer tRNA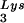 /genomic RNA template in an in vitro reverse transcription reaction, as described for Fig. 1B. Products were analyzed by 1D-PAGE with samples containing equal amounts of genomic RNA. Generation of the standard curve and statistical analyses of the results are as described in the legend to Fig. 2B. (C) Western blots of viral lysates, probed with anti-CA and anti-RT. The results, quantitated by phosphorimaging, are listed as the GagPol/Gag ratios beneath each lane.
/genomic RNA template in an in vitro reverse transcription reaction, as described for Fig. 1B. Products were analyzed by 1D-PAGE with samples containing equal amounts of genomic RNA. Generation of the standard curve and statistical analyses of the results are as described in the legend to Fig. 2B. (C) Western blots of viral lysates, probed with anti-CA and anti-RT. The results, quantitated by phosphorimaging, are listed as the GagPol/Gag ratios beneath each lane.
DISCUSSION
The work presented here indicates a direct relationship between tRNA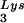 incorporated into the viral population, tRNA
incorporated into the viral population, tRNA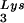 -primed initiation of reverse transcription, and the infectivity of the viral population. The viral population may contain viruses that have packaged primer tRNA
-primed initiation of reverse transcription, and the infectivity of the viral population. The viral population may contain viruses that have packaged primer tRNA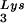 to different extents, with resulting different degrees of tRNA
to different extents, with resulting different degrees of tRNA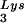 annealing and viral infectivity. This variability could be the result of mutations in viral or cellular proteins involved in this process but could also result from random differences in the amount of GagPol or LysRS incorporated into each virion during assembly. The evolution of a biochemical mechanism for enriching tRNA
annealing and viral infectivity. This variability could be the result of mutations in viral or cellular proteins involved in this process but could also result from random differences in the amount of GagPol or LysRS incorporated into each virion during assembly. The evolution of a biochemical mechanism for enriching tRNA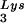 incorporation might help reduce the variability of packaging into viruses.
incorporation might help reduce the variability of packaging into viruses.
The increase in tRNA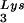 annealing resulting from overexpression of tRNA
annealing resulting from overexpression of tRNA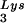 or LysRS is not accompanied by a change in the GagPol/Gag ratio, implying no increase in the incorporation of GagPol (assuming that Gag incorporation has not changed). This result implies that LysRS, rather than GagPol, is a limiting factor for tRNALys packaging, and it explains why the total viral tRNALys remains constant when one of the tRNALys isoacceptors is overexpressed.
or LysRS is not accompanied by a change in the GagPol/Gag ratio, implying no increase in the incorporation of GagPol (assuming that Gag incorporation has not changed). This result implies that LysRS, rather than GagPol, is a limiting factor for tRNALys packaging, and it explains why the total viral tRNALys remains constant when one of the tRNALys isoacceptors is overexpressed.
Seeming to stand counter to the claim of the importance of selective packaging of primer tRNA in virions for obtaining optimum annealing of primer tRNA to the viral template is the work from C. D. Morrow's laboratory. This work indicates that some non-lysyl tRNAs, such as tRNAHis (25) and tRNAMeti (7), can be used as primer tRNAs without their selective packaging. For example, changing the tRNA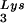 -complementary PBS to a sequence complementary to tRNAHis does not allow the stable use of tRNAHis as a primer, and the PBS reverts back to being complementary to tRNA
-complementary PBS to a sequence complementary to tRNAHis does not allow the stable use of tRNAHis as a primer, and the PBS reverts back to being complementary to tRNA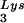 (22). However, if in addition to the PBS the A-rich loop just upstream of the PBS is also made complementary to the anticodon loop of tRNAHis, tRNAHis will be stably used as a primer tRNA, although some other point mutations in the regions near the PBS also occur to facilitate this (25). This mutant virion, termed His-AC-GAC, continues to selectively package tRNALys but does not appear to selectively package the new primer tRNA, tRNAHis (25). Although the replication kinetics of this virus are generally lower than for the wild-type virion with tRNA
(22). However, if in addition to the PBS the A-rich loop just upstream of the PBS is also made complementary to the anticodon loop of tRNAHis, tRNAHis will be stably used as a primer tRNA, although some other point mutations in the regions near the PBS also occur to facilitate this (25). This mutant virion, termed His-AC-GAC, continues to selectively package tRNALys but does not appear to selectively package the new primer tRNA, tRNAHis (25). Although the replication kinetics of this virus are generally lower than for the wild-type virion with tRNA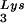 as a primer (25, 26), replication rates of 50% wild type without selective packaging of tRNAHis lead to the conclusion that selective packaging of primer tRNA is not required in the mutant HIV-1. A more general conclusion that might be derived from these observations is that the virus, put under certain conditions requiring the use of a new primer tRNA, can find new ways to utilize low concentrations of the viral tRNA more easily than it can find new ways to selectively package this tRNA. In fact, the normal annealing of tRNAPro to the viral RNA in MuLV does not require selective packaging of this primer (23), and wild-type levels of tRNAPro annealing are found in RT-negative MuLV in which any selective packaging that might occur is abolished (2, 11, 12). Thus, although there may be alternative ways in which retroviruses can develop to achieve optimum primer tRNA annealing to the PBS, wild-type HIV-1 and avian retroviruses appear to achieve this through selective packaging of primer tRNAs into the viruses.
as a primer (25, 26), replication rates of 50% wild type without selective packaging of tRNAHis lead to the conclusion that selective packaging of primer tRNA is not required in the mutant HIV-1. A more general conclusion that might be derived from these observations is that the virus, put under certain conditions requiring the use of a new primer tRNA, can find new ways to utilize low concentrations of the viral tRNA more easily than it can find new ways to selectively package this tRNA. In fact, the normal annealing of tRNAPro to the viral RNA in MuLV does not require selective packaging of this primer (23), and wild-type levels of tRNAPro annealing are found in RT-negative MuLV in which any selective packaging that might occur is abolished (2, 11, 12). Thus, although there may be alternative ways in which retroviruses can develop to achieve optimum primer tRNA annealing to the PBS, wild-type HIV-1 and avian retroviruses appear to achieve this through selective packaging of primer tRNAs into the viruses.
Acknowledgments
This study was supported by grants from the Canadian Institutes for Health Research.
We thank Sandy Fraiberg for assistance in preparation of the manuscript.
REFERENCES
- 1.Faras, A. J., and N. A. Dibble. 1975. RNA-directed DNA synthesis by the DNA polymerase of Rous sarcoma virus: structural and functional identification of 4S primer RNA in uninfected cells. Proc. Natl. Acad. Sci. USA 72:859-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu, W., A. Ortiz-Conde, R. J. Gorelick, S. H. Hughes, and A. Rein. 1997. Placement of tRNA primer on the primer binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 71:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harada, F., G. G. Peters, and J. E. Dahlberg. 1979. The primer tRNA for Moloney murine leukemia virus DNA synthesis: nucleotide sequence and aminoacylation of tRNAPro. J. Biol. Chem. 254:10979-10985. [PubMed] [Google Scholar]
- 4.Harada, F., R. C. Sawyer, and J. E. Dahlberg. 1975. A primer RNA for initiation of in vitro Rous sarcoma virus DNA synthesis: nucleotide sequence and amino acid acceptor activity. J. Biol. Chem. 250:3487-3497. [PubMed] [Google Scholar]
- 5.Huang, Y., J. Mak, Q. Cao, Z. Li, M. A. Wainberg, and L. Kleiman. 1994. Incorporation of excess wild-type and mutant tRNA3Lys into HIV-1. J. Virol. 68:7676-7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang, S., and C. D. Morrow. 1999. Genetic analysis of a unique human immunodeficiency virus type 1 (HIV-1) with a primer binding site complementary to tRNAMeti supports a role for U5-PBS stem-loop RNA structures in initiation of HIV-1 reverse transcription. J. Virol. 73:1818-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khorchid, A., H. Javanbakht, M. A. Parniak, M. A. Wainberg, and L. Kleiman. 2000. Sequences within Pr160gag-pol affecting the selective packaging of tRNALys into HIV-1. J. Mol. Biol. 299:17-26. [DOI] [PubMed] [Google Scholar]
- 9.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leis, J., A. Aiyar, and D. Cobrinik. 1993. Regulation of initiation of reverse transcription of retroviruses, p. 33-47. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase, vol. 1. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 11.Levin, J. G., S. C. Hu, A. Rein, L. I. Messer, and B. I. Gerwin. 1984. Murine leukemia virus mutant with a frameshift in the reverse transcriptase coding region: implications for pol gene structure. J. Virol. 51:470-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levin, J. G., and J. G. Seidman. 1981. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J. Virol. 38:403-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin, J. G., and J. G. Seidman. 1979. Selective packaging of host tRNAs by murine leukemia virus particles does not require genomic RNA. J. Virol. 29:328-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters, G., F. Harada, J. E. Dahlberg, A. Panet, W. A. Haseltine, and D. Baltimore. 1977. Low-molecular-weight RNAs of Moloney murine leukemia virus: identification of the primer for RNA-directed DNA synthesis. J. Virol. 21:1031-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peters, G. G., and J. Hu. 1980. Reverse transcriptase as the major determinant for selective packaging of tRNAs into avian sarcoma virus particles. J. Virol. 36:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prats, A. C., L. Sarih, C. Gabus, S. Litvak, G. Keith, and J. L. Darlix. 1988. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 7:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raba, M., K. Limburg, M. Burghagen, J. Katz, M. Simsek, J. Heckman, U. Rajbhandary, and H. Gross. 1979. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur. J. Biochem. 97:305-318. [DOI] [PubMed] [Google Scholar]
- 19.Sawyer, R. C., and J. E. Dahlberg. 1973. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J. Virol. 12:1226-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiba, K., T. Stello, H. Motegi, T. Noda, K. Musier-Forsyth, and P. Schimmel. 1997. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues Escherichia coli double-defective mutant. J. Biol. Chem. 272:22809-22816. [DOI] [PubMed] [Google Scholar]
- 21.Taylor, J. M. 1977. An analysis of the role of tRNA species as primers for transcription into DNA of RNA tumor virus genomes. Biochim. Biophys. Acta 47:57-71. [DOI] [PubMed] [Google Scholar]
- 22.Wakefield, J. K., A. G. Wolf, and C. D. Morrow. 1995. Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNALys3. J. Virol. 69:6021-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waters, L. C., and B. C. Mullin. 1977. Transfer RNA in RNA tumor viruses. Prog. Nucleic Acid Res. Mol. Biol. 20:131-160. [DOI] [PubMed] [Google Scholar]
- 24.Waters, L. C., B. C. Mullin, T. Ho, and W. K. Yang. 1975. Ability of tryptophan tRNA to hybridize with 35S RNA of avian myeloblastosis virus and prime reverse transcription in vitro. Proc. Natl. Acad. Sci. USA 72:2155-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, Z., S. Kang, A. Le Blanc, S. L. Hajduk, and C. D. Morrow. 1996. Nucleotide sequences within the U5 region of the viral RNA genome are the major determinants for a human immunodeficiency virus type 1 to maintain a primer binding site complementary to tRNAHis. Virology 226:306-317. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Z., S. M. Kang, and C. D. Morrow. 1998. Genetic evidence of the interaction between tRNA(Lys,3) and U5 facilitating efficient initiation of reverse transcription by human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 14:979-988. [DOI] [PubMed] [Google Scholar]