Abstract
Caveolin-1 (Cav-1) is a major protein constituent of caveolae, a type of plasma membrane raft. We observed that coexpression of human Cav-1 with human immunodeficiency virus type 1 (HIV-1) blocked virion production from cells that are ordinarily highly permissive. Further investigation showed that this effect is specific, occurs at low ratios of Cav-1 to HIV-1 DNA, depends on expression of Cav-1 protein, and involves severely impaired expression of HIV-1 proteins. Cav-1 also blocked HIV-2 expression. In contrast, Cav-1 did not inhibit protein expression by a paramyxovirus and did not induce apoptosis or affect cellular morphology, cell viability, or cell cycle progression. Although only small amounts of HIV-1 virions were released from Cav-1-transfected cells, these were fully infectious. Deletion mutagenesis showed that the C-terminal 78 residues were as active as the full-length (178-amino-acid) protein in producing the block. In contrast, the 100 most N-terminal amino acids of Cav-1, which include the previously identified oligomerization and scaffolding domains, were shown to be dispensable. Study of single-amino-acid-exchange mutants of Cav-1 established that palmitoylation was not required. Additional deletion mutants then identified the hydrophobic, membrane-associated domain (residues 101 to 135) as the main determinant. Cellular distribution of wild-type and mutant proteins correlated with ability to block HIV-1 expression. Finally, Cav-2 also blocked HIV-1 expression. These data show that coexpression of caveolins can markedly inhibit expression of HIV proviral DNA and establish that the inhibition is mediated by the hydrophobic, membrane-associated domain.
The lipid raft hypothesis postulates the existence within the plasma membrane bilayer of a heterogeneous population of phase-separated “rafts,” which are conceived as dynamic lateral assemblies of glycosphingolipids, cholesterol, and certain proteins (64, 65). Caveolae, a specialized type of membrane raft present in most cells, were first characterized morphologically as small flask-shaped plasma membrane invaginations (9). Caveolin-1 (Cav-1), initially identified as a 22-kDa substrate for v-src tyrosine kinase in Rous sarcoma virus-transformed fibroblasts (24), localizes to caveolae by immunoelectron microscopy (53) and possesses the ability to drive their biogenesis (22, 28, 29, 53). Two other isoforms (Cav-2 and -3) have subsequently been characterized. A diverse array of functions have been proposed for caveolae and Cav-1, including modulation of signal transduction (33, 67), endocytosis (59), potocytosis (4), and cholesterol trafficking (26, 68, 74, 75), and the functions of the protein remain controversial (45).
In addition, Cav-1 is found in multiple endomembrane compartments within cells in addition to the plasma membrane. Various domains of the protein control its complex, and still controversial, itinerary between these compartments, which includes recycling from the plasma membrane (15, 35, 69). The topology of Cav-1 is distinctive: a 33-amino-acid hydrophobic domain located in the carboxy-terminal half of the 178-amino-acid protein (residues 102 to 134) mediates association of the protein with the lipid bilayer, while both hydrophilic ends protrude into the cytoplasm. Since the hydrophobic domain apparently does not traverse the membrane entirely, the predicted hairpin structure lacks exposure at exocytoplasmic faces of cellular membranes (67). The hydrophobic membrane domain is involved in Cav-1-Cav-2 hetero-oligomerization (17), while residues 61 to 101 (35, 56) and 134 to 154 (35) appear to be important for homo-oligomerization. In CHO cells, deletion of amino- or carboxy-terminal regions flanking the hydrophobic domain results in variable localization of Cav-1 in the Golgi, endoplasmic reticulum (ER), and nuclear membrane rather than at plasma membrane caveolae (35).
Signal transduction molecules have been shown to interact with residues 80 to 101 in the N-terminal region (67). A model has been proposed in which binding to this juxtamembrane domain sequesters key signal transduction molecules, thereby enabling caveolae to regulate diverse signaling events at the cell surface; hence, it has been designated the scaffolding domain (11, 43, 58, 60, 67, 70). Although this hypothesis has received additional support from Cav-1-knockout mice, which lack caveolae and display vascular abnormalities related to defects in nitric oxide and calcium signaling (20, 51), the relatively mild overall phenotypes suggested that redundant or compensatory mechanisms can arise in development (45). In addition to binding to signaling proteins, Cav-1 directly binds cholesterol and mediates cholesterol trafficking through a cytoplasmic complex containing Cav-1, cholesterol, heat shock proteins, and cyclophilin A (74). Palmitoylation of two carboxy-terminal cysteines in Cav-1 is required for formation of this lipid-protein complex (73).
A number of microbial pathogens, including viruses, have been proposed to interact with components of membrane microdomains (61). Simian virus 40 enters cells via caveolae (3, 46), and evidence exists that both human immunodeficiency virus type 1 (HIV-1) (2, 41, 44, 54) and measles virus (37, 76) bud selectively from lipid rafts. Sendai virus and influenza virus virions are also enriched for raft components (1, 27, 55, 66, 77). Nevertheless, raft incorporation is not universal for enveloped RNA viruses, or it can be separable from interactions with raft components; for example, alphavirus and rhabdovirus virions are enriched in cholesterol and glycophospatidylinositol-anchored proteins but do not appear to bud from rafts (34, 57). Cav-1 is expressed in a variety of cells that are susceptible to HIV infection, including macrophages (5, 6, 18, 23, 30, 39, 40, 42) and astrocytes (10), as well as in potential secondary targets such as endothelial cells (14) and mast cells (38, 62). Cav-1 expression is not normally detected in resting peripheral blood T cells (22) but has been found in activated T-cell lines (25). However, no studies have examined potential interactions of HIV-1 and caveolins. The importance of membrane rafts for the assembly of HIV-1 virions, coupled with the known involvement of Cav-1 in cellular processes that include signal transduction, cholesterol trafficking, and virus entry, prompted us to ask whether coexpression of Cav-1 with HIV-1 would influence the assembly and infectivity of this human lentivirus. Initial experiments revealed unexpectedly that cotransfection of Cav-1 with HIV-1 proviral DNA blocked production of virions. We therefore proceeded to further investigate this effect.
MATERIALS AND METHODS
Plasmids.
pYU-2 (gift of G. Shaw, University of Alabama) is a macrophage/CCR5-tropic HIV-1 molecular clone isolated directly from human brain (31). This proviral clone was used in most experiments; HIV-1 NL4-3 and other HIV-1 proviral clones were also used as described in Results. For HIV-2 studies, a full-length molecular plasmid clone of HIV-2 KR (pEP32) was used (49). pCMV is a standard mammalian expression plasmid that contains the promoter-enhancer as well as the first 5′ untranslated exon and intron of the human cytomegalovirus (CMV) immediate-early gene upstream of a polylinker, into which cDNAs for the enhanced green fluorescent protein (eGFP), Cav-1, Cav-1 mutants, and Cav-2 were inserted (71). Only human caveolins were studied. A control, frameshifted version of pCMV.Cav-1 was constructed by Klenow fragment treatment and blunt ligation of the SexA1 site at nucleotide (nt) 149 of the 534-nt Cav-1 open reading frame. pCi (Promega) was also used to express Cav-1, eGFP, and firefly luciferase. Deletion from pCMV of an 896-nt fragment containing the 5′ untranslated exon and intron resulted in pCMV2, which contains only a 91-nt untranslated leader between the TATA box and the site of Cav-1 or eGFP insertion.
Cav-1 mutants and epitope tagging.
A Myc-tagged version of pCMV (pCMV-myc) was constructed by inserting a PCR-generated c-Myc epitope and a stop codon between XbaI and BglII in the polylinker; this plasmid was used to express C-terminally Myc-tagged versions of wild-type Cav-1 (pCMV-myc.Cav-1) or the different truncated mutants. The PCR primers listed in Table 1 were used to generate a panel of Cav-1 fragments, which were subsequently cloned in pCMV-myc. Palmitoylation-minus Cav-1 mutants were generated from pCMV.Cav-1 by site-directed mutagenesis (QuikChange XL; Stratagene) with oligonucleotides listed in Table 1. pCMV.Cav-160-178 and pCMV.Cav-1Δ60-100 were generated by cloning into pCMV the corresponding Myc-tagged cDNAs that were kindly provided by R. G. W. Anderson (University of Texas Southwestern). All mutants were confirmed by DNA sequencing.
TABLE 1.
Oligonucleotides used for mutagenesis of Cav-1
| Mutant | Oligonucleotides |
|---|---|
| Cav-11-60 | 5′-AAATCTAGACCAGCATGTCTGGGGGCAAATACG-3′ |
| 5′-ATATGCGGCCGCTTAGTTGAGGTGTTTAGGGTCGCGG-3′ | |
| Cav-11-78 | 5′-AAATCTAGACCAGCATGTCTGGGGGCAAATACG-3′ |
| 5′-ATATGCGGCCGCTTATGTCCCTTCTGGTTCTGCAATCAC-3′ | |
| Cav-11-101 | 5′-AAATCTAGACCAGCATGTCTGGGGGCAAATACG-3′ |
| 5′-ATATGGGCCCGCGGTAAAACCAGTATTTCGTCACAGTG-3′ | |
| Cav-11-135 | 5′-AAATCTAGACCAGCATGTCTGGGGGCAAATACG-3′ |
| 5′-ATATGGGCCCCTTAATGCATGGTACAACTGCCC-3′ | |
| Cav-1101-135 | 5′-AAATCTAGACCAGCATGCGCTTGCTGTCTGCCCTCTTTGGC-3′ |
| 5′-TATAGGGCCCCTTAATGCATGGTACAACTGCCC-3′ | |
| Cav-1101-178 | 5′-AAATCTAGACCAGCATGCGCTTGCTGTCTGCCCTCTTTGGC-3′ |
| 5′-ATATGGGCCCTATTTCTTTCTGCAAGTTGATGCGG-3′ | |
| Cav-1wt | 5′-AAATCTAGACCAGCATGTCTGGGGGCAAATACG-3′ |
| 5′-ATATGGGCCCTATTTCTTTCTGCAAGTTGATGCGG-3′ | |
| C133A | 5′-CTGCACATCTGGGCAGTTGTACCAGCCATTAAGAGCTTCCTGATTGAGATTC-3′ |
| 5′-GAATCTCAATCAGGAAGCTCTTAATGGCTGGTACAACTGCCCAGATGTGCAG-3′ | |
| C143A | 5′-ATTAAGAGCTTCCTGATTGAGATTCAGGCCACCAGCCGTGTCTATTCCATCTAC-3′ |
| 5′-GTAGATGGAATAGACACGGCTGGTGGCCTGAATCTCAATCAGGAAGCTCTTAAT-3′ | |
| C156A | 5′-TATTCCATCTACGTCCACACCGTCGCTGACCCACTCTTTGAAGCTGTTGG-3′ |
| 5′-CCAACAGCTTCAAAGAGTGGGTCAGCGACGGTGTGGACGTAGATGGAATA-3′ |
Cell culture and transfections.
Human 293T embryonal kidney cells were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% fetal calf serum, penicillin, and streptomycin. Transfections were performed by the calcium phosphate coprecipitation method with a total of 2.4 μg of DNA per well of a six-well plate. Briefly, cells were transfected 24 h after being plated in 2 ml of medium at 0.45 × 106 cells/well. For some experiments, 2 × 106 293T cells were plated in 75-cm2 flasks and cotransfected the next day with 10 μg of HIV-1 proviral DNA plus 10 μg of experimental plasmid or 10 μg of empty vector. Particular attention was paid in all comparisons to keeping total DNA constant between wells by using filler DNA having the same promoter elements. After 14 to 16 h the transfection mix was replaced by fresh culture medium. Cells and cell supernatant were harvested 40 to 48 h after the transfection mix was added. Supernatants and lysates were stored at −80°C.
Virus quantification and titration.
Viral particles were quantitated in cell supernatants by 32P-based reverse transcriptase (RT) assays as previously described (50) or by HIV-1 p24 antigen capture enzyme-linked immunosorbent assay (Coulter). Titers of viruses were determined by endpoint dilution in Magi-CCR5 cells (12).
Cell cycle and apoptosis assays.
293T cells were cotransfected with 1.2 μg of pYU-2 and different concentrations of pCMV.Cav-1 or pCMV empty vector. At 40 h after transfection, cells were harvested with trypsin and used for propidium iodine staining or terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay. Propidium iodine staining was performed as described previously (16). Briefly, cells were resuspended overnight at 4°C in a hypotonic solution containing 50 μg of propidium iodine/ml in 0.1% sodium citrate plus 0.1% Triton X-100. Then cells were washed twice with phosphate-buffered saline (PBS) and analyzed by flow cytometry. TUNEL assay was performed with a Promega Apoptosis Detection System kit according to the manufacturer's instructions. Briefly, cells were fixed in 1% formaldehyde and 70% ethanol, washed in PBS, and incubated for 1 h with terminal deoxynucleotidyltransferase in a reaction buffer containing fluorescein-12-dUTP. The labeling reaction was terminated by adding EDTA, and cells were washed in PBS plus 0.1% Triton X-100, incubated in a propidium iodine solution, and analyzed by flow cytometry.
Flow cytometric analysis of Cav-1 expression.
Cav-1 was detected in transfected cells by indirect immunofluorescence with an anti-Cav-1 monoclonal antibody (MAb), clone 2297 (Transduction Laboratories). Cells were fixed in methanol, permeabilized in 0.1% Triton X-100 in PBS, incubated with the MAb, washed and labeled with R-phycoerythrin-tagged F(ab′)2 rabbit anti-mouse immunoglobulin antibody (Dakopatts, Glostrup, Denmark), and analyzed by flow cytometry (FACScan; Becton Dickinson, Mountain View, Calif.).
Measles-eGFP virus.
Sixteen hours after transfection, 293T cells were infected with a recombinant measles-eGFP virus at a multiplicity of infection of 1.0 in 1 ml of Optimen (Gibco BRL) for 1 h at 37°C. This virus, kindly provided by R. Cattaneo, expresses eGFP in an additional open reading frame upstream of that encoding nucleocapsid (21). Infecting medium was replaced by 2 ml of culture medium, and 6 h later 100 μl of a human serum containing anti-measles virus antibodies was added to the infected cells to prevent syncytium formation. Twenty-four hours later cells were analyzed by flow cytometry.
Immunoblotting and immunofluorescence microscopy.
For Western blotting, cells were lysed in Tris-buffered saline containing 1% Triton X-100 and 1% NP-40, plus a protease inhibitor cocktail (Complete-mini; Boehringer), and sonicated to disrupt aggregates. Proteins (40 μg/lane) were resolved in sodium dodecyl sulfate-12% polyacrylamide gels and transferred to Immobilon P membranes (Millipore). Blocked membranes were incubated overnight at 4°C with anti-Myc MAb (clone 9E10; Covance), anti-α-tubulin MAb (clone B-5-1-2; Sigma), or pooled human anti-HIV serum, diluted 10−3 in Tris-buffered saline-5% nonfat milk plus 0.05% Tween 20. Endogenous Cav-1 was detected with a MAb (Transduction Laboratories clone 2297, 0.25 μg/ml). After washing, membranes were incubated with the appropriate horseradish peroxidase-tagged secondary antibody. Bound antibodies were detected by ECL (Amersham Pharmacia Biotech). For immunofluorescence microscopy, 293T cells cotransfected with Myc-tagged proteins were fixed and permeabilized with paraformaldehyde and methanol and then stained with the anti-Myc MAb and a fluorescein isothiocyanate-labeled secondary antibody. DAPI (4′,6′-diamidino-2-phenylindole) counterstaining was used to identify nuclei.
RESULTS
Cotransfection of Cav-1 blocks HIV production.
A pilot experiment was performed to investigate whether cotransfection of Cav-1 influences HIV-1 virion budding or infectivity. Transfection of 293T human embryonic kidney cells was used for virus production because they do not express detectable Cav-1 by Western blotting (data not shown). In addition, these cells transfect very efficiently and are well established as being highly permissive for producing HIV-1 or vectors when transiently transfected with HIV-1 proviral DNA (13, 47). Because transfected HIV also does not replicate in this CD4-negative cell line, it is used extensively to produce genetically homogeneous viral stocks and to study the productive phase of the viral life cycle (52).
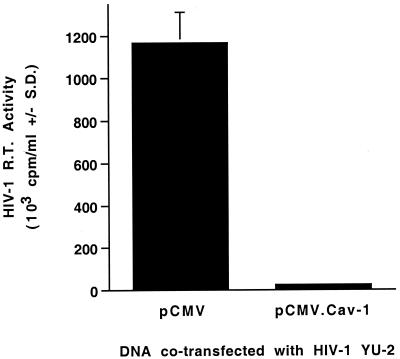
The cells were cotransfected with 10 μg of the full-length HIV-1 molecular clone pYU-2 (31), plus either 10 μg of a Cav-1 expression plasmid (pCMV.Cav-1) or 10 μg of empty vector (pCMV). Supernatant was collected and filtered at 46 h after transfection, and Mg2+-dependent RT activity was determined in sextuplicate. As expected, cotransfection of pYU-2 with the control plasmid pCMV yielded high levels of HIV-1 RT activity (>106 cpm/ml). In contrast, cotransfection of pYU-2 with pCMV.Cav-1 almost completely blocked production of RT activity (Fig. 1).
FIG. 1.
Cav-1 coexpression blocks HIV-1 production. 293T cells (2 × 106) were plated in 75-cm2 flasks and cotransfected the next day with 10 μg of pYU-2 plus pCMV (empty vector) or with 10 μg of pYU-2 plus pCMV.Cav-1 (total of 20 μg of DNA in each transfection) by calcium phosphate coprecipitation. Medium was replaced at 15 h after transfection. Virus supernatant was harvested 31 h later and filtered (0.45-μm pore size). Supernatant RT activities are means ± standard deviations of sextuplicate determinations.
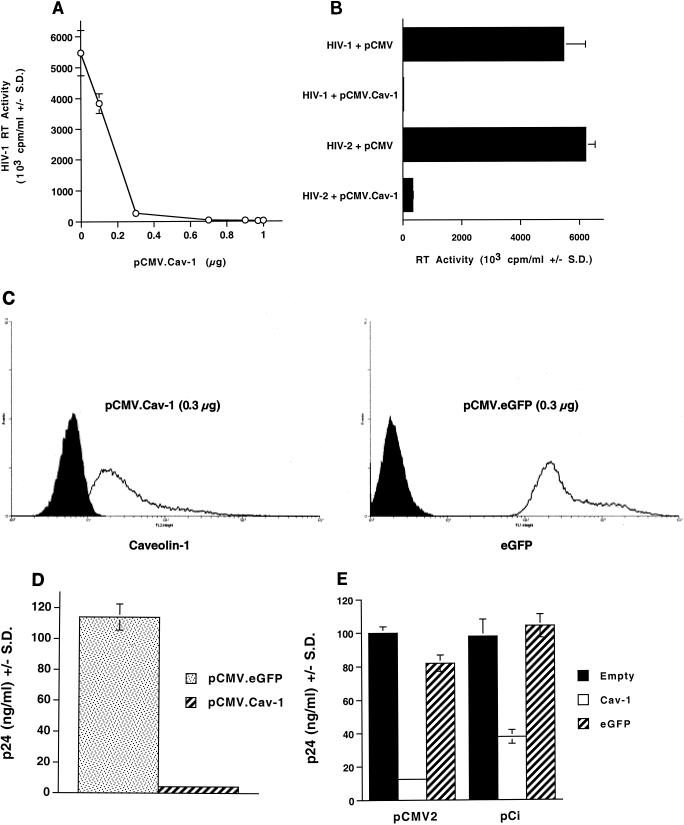
To explore this surprising observation further, we determined that it was highly reproducible, not only with HIV-1 YU-2 but also with a variety of R5-tropic and X4-tropic HIV-1 clones (n = 5; data not shown). We then carried out dose-response analyses. As shown in Fig. 2A, the inhibition was potent. Ninety-six percent inhibition of HIV-1 RT production was seen when 1.2 μg of pYU-2 was cotransfected with as little as 0.3 μg of pCMV.Cav-1 (representing 12.5% of total DNA, which was kept constant at 2.4 μg per well, and 25% of the pYU-2 DNA). An 0.1-μg quantity of pCMV.Cav-1 (4% of total DNA, 8% of the pYU-2 input) produced 30% inhibition of YU-2 RT production. The 0.3-μg input of Cav-1 expression plasmid (one-fourth of the amount of HIV-1 DNA), the lowest ratio that consistently produced near-maximal inhibition, was used in a number of additional experiments.
FIG. 2.
Dose dependence, Cav-1 specificity, and inhibition of HIV-2 production. (A) Dose dependence of inhibition. 293T cells were cotransfected with 1.2 μg of pYU-2 plus different amounts of pCMV.Cav-1 or the empty vector, and RT activity was determined 40 h later. (B) Inhibition of HIV-2 expression. RT activity was determined in supernatants of 293T cells cotransfected with 1.2 μg of either HIV-1 YU-2 or HIV-2 proviral DNA plus 0.7 μg of pCMV.Cav-1 or the control DNA pCMV. (C and D) Overexpression of a control protein does not inhibit HIV-1 production in cotransfected 293T cells. (C) Fluorescence-activated cell sorting histograms of protein expression in two cell populations, each cotransfected with 1.2 μg of pYU-2 and either 0.3 μg of pCMV.Cav-1 (left histogram, anti-Cav-1 MAb labeling) or 0.3 μg of pCMV.eGFP (right histogram, eGFP fluorescence). (D) HIV-1 p24 levels in the supernatants of the cells analyzed by fluorescence-activated cell sorting in panel C. (E) Specific inhibition of HIV-1 production with pCMV2.Cav-1 and pCi.Cav-1. 293T cells were cotransfected with 1.2 μg of YU-2 and 0.3 μg of the indicated plasmids or with empty vector. RT activity (103 cpm/ml) and p24 (nanograms per milliliter) are means ± standard deviations of duplicate or triplicate determinations.
To document that the inhibitory effect required expression of Cav-1 protein and was not occurring through mechanisms operating at the RNA or DNA levels, we introduced a frameshift in the mRNA by blunting a SexA1 site located at nt 149 of the 534-nt Cav-1 open reading frame in pCMV.Cav-1. Cotransfections were carried out with this plasmid following the same design as the experiment depicted in Fig. 2A and showed that the frameshift completely abolished the inhibition. In addition, blockade of virion production by pCMV.Cav-1 was seen whether calcium phosphate, lipofection, or electroporation was used for transfection and virus production or if a different cell line (COS-7) was used (data not shown).
The marked inhibitory effect was also observed with HIV-2, although the effect on HIV-1 was consistently more complete (Fig. 2B). RT activity levels in supernatants of 293T cells cotransfected in parallel with 0.7 μg of pCMV.Cav-1 and 1.2 μg of either HIV-1 or HIV-2 proviral DNA were 0.4 and 5.4% of control, respectively (Fig. 2B). To evaluate the specificity of Cav-1 in producing inhibition, we next determined whether coexpression of control proteins affected HIV-1 particle production in these cells (Fig. 2C and D). 293T cells were cotransfected with 1.2 μg of pYU-2 plus either 0.3 μg of pCMV.Cav-1 or the same expression plasmid encoding eGFP instead of Cav-1 (pCMV.eGFP). Forty hours after transfection, cellular levels of Cav-1 and eGFP were each evaluated by flow cytometric assays (Fig. 2C), and HIV-1 capsid protein (p24 antigen) levels in the cell supernatants were determined by enzyme-linked immunosorbent assay (Fig. 2D). The results showed that eGFP was highly expressed but did not affect HIV p24 production, while Cav-1 markedly inhibited p24 production (Fig. 2C and D). In addition, 85 to 100% transfection efficiency of 293T cells was routinely obtained in our experiments (Fig. 2C). This experiment was repeated four times, with the same result. The same lack of inhibition was also obtained with pCi.eGFP, which encodes eGFP from pCi (Promega), a similarly structured commercial expression plasmid containing the human CMV enhancer-promoter and a synthetic intron, as well as with this plasmid expressing firefly luciferase instead of eGFP (data not shown).
Several different Cav-1 expression plasmids were tested to verify the effect. pCMV, the plasmid into which the Cav-1 cDNA was inserted to create pCMV.Cav-1, contains the promoter-enhancer sequences and first 5′ untranslated exon and first intron of the human CMV immediate-early promoter (63). We also expressed Cav-1 and eGFP from pCi as well as from a version of pCMV from which we have deleted almost all of the 5′ untranslated sequences, including all of the intron, leaving only a 91-nt segment of the original 987-nt leader between the TATA box and the site of cDNA insertion (pCMV2). 293T cells were cotransfected with 1.2 μg of pYU-2 and 0.3 μg of pCMV2.Cav-1 or pCi.Cav-1, and p24 production was compared to that of cells cotransfected with pYU-2 and the respective empty vectors (Fig. 2E). Control transfections were done in parallel with pCMV2 and pCi constructs encoding eGFP instead of Cav-1. Again, inhibition was produced by each of the Cav-1 constructs but not by the eGFP constructs. pCMV2.Cav-1 produced the same inhibition as pCMV.Cav-1, indicating that the CMV 5′ untranslated region was not required. pCi also inhibited pYU-2 expression, although this was slightly less potent at the 0.3-μg input (Fig. 2E).
In summary, these data indicate that cotransfection of Cav-1 under the control of a strong viral promoter blocks expression of HIV-1 proviral DNA in cells that are ordinarily highly permissive for production of this virus.
Virus produced in the presence of Cav-1 is not defective.
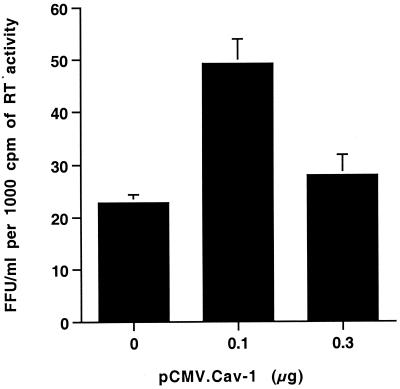
Cav-1 has been demonstrated to mediate cholesterol trafficking from the ER to the plasma membrane through a cytoplasmic, nonvesicle lipid-protein complex containing Cav-1, cholesterol, heat shock proteins, and cyclophilin A (74). Cyclophilin A is specifically incorporated into HIV-1 virions and regulates virion infectivity (7, 8). Therefore, we carried out single-round infectivity analyses to investigate whether involvement of this or other processes would result in a defect in the HIV-1 that was produced in the presence of coexpressed Cav-1 (Fig. 3). Virus supernatants were analyzed from the cells described in Fig. 2A that had been cotransfected with 1.2 μg of pYU-2 plus 0, 0.1, and 0.3 μg of pCMV.Cav-1 (which produced 0, 30, and 96% inhibition of RT production, respectively). The samples were normalized for RT activity in order to equalize particle number, and titers were determined by endpoint dilution on Magi-CCR5 cells (48). Infectivity of virus produced in the presence of Cav-1 did not differ from virus produced in its absence and was even twofold higher for HIV-1 produced from cells transfected with 0.1 μg of pCMV.Cav-1 (Fig. 3). These data suggest that the virions produced in the presence of Cav-1, although lower in quantity, are not defective.
FIG. 3.
Virions budded from cells coexpressing Cav-1 are not defective. The infectious titer of HIV-1 produced by cells cotransfected with 1.2 μg of YU-2 and 0, 0.1, or 0.3 μg of pCMV.Cav-1 (total DNA kept constant at 2.4 μg by using pCMV as filler DNA) was determined by endpoint dilution on Magi-CCR5 cells. Supernatants were normalized for RT activity prior to titration. Values are mean LacZ focus-forming units (FFU) per milliliter per 1,000 cpm of RT activity ± standard deviation of duplicate experiments.
Inhibition of HIV expression is specific to Cav-1 and is not due to cellular toxicity.
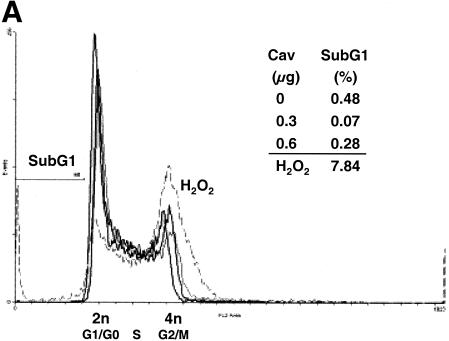
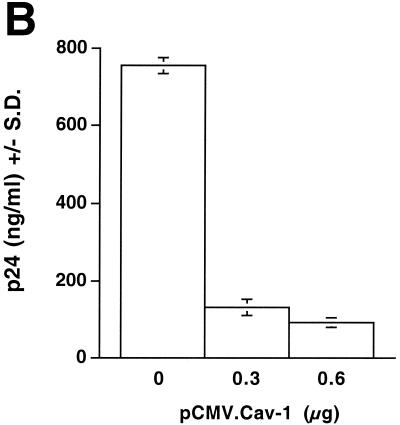
In the experiments described above, transfected cell cultures documented to express Cav-1 with >90% transfection efficiency were morphologically indistinguishable throughout the experiments from control-transfected cells and did not differ in adherence, cell density, trypan blue exclusion, or total protein content per cell (data not shown). Additional experiments were performed to analyze the possibilities of apoptosis and alterations in cell cycle progression in Cav-1-transfected cells. 293T cells were cotransfected with 1.2 μg of HIV YU-2 proviral DNA and different amounts of pCMV.Cav-1 or pCMV. Forty hours later, the cells were subjected to flow cytometry after staining with propidium iodine (Fig. 4A, cell cycle and apoptosis) and also to TUNEL assays (apoptosis). Simultaneously determined supernatant p24 HIV production was high (756 ng/ml) for cells transfected with pCMV (Fig. 4B). Flow cytometry showed that cells displaying a full blockade of HIV-1 p24 production by Cav-1 (Fig. 4B) had neither alterations in their cell cycle profiles nor apoptotic cells (Fig. 4A). The TUNEL assays confirmed the absence of apoptosis (data not shown).
FIG. 4.
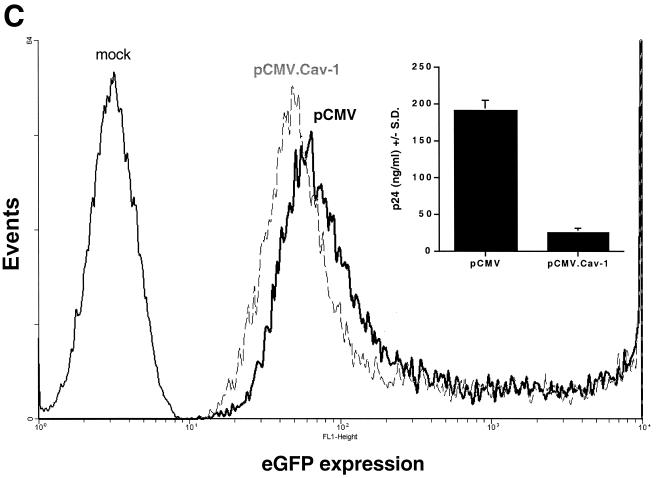
Analyses of apoptosis, cell cycling, and specificity of HIV-1 inhibition in Cav-1-expressing cells. (A and B) 293T cells were cotransfected with 1.2 μg of YU-2 and different amounts of pCMV.Cav-1 or pCMV. Total DNA was kept constant (1.8 μg) by using pCMV as filler DNA. At 40 h, cells were stained with propidium iodine and analyzed by flow cytometry (A), and p24 in their supernatants was quantified (B). Positive control 293T cells were treated with 30 μM H2O2 (dashed line). The inset table gives percentages of cells in sub-G1 phase. Lack of apoptosis was confirmed in separate experiments by TUNEL assays. (C) Fluorescence-activated cell sorting histograms show eGFP expression in 293T cells triply cotransfected with 1.2 μg of YU-2, 1.0 μg of pCi.eGFP, and either 0.3 μg of pCMV.Cav-1 (dashed line) or pCMV (continuous heavy line). p24 levels in the supernatants of the same cells are shown in the inset graph.
In order to further evaluate the specificity for HIV, we performed the converse comparison from that shown in Fig. 2C and D, by comparing the effects of Cav-1 on simultaneous expression of pYU-2 and pCi.eGFP within the same cell (Fig. 4C). Here, 293T cells were triply cotransfected, again by the calcium phosphate method, with 1.2 μg of pYU-2, 1.0 μg of pCi.eGFP, and 0.3 μg of pCMV.Cav-1. Control cells were transfected with the same DNAs, except that 0.3 μg of pCMV was used in place of pCMV.Cav-1. eGFP expression at 40 h after transfection was determined by flow cytometry, and supernatant HIV-1 p24 was quantified. While Cav-1 again severely inhibited HIV p24 production, eGFP expression from the CMV promoter in the same cells was minimally inhibited (Fig. 4C). This experiment was repeated twice, with the same result. In addition, the same differential effect was obtained when the HIV-1 YU-2 provirus and the pCMV.eGFP expression cassette were incorporated within the same plasmid as tandemly arrayed transcription units (data not shown). These results suggested that the inhibitory effect on HIV-1 production was not due to a global perturbation or to nonspecific toxicity to the cell.
Cav-1 blocks expression of HIV proteins but not paramyxovirus proteins.
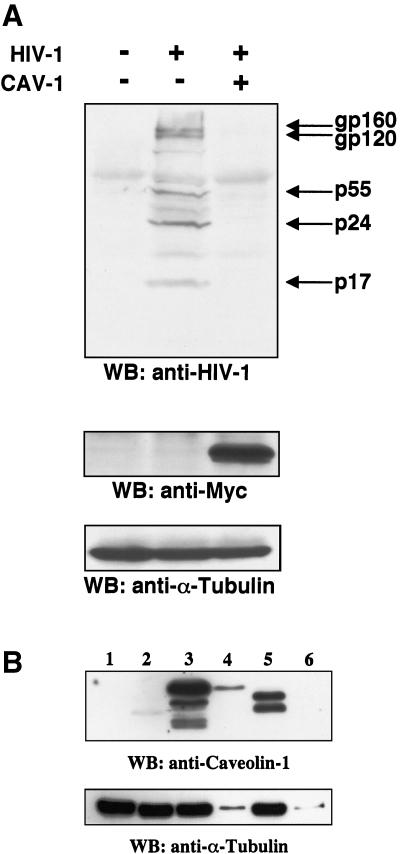
The inhibitory effect of Cav-1 on HIV virion production could occur at any of a number of steps in the productive phase of the viral life cycle—e.g., at the levels of expression and transport of HIV-1 mRNAs, translation, virion assembly, or budding. Interference with assembly or budding might be particularly plausible mechanisms, given the known budding of HIV (2, 41, 44) from glycolipid-enriched membrane raft domains. In order to begin to discriminate between these alternatives, we performed immunoblotting to analyze HIV-1 protein expression. 293T cells were cotransfected with 1.2 μg of pYU-2 and 0.3 μg of pCMV-myc or pCMV-myc.Cav-1 (which expresses Cav-1 with a C-terminal Myc tag). Forty hours later, HIV proteins in cellular lysates were evaluated by Western blotting with pooled HIV-1-infected donor serum, and supernatant p24 levels were determined. The membrane was also reblotted with antibodies to Myc (for caveolin) and α-tubulin (to confirm equivalent protein input). As shown in Fig. 5A, a severe defect in accumulation of HIV-1 proteins was seen within cells coexpressing Cav-1 compared to control cells cotransfected with pCMV-myc. All of the HIV-1 structural proteins were proportionately diminished, while α-tubulin levels were not. The extent of inhibition correlated with a 91% decrease in supernatant p24 production (Fig. 5A legend). To further assess the significance of the results, Western blotting with anti-Cav-1 antibodies was also performed to determine levels of expression in the transfected cells relative to endogenous expression in primary human cells that normally express Cav-1 (primary human vascular smooth muscle cells [VSMCs]) and in a cell line that is known to express some endogenous Cav-1 but is permissive for robust HIV-1 replication (HeLa cells). As shown in Fig. 5B, Cav-1 expression is ample in both primary human VSMCs and transfected 293T cells and is severalfold higher in the latter. Therefore, the inhibitory effect in transiently transfected cells results from relative overexpression of Cav-1. Notably, HIV-1-permissive HeLa cells express very little endogenous Cav-1 compared to the VSMCs.
FIG. 5.
(A) Cav-1 inhibits intracellular HIV-1 structural protein accumulation. 293T cells were cotransfected with 1.2 μg of pYU-2 and 0.3 μg of pCMV-myc or pCMV-myc.Cav-1. HIV protein expression was determined 40 h later in cellular lysates by immunoblotting with pooled HIV-1+ human serum. Equal amounts of protein (40 μg) were loaded in each lane. The same membrane was subsequently immunoblotted with an anti-Myc (for Cav-1) and an anti-α-tubulin MAb. HIV-1 p24 in the supernatants of these cells was <1 (mock), 685 ± 49 (pCMV-myc), and 63 ± 16 (pCMV-myc.Cav-1) ng/ml. (B) Cav-1 expression in the transfected 293T cells used in panel A relative to endogenous levels in other cells. The primary antibody is specific for residues 61 to 71 of Cav-1. Lane 1, pCMV-myc-transfected 293T cells (20 μg of protein); lane 2, HeLa cells (20 μg of protein); lanes 3 and 4, pCMV-myc.Cav-1-transfected 293T cells (20 and 0.8 μg of protein, respectively); lanes 5 and 6, primary human VSMCs (20 and 0.8 μg of protein, respectively). Prominent alpha and beta isoforms can be detected in primary human VSMCs and 293T cells, but only the beta isoform was detectable in HeLa cells; the bands in the 293T lanes are slower migrating because of the Myc tag. WB, Western blotting.
Although these data do not exclude additional effects on viral particle assembly or budding, they demonstrate that the major effect is inhibition of viral protein expression. To further evaluate specificity and to establish whether Cav-1-transfected 293T cells are metabolically healthy and able to support high-level viral protein synthesis, we analyzed the effect of Cav-1 on the expression of proteins by measles virus. We chose this negative-strand RNA virus for comparison because it has a completely cytoplasmic replication cycle, in which mRNAs are generated by a viral polymerase but are translated by the host cell translational apparatus. 293T cells were transfected with 0.6 μg of pCMV or three different amounts of pCMV.Cav-1 (0.1, 0.3, and 0.6 μg) and then infected with a recombinant measles virus that expresses eGFP (21). Twenty-four hours after infection, the cells were immunostained with an anti-Cav-1 MAb and analyzed by flow cytometry for both Cav-1 and eGFP expression. Of pCMV.Cav-1-transfected cells, 82 to 85% were Cav-1 positive (data not shown). The proportion of eGFP-positive cells did not differ significantly between Cav-1− and Cav-1+ cells. Nineteen percent of the control-transfected cells expressed eGFP, whereas 15.3, 17.3, and 18.3% of cells transfected with 0.1, 0.3, and 0.6 μg of pCMV.Cav-1, respectively, were eGFP positive. In addition, the intensity of eGFP expression was similar in both Cav-1+ and Cav-1− cells, and the proportion of double-positive eGFP-Cav-1 cells increased as more pCMV.Cav-1 was transfected (5.0, 9.2, and12.5%). The lack of inhibition of measles virus expression suggested that Cav-1-transfected cells are fully competent for mRNA translation and protein synthesis and provided additional evidence against nonspecific toxicity.
N-terminal regions of Cav-1 do not contribute to inhibition of HIV-1 expression.
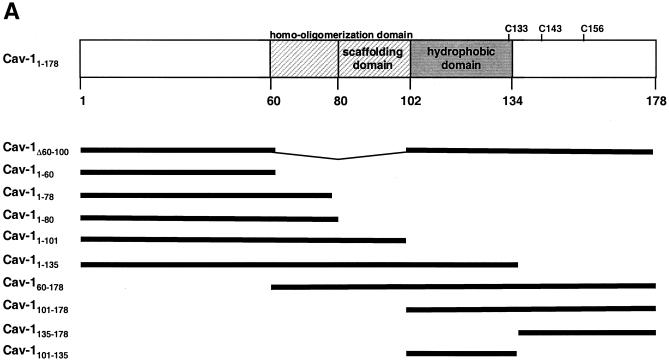
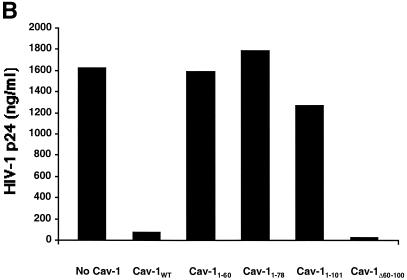
Regions in the amino-terminal half of Cav-1, upstream of the hydrophobic (membrane-anchoring) domain, have been shown by functional analyses of mutant proteins to contain domains involved in caveolar biogenesis and signal transduction (67). In the present study, we derived multiple mutant forms of Cav-1 to map domains involved in inhibiting HIV protein expression (Fig. 6A ). Equivalent expression of each of these Myc-tagged wild-type and mutant caveolins was verified by immunoblotting in cellular lysates of the cells corresponding to the supernatants for which p24 levels are represented in Fig. 6B to D. In all experiments, the same Western blot was also subsequently stripped and reprobed with anti-α-tubulin MAb, which verified both equivalent loading and lack of any effect of Cav-1 or any Cav-1 mutants on cellular α-tubulin expression (data not shown). As an initial step, we found that a Cav-1 mutant protein lacking the N-terminal oligomerization and scaffolding domains (Cav-1Δ60-100) (35) blocked HIV expression to the same extent as the wild-type protein (Fig. 6B). To further evaluate the role of N-terminal functional domains, we constructed and tested a panel of Myc epitope-tagged proteins containing only N-terminal residues 1 to 101 (Cav-11-101), 1 to 78 (Cav-11-78), or 1 to 60 (Cav-11-60). These proteins did not inhibit HIV expression, while Myc-tagged Cav-11-178 (full length) showed full activity (Fig. 6B). Immunoblotting with the cells expressing Cav-11-178 and Cav-11-101, the largest N-terminal fragment that did not inhibit, demonstrated that the lack of inhibition was not due to less mutant protein expression compared to wild type (data not shown). These results suggested that the N-terminal region of the protein, which includes the oligomerization and scaffolding domains, does not play a significant role in the ability of Cav-1 to block HIV expression.
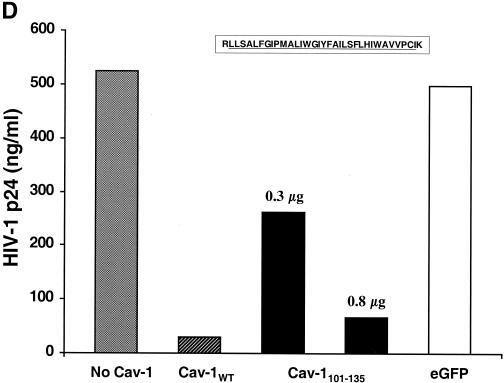
FIG. 6.
Mapping the Cav-1 effect by deletion mutagenesis. Shown are the structures of deletion mutants and effects of amino-terminal and hydrophobic domain deletions. (A) Structures of all Cav-1 deletion mutants used in the present study. Locations of palmitoylated cysteines mutated to alanine in the full-length protein are also shown (latter results described in text). (B) Cav-1 proteins lacking hydrophobic and carboxy-terminal regions do not inhibit HIV expression. (C) Amino-terminal Cav-1 residues proximal to the hydrophobic domain are dispensable. (D) The Cav-1 membrane-anchoring domain is necessary and sufficient to inhibit HIV expression. Residues 101 to 135 of Cav-1 are shown at the top of the panel, and the hydrophobic, putative membrane domain (residues 102 to 134) is underlined. As an additional control, cells in the experiment for panel D were cotransfected with 0.3 μg of pCMV.eGFP and 1.2 μg of pYU-2 (white bar). In panels B to D, equivalent Cav-1 mutant expression was verified by immunoblotting cell lysates with an anti-Myc MAb, and equal loading was verified by reprobing blots with an anti-α-tubulin antibody (data not shown). Unless otherwise indicated (D), 293T cells were cotransfected with 1.2 μg of pYU-2 and 0.3 μg of pCMV-myc (bars labeled “No Cav-1”), pCMV-myc.Cav-1 (bars labeled “Cav-1WT”), or pCMV-expressed Myc-tagged deletion-truncation mutants. p24 production was quantified in the cell supernatants at 40 h.
C-terminal regions of Cav-1 are required for inhibition of HIV-1 expression, but palmitoylation is dispensable.
We next evaluated the effect of Cav-1 mutants containing various portions of the C terminus (Fig. 6C). 293T cells were cotransfected with 1.2 μg of pYU-2 plus either 0.3 μg of pCMV coding for all 178 amino acids of Cav-1 (i.e., pCMV-myc.Cav-1) or two different Myc-tagged mutants containing only residues 60 to 178 (Cav-160-178) or residues 101 to 178 (Cav-1101-178). Both of these mutants were able to block HIV-1 production comparably to the wild-type protein (Fig. 6A). Immunoblotting verified that expression of the smallest active C-terminal fragment (Cav-1101-178) was equivalent to wild type (data not shown). These results suggested that residues 101 to 178 contain an active determinant and again indicated the lack of a role for amino acids N-terminal to the central hydrophobic domain.
Cav-1 is palmitoylated at each of its three cysteine residues, which are located in the carboxy terminus at positions 133, 143, and 156 (19). While these posttranslational modifications are dispensable for localization to caveolae (19), the previously mentioned modulation of intracellular cholesterol trafficking by Cav-1 is dependent on C143 and C156 palmitoylation (73, 74). Membrane cholesterol depletion has been shown elsewhere to decrease the production of infectious HIV-1 (32, 36). We hypothesized that defects in Cav-1 palmitoylation that affect cholesterol binding or other palmitoylation-dependent processes might modulate the inhibitory effect on HIV expression. We constructed a comprehensive panel of palmitoylation-minus mutants by replacing cysteine 133, 143, or 156 (positions shown in Fig. 6A) with alanine by site-directed mutagenesis. All seven possible permutations were constructed (three single mutants, three double mutants, and the triple mutant), and 0.3 μg of each was cotransfected with 1.2 μg of HIV-1 proviral DNA into 293T cells. Supernatants of cells cotransfected with pCMV and YU-2 contained 1,609 ± 28 ng of p24/ml. In contrast, HIV-1 p24 production in 293T cells cotransfected with pCMV.Cav-1 or with each of the seven pCMV.Cav-1 palmitoylation mutants was inhibited 92 to 98%. The triple mutant (C133A + C143A + C156A) produced the greatest inhibition (98%; 34 ± 19 ng of p24/ml). These results indicated definitively that palmitoylation of these residues in Cav-1 is not required and suggested that the palmitoylation-dependent formation of a cholesterol transport complex (73) is also not involved.
The hydrophobic domain of Cav-1 is necessary and sufficient to block HIV expression.
Data obtained with deletion mutants (Fig. 6B and C) indicated that the C-terminal half of Cav-1 was responsible for inhibition of HIV-1 virion production. In addition, a Myc-tagged Cav-1 mutant containing only the C-terminal cytoplasmic domain (residues 135 to 178) was expressed at high levels in cotransfected cells but was unable to block HIV-1 expression (data not shown). These results indicated a role for the hydrophobic, membrane-anchoring domain (residues 101 to 135). We therefore constructed additional truncation mutants to evaluate the role of this domain. A mutant (Cav-11-135), which differs from the inactive Cav-11-101 in containing the hydrophobic domain of the protein, completely blocked virion production in cotransfected cells (>98% inhibition; 28 versus 1,880 ng of p24/ml). Cav-11-135 and the wild-type protein were expressed at similar levels as determined by immunoblotting (data not shown). Finally, we cotransfected cells with 1.2 μg of pYU-2 plus either a pCMV construct encoding a polypeptide consisting only of Myc-tagged Cav-1 residues 101 to 135 (pCMV-myc.Cav-1101-135) or pCMV.eGFP. Cav-1101-135 was able to inhibit HIV expression when 0.3 μg of the expression plasmid was transfected, and at a higher DNA input (0.8 μg, ratio to pYU-2 = 0.66), the blocking activity of Cav-1101-135 was similar to the effect observed with wild-type Cav-1 (Fig. 6D). Moreover, immunoblotting showed that the 0.8-μg input yielded approximately fourfold-lower expression levels of Cav-1101-135 than wild-type Cav-1 (data not shown). This difference in expression level could reflect the lower stability of Cav-1101-135. Taken together, these results demonstrated clearly that the region encompassed by residues 101 to 135 is principally responsible for the effect of Cav-1 on HIV expression.
Blockade of HIV-1 expression correlates with subcellular localization.
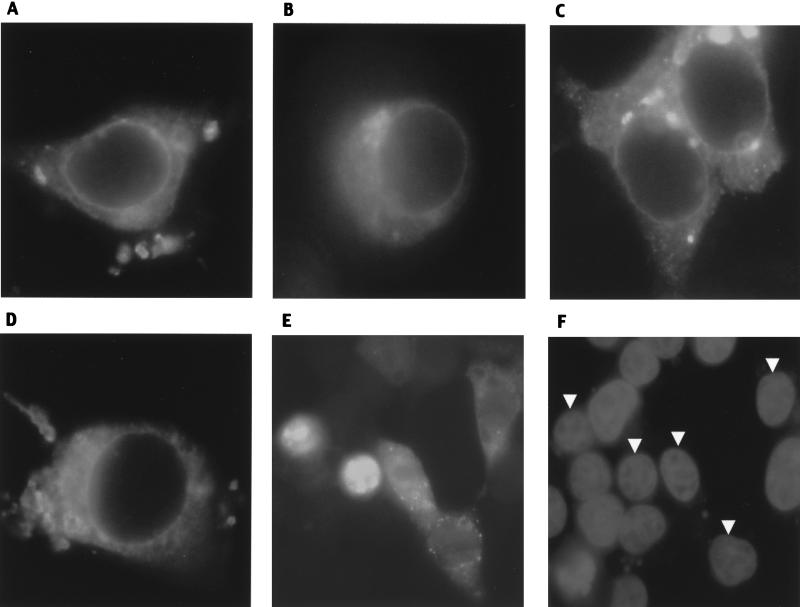
To investigate the subcellular distribution of wild-type and mutant Cav-1 proteins, 293T cells were transfected with Myc-tagged caveolins and analyzed by immunofluorescence with an anti-Myc antibody (Fig. 7). Wild-type Cav-1 (Cav-11-178) was found both at the plasma membrane and intracellularly (Fig. 7A). The same distribution of Cav-1 was seen without the Myc tag, by using an anti-Cav-1 MAb (data not shown). The cellular distribution of Cav-1 suggested localization in plasma membrane, ER, Golgi complex, and nuclear envelope. Similar intracellular distributions were noted with active Cav-11-135, Cav-1101-178, and Cav-1101-135, although these appeared more prominent in the nuclear envelope and Golgi complex (Fig. 7B to D). The similarly active Cav-1Δ60-101 also had a distribution identical to that of Cav-1101-135 (data not shown). All proteins containing the Cav-1 hydrophobic domain—including the mutant containing only this domain (Cav-1101-135)—showed apparent concentration in the nuclear envelope. In contrast, Myc-tagged Cav-11-60, Cav-11-80, Cav-11-101, and Cav-1135-178, which lack the hydrophobic domain and do not block HIV expression, had strikingly different patterns (Fig. 7E and F; also data not shown). No cells transfected with these four mutants showed the discrete nuclear envelope staining seen with variants that contain the hydrophobic domain. Cav-11-101 and Cav-11-80 were localized only to the nucleus in approximately 80% of the transfected cells, while in about 15% both nuclear and cytoplasmic localization occurred. Cav-11-60 and Cav-1135-178 showed diffuse cytoplasmic staining and no nuclear staining (data not shown). Cav-11-101 and Cav-1101-178 both showed punctate cytoplasmic staining that was not seen with Cav-11-135, Cav-1101-135, or wild-type Cav-1.
FIG. 7.
Immunofluorescence of Myc-tagged caveolins in 293T cells. (A) Wild type (Cav-11-178); (B) Cav-11-135; (C) Cav-1101-178; (D) Cav-1101-135; (E and F) Cav-11-101. Panel F shows DAPI (nuclear) staining of cells in panel E, and arrowheads indicate nuclei of transfected cells. Of note, most Cav-11-101-transfected cells (80%) showed the nuclear localization found in the two leftmost cells in panel E; this field, in which a lower percentage has nuclear localization, was selected to show representative examples of the remaining cells in which the protein is also seen in the cytoplasm.
Cav-2 also inhibits HIV virion production.
Cav-1 is one of three known members of the caveolin gene family. Cav-1 and Cav-2 are widely coexpressed in different cell types and form hetero-oligomers, while Cav-3 is expressed principally in muscle. The human Cav-1 and Cav-2 hydrophobic domains exhibit high homology (83%, with 47% amino acid identity), and are also highly conserved from invertebrates to mammals (72). When 293T cells were cotransfected with 1.2 μg of pYU-2 and 0.3 μg of pCMV.Cav-2 or pCMV.Cav-1, supernatant p24 levels were 128 and 70 ng/ml, respectively, compared to 1,627 ng/ml in control cells cotransfected with 0.3 μg of pCMV-myc. In combination with the Cav-1 deletion mapping, the finding that Cav-2 blocked HIV p24 production as potently as Cav-1 suggests that residues conserved between the hydrophobic domains of these isoforms should be considered in future investigations of the molecular basis of the inhibition.
DISCUSSION
This investigation began with the observation that cotransfection of a Cav-1 cDNA potently blocked HIV-1 production from cells that are used routinely to produce high levels of genetically defined HIV-1. Although our data do not exclude effects of this membrane raft-organizing protein on HIV-1 assembly or budding, inhibition of HIV protein expression in cells transfected with Cav-1 correlated directly with the inhibitory effects on p24 and RT production, indicating that the principal block is earlier in the life cycle, at the level of accumulation of viral proteins. This inhibition was eliminated by a frameshift in the Cav-1 mRNA, indicating that protein expression is required and is specific, since cotransfection and equivalent expression of several Cav-1 mutants, as well as control proteins (eGFP and luciferase), in these cells do not block HIV-1 expression. Several lines of evidence also indicate that Cav-1 did not produce nonspecific impairment of cellular function. Appearance, morphology, adherence, cell density, protein content, frequency of cell death (programmed and unprogrammed), and cell cycle progression were unaffected. Furthermore, the Cav-1 inhibitory effect was saturable and potent (reproducibly >90% at low specific DNA/input DNA ratios) and was observed with different Cav-1 expression plasmids.
HIV-1 can replicate efficiently in cells that naturally express Cav-1 (e.g., HeLa cells engineered to express CD4). In contrast to the Cav-1-negative cells studied here, there may be compensatory mechanisms in cells that naturally express Cav-1 that counteract an inhibitory effect on HIV-1. In addition, using very sensitive Western blotting, we found that HeLa cells express very little Cav-1 (Fig. 5B). Although transfected 293T cells moderately overexpress Cav-1 compared to endogenous levels in primary human VSMCs, we have recently found that expression of Cav-1 at physiological levels in 293 cells with a tetracycline-inducible stable cell line also markedly inhibits HIV-1 expression (M. Llano, unpublished data). That cellular adaptation and developmental context are important is supported by Cav-1-knockout mice, which develop to adulthood with relatively mild phenotypes in the total absence of the protein (20, 51); potentially informative conditional knockouts that turn off Cav-1 expression after development have not yet been reported.
HIV-2 was less completely inhibited than HIV-1, and measles virus protein expression was not inhibited at all, further arguing against nonspecific impairment of cellular protein expression. A notable difference between HIV and measles virus is that the latter has a completely cytoplasmic life cycle. Measles virus expression does not depend on nuclear transcription, mRNA splicing or processing, or nuclear export of mRNA, since the negative-strand RNA genome is copied to mRNA by a viral polymerase in the cytoplasm of the infected cells. However, this polycistronic mRNA is then translated by the host protein synthesis machinery. The failure of Cav-1 transfection to affect expression of eGFP by measles virus suggests that actions at the levels of mRNA translation or protein stability are unlikely. Prior to mRNA translation, expression of HIV-1 structural proteins requires a coordinated sequence of events that includes basal transcription and expression of the viral transactivator (Tat), augmentation of transcriptional initiation and/or elongation by Tat acting in concert with cellular factors, production of Rev from initial multiply spliced mRNAs, and Rev-dependent nuclear export of singly spliced and unspliced mRNAs. Complete molecular understanding of the inhibition of HIV-1 expression that we have identified will require additional investigations of these specific components of the life cycle. Given the role of Cav-1 in modulating signal transduction, a primary effect on signaling pathways that modulate HIV-1 expression could be involved. If so, however, this effect is unlikely to be occurring through the sequestration of such molecules in plasmalemma caveolae, since several active Cav-1 mutants, including Cav-11-135, Cav-1101-178, and Cav-1101-135, are defective for transport to the plasma membrane and do not induce caveolae (35) (Fig. 7). Moreover, the scaffolding domain (residues 80 to 101) is entirely absent from a number of active deletion mutants (Cav-1Δ60-100, Cav-1101-178, and Cav-1101-135), and most known interactions between Cav-1 and signaling proteins have been mapped to this domain (43). In addition, the effect observed here appears to be distinguishable from the previously described modulation of intracellular cholesterol trafficking by Cav-1, which is dependent on C143 and C156 palmitoylation (73, 74). Mutants defective for palmitoylation were able to block HIV expression to the same extent as wild-type Cav-1.
The 33-amino-acid hydrophobic region located at residues 102 to 134 was shown by the multiple overlapping deletion mutants to be the principal domain responsible for inhibition of HIV-1 expression. In addition, expression of residues 101 to 135 alone caused the effect. This region is highly homologous between Cav-1 and Cav-2 and is strongly conserved from invertebrates to mammals (72). Few functions have been attributed directly to this domain of Cav-1, except for membrane insertion and for hetero-oligomerization with Cav-2 (17). Indeed, hetero-oligomerization with Cav-2 is the only protein-protein interaction reported for the hydrophobic domain. Our immunofluorescence results in 293T cells are in general consistent with those obtained by others in CHO cells (35). Interestingly, we found that the active mutants and wild-type Cav-1 appear to be present in cytoplasmic locations suggestive of ER and Golgi complex and in the nuclear envelope. The latter location may be relevant to the mechanism of inhibition. The hydrophobicity and size of Cav-1101-135 suggest that this polypeptide should be almost completely embedded in the membranes of these organelles. Nucleocytoplasmic transport of HIV-1 mRNA or another nuclear process may be affected when proteins containing residues 101 to 135 are overexpressed. Such a mechanism would be consistent with the lack of HIV-inhibiting activity of Cav-11-60, Cav-11-80, Cav-11-101, and Cav-1135-178, which did not show discrete nuclear envelope staining. Transfection of Cav-1 may cause the blockade of HIV-1 expression that we have identified here by sequestering a viral or cellular protein required for HIV mRNA synthesis or processing or by disrupting nuclear envelope function. In summary, our data show that caveolins can block HIV expression and identify a previously undisclosed activity of the topologically distinctive membrane-associated domain of this protein family.
Acknowledgments
This work was supported by NIH grant AI47536.
We thank Roberto Cattaneo for providing measles-eGFP virus, laboratory members for helpful scientific discussions, and R. Anderson for two plasmids (Cav-160-178 and Cav-1Δ60-100).
REFERENCES
- 1.Ali, A., R. T. Avalos, E. Ponimaskin, and D. P. Nayak. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aloia, R. C., H. Tian, and F. C. Jensen. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA 90:5181-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, H. A., Y. Chen, and L. C. Norkin. 1996. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol. Biol. Cell 7:1825-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, R. G., B. A. Kamen, K. G. Rothberg, and S. W. Lacey. 1992. Potocytosis: sequestration and transport of small molecules by caveolae. Science 255:410-411. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa, R., S. Abe-Dohmae, M. Asai, J. I. Ito, and S. Yokoyama. 2000. Involvement of caveolin-1 in cholesterol enrichment of high density lipoprotein during its assembly by apolipoprotein and THP-1 cells. J. Lipid Res. 41:1952-1962. [PubMed] [Google Scholar]
- 6.Baorto, D. M., Z. Gao, R. Malaviya, M. L. Dustin, A. van der Merwe, D. M. Lublin, and S. N. Abraham. 1997. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature 389:636-639. [DOI] [PubMed] [Google Scholar]
- 7.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruns, R. R., and G. E. Palade. 1968. Studies on blood capillaries. I. General organization of blood capillaries in muscle. J. Cell Biol. 37:244-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron, P. L., J. W. Ruffin, R. Bollag, H. Rasmussen, and R. S. Cameron. 1997. Identification of caveolin and caveolin-related proteins in the brain. J. Neurosci. 17:9520-9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carman, C. V., M. P. Lisanti, and J. L. Benovic. 1999. Regulation of G protein-coupled receptor kinases by caveolin. J. Biol. Chem. 274:8858-8864. [DOI] [PubMed] [Google Scholar]
- 12.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, B. K., K. Saksela, R. Andino, and D. Baltimore. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 68:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi, D., J. Henry, J. Kelley, R. Thorpe, J. K. Smith, and G. Krishnaswamy. 2000. The effects of HIV infection on endothelial function. Endothelium 7:223-242. [DOI] [PubMed] [Google Scholar]
- 15.Conrad, P. A., E. J. Smart, Y. S. Ying, R. G. Anderson, and G. S. Bloom. 1995. Caveolin cycles between plasma membrane caveolae and the Golgi complex by microtubule-dependent and microtubule-independent steps. J. Cell Biol. 131:1421-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darzynkiewicz, Z., E. Bedner, X. Li, W. Gorczyca, and M. R. Melamed. 1999. Laser-scanning cytometry: a new instrumentation with many applications. Exp. Cell Res. 249:1-12. [DOI] [PubMed] [Google Scholar]
- 17.Das, K., R. Y. Lewis, P. E. Scherer, and M. P. Lisanti. 1999. The membrane-spanning domains of caveolins-1 and -2 mediate the formation of caveolin hetero-oligomers. Implications for the assembly of caveolae membranes in vivo. J. Biol. Chem. 274:18721-18728. [DOI] [PubMed] [Google Scholar]
- 18.de Villiers, W. J., and E. J. Smart. 1999. Macrophage scavenger receptors and foam cell formation. J. Leukoc. Biol. 66:740-746. [DOI] [PubMed] [Google Scholar]
- 19.Dietzen, D. J., W. R. Hastings, and D. M. Lublin. 1995. Caveolin is palmitoylated on multiple cysteine residues. Palmitoylation is not necessary for localization of caveolin to caveolae. J. Biol. Chem. 270:6838-6842. [DOI] [PubMed] [Google Scholar]
- 20.Drab, M., P. Verkade, M. Elger, M. Kasper, M. Lohn, B. Lauterbach, J. Menne, C. Lindschau, F. Mende, F. C. Luft, A. Schedl, H. Haller, and T. V. Kurzchalia. 2001. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science 293:2449-2452. [DOI] [PubMed] [Google Scholar]
- 21.Duprex, W. P., S. McQuaid, B. Roscic-Mrkic, R. Cattaneo, C. McCallister, and B. K. Rima. 2000. In vitro and in vivo infection of neural cells by a recombinant measles virus expressing enhanced green fluorescent protein. J. Virol. 74:7972-7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fra, A. M., E. Williamson, K. Simons, and R. G. Parton. 1995. De novo formation of caveolae in lymphocytes by expression of VIP21-caveolin. Proc. Natl. Acad. Sci. USA 92:8655-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gargalovic, P., and L. Dory. 2001. Caveolin-1 and caveolin-2 expression in mouse macrophages. High density lipoprotein 3-stimulated secretion and a lack of significant subcellular co-localization. J. Biol. Chem. 276:26164-26170. [DOI] [PubMed] [Google Scholar]
- 24.Glenney, J. R., Jr. 1989. Tyrosine phosphorylation of a 22-kDa protein is correlated with transformation by Rous sarcoma virus. J. Biol. Chem. 264:20163-20166. [PubMed] [Google Scholar]
- 25.Hatanaka, M., T. Maeda, T. Ikemoto, H. Mori, T. Seya, and A. Shimizu. 1998. Expression of caveolin-1 in human T cell leukemia cell lines. Biochem. Biophys. Res. Commun. 253:382-387. [DOI] [PubMed] [Google Scholar]
- 26.Ikonen, E., and R. G. Parton. 2000. Caveolins and cellular cholesterol balance. Traffic 1:212-217. [DOI] [PubMed] [Google Scholar]
- 27.Kundu, A., R. T. Avalos, C. M. Sanderson, and D. P. Nayak. 1996. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J. Virol. 70:6508-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurzchalia, T. V., P. Dupree, R. G. Parton, R. Kellner, H. Virta, M. Lehnert, and K. Simons. 1992. VIP21, a 21-kD membrane protein is an integral component of trans-Golgi-network-derived transport vesicles. J. Cell Biol. 118:1003-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurzchalia, T. V., and R. G. Parton. 1999. Membrane microdomains and caveolae. Curr. Opin. Cell Biol. 11:424-431. [DOI] [PubMed] [Google Scholar]
- 30.Lei, M. G., and D. C. Morrison. 2000. Differential expression of caveolin-1 in lipopolysaccharide-activated murine macrophages. Infect. Immun. 68:5084-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao, Z., L. M. Cimakasky, R. Hampton, D. H. Nguyen, and J. E. Hildreth. 2001. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retrovir. 17:1009-1019. [DOI] [PubMed] [Google Scholar]
- 33.Lisanti, M. P., Z. Tang, P. E. Scherer, E. Kubler, A. J. Koleske, and M. Sargiacomo. 1995. Caveolae, transmembrane signalling and cellular transformation. Mol. Membr. Biol. 12:121-124. [DOI] [PubMed] [Google Scholar]
- 34.Lu, Y. E., and M. Kielian. 2000. Semliki Forest virus budding: assay, mechanisms, and cholesterol requirement. J. Virol. 74:7708-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machleidt, T., W. P. Li, P. Liu, and R. G. Anderson. 2000. Multiple domains in caveolin-1 control its intracellular traffic. J. Cell Biol. 148:17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manes, S., G. del Real, R. A. Lacalle, P. Lucas, C. Gomez-Mouton, S. Sanchez-Palomino, R. Delgado, J. Alcami, E. Mira, and A. C. Martinez. 2000. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 1:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marone, G., G. Florio, A. Petraroli, M. Triggiani, and A. de Paulis. 2001. Human mast cells and basophils in HIV-1 infection. Trends Immunol. 22:229-232. [DOI] [PubMed] [Google Scholar]
- 39.Matveev, S., D. R. van der Westhuyzen, and E. J. Smart. 1999. Co-expression of scavenger receptor-BI and caveolin-1 is associated with enhanced selective cholesteryl ester uptake in THP-1 macrophages. J. Lipid Res. 40:1647-1654. [PubMed] [Google Scholar]
- 40.Mulvey, M. A., and S. J. Hultgren. 2000. Cell biology. Bacterial spelunkers. Science 289:732-733. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norkin, L. C., S. A. Wolfrom, and E. S. Stuart. 2001. Association of caveolin with Chlamydia trachomatis inclusions at early and late stages of infection. Exp. Cell Res. 266:229-238. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto, T., A. Schlegel, P. E. Scherer, and M. P. Lisanti. 1998. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J. Biol. Chem. 273:5419-5422. [DOI] [PubMed] [Google Scholar]
- 44.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parton, R. G. 2001. Cell biology. Life without caveolae. Science 293:2404-2405. [DOI] [PubMed] [Google Scholar]
- 46.Parton, R. G., and M. Lindsay. 1999. Exploitation of major histocompatibility complex class I molecules and caveolae by simian virus 40. Immunol. Rev. 168:23-31. [DOI] [PubMed] [Google Scholar]
- 47.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pirounaki, M., N. A. Heyden, M. Arens, and L. Ratner. 2000. Rapid phenotypic drug susceptibility assay for HIV-1 with a CCR5 expressing indicator cell line. J. Virol. Methods 85:151-161. [DOI] [PubMed] [Google Scholar]
- 49.Poeschla, E., J. Gilbert, X. Li, S. Huang, A. Ho, and F. Wong-Staal. 1998. Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. J. Virol. 72:6527-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poeschla, E., F. Wong-Staal, and D. Looney. 1998. Efficient transduction of nondividing cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 51.Razani, B., J. A. Engelman, X. B. Wang, W. Schubert, X. L. Zhang, C. B. Marks, F. Macaluso, R. G. Russell, M. Li, R. G. Pestell, D. Di Vizio, H. Hou, Jr., B. Kneitz, G. Lagaud, G. J. Christ, W. Edelmann, and M. P. Lisanti. 2001. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J. Biol. Chem. 276:38121-38138. [DOI] [PubMed] [Google Scholar]
- 52.Ross, T. M., A. E. Oran, and B. R. Cullen. 1999. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr. Biol. 9:613-621. [DOI] [PubMed] [Google Scholar]
- 53.Rothberg, K. G., J. E. Heuser, W. C. Donzell, Y. S. Ying, J. R. Glenney, and R. G. Anderson. 1992. Caveolin, a protein component of caveolae membrane coats. Cell 68:673-682. [DOI] [PubMed] [Google Scholar]
- 54.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 97:13523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanderson, C. M., R. Avalos, A. Kundu, and D. P. Nayak. 1995. Interaction of Sendai viral F, HN, and M proteins with host cytoskeletal and lipid components in Sendai virus-infected BHK cells. Virology 209:701-707. [DOI] [PubMed] [Google Scholar]
- 56.Sargiacomo, M., P. E. Scherer, Z. Tang, E. Kubler, K. S. Song, M. C. Sanders, and M. P. Lisanti. 1995. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc. Natl. Acad. Sci. USA 92:9407-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 58.Schlegel, A., D. Volonte, J. A. Engelman, F. Galbiati, P. Mehta, X. L. Zhang, P. E. Scherer, and M. P. Lisanti. 1998. Crowded little caves: structure and function of caveolae. Cell. Signal. 10:457-463. [DOI] [PubMed] [Google Scholar]
- 59.Schnitzer, J. E., P. Oh, and D. P. McIntosh. 1996. Role of GTP hydrolysis in fission of caveolae directly from plasma membranes. Science 274:239-242. [DOI] [PubMed] [Google Scholar]
- 60.Shaul, P. W., and R. G. Anderson. 1998. Role of plasmalemmal caveolae in signal transduction. Am. J. Physiol. 275:L843-L851. [DOI] [PubMed] [Google Scholar]
- 61.Shin, J. S., and S. N. Abraham. 2001. Co-option of endocytic functions of cellular caveolae by pathogens. Immunology 102:2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shin, J. S., Z. Gao, and S. N. Abraham. 2000. Involvement of cellular caveolae in bacterial entry into mast cells. Science 289:785-788. [DOI] [PubMed] [Google Scholar]
- 63.Simari, R. D., H. San, M. Rekhter, T. Ohno, D. Gordon, G. J. Nabel, and E. G. Nabel. 1996. Regulation of cellular proliferation and intimal formation following balloon injury in atherosclerotic rabbit arteries. J. Clin. Investig. 98:225-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 65.Simons, K., and G. van Meer. 1988. Lipid sorting in epithelial cells. Biochemistry 27:6197-6202. [DOI] [PubMed] [Google Scholar]
- 66.Skibbens, J. E., M. G. Roth, and K. S. Matlin. 1989. Differential extractability of influenza virus hemagglutinin during intracellular transport in polarized epithelial cells and nonpolar fibroblasts. J. Cell Biol. 108:821-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smart, E. J., G. A. Graf, M. A. McNiven, W. C. Sessa, J. A. Engelman, P. E. Scherer, T. Okamoto, and M. P. Lisanti. 1999. Caveolins, liquid-ordered domains, and signal transduction. Mol. Cell. Biol. 19:7289-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smart, E. J., Y. Ying, W. C. Donzell, and R. G. Anderson. 1996. A role for caveolin in transport of cholesterol from endoplasmic reticulum to plasma membrane. J. Biol. Chem. 271:29427-29435. [DOI] [PubMed] [Google Scholar]
- 69.Smart, E. J., Y. S. Ying, P. A. Conrad, and R. G. Anderson. 1994. Caveolin moves from caveolae to the Golgi apparatus in response to cholesterol oxidation. J. Cell Biol. 127:1185-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song, K. S., S. Li, T. Okamoto, L. A. Quilliam, M. Sargiacomo, and M. P. Lisanti. 1996. Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent-free purification of caveolae microdomains. J. Biol. Chem. 271:9690-9697. [DOI] [PubMed] [Google Scholar]
- 71.Stephan, D. J., Z. Y. Yang, H. San, R. D. Simari, C. J. Wheeler, P. L. Felgner, D. Gordon, G. J. Nabel, and E. G. Nabel. 1996. A new cationic liposome DNA complex enhances the efficiency of arterial gene transfer in vivo. Hum. Gene Ther. 7:1803-1812. [DOI] [PubMed] [Google Scholar]
- 72.Tang, Z., T. Okamoto, P. Boontrakulpoontawee, T. Katada, A. J. Otsuka, and M. P. Lisanti. 1997. Identification, sequence, and expression of an invertebrate caveolin gene family from the nematode Caenorhabditis elegans. Implications for the molecular evolution of mammalian caveolin genes. J. Biol. Chem. 272:2437-2445. [DOI] [PubMed] [Google Scholar]
- 73.Uittenbogaard, A., and E. J. Smart. 2000. Palmitoylation of caveolin-1 is required for cholesterol binding, chaperone complex formation, and rapid transport of cholesterol to caveolae. J. Biol. Chem. 275:25595-25599. [DOI] [PubMed] [Google Scholar]
- 74.Uittenbogaard, A., Y. Ying, and E. J. Smart. 1998. Characterization of a cytosolic heat-shock protein-caveolin chaperone complex. Involvement in cholesterol trafficking. J. Biol. Chem. 273:6525-6532. [DOI] [PubMed] [Google Scholar]
- 75.van Meer, G. 2001. Caveolin, cholesterol, and lipid droplets? J. Cell Biol. 152:F29-F34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vincent, S., D. Gerlier, and S. N. Manie. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]