Abstract
Hepatitis A virus (HAV) is a positive-stranded RNA virus in the genus Hepatovirus in the family Picornaviridae. So far, analysis of the genetic variability of HAV has been based on two discrete regions, the VP1/2A junction and the VP1 N terminus. In this report, we determined the nucleotide and deduced amino acid sequences of the complete VP1 gene of 81 strains from France, Kosovo, Mexico, Argentina, Chile, and Uruguay and compared them with the sequences of seven strains of HAV isolated elsewhere. Overall strain variation in the complete VP1 gene was found to be as high as 23.7% at the nucleotide level and 10.5% at the amino acid level. Different phylogenetic methods revealed that HAV sequences form five distinct and well-supported genetic lineages. Within these lineages, HAV sequences clustered by geographical origin only for European strains. The analysis of the complete VP1 gene allowed insight into the mode of evolution of HAV and revealed the emergence of a novel variant with a 15-amino-acid deletion located on the VP1 region where neutralization escape mutations were found. This could be the first antigenic variant of HAV so far identified.
Human hepatitis A virus (HAV) is a hepatotropic member of the Picornaviridae family (31, 33). The clinical manifestations of HAV infection in humans can vary greatly, ranging from asymptomatic infection, commonly seen in young children, to fulminant hepatitis, which in some cases can result in death (51). HAV is transmitted primarily by the fecal-oral route, and epidemics are common in regions where sanitation is poor.
The virion is nonenveloped with a capsid composed of three main polypeptides (VP1, VP2, and VP3). There is only one serotype of HAV identified thus far, and the only known antigenic variants are HAV strains collected from Old World monkeys (37, 58). Monoclonal antibody studies suggest that there are a limited number of antigenic epitopes which are closely grouped at the surface of the virus (42, 43, 54).
Genetic variants of HAV have been identified by sequencing of selected genome regions, including the VP3 C terminus (24), the VP1 amino terminus (2, 3, 7, 12, 14, 16, 47, 50), and the VP1/2A junction (5, 7, 8, 12, 15, 17, 24, 39, 41, 47, 49, 56).
These three regions, i.e., the VP3 C terminus, the VP1 amino terminus, and the VP1/2A junction, present some genetic differences. The VP3 C-terminal region is relatively conserved, while the VP1/2A junction is more variable and can be used to distinguish one strain from another (46, 47, 49, 50). The VP1 amino terminus presents an intermediate variability between the two regions cited above. On the basis of the genetic variability observed within the putative VP1/2A junction, seven HAV genotypes have been identified (49). However, an extensive study of South American HAV strains revealed that the VP1 amino terminus contains more-informative variable positions than the VP1/2A junction (12).
To gain insight into the molecular epidemiology of HAV, we investigated the genetic variability of HAV strains recovered from different countries using the complete sequences of the VP1 gene. The sequences of 81 HAV isolates from France, Kosovo, Mexico, and three other South American countries have been determined. The analysis of these sequences allowed us to estimate the mode of HAV evolution, to determine the multiple genotypes cocirculating during epidemic outbreaks, and to document the emergence of a novel variant which could not be detected using either the VP1 amino-terminal region or the VP1/2A junction.
MATERIALS AND METHODS
Virus strains.
HAV strains were collected from different geographical areas. The source, origin, year of isolation, and epidemiological data from these and other HAV strains whose sequences were taken from GenBank and used in our analysis are listed in Table 1. The French isolates were obtained from an epidemic outbreak associated with shellfish consumption which occurred north of Bretagne during the winter of 1999 (14); these isolates were taken from sludge samples obtained from a wastewater treatment plant in Nantes (France) and from a virus collection of the Unité de Virologie at the Centre Hôspitalier Universitaire, Nantes, France, from 1983 to 2001. Seven serum samples from patients with HAV during an epidemic outbreak during the Kosovo war (1998 to 1999) were collected and processed at the Hôpital Val de GrÂce, Paris, France.
TABLE 1.
HAV strains examined in this study
| Strain | Yr | Countrya | Specimen | Epidemiol- ogical datab | GenBank accession no. |
|---|---|---|---|---|---|
| Arg-1 | 1999 | Argentina | Stool | Outbreak | AJ437151 |
| Arg-5 | 1999 | Argentina | Serum | Outbreak | AJ437152 |
| Arg-6 | 1999 | Argentina | Stool | Outbreak | AJ437153 |
| Arg-8 | 2000 | Argentina | Stool | Outbreak | AJ437154 |
| Arg-11 | 1999 | Argentina | Stool | Outbreak | AJ437155 |
| Arg-15 | 2000 | Argentina | Serum | Outbreak | AJ437156 |
| Arg-23 | 1999 | Argentina | Serum | Outbreak | AJ437157 |
| Arg-27 | 1999 | Argentina | Serum | Outbreak | AJ437158 |
| Arg-28 | 2000 | Argentina | Serum | Outbreak | AJ437159 |
| Chile-5 | 1999 | Chile | Stool | Outbreak | AJ437160 |
| Chile-6 | 1999 | Chile | Stool | Outbreak | AJ437161 |
| Chile-9 | 1999 | Chile | Stool | Outbreak | AJ437163 |
| Chile-J | 1999 | Chile | Stool | Outbreak | AJ437164 |
| Chato-Mex | 2000 | Mexico (France)c | Stool | Sporadic | AJ437179 |
| Uru-1 | 1999 | Uruguay | Serum | Outbreak | AJ437165 |
| Uru-3 | 1999 | Uruguay | Serum | Outbreak | AJ437166 |
| Uru-4 | 1999 | Uruguay | Serum | Outbreak | AJ437167 |
| Uru-5 | 1999 | Uruguay | Serum | Outbreak | AJ437168 |
| Uru-6 | 1999 | Uruguay | Serum | Outbreak | AJ437169 |
| Uru-8 | 1999 | Uruguay | Serum | Outbreak | AJ437170 |
| Uru-9 | 1999 | Uruguay | Serum | Outbreak | AJ437171 |
| Uru-12 | 1999 | Uruguay | Serum | Outbreak | AJ437172 |
| Uru-13 | 1999 | Uruguay | Serum | Outbreak | AJ437173 |
| Uru-14 | 1999 | Uruguay | Serum | Outbreak | AJ437174 |
| Uru-17 | 1999 | Uruguay | Serum | Outbreak | AJ437175 |
| Uru-20 | 2000 | Uruguay | Serum | Outbreak | AJ437176 |
| Uru-21 | 2000 | Uruguay | Serum | Outbreak | AJ437177 |
| UruK | 1999 | Uruguay | Serum | Outbreak | AJ437178 |
| 1K | 1998 | Kosovo | Serum | Outbreak | AJ437180 |
| 2K | 1998 | Kosovo | Serum | Outbreak | AJ437181 |
| 3K | 1998 | Kosovo | Serum | Outbreak | AJ437182 |
| 7K | 1998 | Kosovo | Serum | Outbreak | AJ437183 |
| 8K | 1998 | Kosovo | Serum | Outbreak | AJ437184 |
| 9F83 | 1983 | France | Serum | Sporadic | AJ437252 |
| 2F84 | 1984 | France | Serum | Sporadic | AJ437244 |
| 19F85 | 1985 | France | Serum | Sporadic | AJ437240 |
| 20F85 | 1985 | France | Serum | Sporadic | AJ437241 |
| 22F85 | 1985 | France | Serum | Sporadic | AJ437242 |
| YF87 | 1987 | France | Serum | Sporadic | AJ437243 |
| 9F88 | 1988 | France | Serum | Sporadic | AJ437185 |
| 13F89 | 1989 | France | Serum | Sporadic | AJ437245 |
| 23F90 | 1990 | France | Serum | Sporadic | AJ437253 |
| UF93 | 1993 | France | Serum | Sporadic | AJ437186 |
| TF93 | 1993 | France | Serum | Sporadic | AJ437246 |
| WF93 | 1993 | France | Serum | Sporadic | AJ437254 |
| 14F94 | 1994 | France | Serum | Sporadic | AJ437247 |
| 9F94 | 1994 | France (Morocco) | Serum | Sporadic | AJ437317 |
| 10F94 | 1994 | France | Serum | Sporadic | AJ437255 |
| 12F94 | 1994 | France | Serum | Sporadic | AJ437248 |
| SF94 | 1994 | France | Serum | Sporadic | AJ437187 |
| 1F95 | 1995 | France | Serum | Sporadic | AJ437188 |
| 2F95 | 1995 | France | Serum | Sporadic | AJ437256 |
| 3F95 | 1995 | France | Serum | Sporadic | AJ437189 |
| 5F96 | 1996 | France | Serum | Sporadic | AJ437190 |
| FF97 | 1997 | France | Serum | Sporadic | AJ437249 |
| EF97 | 1997 | France | Serum | Sporadic | AJ437218 |
| CF97 | 1997 | France | Serum | Sporadic | AJ437257 |
| HF97 | 1997 | France | Serum | Sporadic | AJ438160 |
| GF97 | 1997 | France | Serum | Sporadic | AJ438161 |
| AF98 | 1998 | France | Serum | Sporadic | AJ438162 |
| CF98 | 1998 | France | Serum | Sporadic | AJ438163 |
| LF98 | 1998 | France | Serum | Sporadic | AJ437251 |
| P18-99 | 1999 | France | Serum | Outbreak | AJ438164 |
| P14-99 | 1999 | France | Serum | Outbreak | AJ438165 |
| P21-99 | 1999 | France | Serum | Outbreak | AJ438166 |
| P20-99 | 1999 | France | Serum | Outbreak | AJ438167 |
| P27-99 | 1999 | France | Serum | Outbreak | AJ437316 |
| 4F-99 | 1999 | France | Serum | Sporadic | AJ437228 |
| 6F99 | 1999 | France | Serum | Sporadic | AJ437229 |
| 7F99 | 1999 | France | Serum | Sporadic | AJ437250 |
| 10F99 | 1999 | France | Serum | Sporadic | AJ437230 |
| 11F99 | 1999 | France | Serum | Sporadic | AJ437231 |
| 6F-00 | 2000 | France | Serum | Sporadic | AJ437236 |
| 7F-00 | 2000 | France | Serum | Sporadic | AJ437314 |
| Boue2 | 2000 | France | Sludge | Sporadic | AJ437315 |
| Boue4 | 2000 | France | Sludge | Sporadic | AJ437232 |
| Boue6 | 2000 | France | Sludge | Sporadic | AJ437233 |
| Boue9 | 2000 | France | Sludge | Sporadic | AJ437234 |
| AF-01 | 2001 | France | Serum | Outbreak | AJ437237 |
| BF-01 | 2001 | France | Serum | Outbreak | AJ437238 |
| CF-01 | 2001 | France | Serum | Outbreak | AJ437239 |
| HM-175 | 1976 | Australia | Sporadic | M14707 | |
| AGM-27 | 1985 | Kenya (USSR) | Liver homogenate | Sporadic | D00926 |
| MBB | 1978 | N. Africa (Germany) | Sporadic | M20273 | |
| GA76 | 1976 | US | Sporadic | M66695 | |
| PA21 | 1980 | Panama | Sporadic | M34084 | |
| CY145 | 1988 | Philippines US | Liver homogenate | Sporadic | M59286 |
| SLF88 | 1988 | Sierra Leona | Liver homogenate | Sporadic | AY032861 |
The country or autonomous region where the strain was collected is shown. US, United States.
Strains were from outbreaks or sporadic cases of HAV infection.
This strain was isolated in France from a Mexican patient.
South American strains were collected from three countries. Fourteen HAV-positive serum samples were collected at Pereira Rossell Hospital and Asociacion Espanola Primera de Socorros Mutuos during an epidemic outbreak that occurred in Montevideo, Uruguay, from September 1999 to February 2000. Over the same period of time, four stool samples from Chilean patients were collected at Hospital Regional in Valdivia, and nine stool and serum samples from Argentinian patients were collected at Hospital San Juan de Dios in Buenos Aires (Table 1). In September 2000, stool samples were isolated in Nantes, France, from a recently arrived Mexican student.
RNA extraction and amplification.
Viral RNA was extracted from 140 μl of serum or 400 μl of a stool suspension (0.3 g of stool diluted in 2 ml of water) using the QIAmp viral extraction kit (Qiagen) or the RNeasy plant minikit (Qiagen), respectively. HAV RNA was extracted from sludge samples as previously described (35). Briefly, viral RNA was extracted from the samples, and the amount obtained was quantified by a real-time reverse transcriptase PCR (RT-PCR) method. Typically, there were approximately 6.38 × 105 copies of viral RNA/ml (13). The primers used to reverse transcription and sequencing are listed in Table 2. From each viral RNA, an amplicon, encompassing the 3′end of VP3, all of VP1, and the 5′ end of 2A was amplified by using primers HAV1 and HAV2 (Table 2) in a reaction driven by the SuperScript One-Step RT-PCR with Platinum Taq kit (Invitrogen). RT-PCR was performed with a one-tube reaction mixture (25 μl) containing 5 μl of viral RNA, 20 pmol of each primer, a 100 μM concentration of each deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 2 mM MgSO4, 20 IU of RNase inhibitor (Promega), and 1 μl of the RT/Platinum Taq Mix (Invitrogen). VP1-specific cDNA was synthesized by incubation of the reaction mixture for 45 min at 42°C and 5 min at 94°C, and it was amplified by 35 cycles, with 1 cycle consisting of 30 s at 94°C, 45 s at 50°C, and 1 min at 68°C. For some samples, the complete VP1 gene was amplified as two overlapping fragments with internal primers. The primers used either to amplify or to sequence the entire VP1 genes are listed in Table 2. The products were purified and sequenced using the Big Dye DNA sequencing kit (Perkin-Elmer) and a DNA sequencer (model 373; Perkin-Elmer). To distinguish natural heterogeneity from possible technical artifacts, both strands of each DNA fragment were sequenced. When discrepancies were observed, the procedure was repeated three times using a different set of primers (Table 2).
TABLE 2.
Oligonucleotide primers used in RT-PCR and sequencing
| Primer | Sequence | Positionsa |
|---|---|---|
| HAV1 | GTTTTGCTCCTCTTTATCATGCTATG | 2167-2192 |
| HAV2 | AGTCACACCTCTCCAGGAAAACTT | 3308-3285 |
| HAV3 | TTCATTATTTCATGCTCCTC | 3286-3267 |
| HAV4 | ACAAATAACAACTAAAAGACAA | 2955-2934 |
| HAV5 | TTGTCTTTTAGTTGTTATTTGTC | 2934-2956 |
| HAV6 | AGGAAATGTCTCAGGTACTTTCTTTG CTAAAACTG | 2414-2380 |
| HAV7 | CTGGAGTGAACCAGGCCATGCCATC | 2691-2667 |
| HAV8 | AGAGTCATATAGATTGCAGG | 3106-3125 |
| HAV9 | GAGCACTGGATGGTTTGGG | 2845-2863 |
| HAV10 | TTGTTCTTTAATTTCCTG | 3674-3657 |
Positions relative to the genome of HAV strain HM-175 (M14707).
Sequence analysis.
The entire VP1 nucleotide sequences were aligned using the CLUSTAL W program (57). Matrix distances for the Kimura two-parameter model were then generated (18) and used to compute neighbor-joining phylogenetic trees. The robustness of each node was assessed by bootstrap resampling (1,000 pseudoreplicates). These methods were implemented by using software from the MEGA program (28).
Substitution rate analysis.
The substitution rate along the VP1 gene was measured using a sliding window by the procedure of Alvarez-Valin et al. (1). Pairwise nucleotide distances (synonymous and nonsynonymous) within each window were estimated by the method of Comeron (11) as implemented in the computer program k estimator, where k is the number of nucleotide substitutions between sequences. For those windows where this method could not be applied, the Jukes-Cantor method (25) was used for correction for multiple hits. The window had a size of 30 codons and a movement of 3.
Nucleotide sequence accession numbers.
The complete VP1 sequences obtained from the 81 HAV strains were deposited in GenBank, and their accession numbers are shown in Table 1.
RESULTS
VP1 and picornavirus phylogeny.
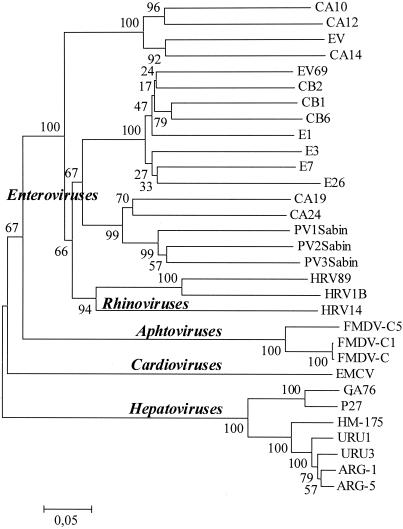
To assess the utility of the complete VP1 protein to infer the relationships among HAVs and other human picornaviruses, a phylogenetic tree was constructed with representative strains from the Picornaviridae family, taken from databases using p-distance and the neighbor-joining method. The results of this study are shown in Fig. 1. As can be seen, the enteroviruses clustered into four major lineages. This result is consistent with previous phylogeny studies (22, 40, 44). Moreover, human rhinoviruses form a cluster among the human enterovirus group, consistent with the results of previous phylogeny studies (40). Each lineage was very strongly supported with bootstrap values from 94 to 100% (Fig. 1). Aphthoviruses (foot-and-mouth disease virus), cardioviruses, and hepatoviruses were assigned to different clusters, which were also strongly supported (supported by a bootstrap value of 100%).
FIG. 1.
Consensus phylogenetic tree constructed for representative human picornaviruses. The tree was constructed by neighbor-joining and p-distance methods. The numbers at nodes represent the percentages of 500 bootstrap pseudoreplicates. The bar indicates genetic distance. Abbreviations: CA, coxsackie A virus; EV, enterovirus; CB, coxsackie B virus; E, echovirus; FMDV, foot-and-mouth disease virus; EMCV, encephalomyocarditis virus.
Phylogenetic analysis of HAV strains.
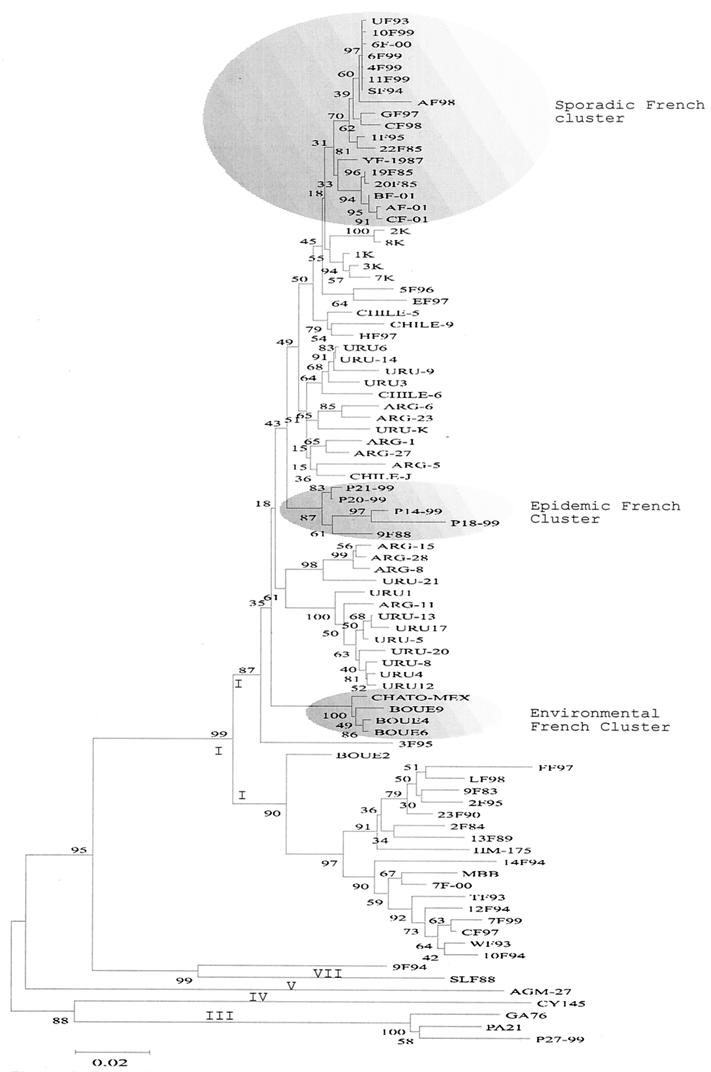
The complete VP1 nucleotide sequences (900 nucleotides) for 81 HAV strains from Argentina, Chile, Mexico, Uruguay, Kosovo, and France isolated from 1983 to 2001 were determined and aligned with those of 10 isolates of HAV taken from the database (Table 1). Phylogenetic trees were generated using Kimura two-parameter distance and the neighbor-joining method (Fig. 2). These results revealed the existence of five genetic groups, strongly supported by bootstrap values.
FIG. 2.
Phylogenetic analysis of the complete VP1 region using the two-parameter model of Kimura. The numbers at nodes indicate bootstrap percentages after 1,000 replications of bootstrap sampling. Genotypes and subgenotypes are indicated at nodes. The bar indicates genetic distance.
The majority of the human HAV strains included in these studies belong to genotype I. This genotype is subdivided into two well-identified subgenotypes, namely, IA and IB (Fig. 2) Genotype IA, supported by a bootstrap value of 87%, includes strains from Argentina, Chile, Mexico, Uruguay, Kosovo, and France. Nevertheless, genetic heterogeneity was observed within the main IA cluster. The South American strains clustered (even those from the same country) in different branches, and no geographical cluster was found.
Interestingly, different clusters of strains isolated in France were well identified within the main subgenotype IA cluster. While one of these clusters contains strains isolated during an epidemic outbreak north of Bretagne (France), another contains environmental strains isolated from sludge samples recovered near the Loire region (Nantes, France). Remarkably, virus causing sporadic cases clustered separately from the two French clusters cited above (sporadic and epidemic French clusters in Fig. 2). Nevertheless, bootstrap values did not allow us to establish definitive genetic relationships among these strains. Genotype IB, also strongly supported by bootstrap values, contains strains from France which cluster with the genotype IB reference strain, HM-175. Within this genotype, two distinct lineages were found. One cluster included HAV strains isolated from sporadic cases of infection, while the other one was made up of the only isolate of this subgenotype found in the environment (Boue2 strain).
Genotype IIIA comprises the simian prototype strain from Panama (PA21 [29]), one strain isolated in the United States (26), and the only genotype IIIA strain isolated in France thus far (14).
Genotypes IV and V are represented by strains recovered from Old World monkey species (37, 58). The VP1 sequences from genotypes IV and V were obtained from databases and were included in this analysis. These two genotypes also clustered separately (Fig. 2).
The VP1 sequence from the only example of genotype VII identified so far was recently published (9) and was included in our analysis. This strain was isolated from an African (Sierra Leone) woman who developed fulminant hepatitis.
Surprisingly, strain 9F94 clustered separately from all other strains, even though it was genetically closer to genotype VII than to any other genotype (Fig. 2).
These findings were further confirmed by using both the Tamura-Nei (55) and Jukes-Cantor distances (25) (data not shown).
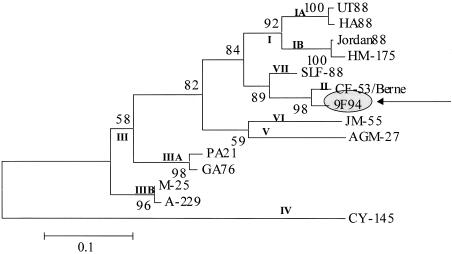
Because the complete VP1 sequences from genotypes II, IIIB, and VI were not available and could not be included in this study, phylogenetic analysis using the VP1/2A region was performed to ascertain whether strain 9F94 was related to any of these genotypes or subgenotypes (Fig. 3). As can be seen, the 9F94 strain has a close genetic relationship with the only identified genotype II strain of HAV, the CF-53/Berne strain (Fig. 3).
FIG. 3.
Neighbor-joining phylogenetic tree of the VP1/2A region using the two-parameter model of Kimura. Genotypes and subgenotypes are shown at nodes for strains reported previously. The numbers at the nodes indicate bootstrap percentages after 1,000 replications of bootstrap sampling. The bar indicates genetic distance.
Interlineage and intralineage sequence diversity.
Sequence differences within and between HAV types and subtypes are shown in Table 3. In the VP1 region, there is a great level of overall genetic diversity within HAV strains, approaching a maximum of 23.5% at the nucleotide level and 10.5% at the amino acid level.
TABLE 3.
Sequence differences observed between HAV strains using the complete VP1 gene
| Comparison of strains | Nucleotide difference (%)a
|
Amino acid difference (%)a
|
||||
|---|---|---|---|---|---|---|
| Range | Mean | SD | Range | Mean | SD | |
| Among all strains | 0-23.5 | 7.47 | 5.33 | 0-10.5 | 1.82 | 1.86 |
| Within genotype I | 0-11.1 | 5.54 | 2.74 | 0-5.6 | 1.22 | 0.9 |
| Within subgenotype IA | 0-8.5 | 3.86 | 1.42 | 0-3.9 | 0.96 | 0.7 |
| Within subgenotype IB | 0.9-9.9 | 4.77 | 1.79 | 0-5.6 | 1.3 | 1.4 |
| Within subgenotype IIIA | 4-5.7 | 4.66 | 0.91 | 1.8-2.8 | 2.36 | 0.51 |
| Between subgenotypes IA and IB | 5.1-11.1 | 9.27 | 1.73 | 0-5.6 | 1.74 | 0.98 |
| Between genotype I and subgenotype IIIA | 18.4-21.4 | 19.87 | 0.57 | 4.6-9.1 | 5.97 | 0.76 |
| Between subgenotypes IA and IIIA | 18.6-21.3 | 19.85 | 0.59 | 4.6-7.7 | 5.91 | 0.65 |
| Between subgenotypes IB and IIIA | 18.4-21.4 | 19.95 | 0.67 | 4.6-9.1 | 6.15 | 1.08 |
| Between subgenotype IIIA and II | 19.9-21.2 | 20.57 | 0.65 | 7-8.1 | 7.73 | 0.63 |
| Between subgenotypes IIIA and IV | 19.6-20.8 | 20.36 | 0.66 | 7.4-9.1 | 8.3 | 0.85 |
| Between subgenotype IIIA and V | 22.1-23.5 | 22.97 | 0.75 | 7-8.8 | 8.2 | 1.04 |
| Between subgenotypes IIIA and VII | 19.8-20.1 | 19.9 | 0.17 | 4.2-5.3 | 4.93 | 0.63 |
| Between genotype I and genotype IV | 19.4-21.5 | 20.18 | 0.39 | 7-10.5 | 7.65 | 0.62 |
| Between subgenotype IA and genotype IV | 19.4-21.2 | 20.18 | 0.39 | 7-9.1 | 7.57 | 0.49 |
| Between subgenotype IB and genotype IV | 19.8-21.5 | 20.31 | 0.55 | 7-10.5 | 7.92 | 0.96 |
| Between genotypes I and V | 18.2-21.4 | 19.01 | 0.46 | 5.6-8.8 | 6.65 | 0.59 |
| Between subgenotype IA and V | 18.2-20 | 18.9 | 0.36 | 6-8.1 | 6.74 | 0.44 |
| Between subgenotype IB and V | 18.6-20.7 | 19.47 | 0.55 | 5.6-8.8 | 6.27 | 0.95 |
| Between genotypes I and II | 12.6-16.7 | 14.07 | 0.88 | 2.1-5.3 | 2.72 | 0.61 |
| Between subgenotype IA and II | 12.6-15.1 | 13.78 | 0.52 | 2.1-4.2 | 2.6 | 0.5 |
| Between subgenotype IB and II | 12.6-16.7 | 15.28 | 1.05 | 2.8-5.3 | 3.2 | 0.75 |
| Between genotypes I and VII | 14-17.8 | 15.25 | 0.6 | 0.7-4.2 | 1.37 | 0.67 |
| Between subgenotype IA and genotype VII | 14-15.9 | 14.41 | 0.41 | 0.7-2.8 | 1.22 | 0.52 |
| Between subgenotype IB and genotype VII | 15-17.8 | 1.05 | 1.05 | 1.1-4.2 | 1.95 | 0.2 |
| Between genotypes II and IV | 21.4 | NA | NA | 9.5 | NA | NA |
| Between genotypes II and V | 20.1 | NA | NA | 8.4 | NA | NA |
| Between genotypes II and VII | 10.6 | NA | NA | 2.8 | NA | NA |
| Between genotypes IV and V | 22 | NA | NA | 5.3 | NA | NA |
| Between genotypes IV and VII | 21.5 | NA | NA | 7.7 | NA | NA |
| Between genotypes V and VII | 20.1 | NA | NA | 7 | NA | NA |
Uncorrected p distances multiplied by 100. NA, not applicable because only one complete VP1 sequence from genotypes II, IV, V, and VII was available.
Within lineages, differences are less than 11.1% at the nucleotide level and 5.6% at the amino acid level. Genetic variations between genotypes range from 10.6 to almost 23.5% at the nucleotide level and 0.7 to almost 10.5% to the amino acid level.
Amino acid sequence diversity.
Despite the extensive nucleotide variation in the HAV strains, the deduced amino acid sequences for all 81 HAV strains showed a high degree of sequence homology (89.5 to 100%) within the VP1 protein. The majority of differences involve synonymous mutations with only few changes resulting in amino acid changes.
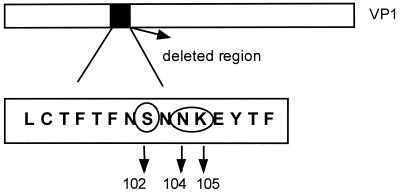
Surprisingly, the Uru-3 VP1 protein contains a 45-nucleotide deletion (positions 2488 to 2532 numbering as for wild-type strain HM-175 [M14707] [10]), resulting in a 15-amino-acid deletion (Fig. 4). Some of the deleted amino acids have been previously described as part of the immunodominant antigenic site of HAV (42).
FIG. 4.
Scheme representing the 15-amino-acid deletion within the VP1 protein of the Uru-3 strain. Neutralization escape mutations are circled.
Within-gene covariation between synonymous and nonsynonymous substitutions.
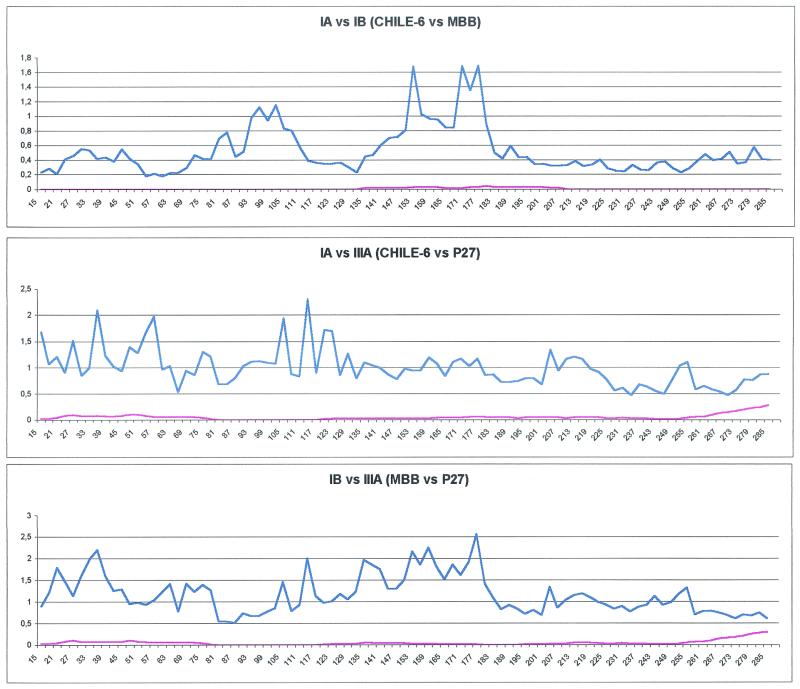
Figure 5 shows the variation in the rates of synonymous and nonsynonymous substitutions within the HAV VP1 region. We have compared strains of three different subgenotypes (Chile-6, IA; MBB, IB; P27, IIIA). Synonymous distances are significantly higher than nonsynonymous ones for almost all pairwise comparisons (Fig. 5). As a consequence, the synonymous distance/nonsynonymous distance (ka/ks) ratio is very low along the whole sequences; this has usually been associated with purifying selection acting at the level of amino acid conservation.
FIG. 5.
Profiles of synonymous (blue line) and nonsynonymous (red line) distances between different HAV genetic lineages. Sequences from strains Chile-6 (subgenotype IA) and MBB (subgenotype IB) (A), strains Chile-6 (subgenotype IA) and P27 (subgenotype IIIA) (B), and strains MBB (subgenotype IB) and P27 (subgenotype IIIA) (C). The x axis depicts the window number, and the y axis depicts distance.
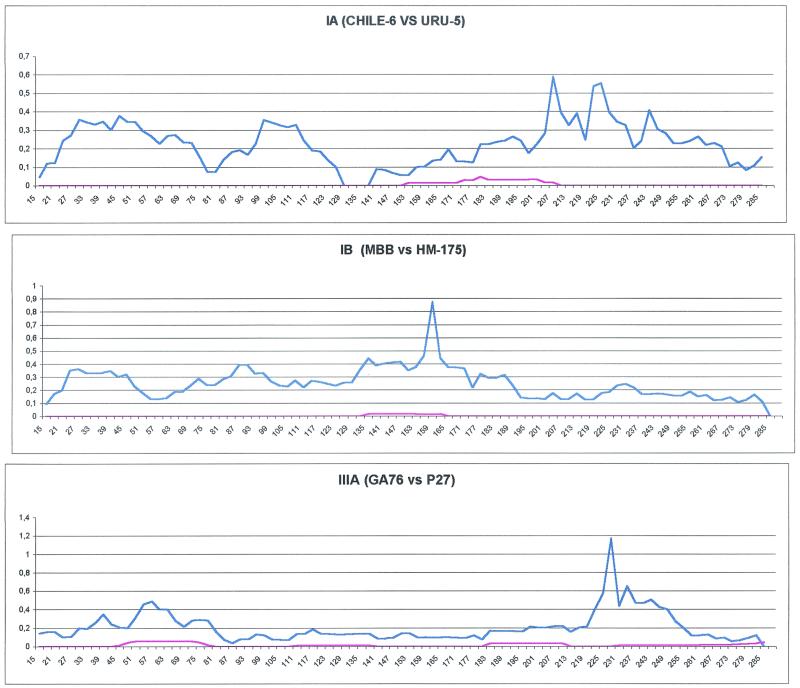
To obtain a clearer picture of the evolutionary mechanisms underlying the changes in VP1, we analyzed the processes of divergence within phylogenetically independent lineages (by the method of Alvarez-Valin et al. [1]). Therefore, we obtained profiles of synonymous and nonsynonymous distances for pairs of strains of types IA, IB, and IIIA (shown in the phylogenetic tree presented in Fig. 2). The results are shown in Fig. 6.
FIG. 6.
Profiles of synonymous (blue line) and nonsynonymous (red line) distances within different HAV genetic lineages. Sequences from strains Uru-5 and and Chile-6 (subgenotype IA) (A), strains HM-175 and MBB (subgenotype IB) (B), and strains GA76 and P27 (subgenotype IIIA) (C). For further details, see the legend to Fig. 5.
Although comparison of the synonymous substitution profiles in the three different genetic lineages revealed quite different patterns and no significant association between the ks values was observed, the profiles obtained for the three pairs show that nonsynonymous substitutions revealed extremely low rates all over the gene.
DISCUSSION
VP1 Picornaviridae phylogeny.
VP1 is the major surface-accessible protein in the mature picornavirus virion (21, 30, 42, 43). Monoclonal antibody-resistant mutants have shown that a number of amino acids within the VP1 protein contribute to the major immunodominant site of HAV (42, 43, 54). Most escape mutations in other picornaviruses are similarly located on loops connecting β-strands within an eight-strand antiparallel β-barrel structure assumed by other picornaviral proteins (42, 43). Therefore, the use of VP1 protein to establish the genetic relationships in the family Picornaviridae, using complete VP1 sequences, will be extremely useful for strain characterization and molecular evolution studies. Using this approach, it was possible to establish a well-defined phylogeny for the family Picornaviridae with very strong statistical and phylogenetic support (Fig. 1).
New classification of HAV based on the VP1 region.
Genetic characterization of HAV was based on the traditional criteria applied to poliovirus (45). It was based upon the percentage identity within the putative VP1/2A junction (168 nucleotides) (49), and seven different genotypes, designated I to VII were determined. Four of these genotypes have been associated with human disease (I, II, III, and VII).
Recently, evolution and phylogenetic analysis of different members of the Picornavirus family took into account the complete VP1 gene (6, 20, 32, 38, 40).
In this study, which includes virus strains recovered from six different countries, for the first time, the complete VP1 protein sequence (900 nucleotides) was used to determine the molecular epidemiology and evolution of HAV strains isolated from 1983 to 2001. The results revealed the existence of five genetic groups or genotypes. Each genetic group was strongly supported by bootstrap values. The bootstrap values observed for the phylogenetic trees (Fig. 2) allowed us to differentiate among the different genotypes and subgenotypes and even established some definitive relationships among strains within each subgenotype.
How many genotypes of HAV?
So far, HAV strains have been classified into seven distinct genotypes by the method of Robertson et al. (49), who considered only 168 bases of the VP1/2A junction and/or the first 148 bases of the VP1 gene (VP1 N-terminal region). By using this approach, viruses in three of these genotypes (I, II, and VII) were recovered from infected humans, while one genotype (III) contained viruses isolated from humans and owl monkeys. Genotypes I and III comprised the vast majority of human strains studied, while genotypes II and VII were each represented by only a single strain. Unexpectedly, phylogenetic analysis using the complete VP1 region revealed the presence of five distinct genetic groups, all of them supported by high bootstrap values (Fig. 2). This was surprising, since the only sequence not included in our analysis (not available at present) was from genotype VI (JM-55 strain). Strain 9F94, which clustered separately from all other strains included in this analysis (Fig. 1), was shown to be closely related to strain CF-53/Berne (genotype II) when the available sequences of VP1/2A region are studied (Fig. 3) and has the same genetic lineage as SLF88 strain (genotype VII). Moreover, sequence differences between HAV genotypes revealed that the least variation observed was found between genotypes II and VII (Table 3).
Considering all these observations together, it was tempting to speculate that this two genotypes may just be one or two subgenotypes of the same type, the type being genotypes II and VII described by Robertson et al. (49).
Further studies are needed to test this hypothesis. Since the complete sequence of the SLF88 strain has been recently published (9), the determination of the complete sequence of strain 9F94 might help to clarify this issue.
HAV variant strain.
In the course of this study, a second, unexpected finding emerged: we were able to detect a 45-nucleotide deletion within the VP1 gene of the Uru-3 strain, resulting in a 15-amino-acid deletion (Fig. 4).
The only VP1 deletion (18 nucleotides) reported so far was found in strains adapted to grow in cell culture (4, 19, 48). As these adapted strains grow in FrhK/4 cells, this deletion appears to be related to the adaptation of the virus to that particular cell line. Since the Uru-3 strain was directly amplified from serum samples without previous passage in cell culture, mutations induced by adaptation to growth in cell culture may be ruled out. Moreover, the Uru-3 45-nucleotide deletion mapped in a VP1 region far removed from the 18-nucleotide deletions associated with the adaptation to grow in cell culture.
The VP1 Uru-3 deletion is intriguing, since there is no evidence of antigenic variation among human HAV strains detected by an immunological method. The only known antigenic variants of HAV are strains collected from Old World monkeys (genotypes IV and V) (37, 58). These variants presented two amino acid mutations (Ser102 of VP1 and Asp70 of VP3) which have been identified as part of the immunodominant region in human HAV using escape mutants to monoclonal antibody K24F2 (42).
Neutralization escape mutations for the HAV strain HM-175 were identified at Asp70 and Gln74 of the VP3 protein and at Ser102, Val171, and Lys221 of the VP1 protein (42, 43), and those for strain HAS15 were identified at Pro65, Asp70, and Ser71 of the VP3 protein and at Asn104, Lys105, and Gln232 of the VP1 protein (36). The deleted region in the Uru-3 strain contains three amino acids (Ser102, Asn104, and Asn105) which were reported to be able to induce a escape response in neutralization experiments (Fig. 4). Moreover, these residues align with recognized immunogenic sites in human rhinovirus 14 (HRV14) (52) and poliovirus type 3 (PV3) (21, 34). This results suggest that this residues are part of an immunogenic site that is analogous to neutralization immunogenic sites found in other picornaviruses (HRV14 and PV3). Therefore, it is possible that the deletion found in this strain would alter the antigenic structure of this virus. This observation suggests that this strain may be the first antigenic variant of HAV found in humans. Although the deletion of 45 nucleotides would conserve the reading frame, we cannot rule out the possibility that this strain is a defective virus.
The construction of chimeric full-length cDNA clones carrying the sequences coding for the capsid protein of Uru-3, followed by cell culture growing experiments and monoclonal antibody studies, may allow us to address this issue.
Since all the data obtained from escape mutant studies performed thus far (27, 42, 43) suggested that the immunodominant antigenic site is formed for the VP3 and VP1 proteins and only the VP1 region was examined in this study, mutations located in another parts of the genome may complement those located in the VP1 protein. The possibility that mutation in one of these sites could affect antibody binding cannot be ruled out until the HAV structure is resolved.
Mode of evolution and substitution rates in HAV.
The different patterns between the intragenic distributions of synonymous substitutions in the HAV VP1 protein (also observed among different genetic groups [Fig. 5]) suggest that synonymous divergence could be random in the VP1 gene. The distribution of nonsynonymous substitutions shows a complete different situation, with extremely low rates of substitutions compared to those of synonymous substitutions. This suggests that the pattern of divergence observed for HAV VP1 is probably due to selective forces that do not allow amino acid replacements, despite the relative high rates of synonymous substitutions observed all over the gene. These results show that both kinds of nucleotide substitutions are undergoing quite different modes of change. While synonymous divergence is expected to follow a neutral mode of evolution (Fig. 5 and 6) (even though the well-documented existence of cis-acting regulatory elements within the open reading frames of picornaviruses suggest that further work will be needed to qualify this statement), negative selection appears to be the main force shaping the pattern of nonsynonymous substitutions, selecting against most replacement changes in all protein regions, giving a quite conserved protein. In contrast, the antigenic sites of multiple serotype viruses, such as the hemagglutinin gene of influenza virus (23), the complete capsid region of serotypes A and C of foot-and-mouth disease virus (20), and the VP3 region of human immunodeficiency virus (53), were subject to positive selection. As a consequence, the mode of evolution of HAV appears, at least in part, to contribute to explain the presence of only one serological group of HAV so far.
Acknowledgments
We thank anonymous reviewers for constructive comments and suggestions. We thank Michel Pletchette from the European Commission for helpful advice. We also thank Lorica Smith and Nicolas Le Roux for technical assistance, Pascaline Guerin and Bernard Besse for technical support, and Marie Martine Hallet for critical reading of our manuscript. Mauro Costa-Mattioli thanks Ana Maria Renna for unconditional support.
This work was supported in part by the Commission of the European Communities through contract IC18-CT98-0378 (DG12-CEOR) and the Project UNESCO 00URU606 (Ana Maria Renna).
REFERENCES
- 1.Alvarez-Valin, J. F. Tort, and G. Bernardi. 2000. Nonrandom spatial distribution of synonymous substitutions in the GP63 gene from Leishmania. Genetics 155:1683-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apaire-Marchais, V., B. H. Robertson, V. Aubineau-Ferre, M. G. Le Roux, F. Leveque, L. Schwartzbrod, and S. Billaudel. 1995. Direct sequencing of hepatitis A virus strains isolated during an epidemic in France. Appl. Environ. Microbiol. 61:3977-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arauz-Ruiz, P., L. Sundqvist, Z. Garcia, L. Taylor, K. Visona, H. Norder, and L. O. Magnius. 2001. Presumed common source outbreaks of hepatitis A in an endemic area confirmed by limited sequencing within the VP1 region. J. Med. Virol. 65:449-456. [PubMed] [Google Scholar]
- 4.Beneduce, F., G. Pisani, M. Divizia, A. Pana, and G. Morace. 1995. Complete nucleotide sequence of a cytopathic hepatitis A virus strain isolated in Italy. Virus Res. 36:299-309. [DOI] [PubMed] [Google Scholar]
- 5.Bosch, A., G. Sanchez, F. Le Guyader, H. Vanaclocha, L. Haugarreau, and R. M. Pinto. 2001. Human enteric viruses in coquina clams associated with a large hepatitis A outbreak. Water Sci. Technol. 43:61-65. [PubMed] [Google Scholar]
- 6.Brown, B. A., M. S. Oberste, J. P. Alexander, Jr., M. L. Kennett, and M. A. Pallansch. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969-9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruisten, S. M., J. E. van Steenbergen, A. S. Pijl, H. G. Niesters, G. J. van Doornum, and R. A. Coutinho. 2001. Molecular epidemiology of hepatitis A virus in Amsterdam, The Netherlands. J. Med. Virol. 63:88-95. [PubMed] [Google Scholar]
- 8.Byun, K. S., J. H. Kim, K. J. Song, L. J. Baek, J. W. Song, S. H. Park, O. S. Kwon, J. E. Yeon, J. S. Kim, Y. T. Bak, and C. H. Lee. 2001. Molecular epidemiology of hepatitis A virus in Korea. J. Gastroenterol. Hepatol. 16:519-524. [DOI] [PubMed] [Google Scholar]
- 9.Ching, K. Z., T. Nakano, L. E. Chapman, A. Demby, and B. H. Robertson. 2002. Genetic characterization of wild-type genotype VII hepatitis A virus. J. Gen. Virol. 83:53-60. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, J. I., B. Rosenblum, J. R. Ticehurst, R. J. Daemer, S. M. Feinstone, and R. H. Purcell. 1987. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc. Natl. Acad. Sci. USA 84:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comeron, J. M. 1995. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J. Mol. Evol. 41:1152-1159. [DOI] [PubMed] [Google Scholar]
- 12.Costa-Mattioli, M., V. Ferre, S. Monpoeho, L. Garcia, R. Colina, S. Billaudel, I. Vega, R. Perez-Bercoff, and J. Cristina. 2001. Genetic variability of hepatitis A virus in South America reveals heterogeneity and co-circulation during epidemic outbreaks. J. Gen. Virol. 82:2647-2652. [DOI] [PubMed] [Google Scholar]
- 13.Costa-Mattioli, M., S. Monpoeho, E. Nicand, M. H. Aleman, S. Billaudel, and V. V. Ferre. 2002. Quantification and duration of viraemia during hepatitis A infection as determined by real-time RT-PCR. J. Viral Hepat. 9:101-106. [DOI] [PubMed] [Google Scholar]
- 14.Costa-Mattioli, M., S. Monpoeho, C. Schvoerer, B. Besse, M. H. Aleman, S. Billaudel, J. Cristina, and V. Ferre. 2001. Genetic analysis of hepatitis A virus outbreak in France confirms the co-circulation of subgenotypes Ia, Ib and reveals a new genetic lineage. J. Med. Virol. 65:233-240. [DOI] [PubMed] [Google Scholar]
- 15.de Paula, V. S., M. L. Baptista, E. Lampe, C. Niel, and A. M. Gaspar. 2002. Characterization of hepatitis A virus isolates from subgenotypes IA and IB in Rio de Janeiro, Brazil. J. Med. Virol. 66:22-27. [DOI] [PubMed] [Google Scholar]
- 16.De Serres, G., T. L. Cromeans, B. Levesque, N. Brassard, C. Barthe, M. Dionne, H. Prud'homme, D. Paradis, C. N. Shapiro, O. V. Nainan, and H. S. Margolis. 1999. Molecular confirmation of hepatitis A virus from well water: epidemiology and public health implications. J. Infect. Dis. 179:37-43. [DOI] [PubMed] [Google Scholar]
- 17.Diaz, B. I., C. A. Sariol, A. Normann, L. Rodriguez, and B. Flehmig. 2001. Genetic relatedness of Cuban HAV wild-type isolates. J. Med. Virol. 64:96-103. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1993. PHYLIP: phylogeny inference package, 3.5th ed. University of Washington, Seattle, Wash.
- 19.Graff, J., O. C. Richards, K. M. Swiderek, M. T. Davis, F. Rusnak, S. A. Harmon, X. Y. Jia, D. F. Summers, and E. Ehrenfeld. 1999. Hepatitis A virus capsid protein VP1 has a heterogeneous C terminus. J. Virol. 73:6015-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haydon, D. T., A. D. Bastos, N. J. Knowles, and A. R. Samuel. 2001. Evidence for positive selection in foot-and-mouth disease virus capsid genes from field isolates. Genetics 157:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 22.Huttunen, P., J. Santti, T. Pulli, and T. Hyypia. 1996. The major echovirus group is genetically coherent and related to coxsackie B viruses. J. Gen. Virol. 77:715-725. [DOI] [PubMed] [Google Scholar]
- 23.Ina, Y., and T. Gojobori. 1994. Statistical analysis of nucleotide sequences of the hemagglutinin gene of human influenza A viruses. Proc. Natl. Acad. Sci. USA 91:8388-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen, R. W., G. Siegl, and S. M. Lemon. 1990. Molecular epidemiology of human hepatitis A virus defined by an antigen-capture polymerase chain reaction method. Proc. Natl. Acad. Sci. USA 87:2867-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jukes, T., and C. Kantor. 1969. Evolution of protein molecules. Academic Press, New York, N.Y.
- 26.Khanna, B., J. E. Spelbring, B. L. Innis, and B. H. Robertson. 1992. Characterization of a genetic variant of human hepatitis A virus. J. Med. Virol. 36:118-124. [DOI] [PubMed] [Google Scholar]
- 27.Khudyakov, Y. E., E. N. Lopareva, D. L. Jue, S. Fang, J. Spelbring, K. Krawczynski, H. S. Margolis, and H. A. Fields. 1999. Antigenic epitopes of the hepatitis A virus polyprotein. Virology 260:260-272. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 29.Lemon, S. M., J. W. LeDuc, L. N. Binn, A. Escajadillo, and K. G. Ishak. 1982. Transmission of hepatitis A virus among recently captured Panamanian owl monkeys. J. Med. Virol. 10:25-36. [DOI] [PubMed] [Google Scholar]
- 30.Mateu, M. G., J. A. Camarero, E. Giralt, D. Andreu, and E. Domingo. 1995. Direct evaluation of the immunodominance of a major antigenic site of foot-and-mouth disease virus in a natural host. Virology 206:298-306. [DOI] [PubMed] [Google Scholar]
- 31.Matthews, R. E. 1979. The classification and nomenclature of viruses. Summary of results of meetings of the International Committee on Taxonomy of Viruses in The Hague, September 1978. Intervirology 11:133-135. [DOI] [PubMed] [Google Scholar]
- 32.McMinn, P., K. Lindsay, D. Perera, H. M. Chan, K. P. Chan, and M. J. Cardosa. 2001. Phylogenetic analysis of enterovirus 71 strains isolated during linked epidemics in Malaysia, Singapore, and Western Australia. J. Virol. 75:7732-7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melnick, J. L. 1982. Classification of hepatitis A virus as enterovirus type 72 and of hepatitis B virus as hepadnavirus type 1. Intervirology 18:105-106. [DOI] [PubMed] [Google Scholar]
- 34.Minor, P. D., A. John, M. Ferguson, and J. P. Icenogle. 1986. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J. Gen. Virol. 67:693-706. [DOI] [PubMed] [Google Scholar]
- 35.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferre. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nainan, O. V., M. A. Brinton, and H. S. Margolis. 1992. Identification of amino acids located in the antibody binding sites of human hepatitis A virus. Virology 191:984-987. [DOI] [PubMed] [Google Scholar]
- 37.Nainan, O. V., H. S. Margolis, B. H. Robertson, M. Balayan, and M. A. Brinton. 1991. Sequence analysis of a new hepatitis A virus naturally infecting cynomolgus macaques (Macaca fascicularis). J. Gen. Virol. 72:1685-1689. [DOI] [PubMed] [Google Scholar]
- 38.Norder, H., L. Bjerregaard, and L. O. Magnius. 2001. Homotypic echoviruses share aminoterminal VP1 sequence homology applicable for typing. J. Med. Virol. 63:35-44. [PubMed] [Google Scholar]
- 39.Normann, A., M. Pfisterer-Hunt, S. Schade, J. Graff, R. L. Chaves, P. Crovari, G. Icardi, and B. Flehmig. 1995. Molecular epidemiology of an outbreak of hepatitis A in Italy. J. Med. Virol. 47:467-471. [DOI] [PubMed] [Google Scholar]
- 40.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pina, S., M. Buti, R. Jardi, P. Clemente-Casares, J. Jofre, and R. Girones. 2001. Genetic analysis of hepatitis A virus strains recovered from the environment and from patients with acute hepatitis. J. Gen. Virol. 82:2955-2963. [DOI] [PubMed] [Google Scholar]
- 42.Ping, L. H., R. W. Jansen, J. T. Stapleton, J. I. Cohen, and S. M. Lemon. 1988. Identification of an immunodominant antigenic site involving the capsid protein VP3 of hepatitis A virus. Proc. Natl. Acad. Sci. USA 85:8281-8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ping, L. H., and S. M. Lemon. 1992. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J. Virol. 66:2208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pulli, T., P. Koskimies, and T. Hyypia. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 212:30-38. [DOI] [PubMed] [Google Scholar]
- 45.Rico-Hesse, R., M. A. Pallansch, B. K. Nottay, and O. M. Kew. 1987. Geographic distribution of wild poliovirus type 1 genotypes. Virology 160:311-322. [DOI] [PubMed] [Google Scholar]
- 46.Robertson, B., and O. Naiman. 1997. Genetic and antigenic variants of hepatitis A virus. Edizioni Minerva Medica, Turin, Italy.
- 47.Robertson, B. H., F. Averhoff, T. L. Cromeans, X. Han, B. Khoprasert, O. V. Nainan, J. Rosenberg, L. Paikoff, E. DeBess, C. N. Shapiro, and H. S. Margolis. 2000. Genetic relatedness of hepatitis A virus isolates during a community-wide outbreak. J. Med. Virol. 62:144-150. [PubMed] [Google Scholar]
- 48.Robertson, B. H., V. K. Brown, and B. Khanna. 1989. Altered hepatitis A VP1 protein resulting from cell culture propagation of virus. Virus Res. 13:207-212. [DOI] [PubMed] [Google Scholar]
- 49.Robertson, B. H., R. W. Jansen, B. Khanna, A. Totsuka, O. V. Nainan, G. Siegl, A. Widell, H. S. Margolis, S. Isomura, K. Ito, et al. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J. Gen. Virol. 73:1365-1377. [DOI] [PubMed] [Google Scholar]
- 50.Robertson, B. H., B. Khanna, O. V. Nainan, and H. S. Margolis. 1991. Epidemiologic patterns of wild-type hepatitis A virus determined by genetic variation. J. Infect. Dis. 163:286-292. [DOI] [PubMed] [Google Scholar]
- 51.Ross, B. C., and D. A. Anderson. 1991. Characterization of hepatitis A virus capsid proteins with antisera raised to recombinant antigens. J. Virol. Methods 32:213-220. [DOI] [PubMed] [Google Scholar]
- 52.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, et al. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 53.Seibert, S. A., C. Y. Howell, M. K. Hughes, and A. L. Hughes. 1995. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 12:803-813. [DOI] [PubMed] [Google Scholar]
- 54.Stapleton, J. T., and S. M. Lemon. 1987. Neutralization escape mutants define a dominant immunogenic neutralization site on hepatitis A virus. J. Virol. 61:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 56.Taylor, M. B. 1997. Molecular epidemiology of South African strains of hepatitis A virus: 1982-1996. J. Med. Virol. 51:273-279. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsarev, S. A., S. U. Emerson, M. S. Balayan, J. Ticehurst, and R. H. Purcell. 1991. Simian hepatitis A virus (HAV) strain AGM-27: comparison of genome structure and growth in cell culture with other HAV strains. J. Gen. Virol. 72:1677-1683. [DOI] [PubMed] [Google Scholar]