Abstract
Human immunodeficiency virus type 1 is characterized by extensive genetic heterogeneity. Having previously demonstrated that, in the peripheral blood, the initial viral population is more homogeneous than at subsequent stages of infection, we have extended our studies to tissue samples, allowing comparisons between viral populations in peripheral blood and tissues during both the acute and chronic stages of infection. We found that homogeneity in gp120 sequences during the acute infection phase is not just restricted to the peripheral blood but also extends to other tissue compartments. However, in chronically infected individuals, heterogeneous and distinct viral populations were found in different compartments. We therefore conclude that the dominant and homogeneous viral population observed during the acute infection phase is likely to infiltrate lymphoid tissues and form the genetic bases for subsequent diversification. It is therefore likely that the compartmentalization of viral sequences observed in chronically infected patients reflects a gradual diversification of a common dominant viral variant rather than the preferential migration of distinct viral populations to different tissue compartments at the beginning of infection.
A striking feature of human immunodeficiency virus type 1 (HIV-1) infection in vivo is the rapid generation and turnover of viral variants, resulting in a high degree of sequence diversity both within and between infected individuals (1, 6, 8, 11, 18, 23, 26, 27, 29, 39, 47-49, 53). While the time course of the disease seems slow, the generation of different viral variants in vivo is rapid and persistent throughout the course of infection. It is generally believed that HIV-1 infection begins with a relatively homogeneous population (7, 25, 29, 33, 45, 56, 57). However, as the disease progresses, divergent viral variants with distinct genetic sequences emerge, probably as a result of selective pressures imposed by the immune system, therapeutic regimens, or preferential tropism for certain target cells (3-5, 17, 26, 36, 38, 42, 56). Thus, an increasingly complex and heterogeneous viral population is almost exclusively found in chronically rather than acutely infected patients. Over time, these viral variants often result in the formation of distinct quasispecies and are capable of localizing themselves in various tissue compartments (9, 10, 15, 17, 21, 22, 31, 32, 43-46, 58). Understanding more about the unique nature of viral diversification, dissemination, and compartmentalization during the acute and chronic phases of infection is critical to our understanding of viral pathogenesis and the development of an effective drug regimen against those variants residing in various tissue compartments.
Numerous reports have provided evidence of distinct HIV-1 variants in vivo in chronically infected patients, as indicated by their envelope sequences. Viral variants recovered from the brain, lungs, lymph nodes, spleen, gastrointestinal and genital tracts, and peripheral blood mononuclear cells (PBMC) may all be distinct from one another (2, 9, 10, 12, 14, 15, 17, 20-22, 24, 30-32, 34, 35, 37, 43-46, 58). Even within a single organ, subpopulations of HIV-1 appear to exist. Viral variants residing within the white pulp are genetically distinct from those in the red pulp within the same spleen section (17, 19). Viral variants residing in different parts of the brain, such as the frontal lobe, basal ganglia, medial temporal lobe, and nonmedial temporal lobe, are also genetically different (37). These findings suggest that very little genetic flow occurs not only between different tissue compartments but also between the different subcompartments within a given tissue. A similar observation has also been made within HIV-2-infected patients (35).
Two distinct hypotheses have been put forward to explain these organ- or tissue-specific viral populations that arise during the course of HIV-1 infection. The first seeks to explain the quasispecies compartmentalization by selective migration. That is, viral variants best suited for a particular tissue, because of either their preferential cell tropism or their ability to escape immune recognition, will actively migrate to that location upon infection. The second theory is based on the parallel evolution of viral variants. Upon infection, a homogeneous viral population infiltrates all tissues. However, over the course of infection, tissue-specific selection pressures favor minor independent variants, which will actively replicate in accordance with their unique environment and thereby produce a subpopulation distinct from those of neighboring compartments.
Our present study sought to evaluate these two hypotheses by studying variation in the gp120 sequences in tissues and peripheral blood from both acutely and chronically infected individuals. We believe that this is the first study of this kind to evaluate the genetic diversity and relatedness of HIV-1 in both peripheral blood and tissues during the acute infection phase. Our results show that homogeneity in gp120 sequences during the acute infection phase is not just restricted to the peripheral blood but also extends to various tissue compartments. In contrast, in chronically infected individuals, distinct viral populations were found in different compartments. We therefore conclude that the dominant homogeneous viral population observed during the acute infection phase is likely to infiltrate lymphoid tissues and thereby form the genetic basis for further diversification during the subsequent stages of infection. The unique selection pressures that individual compartments are believed to experience will encourage the best-suited local variant to replicate with a higher frequency over the course of the infection. The compartmentalization of viral sequences observed in chronically infected patients therefore reflects a gradual diversification from a common dominant viral variant rather than the preferential migration of distinct viral populations to different tissue compartments at the beginning of the infection.
MATERIALS AND METHODS
Patients.
Eleven patients ranging in age from 30 to 54 years were recruited for the study (Table 1). Five of them were enrolled in treatment protocols during the acute infection phase and selected for this study because of their complete suppression of HIV-1 in plasma under the prior treatment and their full compliance with the prescribed antiretroviral therapy. Acute infection in our cohort was defined as having a detectable viral load in the plasma but having either no HIV-1-specific antibodies or an evolving antibody responses on a Western blot assay. Most of these acutely infected patients demonstrated clinical symptoms consistent with acute infection. All acutely infected individuals were enrolled within 90 days of documented seroconversion. The remaining six patients were enrolled in treatment protocols at least 90 days after infection and were therefore defined as chronically infected. At baseline, these patients had a mean CD4 lymphocyte count of 308 μl of blood and an average viral load of 123,071 RNA copies per ml of plasma. The level of HIV-1 in plasma was measured by a commercial reverse transcriptase PCR assay (Amplicor Ultrasensitive HIV-1 Monitor assay; Roche Molecular Diagnostic Systems, Branchburg, N.J.), which has a limit of detection of 50 HIV-1 RNA copies per ml of plasma. The antiretroviral regimen consisted of zidovudine (AZT) and lamivudine (3TC), along with either indinavir (Ind), nelfinavir, both ritonavir and saquinavir, or both abacavir (ABC) and amprenavir (Amp) (Table 1).
TABLE 1.
Demographic, clinical, and virologic profiles of study subjects
| Patient no. | Age (yr)/gendera/risk factor | Baseline plasma HIV-1 RNA (no. of copies/ml)b | Baseline CD4 count (no. of cells/mm3) | Treatment regimenc | Infection phase at initiation of therapyd | Tissue sample collectede |
|---|---|---|---|---|---|---|
| 1 | 33/M/homosexuality | 611,566 | 432 | AZT/3TV/Ind | Acute | Tonsil |
| 2 | 37/M/homosexuality | 9,806 | 387 | AZT/3TV/Ind | Acute | Tonsil |
| 3 | 33/M/homosexuality | 49,737 | 544 | AZT/3TV/Ind | Acute | Tonsil |
| 4 | 38/M/homosexuality | 38,940 | 290 | AZT/3TC/Rit/Saq | Acute | Rectum |
| 5 | 47/M/homosexuality | 287,070 | 284 | AZT/3TC/ABC/Amp | Acute | Rectum |
| 6 | 42/M/homosexuality | 19,013 | 314 | AZT/3TC/ABC/Amp | Chronic | Rectum |
| 7 | 30/M/homosexuality | 37,497 | 355 | AZT/3TC/ABC/Amp | Chronic | Rectum |
| 8 | 32/M/homosexuality | 18,124 | 392 | AZT/3TC/ABC/Amp | Chronic | Rectum |
| 9 | 48/M/homosexuality | 54,783 | 302 | AZT/3TC/ABC/Amp | Chronic | Rectum |
| 10 | 54/M/homosexuality | 98,824 | 75 | AZT/3TC/Nel | Chronic | Rectum |
| 11 | 30/M/homosexuality | 128,425 | 12 | AZT/3TC/ABC/Amp | Chronic | Rectum |
M, male.
Determined by the Amplicor assay.
Rit, ritonavir; Saq, saquinavir; Nel, nelfinavir.
Acute, treatment initiation within 90 days of documented seroconversion.
All rectal samples were collected before initiation of therapy, whereas tonsil samples were collected 12 to 24 months after initiation of therapy.
Peripheral blood and tissue samples.
Informed consent was obtained from all of the subjects enrolled in this study in accordance with the guidelines of the Committee for the Protection of Human Subjects from Research Risks at The Rockefeller University prior to any endoscopic or biopsy procedures. Whole-blood samples and flexible sigmoidoscopic biopsies from noninflamed rectal tissue were collected simultaneously before the initiation of highly active antiretroviral therapy. The endoscopic mucosal specimens collected in the study contained primarily the surface epithelium and lamina propria. Tonsillar biopsies were collected from acutely infected patients 12 to 24 months after initiation of therapy. PBMC were isolated from the whole blood by Ficoll-Hypaque separations (52, 55). Tissue samples were minced with scissors and then treated with 0.5 mg of collagenase (Sigma Chemical Company, St. Louis, Mo.) per ml at 37°C until the tissue was completely digested. Cell suspensions were obtained by filtration of collagenase-treated tissue samples through a 100-μm-pore-size nylon cell strainer (Becton Dickinson, San Jose, Calif.). Isolated PBMC and tissue cells were then used for viral isolation and genomic DNA extraction (see below). Viral RNA was extracted from the plasma; this was followed by cDNA synthesis and PCR amplification in accordance with previously published protocols (52, 55).
Virus isolation from clinical samples.
Approximately 5 × 106 of patients' PBMC or cells isolated from rectal biopsies were cocultured with 5 × 106 phytohemagglutinin-stimulated normal donor PBMC as previously described (51, 54). The culture supernatant was monitored for p24 production on days 0, 4, 7, 10, and 14 with a commercial enzyme immunoassay (Abbott Laboratories, Abbott Park, Ill.). A culture was considered positive if the p24 value was above a cutoff of 40 pg/ml. Virus titers were determined on PBMC to determine the 50% tissue culture-infective dose (TCID50).
Determination of viral phenotype and coreceptor usage.
The phenotype of viruses was determined by testing their ability to generate syncytia in MT-2 cells as described previously (51, 54). In brief, MT-2 cells were cultured in RPMI 1640 medium containing 10% fetal calf serum, L-glutamine, and antibiotics and split twice a week. For infection assays, 2 × 105 cells were incubated with 100 TCID50 of each viral isolate for 4 h. Unbound viruses were removed by two washes in culture medium. The culture supernatant was tested for HIV-1 p24 antigen on days 0, 4, and 7 postinfection. MT-2 cell cultures were evaluated by light microscopy for syncytium formation.
U87MG.CD4 cell lines stably transfected with the CCR1, CCR2b, CCR3, CCR5, or CXCR4-encoding gene and HOS.CD4 cells stably expressing BOB/gpr15 and Bonzo/STRL33 were kindly provided by D. Littman (Skirball Institute for Molecular Medicine, New York, N.Y.) (51, 54). These cells were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum, L-glutamine, and antibiotics (1 μg of puromycin [Sigma Chemical Company)] per ml, 100 μg of neomycin [G418; Sigma Chemical Company] per ml, and 250 μg of hygromycin [Sigma Chemical Company] per ml) and split twice a week. For infection experiments, 104 cells were incubated with 100 TCID50 of each isolate for 4 h and unbound virus was removed by three washes in culture medium. The cultures were examined microscopically for the formation of multinucleated foci, and the supernatant was analyzed for the presence of p24 antigen.
PCR amplification, length polymorphism, sequencing, and sequence analysis of HIV-1 gp120.
The entire procedure for PCR amplification, length polymorphism, sequencing, and sequence analysis of HIV-1 gp120 has been previous described in detail (52). In brief, viral RNA was extracted from plasma and cDNA was synthesized (52). Proviral DNA was extracted from both PBMC and tissue samples (55). Our genetic analyses were focused on the V1-V2 and V4-V5 domains of the viral envelope glycoprotein. These regions contain not only frequent base substitutions but also extensive length polymorphisms. The pattern and profile of length polymorphism in the V1-V2 and V4-V5 regions can therefore be used as a marker for genetic comparisons between viral populations (52). The primer sequences, PCR and electrophoresis conditions, and analyses using GeneScan programs (PE Biosystems) were performed as described previously (52). Similarly, generation of single proviral DNA and viral cDNA, nucleotide sequencing with an automated sequencer (ABI Prism 377), and sequence analyses with the ClustalW program and the PHYLIP package were carried out as described in our previous reports (52, 55). All RNA and DNA extractions and subsequent amplification reactions were carried out with appropriate negative controls in parallel to detect contamination at each step of the procedure.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the C2-V3 region sequences described here are AY120369 through AY120564.
RESULTS
Comparison of length polymorphisms in the surface envelope glycoprotein of HIV-1 samples from peripheral blood and tissues during the acute and chronic phase of infection.
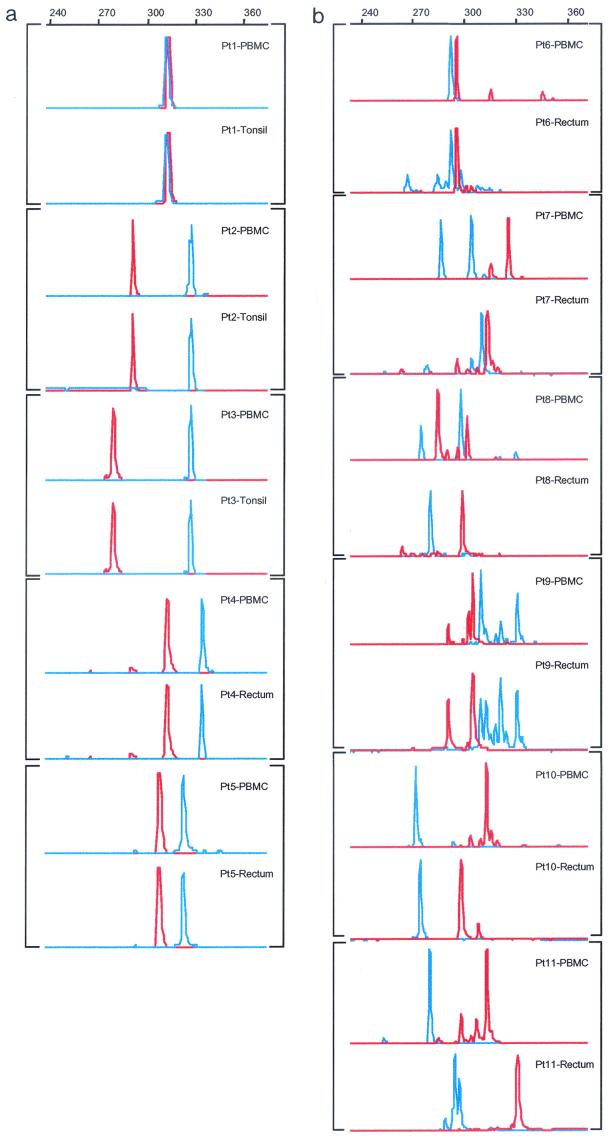
To compare the genetic relationship between peripheral blood and various tissues, we used a fluorescence technique based on the extensive length polymorphism in the V1-V2 and V4-V5 regions of HIV-1 gp120, as reported previously (52). This technique is able to detect a single nucleotide deletion or insertion within PCR products, thereby offering a quick and reliable technique for studying the pattern and profile of length polymorphism at the population level (52). As shown in Fig. 1a, single and uniform peaks of V1-V2 and V4-V5 products were found in both the PBMC and tissues of all five acutely infected individuals (Pt1 to Pt5), although the lengths varied from patient to patient. More importantly, the length polymorphism profiles of PBMC and tissues are identical, suggesting that both compartments bear homogeneous and identical viral populations during the acute infection phase. It is worth noting that although the tonsillar samples were collected from patients 1, 2, and 3 12 to 24 months after the initiation of highly active antiretroviral therapy, the finding of a length polymorphism profile identical to that of PBMC during the acute infection phase suggests that virus replication was well suppressed by the combination therapy and that there was no significant viral evolution in tissues during the study period. This notion is supported by the sustained absence of plasma viremia in these patients without a single viral blip above the detection limit during the study period (data not shown). Furthermore, the length polymorphism profile in plasma during the acute infection phase is identical to that of PBMC and tissues (data not shown), which suggests that a relatively homogeneous viral population dominates in peripheral blood and probably in all tissues.
FIG.1.
Length polymorphism of the proviruses in PBMC in comparison with those isolated from tonsillar or rectal samples during the acute (a) or chronic (b) phase of HIV-1 infection. V1-V2 region, red; V4-V5 region, blue. The size of each peak is indicated by the scale at the top of each panel.
Contrary to what has been observed in acutely infected patients, multiple peaks with different lengths were identified in viral populations from both PBMC and rectal tissues for both V1-V2 and V4-V5 products in all six chronically infected patients (Pt6 to Pt11) (Fig. 1b). In addition, the length polymorphism patterns in both the V1-V2 and V4-V5 regions differed significantly between PBMC and rectal tissue, indicating that distinct viral populations reside in the different tissue compartments (Fig. 1b).
Comparison of sequences in the surface envelope glycoprotein of HIV-1 between peripheral blood and tissues during the acute and chronic phases of infection.
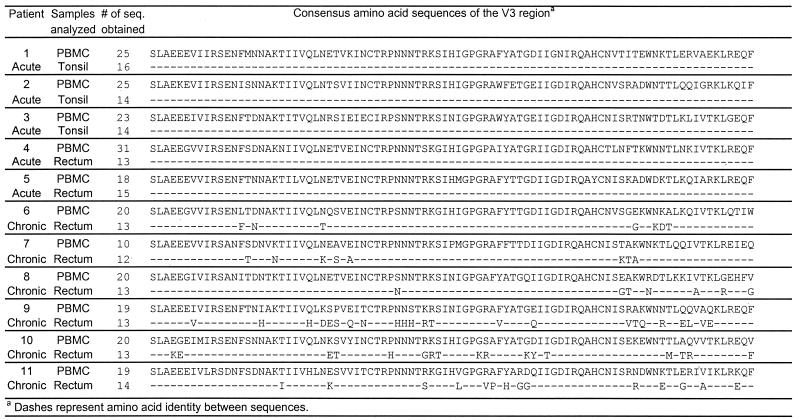
To analyze viral populations in both peripheral blood and tissues in greater detail, extensive sequencing was conducted in all five acutely infected and six chronically infected individuals. A total of 380 sequences from the C2-V3 region were obtained from both PBMC and tissues, 230 of which were from PBMC and the remaining 150 of which were from tissues. All of these sequences were obtained as single molecules through limiting dilution prior to PCR to avoid errors generated during the amplification step (40). The most notable result of the sequencing is the confirmation of the results obtained by length polymorphism analysis. Figure 2 summarizes the consensus sequences obtained from PBMC, tonsillar, and rectal samples. In five acutely infected individuals (patients 1 to 5), the sequences from PBMC and tonsil or rectum tissue are identical, consistent with the results obtained from the length polymorphism studies (see above). In contrast, significant differences were found between PBMC and rectal samples in all chronically infected patients (patients 6 to 11). In patients 6, 7, and 8, large numbers of amino acid changes were found outside the V3 loop; in patients 9, 10, and 11, changes were also found inside the V3 loop sequence. The impact of these changes on the viral replication kinetics, phenotype, and coreceptor usages is relatively minimal (see below). An analysis of the genetic diversity within and between different compartments has also been conducted. It has been found that the average genetic diversity in the peripheral blood (0.38% ± 0.38%) is significantly smaller (P < 0.5) that that in tissues (0.58% ± 0.58%). In addition, the average genetic distance between sequences from the peripheral blood and tissue is approximately 6.3% ± 3.2%, which is roughly 10- to 20-fold greater than that within the same compartment. The average interpatient diversity, however, is about 15 to 20%.
FIG. 2.
Comparison of deduced amino acid sequences of the V3 region between peripheral blood and tissues. # of seq., number of sequences.
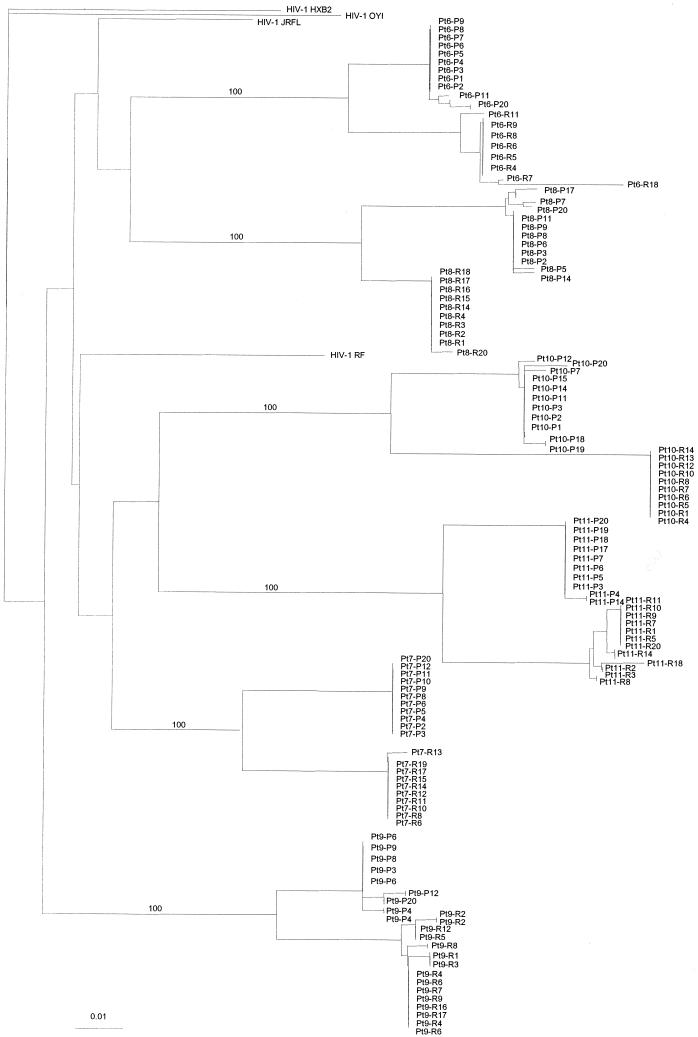
Further analyses of sequences obtained from the chronically infected individuals were conducted by using several phylogenetic approaches. Figure 3 depicts the neighbor-joining trees generated for all six patients with sequences from both PBMC and rectal tissue. Individual sequences are represented by patient codes. For clarity of tree presentation, only selected sequences are shown for each patient. The horizontal branches reflect the relative genetic distance between different sequences. The most obvious and consistent result of these analyses is that sequences from each individual clustered together without contamination between patients or with HIV-1 laboratory strains such as HXB2, OYI, JRFL, and RF (Fig. 3). In addition, sequences from PBMC and rectal tissue are more similar within each compartment, as reflected by their tight clustering on the phylogenetic tree (Fig. 3). In contrast, sequences obtained from different compartments are significantly different, as demonstrated by the distinct clustering of sequences separated by a long branch length on the tree (Fig. 3). This observation supports the findings of length polymorphism analysis that there are significant differences between peripheral blood and rectal tissue in all six chronically infected patients.
FIG. 3.
Unrooted neighbor-joining tree (Kimura two-parameter model) depicting the evolutionary relationship between viruses from PBMC and tissues of six chronically infected patients. Individual sequences are represented by patient codes at the ends of braches, which are drawn to scale so that the relatedness between different sequences can be readily assessed. P; PBMC; R; rectum. For each individual, only a selected number of sequences are shown in the tree for clarity of presentation. Laboratory HIV-1 strains such as HXB2, OYI, JRFL, and RF were included as controls.
Isolation and phenotypic characterization of viruses from the PBMC and rectums of chronically infected individuals.
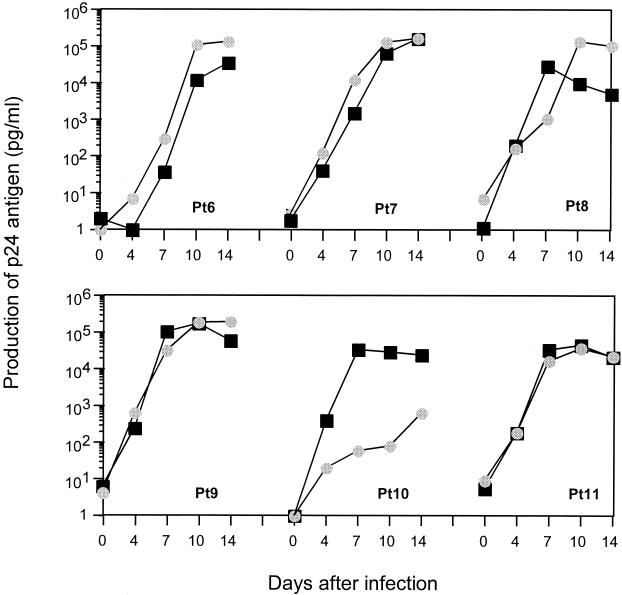
To study whether the genetic differences observed between virus samples obtained from peripheral blood and rectal tissue have differing viral phenotypes, we isolated viruses from all six chronically infected patients by using the same PBMC and rectal cells as were used for the genetic analysis. We then characterized viral phenotypic properties by several means. Firstly, we tested the ability of viral isolates to replicate in PBMC. Approximately 100 TCID50 of each isolate was used to inoculate 2 × 106 phytohemagglutinin-stimulated PBMC, and the production of p24 antigen in the culture supernatant was measured on days 0, 4, 7, 10, and 14. All viral isolates, except one from the PBMC of patient 10, replicated in PBMC with similar kinetics, without significant differences between those from peripheral blood and those from rectal cells (Fig. 4).
FIG. 4.
Replication kinetics of primary viruses isolated from peripheral blood (squares) in comparison with those from rectal samples (circles) before initiation of therapy. Production of p24 antigen in the culture supernatant was measured on days 0, 4, 7, 10, and 14.
To assess the ability of virus to generate syncytia in MT-2 cells and to evaluate their coreceptor usage, isolates were inoculated into MT-2 cells and U87MG.CD4 cells stably transfected with the chemokine receptor CCR1, CCR2b, CCR3, CCR5, or CXCR4. All viral isolates failed to generate syncytia in an MT-2 cell culture and failed to use coreceptors other than CCR5 (data not shown). All of these isolates therefore fit into the category of R5 or non-syncytium-inducing viruses.
DISCUSSION
In this work, we have carried out an extensive investigation of the genetic polymorphism in the gp120 sequences of HIV-1 in peripheral blood and tissues during the acute and chronic infection phases. We believe that this is the first study of this kind to evaluate the genetic diversity and relatedness of HIV-1 in both peripheral blood and tissue during the acute infection phase. Viral sequences from both peripheral blood and tissues were analyzed by using multiple techniques. The profiles of length polymorphism in the V1-V2 and V4-V5 regions of gp120 gave a single and uniform peak in both the peripheral blood and tissues of all acutely infected individuals. More importantly, these profiles are identical in all tissues during the acute infection phase. This inferred homogeneity was confirmed by sequence analysis. This finding suggests that homogeneity of viral sequences during the acute infection phase is not just restricted to the peripheral blood but extends to most tissue compartments. Explosive viral replication during the acute infection phase therefore populates all body compartments with a dominant viral variant, discounting the likelihood that different variants preferentially target different tissue compartments during the acute infection phase. This observation is consistent with previous findings based on peripheral blood sampling (7, 25, 29, 56, 57) that during the acute infection phase, the viral population is relatively homogeneous compared with that in the later stages of infection.
In contrast to what was observed in acutely infected patients, multiple peaks with different lengths of both the V1-V2 and V4-V5 regions were identified in peripheral blood and tissues in chronically infected patients. Significant differences between the peripheral blood and tissues in the profile of length polymorphism and sequences were also found in these patients, indicating that distinct viral populations reside in the different compartments. In the context of what we have observed in acutely infected individuals, this finding suggests that the compartmentalization of viral sequences observed here and elsewhere (2, 9, 10, 12, 14, 15, 17, 20-22, 24, 30-32, 34, 35, 37, 43-46, 58) must have occurred at a later point in infection. Furthermore, phenotypic characterization of viruses isolated simultaneously from the peripheral blood and rectal samples of the six chronically infected patients showed them to be similar in replication kinetics and coreceptor usage despite the significant genetic differences between their gp120 sequences. It is probable that the observed amino acid differences do not have a significant impact on these viral phenotypic characters or that current assays are not sensitive enough to discern those differences. In any case, homogeneity in viral sequences during the acute infection phase in both peripheral blood and tissues strongly suggests that a dominant variant has infiltrated all of the tissue compartments. Once this homogeneous viral population populates a particular tissue compartment, it subsequently diversifies and becomes distinct from the virus in other compartments, most likely as a result of the different selective environments in different tissue compartments. The different viral populations in the peripheral blood and tissues of chronically infected patients are therefore more likely to reflect gradual diversification from a common dominant viral variant rather than preferential targeting of distinct viral populations at the beginning of the infection.
One may only speculate as to the mechanism underlying the homogeneous viral population that exists during the acute infection phase. The simple laws of population genetics dictate that when even a highly complex mixture of species expands to fill a new environment by uncontrolled replication, genetic polymorphism is lost, sometimes completely (13, 16, 41, 50). These founder effects are a natural consequence of uncontrolled expansion, are stochastic in nature, and (perhaps surprisingly) do not require any selective force to deliver a monotypic population (13, 16, 41, 50). It is also possible that homogeneity is a result of selective constraints imposed on the incoming viral population either by replication competition among different variants or by the external environment in the form of immune surveillance and the availability of target cells. As a result, the fastest-growing and/or best-suited viral variant comes to dominate the entire human body during the acute infection phase. Part of these selection pressures could act on the envelope gene, which could help explain the observation that the majority of the viruses isolated during the acute infection phase use CCR5 as a coreceptor. However, selective pressure on the envelope-encoding gene alone cannot explain the whole story, in particular in those acutely infected women in whom relatively divergent rather than homogeneous viral populations have been identified during the acute infection phase (28). Differences in the hormonal regulation or the availability, type, and number of target cells between the male rectal and female vaginal tracts may offer some explanations for the observed differences. It is probable that both viral and host factors play critical roles in the emergence of a homogeneous population during the acute infection phase.
In summary, our study demonstrates that homogeneity in gp120 sequences during the acute infection phase is not restricted to the peripheral blood but extends to the tonsils and rectum and possibly to all tissue compartments. The dominant viral strain during the acute infection phase is therefore likely to form the genetic basis for the subsequent diversification of HIV-1 in various tissue compartments during the later stages of infection. Our previous findings that viral populations continue to diverge from the initially dominant population during the course of infection (18, 53) strongly support this notion. We conclude that the compartmentalization of viral sequences observed in chronically infected patients reflects a gradual diversification from a common dominant viral variant at seroconversion rather than a variety of distinct viral populations migrating to different tissue compartments at the start of the infection. Finally, our findings here and elsewhere argue strongly for early treatment intervention before these viruses become divergent and hence more likely to develop both drug resistance mutations and other mutations associated with immune evasion.
Acknowledgments
U87MG.CD4 cell lines and HOS.CD4 cells were kindly provided by D. Littman, Skirball Institute for Molecular Medicine, New York, N.Y. Thanks to P. Balfe for critical reading of the manuscript.
This work was supported by National Institutes of Health grants AI46964, AI40387, and AI 41534 and by the General Clinical Research Center of Rockefeller University (M01-RR00102).
REFERENCES
- 1.Balfe, P., P. Simmonds, C. A. Ludlam, J. O. Bishop, and A. J. Brown. 1990. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J. Virol. 64:6221-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, S. W., A. Barboza, C. M. Wilcox, C. E. Forsmark, and J. A. Levy. 1991. Characterization of human immunodeficiency virus type 1 strains recovered from the bowel of infected individuals. Virology 182:802-809. [DOI] [PubMed] [Google Scholar]
- 3.Bonhoeffer, S., E. C. Holmes, and M. A. Nowak. 1995. Causes of HIV diversity. Nature 376:125.. [DOI] [PubMed] [Google Scholar]
- 4.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 5.Cheng-Mayer, C., D. Seto, M. Tateno, and J. A. Levy. 1988. Biologic features of HIV-1 that correlate with virulence in the host. Science 240:80-82. [DOI] [PubMed] [Google Scholar]
- 6.Cichutek, K., H. Merget, S. Norley, R. Linde, W. Kreuz, M. Gahr, and R. Kurth. 1992. Development of a quasispecies of human immunodeficiency virus type 1 in vivo. Proc. Natl. Acad. Sci. USA 89:7365-7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cichutek, K., S. Norley, R. Linde, W. Kreuz, M. Gahr, J. Lower, G. von Wangenheim, and R. Kurth. 1991. Lack of HIV-1 V3 region sequence diversity in two haemophiliac patients infected with a putative biologic clone of HIV-1. AIDS 5:1185-1187. [DOI] [PubMed] [Google Scholar]
- 8.Coffin, J. M. 1992. Genetic diversity and evolution of retroviruses. Curr. Top. Microbiol. Immunol. 176:143-164. [DOI] [PubMed] [Google Scholar]
- 9.Delassus, S., R. Cheynier, and S. Wain-Hobson. 1992. Nonhomogeneous distribution of human immunodeficiency virus type 1 proviruses in the spleen. J. Virol. 66:5642-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwart, E. L., J. I. Mullins, P. Gupta, G. H. Learn, Jr., M. Holodniy, D. Katzenstein, B. D. Walker, and M. K. Singh. 1998. Human immunodeficiency virus type 1 populations in blood and semen. J. Virol. 72:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart, E. L., H. W. Sheppard, B. D. Walker, J. Goudsmit, and J. I. Mullins. 1994. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J. Virol. 68:6672-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dittmar, M. T., G. Simmons, Y. Donaldson, P. Simmonds, P. R. Clapham, T. F. Schulz, and R. A. Weiss. 1997. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J. Virol. 71:5140-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobzhansky, T. 1951. Genetics and the origin of species. Columbia University Press, New York, N.Y.
- 14.Donaldson, Y. K., J. E. Bell, E. C. Holmes, E. S. Hughes, H. K. Brown, and P. Simmonds. 1994. In vivo distribution and cytopathology of variants of human immunodeficiency virus type 1 showing restricted sequence variability in the V3 loop. J. Virol. 68:5991-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epstein, L. G., C. Kuiken, B. M. Blumberg, S. Hartman, L. R. Sharer, M. Clement, and J. Goudsmit. 1991. HIV-1 V3 domain variation in brain and spleen of children with AIDS: tissue-specific evolution within host-determined quasispecies. Virology 180:583-590. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, R. A. 1958. The genetic theory of natural selection. Dover, New York, N.Y.
- 17.Gratton, S., R. Cheynier, M. J. Dumaurier, E. Oksenhendler, and S. Wain-Hobson. 2000. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc. Natl. Acad. Sci. USA 97:14566-14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes, E. C., L. Q. Zhang, P. Simmonds, C. A. Ludlam, and A. J. Brown. 1992. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc. Natl. Acad. Sci. USA 89:4835-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosmalin, A., A. Samri, M. J. Dumaurier, Y. Dudoit, E. Oksenhendler, M. Karmochkine, B. Autran, S. Wain-Hobson, and R. Cheynier. 2001. HIV-specific effector cytotoxic T lymphocytes and HIV-producing cells colocalize in white pulps and germinal centers from infected patients. Blood 97:2695-2701. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, E. S., J. E. Bell, and P. Simmonds. 1997. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17gag and env genes. J. Virol. 71:1272-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itescu, S., P. F. Simonelli, R. J. Winchester, and H. S. Ginsberg. 1994. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc. Natl. Acad. Sci. USA 91:11378-11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keys, B., J. Karis, B. Fadeel, A. Valentin, G. Norkrans, L. Hagberg, and F. Chiodi. 1993. V3 sequences of paired HIV-1 isolates from blood and cerebrospinal fluid cluster according to host and show variation related to the clinical stage of disease. Virology 196:475-483. [DOI] [PubMed] [Google Scholar]
- 23.Korber, B. T., E. E. Allen, A. D. Farmer, and G. L. Myers. 1995. Heterogeneity of HIV-1 and HIV-2. AIDS 9(Suppl. A):S5-S18. [PubMed] [Google Scholar]
- 24.Korber, B. T., K. J. Kunstman, B. K. Patterson, M. Furtado, M. M. McEvilly, R. Levy, and S. M. Wolinsky. 1994. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 68:7467-7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuiken, C. L., V. V. Lukashov, E. Baan, J. Dekker, J. A. Leunissen, and J. Goudsmit. 1996. Evidence for limited within-person evolution of the V3 domain of the HIV-1 envelope in the Amsterdam population. AIDS 10:31-37. [DOI] [PubMed] [Google Scholar]
- 26.Kuiken, C. L., G. Zwart, E. Baan, R. A. Coutinho, J. A. van den Hoek, and J. Goudsmit. 1993. Increasing antigenic and genetic diversity of the V3 variable domain of the human immunodeficiency virus envelope protein in the course of the AIDS epidemic. Proc. Natl. Acad. Sci. USA 90:9061-9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaRosa, G. J., J. P. Davide, K. Weinhold, J. A. Waterbury, A. T. Profy, J. A. Lewis, A. J. Langlois, G. R. Dreesman, R. N. Boswell, P. Shadduck, et al. 1990. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science 249:932-935. [DOI] [PubMed] [Google Scholar]
- 28.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 29.McNearney, T., Z. Hornickova, R. Markham, A. Birdwell, M. Arens, A. Saah, and L. Ratner. 1992. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc. Natl. Acad. Sci. USA 89:10247-10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overbaugh, J., R. J. Anderson, J. O. Ndinya-Achola, and J. K. Kreiss. 1996. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res. Hum. Retrovir. 12:107-115. [DOI] [PubMed] [Google Scholar]
- 31.Panther, L. A., L. Tucker, C. Xu, R. E. Tuomala, J. I. Mullins, and D. J. Anderson. 2000. Genital tract human immunodeficiency virus type 1 (HIV-1) shedding and inflammation and HIV-1 env diversity in perinatal HIV-1 transmission. J. Infect. Dis. 181:555-563. [DOI] [PubMed] [Google Scholar]
- 32.Poss, M., H. L. Martin, J. K. Kreiss, L. Granville, B. Chohan, P. Nyange, K. Mandaliya, and J. Overbaugh. 1995. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J. Virol. 69:8118-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poss, M., and J. Overbaugh. 1999. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 73:5255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy, R. T., C. L. Achim, D. A. Sirko, S. Tehranchi, F. G. Kraus, F. Wong-Staal, and C. A. Wiley. 1996. Sequence analysis of the V3 loop in brain and spleen of patients with HIV encephalitis. AIDS Res. Hum. Retrovir. 12:477-482. [DOI] [PubMed] [Google Scholar]
- 35.Sankale, J. L., R. S. De La Tour, R. G. Marlink, R. Scheib, S. Mboup, M. E. Essex, and P. J. Kanki. 1996. Distinct quasi-species in the blood and the brain of an HIV-2-infected individual. Virology 226:418-423. [DOI] [PubMed] [Google Scholar]
- 36.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapshak, P., D. M. Segal, K. A. Crandall, R. K. Fujimura, B. T. Zhang, K. Q. Xin, K. Okuda, C. K. Petito, C. Eisdorfer, and K. Goodkin. 1999. Independent evolution of HIV type 1 in different brain regions. AIDS Res. Hum. Retrovir. 15:811-820. [DOI] [PubMed] [Google Scholar]
- 38.Shpaer, E. G., and J. I. Mullins. 1993. Rates of amino acid change in the envelope protein correlate with pathogenicity of primate lentiviruses. J. Mol. Evol. 37:57-65. [DOI] [PubMed] [Google Scholar]
- 39.Simmonds, P., P. Balfe, C. A. Ludlam, J. O. Bishop, and A. J. Brown. 1990. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J. Virol. 64:5840-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmonds, P., P. Balfe, J. F. Peutherer, C. A. Ludlam, J. O. Bishop, and A. J. Brown. 1990. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J. Virol. 64:864-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, D. T., A. J. F. Griffiths, J. H. Miller, and R. C. Lweontin. 1989. An introduction to genetic analysis. W. H. Freeman & Co., New York, N.Y.
- 42.Tersmette, M., J. M. Lange, R. E. de Goede, F. de Wolf, J. K. Eeftink-Schattenkerk, P. T. Schellekens, R. A. Coutinho, J. G. Huisman, J. Goudsmit, and F. Miedema. 1989. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet i:983-985. [DOI] [PubMed]
- 43.van der Hoek, L., J. Goudsmit, J. Maas, and C. J. Sol. 1998. Human immunodeficiency virus type 1 in faeces and serum: evidence against independently evolving subpopulations. J. Gen. Virol. 79:2455-2459. [DOI] [PubMed] [Google Scholar]
- 44.van der Hoek, L., C. J. Sol, J. Maas, V. V. Lukashov, C. L. Kuiken, and J. Goudsmit. 1998. Genetic differences between human immunodeficiency virus type 1 subpopulations in faeces and serum. J. Gen. Virol. 79:259-267. [DOI] [PubMed] [Google Scholar]
- 45.van der Hoek, L., C. J. Sol, F. Snijders, J. F. Bartelsman, R. Boom, and J. Goudsmit. 1996. Human immunodeficiency virus type 1 RNA populations in faeces with higher homology to intestinal populations than to blood populations. J. Gen. Virol. 77:2415-2425. [DOI] [PubMed] [Google Scholar]
- 46.van't Wout, A. B., L. J. Ran, C. L. Kuiken, N. A. Kootstra, S. T. Pals, and H. Schuitemaker. 1998. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J. Virol. 72:488-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wain-Hobson, S. 1992. Human immunodeficiency virus type 1 quasispecies in vivo and ex vivo. Curr. Top. Microbiol. Immunol. 176:181-193. [DOI] [PubMed] [Google Scholar]
- 48.Wolfs, T. F., J. J. de Jong, H. Van den Berg, J. M. Tijnagel, W. J. Krone, and J. Goudsmit. 1990. Evolution of sequences encoding the principal neutralization epitope of human immunodeficiency virus 1 is host dependent, rapid, and continuous. Proc. Natl. Acad. Sci. USA 87:9938-9942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 50.Wright, S. 1977. Evolution and the genetics of populations. Vol. 3. Experimental results and evolutionary deductions. University of Chicago Press, Chicago, Ill.
- 51.Zhang, L., C. D. Carruthers, T. He, Y. Huang, Y. Cao, G. Wang, B. Hahn, and D. D. Ho. 1997. HIV type 1 subtypes, coreceptor usage, and CCR5 polymorphism. AIDS Res. Hum. Retrovir. 13:1357-1366. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, L., C. Chung, B. S. Hu, T. He, Y. Guo, A. J. Kim, E. Skulsky, X. Jin, A. Hurley, B. Ramratnam, M. Markowitz, and D. D. Ho. 2000. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J. Clin. Investig. 106:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, L., R. S. Diaz, D. D. Ho, J. W. Mosley, M. P. Busch, and A. Mayer. 1997. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J. Virol. 71:2555-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, L., T. He, Y. Huang, Z. Chen, Y. Guo, S. Wu, K. J. Kunstman, R. C. Brown, J. P. Phair, A. U. Neumann, D. D. Ho, and S. M. Wolinsky. 1998. Chemokine coreceptor usage by diverse primary isolates of human immunodeficiency virus type 1. J. Virol. 72:9307-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, D. A. Cooper, and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]