Abstract
The 2B protein of enteroviruses is the viral membrane-active protein that is responsible for the modifications in host cell membrane permeability that take place in enterovirus-infected cells. The 2B protein shows structural similarities to the group of lytic polypeptides, polypeptides that permeate membranes either by forming multimeric membrane-integral pores or, alternatively, by lying parallel to the lipid bilayer and disturbing the curvature and symmetry of the membrane. Our aim is to gain more insight into the molecular architecture of the 2B protein in vivo. In this study, the possible existence of multimers of the coxsackie B3 virus 2B protein in single living cells was explored by fluorescence resonance energy transfer (FRET) microscopy. FRET between fusion proteins 2B-ECFP and 2B-EYFP (enhanced cyan and yellow fluorescent variants of green fluorescent protein) was monitored by using spectral imaging microscopy (SPIM) and fluorescence lifetime imaging microscopy (FLIM). Both techniques revealed the occurrence of intermolecular FRET between 2B-ECFP and 2B-EYFP, providing evidence for the formation of protein 2B homomultimers. Putative models for the mode of action of the membrane-active 2B protein and the formation of membrane-integral pores by 2B multimers are discussed.
A large number of human pathogenic viruses are cytolytic. During their replication cycle they gradually modify host cell membrane permeability by mechanisms that are as yet poorly understood (reviewed in reference 8). Enteroviruses (e.g., poliovirus, coxsackievirus, and echovirus) are small cytolytic human viruses that replicate their RNA genomes at virus-induced membrane vesicles that accumulate in the cytoplasm of the infected cell (reviewed in reference 6). During infection, enteroviruses gradually modify host cell membrane permeability. Early in infection, Ca2+ is released from intracellular stores (endoplasmic reticulum [ER] and Golgi complex) and gradients of cations maintained by the plasma membrane are disrupted (8, 34). Later, small low-molecular-weight compounds can also pass the plasma membranes of infected cells (8). Opposed to the traditional idea that the membrane modifications are due to either the bulk of viral gene expression or the accumulation of virus particles is the new concept that cytolytic viruses manipulate host cell membranes in a controlled manner by producing membrane-active (i.e., membrane-permeabilizing) proteins. Consistent with this concept, we and others have shown that individual expression of protein 2B (or its precursor protein 2BC) in mammalian cells leads to the release of Ca2+ from intracellular stores (33) and an increased plasma membrane permeability to both extracellular Ca2+ and hygromycin B, a small normally nonpermeative translation inhibitor (1, 12, 34, 35).
The 2B protein is a small hydrophobic protein (99 amino acids) that in infected cells is localized at the virus-induced, secretory-pathway-derived membrane vesicles at which viral RNA replication takes place (6). The 2B protein plays a functional role (as part of the 2BC precursor) in the accumulation of these membrane vesicles. Mutations in the 2B protein abolish viral RNA replication (5, 21, 32, 33, 36), suggesting that the ability of the 2B protein to modify secretory pathway membranes is required to create the membranous replication complex required for the replication of the viral RNA. How the ability of the 2B protein to disturb secretory-pathway membranes leads to an increased permeability of the plasma membrane is as yet unknown.
The mechanism by which the enterovirus 2B protein modifies membrane permeability is largely unknown. All enterovirus 2B proteins contain two hydrophobic domains (Fig. 1A), one of which is predicted to form a cationic amphipathic α-helix (Fig. 1B) (32, 33). This cationic amphipathic α-helix is a major determinant for the membrane-active function of the 2B protein (34, 35) and displays characteristics typical of the family of the so-called membrane-lytic peptides (25, 33). It has been proposed that these cytolytic peptides, which are ubiquitous in nature and which constitute a defense system against invading microorganisms in many organisms, permeate membranes either by forming multimeric membrane-integral pores or, alternatively, by lying parallel to the lipid bilayer and increasing membrane curvature (reviewed in references 4 and 28).
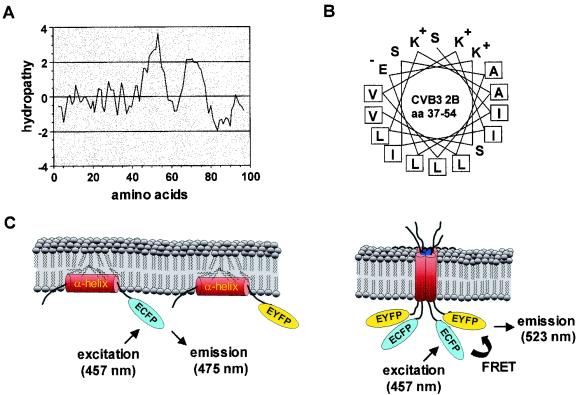
FIG. 1.
(A) Hydropathy plot of CBV3 protein 2B (99 amino acids) by using a hydropathy window size of nine residues. Positive hydropathy indicates a hydrophobic nature. Negative hydropathy indicates a hydrophilic nature. (B) Helical-wheel representation of the putative cationic amphipathic α-helix formed by amino acids (aa) 37 to 54 of CBV3 protein 2B. Positively and negatively charged residues are indicated. Hydrophobic residues are boxed. (C) FRET between 2B-ECFP and 2B-EYFP can occur when these fluorophores are brought in close proximity through an intermolecular 2B-2B interaction. The occurrence of FRET leads to (i) a decreased ECFP fluorescence and increased EYFP fluorescence as the result of sensitized emission and (ii) a decreased ECFP fluorescence lifetime.
In this study, we investigated the possible existence of membrane-integral multimers of protein 2B of the coxsackie B3 virus (CBV3) in living cells by fluorescence resonance energy transfer (FRET) microscopy (15, 23, 26). FRET is the phenomenon by which a donor fluorophore transfers its excited-state energy to an absorbing chromophore in a nonradiative manner. The efficiency of FRET strongly depends on the molecular proximity between the donor and the acceptor. One of the applications of the FRET technology is the investigation of the spatial distribution and assembly of protein complexes in living cells in a noninvasive and nondestructive manner. Fluorophores ECFP and EYFP, enhanced cyan and yellow fluorescent chromophore-mutated variants of green fluorescent protein (GFP), show an excellent spectral overlap and therefore form an appropriate donor-acceptor FRET pair, with a R0 value (i.e., the distance at which 50% of the excited donor molecules transfer energy by FRET) of ∼50 Å (31). Because this distance is in the same order of magnitude as protein dimensions, the occurrence of FRET is indicative of a direct protein-protein interaction between ECFP and EYFP fusion proteins. FRET results in a reduced fluorescence intensity of the donor (ECFP) and an increased fluorescence intensity of the acceptor (EYFP), as can be detected by spectral imaging microscopy (SPIM). Moreover, FRET results in a decrease in fluorescence lifetime (the average time that the molecule spends in the excited state) of the donor (10). Unlike fluorescence intensity, the fluorescence lifetime is not dependent on the concentration of the chromophore, direct absorption of donor fluorescence intensity, moderate photobleaching, or light path length, conditions that are difficult to control inside a cell. Therefore, production of high-resolution micrographs and fluorescence lifetime imaging microscopy (FLIM) analysis of the fluorescence lifetime on a pixel-by-pixel basis, in which each pixel represents the local fluorescence lifetime, constitute a very reliable tool to obtain quantitative information about the occurrence and efficiency of FRET in single living cells (3, 13). The FRET technology has recently been applied to demonstrate homomultimerization reactions in living cells (20, 27, 30, 37). Here, we report the analysis of the possible existence of protein 2B homomultimers by FRET microscopy (Fig. 1C). For this purpose, 2B proteins fused either with ECFP or EYFP were coexpressed and assayed in vivo for FRET by SPIM and FLIM to monitor intermolecular 2B-2B protein interactions.
MATERIALS AND METHODS
Plasmids.
For the construction of plasmids p2B-ECFP and p2B-EYFP, we made use of a previously constructed p2B-EGFP plasmid. This plasmid was constructed by amplification of the CBV3 2B coding sequence by PCR with forward primer 5′-GCTAGCGTCGACGCCACCATGGGAGTGAAGGACTATGTGGAA-3′ (restriction sites are in italics and the start codon is underlined) and reverse primer 5′-CCAGCTGGATCCTTGGCGTTCAGCCATAGG-3′ and cutting the PCR product with SalI and BamHI and cloning it into plasmid pEGFP-N3 (Clontech) cut with the same enzymes. The sequence of the 2B insert was confirmed by sequence analysis. To construct plasmids p2B-ECFP and p2B-EYFP, first unique AgeI and EcoRV sites were introduced between the 2B coding region and the enhanced GFP (EGFP) coding region. Plasmid p2B-ECFP was generated by deleting the EGFP coding sequence with AgeI and SspBI and replacing it with the ECFP coding sequence cut out of plasmid pECFP-N1 (Clontech) with the same enzymes. In plasmid p2B-ECFP, the 2B and ECFP coding sequences are separated by the coding sequence for a seven-amino-acid linker (GSPPVAT). Plasmid p2B-EYFP was generated by deleting the EGFP coding sequence with EcoRV and SspBI and replacing it with the EYFP coding sequence (containing mutations V68L and Q69K, which reduce the pH sensitivity of the EYFP protein) cut out of plasmid yellow cameleon 2.1 (22) (a kind gift from R. Y. Tsien, Howard Hughes Medical Institute, University of California, San Diego) with enzymes EcoICRI and SspBI. In plasmid p2B-EYFP, the 2B and EYFP coding sequences are separated by the coding sequence for a seven-amino-acid linker (GSPPVDL).
Plasmids p2BC-ECFP and p2BC-EYFP and plasmids p2C-ECFP and p2C-EYFP were constructed by deleting the 2B-encoding sequence from plasmids p2B-ECFP and p2B-EYFP with SalI and BamHI and replacing it with the sequence encoding 2BC and 2C. Thereby, the 2BC and 2C proteins and the ECFP and EYFP proteins are separated by the same seven-amino-acid linkers as those described for the 2B-ECFP and 2B-EYFP proteins (GSPPVAT and GSPPVDL, respectively). The 2BC coding sequence was amplified by PCR with the 2B forward primer and reverse primer 5′-GTCGACGGATCCCTGGAACAGTGCCTCAAG-3′. The 2C coding sequence was amplified by PCR with forward primer 5′-GGGGGGGTCGACGCCACCATGGGGAACAATAGCTGGCTTAAGAAA-3′ and the 2BC reverse primer. The PCR products were cut with SalI and BamHI and cloned upstream of the ECFP and EYFP coding sequences. In the 2C protein, an additional glycine residue is inserted after the starting methionine in order to maintain an optimal Kozak sequence. The sequences of the 2BC and 2C inserts were confirmed by sequence analysis.
Plasmids p2B[1-85]ECFP and p2B[1-85]EYFP contain a fusion of the coding sequence for the N-terminal 85 amino acids of 2B [2B (1-85)] and the coding sequences for the ECFP and EYFP proteins, respectively. In these plasmids the coding sequences for 2B[1-85] and ECFP or EYFP are separated by the coding sequences for six-amino-acid linkers (DPPVAT and DPPVPL, respectively).
The linkers between viral proteins 2B, 2B[1-85], 2BC, and 2C and the fluorescent proteins contain a number of prolines (which may somehow influence, positively or negatively, the orientation between the viral proteins and the fluorescent protein, but this cannot be predicted) and differ in two amino acids from the linkers in the ECFP and the EYFP fusion proteins. It is unlikely, however, that these features interfered with the outcome of the FRET experiments because it is the distance between the fluorescent protein and the viral protein that is of importance rather than the amino acid composition of the linker. In fact, in Results FRET reactions are demonstrated, indicating that neither the composition of the linkers nor the distance relative to the viral proteins was detrimental to the occurrence of FRET.
Plasmids p2B-myc, p2BC-myc, and p2C-myc were constructed by replacing the EYFP coding sequence from plasmids p2B-EYFP, p2BC-EYPF, and p2C-EYFP, respectively, with a DNA fragment encoding the myc tag (followed by a stop codon).
Plasmid pEYFP-2B was constructed by amplification of the 2B coding sequence by PCR with the 2B forward primer and reverse primer 5′-AAGCCACCCGGGCTATTGGCGTTCAGCCATAGG-3′ and cutting the PCR product with SalI and SmaI and cloning it into plasmid pEYFP-C1 (containing mutations V68L and Q69K) cut with the same enzymes. The sequence of the 2B insert was confirmed by sequence analysis.
Plasmid pEYFP-ECFP was constructed by cloning the ECFP coding sequence, cut out of pECFP-N1 (Clontech) with XhoI and DraI, into pEYFP-C1 (containing mutations V68L and Q69K) cut with SalI and SmaI. The resulting plasmid contains an in frame fusion of the EYFP and ECFP coding sequences separated by the coding sequence for a 35-amino-acid linker region.
Plasmid pECFP-ER (Clontech) encodes an ECFP fusion protein with the calreticulin signal sequence at its amino terminus and a KDEL retrieval sequence at its carboxy terminus. The ECFP-ER protein is a soluble protein that localizes in the lumen of the ER. Plasmid pECFP-Golgi (Clontech) encodes an ECFP fusion protein that contains the N-terminal 81 amino acids of human β-1,4-galactosyltransferase at its amino terminus. This β-1,4-galactosyltransferase domain contains the membrane anchor that targets the ECFP fusion protein to the transmedial region of the Golgi complex.
Transfections.
Buffalo green monkey (BGM) cells were grown to 70% confluence on coverslips in 24-well tissue culture plates in minimal essential medium. Cells were transfected with a total of 3 μg of plasmid DNA by using FuGENE 6 transfection reagent according to the instructions of the manufacturer (Roche). When cells were cotransfected with two plasmids, a ratio of 1:2 (ECFP-EYFP construct) was used because an excess of the EYFP fluorescence acceptor relative to the ECFP fluorescence donor increases the occurrence of FRET. For each transfection, 3 μl of FuGENE 6 reagent was added to 100 μl of serum-free medium and the mixture was incubated for 5 min at room temperature. The mixture was added drop-wise to the plasmid DNA and incubated at room temperature for 15 min. After this incubation, the FuGENE 6 reagent-DNA mixture was added drop-wise to the cells. The cells were grown at 37°C until further analysis. Living cells were assayed by microscopical analysis (confocal laser scanning microscopy [CLSM], SPIM, and FLIM) at 40 h posttransfection at room temperature in HEPES-buffered, phenol red-free and serum-free medium.
Hygromycin B permeability analysis.
Membrane permeability was assayed by the analysis of protein synthesis in the presence or absence of hygromycin B (hygB), a small inhibitor of translation that under physiological conditions poorly passes the membrane. BGM cells were transfected with plasmids encoding EYFP, 2B-EYFP, or EYFP-2B as described above. At 40 h posttransfection, cells were pulse labeled with 25 μCi of [35S]methionine (Amersham) in methionine- and serum-free minimal essential medium in the presence or absence of hygB (500 μg per ml). The EYFP proteins were immunoprecipitated with anti-EGFP polyclonal antisera (raised against a recombinant glutathione S-transferase-EGFP fusion protein) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography.
CLSM.
Confocal images were taken with an LSM510 confocal laser scanning microscope system (Carl-Zeiss Gmbh) based on an Axiovert inverted microscope equipped with an argon ion laser and with a 40× oil immersion objective with a numerical aperture (NA) of 1.3. Excitation was provided by the 457- and 514-nm Ar laser lines controlled by an acoustic-optical tunable filter. ECFP was excited at 457 nm and selectively detected by using an HFT457/514 main dichroic splitter, a NFT 515 dichroic splitter acting as a short-pass filter, and a band-pass 470- to 500-nm filter. EYFP was excited at 514 nm with the same HFT457/514 main dichroic splitter, the same NFT 515 dichroic splitter acting as a long-pass filter, and a band-pass 535- to 590-nm filter. Pinholes were set at 1 airy disk unit for both ECFP and EYFP confocal detection channels. Operating in a multitracking mode allowed cross talk-free ECFP and EYFP images to be collected without changing filters.
SPIM.
SPIM was performed using a Leica DMR/BE upright epifluorescence microscope equipped with Chromex (Albuquerque, N.Mex.) 250IS imaging spectrograph coupled to a Photometrics (Tucson, Ariz.) CH250 charge-coupled device camera incorporating a back-illuminated SIT502 chip with 512 by 512 24-μm square pixels. For SPIM, excitation was provided by a 100-W Hg arc lamp whose 435-nm line was selected by inserting an Omega (Brattleboro, Vt.) 435DF10 band-pass filter in the excitation light path. The excitation light was reflected onto the sample by an Omega 430DCLP dichroic mirror. A Leica 20× HR-Fluotar air objective (NA = 0.5) was employed. Residual excitation light was rejected by using a Schott (Mainz, Germany) GG455 long-pass filter. In the spectrograph a 150-groove/mm grating with a central wavelength of 500 nm was used. Details of the setup have been described previously (19, 27). Single cells expressing specific ECFP and/or EYFP fusion proteins were positioned by aligning them across the entrance slit of the spectrograph (set at 200 μm wide corresponding to a line 10 μm wide in the object plane). Acquisition time was 1 to 5 s. Regions of the image spectrum corresponding to intracellular locations of labeled cells were distance averaged (typically 10 to 100 rows of pixels), and the resulting fluorescence spectra were corrected for background fluorescence and camera bias by background subtraction using an extracellular region just next to the plasma membrane region from the same spectral image. The resulting spectra were not corrected for the spectral instrument response, yielding a slightly underestimated intensity in the blue edge of the spectra.
FLIM.
Frequency domain FLIM (14, 17, 18, 29) was used to measure the ECFP fluorescence lifetime. The instrument is described in detail elsewhere (13, 18). For exciting ECFP, the sample was continuously excited with a 457-nm argon ion laser line (Coherent Innova 70 spectrum CW Ar/Kr laser) sinusoidally modulated at 40.000 MHz. The ECFP fluorescence was selectively imaged with an Omega 470 DCLP dichroic mirror and an Omega 487RDF42 band-pass emission filter. A 63× 1.4-NA oil immersion objective was used. Twenty phase images (1 to 3 s each) were taken (10 with increasing phase and 10 with decreasing phase, allowing correction for photobleaching). The emitted fluorescence light is sinusoidally modulated at the same frequency as the excitation light but displays a shift in phase (Δφ) and a reduction in the relative modulation depth (M) due to the time difference between excitation and emission (i.e., the fluorescence lifetime of the ECFP protein). The phase shift and relative modulation depths can be used to calculate the respective phase and modulation lifetimes (τφ and τM, respectively). Reference phase settings and modulation were calibrated approximately every 30 min by measuring a glass microcuvette filled with an erythrosine-B solution in water (single-component fluorescence decay with a lifetime of 0.08 ns). Image analysis for calculating lifetime images on a pixel-by-pixel basis was performed on a Silicon Graphics Instruments workstation with software described previously (17). Fluorescence lifetime τφ was calculated according to the equation τφ = ω−1tan Δφ. Fluorescence lifetime τM was calculated from the equation τM = ω−1(M−2-1)1/2.
RESULTS
Construction and characterization of fluorescent fusion proteins.
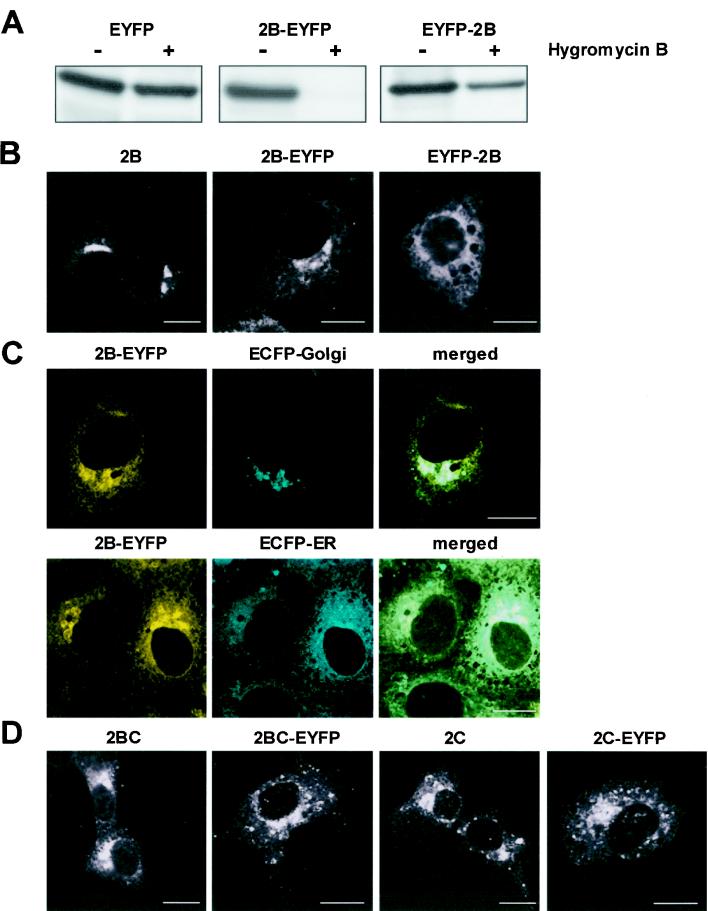
The fusion of a fluorescent protein at the amino terminus or carboxy terminus of a protein may affect its biological function. Therefore, EYFP was fused at both ends of the 2B protein and the corresponding constructs (2B-EYFP and EYFP-2B) were tested for their membrane-active function. For this, BGM cells were transiently transfected with plasmids encoding EYFP (control), 2B-EYFP, or EYFP-2B and assayed for increased membrane permeability to hygromycin B, a small translation inhibitor that normally passes the membrane poorly. Expression of the EYFP protein had little effect on membrane permeability, demonstrating the relative impermeability of the cellular membranes to this translation inhibitor (Fig. 2A). The 2B-EYFP fusion protein retained its membrane-active function, as indicated by the severe inhibition of translation in the presence of hygromycin B. The membrane-active function of the EYFP-2B fusion protein, however, appeared to be impaired. Furthermore, CLSM analysis revealed that the 2B-EYFP fluorescence pattern most closely resembled the localization of epitope-tagged 2B protein as detected by immunocytochemistry (Fig. 2B). The intracellular localization of the 2B-EYFP protein was further investigated by cotransfection with ECFP fusion proteins targeted to the Golgi complex and the ER. Colocalization analysis was performed in such a way that there was no cross talk between ECFP and EYFP fluorescence signals. Figure 2C shows that 2B-EYFP colocalized with the ER network and the Golgi complex, consistent with the localization described for the closely related poliovirus 2B protein (7, 24). Thus, fusion of a fluorescent protein at the carboxy terminus of the 2B protein does not interfere with the membrane-active function and intracellular localization of the 2B protein.
FIG. 2.
Functional characterization and localization of fluorescent fusion proteins. (A) Analysis of the membrane-active function of the 2B-EYFP and EYFP-2B proteins. BGM cells were transiently transfected with either EYFP, 2B-EYFP, or EYFP-2B and pulse labeled for 30 min with [35S]methionine in the absence (−) or presence (+) of hygromycin B, a small normally nonpermeative translation inhibitor, at 40 h posttransfection. The EYFP proteins were immunoprecipitated using anti-EGFP polyclonal antisera and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (B) CLSM localization of (epitope-tagged) 2B protein, 2B-EYFP, and EYFP-2B. Left image, immunocytochemical localization of 2B-myc, as detected by the mouse monoclonal anti-c-myc (clone 9E10) antibody. An identical localization pattern was observed with a FLAG-2B protein (data not shown). (C) Colocalization of the 2B-EYFP protein with the ER and Golgi complex. BGM cells were cotransfected with 2B-EYFP- and ECFP-ER- or ECFP-Golgi-encoding plasmids and examined by CLSM at 40 h posttransfection. (D) CLSM localization of 2BC-myc, 2BC-EYFP, 2C-myc, and 2C-EYFP. Bars, 10 μm.
The efficiency of FRET depends on the distance between the ECFP and EYFP fluorophores. The distance between these fluorophores and the molecular backbone to which they are attached can influence the extent of FRET. Therefore, we also fused the ECFP and EYFP molecules to a carboxy-terminal-truncated 2B protein, lacking the last 14 amino acids (protein 2B[1-85]). Deletion of the last 14 amino acids had no adverse effects on either the membrane-active function or the localization of the 2B protein (data not shown).
We also generated fusions between ECFP and EYFP and the 2BC protein (the precursor protein of the 2B protein) and the 2C protein. Expression of both the 2BC and the 2C proteins in mammalian cells results in a massive accumulation of membrane vesicles with which these proteins are associated (2, 9). Fusion of fluorescent proteins to the carboxy termini of the 2BC and 2C proteins had little or no effect on their intracellular localization (Fig. 2D), and the 2BC-EYFP fusion protein retained its membrane-active function (data not shown).
Expression and (co)localization of fluorescent proteins.
BGM cells were transiently (co)transfected with plasmids driving expression of fluorescent fusion proteins and were investigated for (i) the expression and (co)localization of fluorescent proteins and (ii) the occurrence of FRET. All experiments were performed between 40 and 44 h posttransfection, which is the time point at which the 2B-EYFP fusion protein exerts its membrane-active function (Fig. 2A).
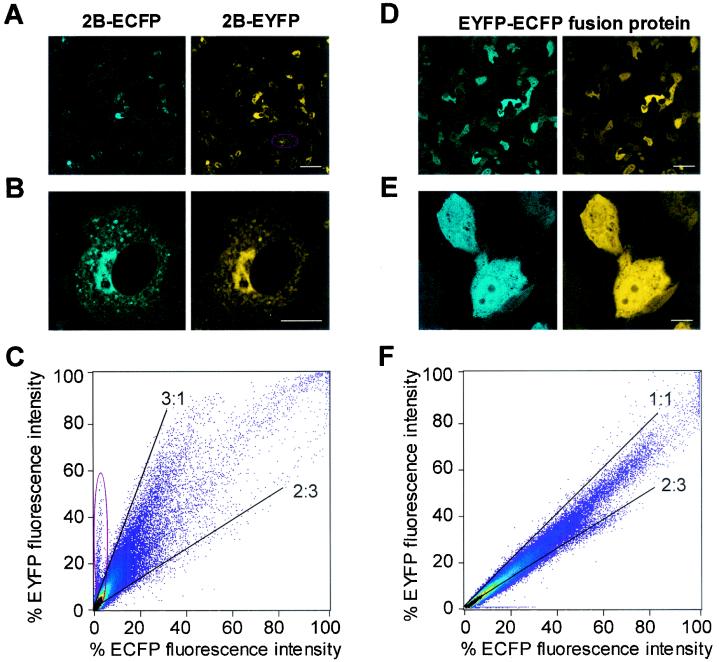
CLSM analysis of transiently transfected cells showed protein expression in about 30% of the cells. Upon cotransfection, the vast majority of the cells efficiently expressed both proteins. Figure 3A and B show overview images and detailed images of cells cotransfected with plasmids encoding 2B-ECFP and 2B-EYFP. These images, as well as the two-dimensional fluorescence intensity histogram quantitatively correlating ECFP and EYFP fluorescence intensities (Fig. 3C), demonstrate that (i) all transfected cells coexpressed both proteins and (ii) the expression ratios of both proteins in individual cells were more or less similar. The EYFP/ECFP intensity ratios varied between 3:1 and 2:3 (with these confocal settings) for the vast majority of the cells. Only in a few cells were aberrant expression ratios found (i.e., see the cell marked in Fig. 3A).
FIG. 3.
Expression and localization of fluorescent proteins. (A and B) Confocal fluorescence overview image of cells cotransfected with 2B-ECFP- and 2B-EYFP-encoding plasmids at low magnification (A) and high magnification (B). (C) 2D histogram comparing 2B-ECFP and 2B-EYFP fluorescence intensities of the image shown in panel A. The slopes of the two lines indicate that for most cells when these confocal settings are used, the intensity ratios were between 3:1 and 2:3. Ovals (A and C), aberrant cell almost lacking 2B-ECFP expression. (D and E) Confocal fluorescence images of cells expressing the EYFP-ECFP fusion protein at low magnification (D) and high magnification (E). (F) 2D histogram comparing ECFP and EYFP fluorescence intensities of the image shown in panel D. Bars, 50 (A and D) and 10 μm (B and E).
Figure 3D and E show overview images and detailed images of cells transfected with the plasmid encoding an EYFP-ECFP fusion protein (a fusion construct that was generated as positive control for the FRET experiments). Cells expressing the EYFP-ECFP fusion protein showed a perfect colocalization of ECFP and EYFP fluorescence signals in the cytosol and nucleus, a localization that is similar to that observed with the ECFP protein alone (data not shown). The perfect colocalization is expected given the fact that ECFP and EYFP are fused into one protein structure. As a result the intensity ratio, as illustrated in Fig. 3F, varies only between 1:1 and 2:3; this ratio is much better defined than the ratios observed in the cotransfection of 2B-ECFP and 2B-EYFP (Fig. 3C).
Analysis of molecular interactions by SPIM.
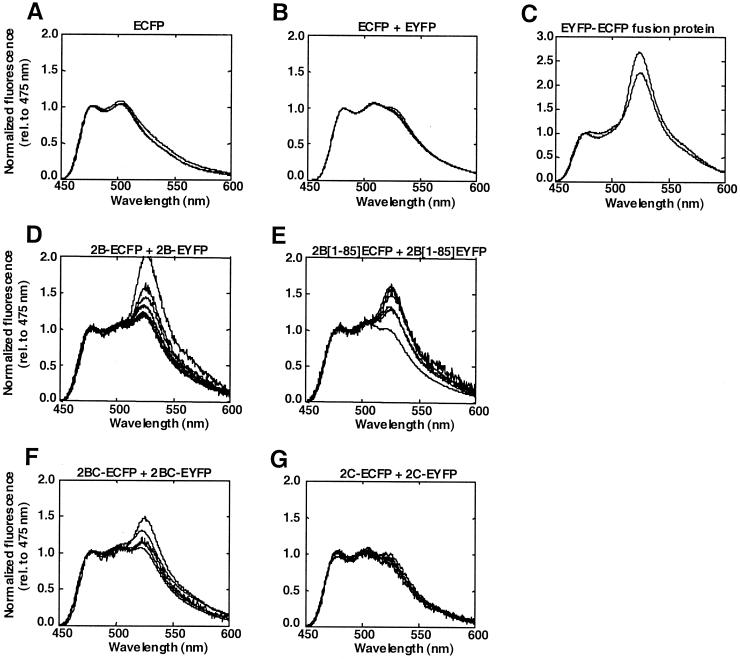
To investigate molecular interactions between the fluorescent fusion proteins in vivo, we investigated the occurrence of FRET by SPIM. FRET between ECFP and EYFP fusion proteins leads to the quenching of the ECFP donor and sensitized emission of the EYFP acceptor. With SPIM, fluorescence emission spectra can be recorded from living cells with spatial resolution along one dimension (15, 16). Increased ratios of the fluorescence emitted at 523 nm (EYFP) to that emitted at 475 nm (ECFP) (523/475 ratios) suggest the occurrence of FRET. As shown in Fig. 4A, the spectra of cells expressing ECFP alone display two peaks, one at 475 nm and one at 505 nm. The spectra of cells coexpressing (unfused) ECFP and EYFP contain an additional peak at 523 nm (Fig. 4B). Because EYFP slightly absorbs at the employed excitation wavelength (430 nm), a small contribution of directly excited EYFP fluorescence is expected in the emission spectrum. For comparative reasons all single-cell spectra were normalized to the fluorescence observed at 475 nm. Consequently, for FRET the reduction in the ECFP intensity is not visible in these normalized spectra but will contribute to a “further increased” EYFP fluorescence at 523 nm. From Fig. 4B it can be concluded that the majority of the fluorescence in cells coexpressing ECFP and EYFP can be attributed to ECFP. Cells that coexpressed the ECFP and EYFP proteins showed 523/475 ratios that varied between 0.95 and 1.0 (Fig. 4B). We did not test the combination of the 2B-ECFP and the EYFP proteins as a negative control, because the nonfused EYFP was expressed at much higher levels than the 2B-ECFP protein (thereby causing an abnormally high EYFP/ECFP expression ratio, which disturbs the SPIM analysis). Cells that expressed the EYFP-ECFP fusion protein (i.e., the positive control) showed a dramatic increase in EYFP fluorescence (523/475 ratio of on average 2.4), indicating substantial sensitized emission due to the occurrence of intramolecular FRET (Fig. 4C).
FIG. 4.
SPIM analysis of individual cells expressing ECFP (n = 3) (A), ECFP and EYFP (n = 4) (B), the EYFP-ECFP fusion protein (n = 2) (C), 2B-ECFP and 2B-EYFP (n = 9) (D), 2B[1-85]ECFP and 2B[1-85]EYFP (n = 6) (E), 2BC-ECFP and 2BC-EYFP (n = 5) (F), or 2C-ECFP and 2C-EYFP (n = 5) (G). The y axis represents the fluorescence emission spectra normalized to the first emission peak of the ECFP spectrum (as measured at 475 nm) for comparative reasons. The occurrence of sensitized emission, i.e., an increase in the ratio of EYFP (523 nm) to ECFP (475 nm), is indicative of the occurrence of FRET.
Cells that coexpressed the 2B-ECFP and 2B-EYFP fusion proteins showed increased 523/475 ratios. The spectra of a number of randomly chosen cells (n = 9) that coexpressed the 2B-ECFP and 2B-EYFP fusion proteins are shown in Fig. 4D. A large portion of the cells (n = 4) showed an increased 523/475 ratio of 1.2. The other cells showed further increases in the 523/475 ratio which varied between 1.3 (n = 2), 1.45 (n = 1), and 1.6 (n = 1), whereas one cell showed an enormous increase in 523/475 ratio (2.1). Thus, cells that coexpress 2B-ECFP and 2B-EYFP have significantly higher 523/475 ratios than control cells expressing ECFP and EYFP under similar conditions. This suggests the occurrence of intermolecular FRET due to a 2B-2B interaction.
Increased 523/475 ratios were also observed in cells coexpressing 2B[1-85]ECFP and 2B[1-85]EYFP (Fig. 4E). The overall efficiency of FRET between 2B[1-85]ECFP and 2B[1-85]EYFP was not significantly different from that observed in cells expressing 2B-ECFP and 2B-EYFP, indicating that (i) the distance between 2B and ECFP or EYFP is not a limiting feature for the efficiency of FRET and (ii) the C-terminal region of the 2B protein is not required for the 2B-2B interaction. Increased 523/475 ratios were also observed, although to a lesser extent, in cells coexpressing 2BC-ECFP and 2BC-EYFP (Fig. 4F), but not in cells coexpressing 2C-ECFP and 2C-EYFP (Fig. 4F). These data further stress the occurrence of FRET due to an intermolecular 2B-2B interaction.
Taken together, these data suggest that, in cells coexpressing 2B-ECFP and 2B-EYFP, sensitized emission of the acceptor fluorophore occurs as result of intermolecular FRET due to a 2B-2B interaction. However, the possibility that the variations in the 523/475 ratios observed in cells transfected with the 2B-ECFP and 2B-EYFP plasmids are due to abnormally high 2B-EYFP-to-2B-ECFP expression ratios in some of the cells cannot be excluded. This possibility seems unlikely from the data shown in Fig. 3A and B, which demonstrated that nearly all cotransfected cells expressed the 2B-ECFP and 2B-EYFP fusion proteins to more or less similar levels. To further investigate the occurrence of FRET, FLIM was performed.
Analysis of protein 2B homomultimerization reactions by FLIM.
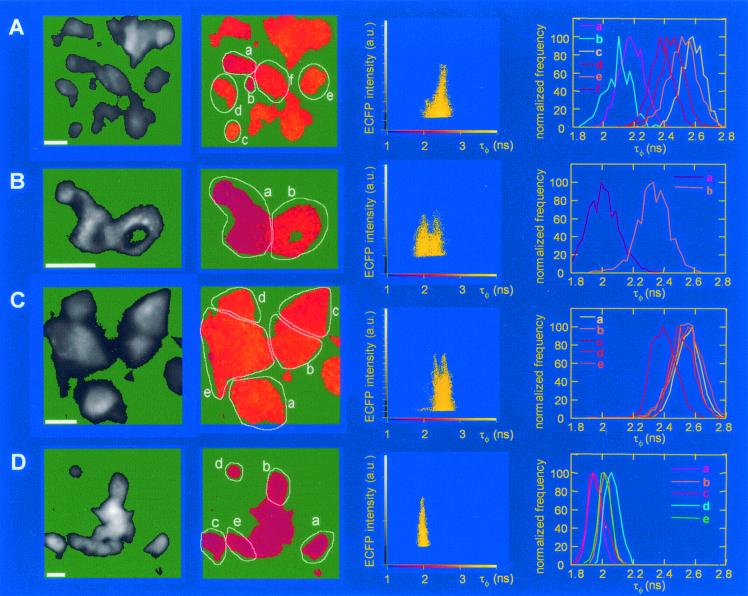
Frequency domain FLIM was used to investigate the occurrence of FRET between the 2B-ECFP and 2B-EYFP fusion proteins. FLIM is a more reliable, but less sensitive, technique to monitor FRET (30). FRET causes a reduction in the fluorescence lifetime of ECFP. Frequency domain FLIM was used to obtain two separate determinations for the fluorescence lifetime, τφ and τM, at each pixel of an image from the phase shift and demodulation of the fluorescence emission, respectively. We show only images of τφ. Comparison of the fluorescence lifetime modulation τM values yielded similar relative changes (Table 1). The τM values are higher than the τφ values, as expected for multiexponential complex fluorescence decay (14).
TABLE 1.
Results of FLIM analysis
| (Co)transfection (Fig. 5 panel) | τφ ± SD (ns)a | τM ± SD (ns)b | Res (%)c |
|---|---|---|---|
| 2B-ECFP + 2B-EYFP (A)d | 2.42 ± 0.13 | 2.99 ± 0.13 | 1.7 |
| 2B-ECFP + 2B-EYFP (B) | 2.13 ± 0.19 | 2.66 ± 0.16 | 1.9 |
| 2B-ECFP + EYFP (C) | 2.53 ± 0.11 | 3.03 ± 0.10 | 1.7 |
| EYFP-ECFP fusion (D) | 1.95 ± 0.05 | 2.60 ± 0.03 | 1.5 |
Fluorescence lifetime calculated from the phase shift Δφ (τφ = 1/ωtan Δφ). SD, standard deviation in the τφ image corrected for noise in the fit procedure (17).
Fluorescence lifetime calculated from the modulation (M) with τM = (1/ω) (M−2 − 1)1/2. SD, standard deviation in the τM image corrected for noise in the fit procedure (17).
The quality of the data is indicated by the low average difference between calculated (lc) and observed (lo) fluorescence intensity in each phase image: Res2 = (lo − lc)2/lc2 (17).
Note that the τφ and the τM values shown in Fig. 5A differ from those in Fig. 5B, because in Fig. 5A an overview of the cell population is shown (of which only a limited number exhibit a reduced fluorescence lifetime), whereas Fig. 5B shows only two cells (of which one exhibits a major reduction in fluorescence lifetime, whereas the other one exhibits a minor decrease in fluorescence lifetime).
The ECFP fluorescence lifetime was recorded in cells that (co)expressed either (i) 2B-ECFP and 2B-EYFP, (ii) 2B-ECFP and EYFP (negative control), or (iii) the EYFP-ECFP fusion protein (positive control). ECFP fluorescence intensity images, ECFP fluorescence lifetime τφ images, and temporal histograms of the ECFP fluorescence lifetime τφ values of individual cells are shown in Fig. 5. The fluorescence lifetime τφ of the 2B-ECFP protein when expressed together with the unfused EYFP protein varied between 2.4 and 2.6 ns (Fig. 5C, Table 1). A similar fluorescence lifetime τφ was observed in cells that expressed the 2B-ECFP protein alone (data not shown). Cells transfected with the EYFP-ECFP fusion protein all displayed a reduced ECFP fluorescence lifetime (τφ, 1.95 to 2.05 ns) (Fig. 5D, Table 1), consistent with the SPIM data (Fig. 4C) and the occurrence of intramolecular FRET.
FIG. 5.
FLIM analysis of the ECFP fluorescence in living BGM cells coexpressing either 2B-ECFP and 2B-EYFP (A and B), 2B-ECFP and EYFP (C), or the EYFP-ECFP fusion protein (D). Only the ECFP fluorescence (lifetime) of the fusion proteins is monitored by use of a cyan fluorescent protein-specific emission filter. The first (left) column of micrographs represents the ECFP fluorescence intensity distribution in different cells. The second column of micrographs represents ECFP fluorescence lifetime (τφ) images calculated from the fluorescence intensity image shown in the first column. The lifetime images were calculated on a pixel-by-pixel basis (17), and they are pseudocolored according to the lifetime scale indicated in the horizontal axis of the 2D histograms in the third column. Blue-red colors in the τφ image depict low fluorescence lifetimes (FRET), and orange-yellow colors in the τφ image depict higher fluorescence lifetimes (no FRET). The third column represents a 2D histogram depicting the correlation between the ECFP fluorescence intensity (y axis) and the τφ ECFP fluorescence lifetime (x axis) for the images in the first two columns on a pixel-by-pixel basis. Clearly pixels with equal fluorescence intensity can have different lifetimes. The fourth column of images depicts the fluorescence lifetime distribution in the region of interest (cells) indicated in the second column of τφ micrographs. The lifetime heterogeneity, which demonstrates FRET heterogeneity, is observed only for the 2B-ECFP and 2B-EYFP cotransfections (A and B). Bars in the fluorescence intensity images, 20 μm in the object plane.
The fluorescence lifetime τφ values recorded in cells that coexpressed the 2B-ECFP and 2B-EYFP fusion proteins are shown in Fig. 5A and B. Figure 5A shows that in some of the cells, but not in all, a reduced 2B-ECFP fluorescence lifetime (τφ, 2.1 to 2.2 ns) was observed. The observation that not all cells displayed a reduced 2B-ECFP fluorescence reflects a cell-to-cell heterogeneity. Because the reduction in ECFP fluorescence lifetime is rather small, only for a relatively extensive intermolecular interaction between 2B-ECFP and 2B-EYFP will a reduction in ECFP fluorescence lifetime be observed. In cells in which only low levels of FRET occur (i.e., cells that exhibit only minor increases in the 523/475 ratio in the SPIM analysis), the reduction in the 2B-ECFP fluorescence lifetime is too small to detect. The variation in the ECFP fluorescence lifetime in cells coexpressing 2B-ECFP and 2B-EYFP is further illustrated in Fig. 5B, which shows two cells with ECFP fluorescence lifetime τφ values of 2.0 and 2.3 ns, respectively. In some other cells coexpressing 2B-ECFP and 2B-EYFP, an ECFP fluorescence lifetime τφ as low as 1.85 ns was observed (data not shown). Figure 5B further illustrates that there is no correlation between the occurrence and extent of FRET on the one hand and the morphology of the cells or the level of protein expression on the other hand, indicating that FRET is not a mere consequence of high expression levels or altered cellular conditions.
Table 1 shows the average fluorescence lifetime τφ and τM values of the cells shown in Fig. 5. The differences in the average fluorescence lifetimes (both τφ and τM) of the cells coexpressing 2B-ECFP and 2B-EYFP shown in Fig. 5A and B, respectively, again demonstrate the importance of analyzing cells at the single-cell level. This table also shows that the heterogeneity of the donor ECFP fluorescence lifetime (both τφ and τM), as quantified by the standard deviations in their values across the images, is much higher for 2B-ECFP and 2B-EYFP coexpression (0.13 to 0.19 ns) than for expression of the EYFP-ECFP fusion protein (0.03 to 0.05 ns) or for coexpression of 2B-ECFP and EYFP (0.10 to 0.11 ns). This quantitatively demonstrates that the observed lifetime heterogeneity for 2B-ECFP and 2B-EYFP expression reflects the heterogeneity in FRET efficiency that is intrinsic to the 2B fusion protein and its homomultimerization. Interestingly, the very small lifetime heterogeneity observed for the EYFP-ECFP fusion protein (see also the sharp temporal histograms in Fig. 5C) correlates very well with the observed narrow range of the ratio of ECFP fluorescence intensity to EYFP fluorescence intensity as shown in the confocal two-dimensional (2D) histogram (Fig. 3F).
Taken together, both FLIM and SPIM (from which the data were recorded in separate experiments) provide evidence for the occurrence of intermolecular FRET through the formation of homomultimers of the 2B protein.
DISCUSSION
Unraveling the mechanism by which a viral membrane-active protein manipulates host cell membranes is of major importance for a better understanding of virus-induced cytopathogenesis. The 2B protein of enterovirus is responsible for the modifications in host cell membrane permeability that take place in enterovirus-infected cells. The 2B protein is a small hydrophobic protein that is localized at secretory-pathway membranes. Our aim is to gain more insight into the molecular mechanism and the molecular architecture adopted by the 2B protein to permeate host cell membranes. In this study, we probed the existence of multimers of the CBV3 2B protein by FRET microscopy. FRET was assayed in living cells that coexpressed biologically active 2B-ECFP and 2B-EYFP fusion proteins by both SPIM and FLIM, at a time point at which the 2B fusion proteins exerted their membrane-active function.
FRET between ECFP and EYFP fusion proteins leads to the quenching of the ECFP donor and sensitized emission of the EYFP acceptor. Moreover, FRET results in a decrease in the fluorescence lifetime of the ECFP donor. Using SPIM, we found that all cells coexpressing 2B-ECFP and 2B-EYFP displayed significantly higher 523/475 ratios than cells expressing ECFP and EYFP under similar conditions. The SPIM ratio analysis is much more accurate than conventional ratiometric methods as background components are spectrally subtracted and also Raman scattering is corrected for (19). In addition, the shape of the spectrum provides an additional check that indeed the fluorescence ratios reflect ECFP-to-EYFP fluorescence emission ratios and not other autofluorescing species with other spectral signatures. FLIM analysis of the fluorescence lifetime on a pixel-by-pixel basis revealed reductions in the ECFP fluorescence lifetime in cells coexpressing 2B-ECFP and 2B-EYFP, providing further evidence for the occurrence of intermolecular FRET between the 2B-ECFP and 2B-EYFP proteins due to homomultimerization reactions of the 2B protein. The occurrence of FRET between the 2B-ECFP and 2B-EYFP proteins indicates a close proximity of 2B-2B proteins. In some cells, a reduction in ECFP fluorescence lifetime τφ from 2.5 to 2.0 ns (Fig. 5B) or even to 1.85 ns (data not shown) was observed, demonstrating a FRET efficiency that varies between 20 and 25%. Given the R0 value for ECFP and EYFP (50 Å), this indicates an average proximity of ECFP and EYFP in the 2B-2B protein complex of ∼6 nm (for a 1:1 stoichiometry in the complex). In other cells, a smaller reduction in ECFP fluorescence lifetime was observed (τφ, 2.2 to 2.3 ns; Fig. 5A and B). This heterogeneity may reflect the difference in composition of the protein 2B homomultimeric complex, as the FRET efficiency in a multimer consisting of one 2B-ECFP protein and several 2B-EYFP proteins is more extensive than the FRET efficiency in a protein 2B complex of reverse composition. Heterogeneity in the FRET efficiency between ECFP- and EYFP-tagged homomultimers was recently also observed by Zacharias et al. (37)
Using SPIM, we also obtained evidence for homomultimerization reactions of the 2BC protein (the precursor of the 2B protein) but not of the 2C protein. This finding provides further evidence for the ability of the 2B protein to form homomultimers. The level of sensitized emission between proteins 2BC-ECFP and 2BC-EYFP was lower than that between proteins 2B-ECFP and 2B-EYFP, possibly because of the increased distance between the ECFP and EYFP fluorophores. The results of this study are in agreement with recent results that demonstrated homomultimerization reactions of the CVB3 2B and 2BC proteins, but not of the 2C protein, in a mammalian two-hybrid system (11). The two-hybrid system is a complex and artificial method that requires translocation of the bait and prey to the nucleus in order to activate transcription of a reporter. The results of this FRET microscopy study not only confirm our mammalian two-hybrid results but also extend these observations by providing evidence for homomultimerization reactions in living cells of proteins at their “normal” intracellular localization (i.e., at secretory-pathway membranes).
How do these observations aid our understanding of the molecular architecture of the 2B protein? The 2B protein contains a cationic amphipathic α-helix motif that is typical for the group of the lytic polypeptides (33). The most extensively studied lytic peptides are the antimicrobial peptides, amphipathic α-helical peptides produced by animals as well as plants to defend themselves against invading pathogenic microorganisms. Two distinct molecular models of action have emerged from structural and functional studies on these antimicrobial peptides (4, 28). According to the first model, the barrel stave model (also referred to as the helical-bundle model), the amphipathic α-helices form aqueous pores or channels by forming multimeric transmembrane bundles, which expose their hydrophobic sides to the lipid bilayer while their hydrophilic faces point inward to form the aqueous interior. Progressive recruitment of monomers may occur and cause a gradual increase in pore size. A crucial step in this mechanism is the formation of multimers on the surface of the membrane before the peptide is inserted, since it is energetically unfavorable for a single amphipathic α-helix to traverse the membrane as a monomer. According to the second model, the carpet model, the amphipathic α-helical peptides are located at the membrane plane as monomers, with their hydrophobic backbone inserted in the lipid bilayer and their positively charged amino acids interacting with the phospholipid head groups. The peptides remain in contact with the phospholipid head groups throughout the entire membrane permeabilization process. Membrane disruption is supposed to occur at certain high local threshold concentrations of membrane-bound peptides, which are thought to disturb the membrane by increasing the membrane curvature and disturbing the lipid bilayer organization. It has been suggested that breakage of the membrane is preceded by the formation in the membrane of transient pores (so-called toroidal or wormhole pores), enabling the passage of ions and low-molecular-weight molecules prior to complete lysis of the membrane (28).
The occurrence of FRET between 2B-ECFP and 2B-EYFP and the calculated proximity of 6 nm, which is in the order of protein dimensions, support the hypothesis that homomultimers of protein 2B build membrane-integral pores and thereby account for the alterations in host cell membrane permeability (Fig. 1C). As described above, we recently obtained evidence for homomultimerization reactions of the 2B and 2BC proteins using a mammalian two-hybrid system (11). Upon testing mutant 2B and 2BC proteins, we found that the homomultimerization reactions were dependent on determinants located in both the amphipathic α-helix and the second hydrophobic domain of the 2B protein (11). The N terminus of the 2B protein (amino acids 1 to 30) was found to be dispensable for the homomultimerization reactions of the 2B and 2BC proteins (11). In this study, we found that the C terminus of the 2B protein (amino acids 86 to 99) is also dispensable for the 2B homomultimerization reaction. Based on these observations, it is tempting to speculate that both the amphipathic α-helix and the second hydrophobic domain traverse the lipid bilayer and that cooperative interactions between these domains are required for the formation of a multimeric membrane-integral pore structure. Such a membrane topology is consistent with the need for a cytosolic localization of both the amino terminus and the carboxy terminus of the 2B protein because of the need for proteolytic processing at the 2A-2B and 2B-2C cleavage sites by the viral 3C protein, a cytosolic protease. This membrane topology is also consistent with previous observations that mutations in the second hydrophobic domain can abolish the membrane-active function of the 2B protein (34, 35) and that the α-helix in the 2B protein of CBV3 cannot be functionally exchanged with the corresponding domain of the poliovirus 2B protein or with another structurally related cationic amphipathic α-helix (33, 36).
In conclusion, the results of this study indicate that the membrane-active enterovirus 2B protein is able to form multimeric transmembrane pores in endomembranes of living cells. Further research is required to clarify the relationship between the membrane-active function of the 2B protein and its contribution to the accumulation of the membrane vesicles at which viral RNA replication takes place in infected cells. Moreover, further investigation is required to solve the intriguing mechanism by which a viral membrane-active protein that is localized at ER and Golgi membranes can modify plasma membrane permeability. Answers to these questions will provide important insight into the molecular mechanism by which a cytolytic virus manipulates host cell membranes and causes cell lysis.
Acknowledgments
We are grateful to R. Tsien (Howard Hughes Medical Institute, University of California, San Diego) for the kind gift of plasmid yellow cameleon 2.1.
T. W. J. Gadella, Jr., was supported by the Royal Netherlands Academy of Sciences (KNAW). This work was partly supported by grants from the European Communities (INTAS 2012).
REFERENCES
- 1.Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134-23137. [DOI] [PubMed] [Google Scholar]
- 2.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 3.Bastiaens, P. I. H., and A. Squire. 1999. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9:48-52. [DOI] [PubMed]
- 4.Bechinger, B. 1999. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta 1462:157-183. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, H. D., P. Sarnow, and D. Baltimore. 1986. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J. Virol. 60:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienz, K., D. Egger, and T. Pfister. 1994. Characteristics of the poliovirus replication complex. Arch. Virol. Suppl. 9:147-157. [DOI] [PubMed] [Google Scholar]
- 7.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39-48. [DOI] [PubMed] [Google Scholar]
- 8.Carrasco, L. 1995. Modification of membrane permeability by animal viruses. Adv. Virus Res. 45:61-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 10.Clegg, R. M. 1996. Fluorescence resonance energy transfer, p. 179-252. In X. F. Wang and B. Herman (ed.), Fluorescence imaging spectroscopy and microscopy. John Wiley and Sons, New York, N.Y.
- 11.de Jong, A. S., I. W. J. Schrama, P. H. G. M. Willems, J. M. D. Galama, W. J. G. Melchers, and F. J. M. van Kuppeveld. 2002. Multimerization reactions of the coxsackievirus proteins 2B, 2C, and 2BC: a mammalian two-hybrid analysis. J. Gen. Virol. 83:783-793. [DOI] [PubMed] [Google Scholar]
- 12.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadella, T. W. J., Jr. 1999. Fluorescence lifetime imaging microscopy (FLIM): instrumentation and applications, p. 467-489. In W. T. Mason (ed.), Fluorescent and luminescent probes for biological activity. A practical guide to technology for quantitative real-time analysis, 2nd ed. Academic Press, New York, N.Y.
- 14.Gadella, T. W. J., Jr., T. M. Jovin, and R. M. Clegg. 1993. Fluorescence lifetime imaging microscopy (FLIM)—spatial resolution of microstructures on the nanosecond time-scale. Biophys. Chem. 48:221-239. [Google Scholar]
- 15.Gadella, T. W. J., Jr., G. N. van der Krogt, and T. Bisseling. 1999. GFP-based FRET microscopy in living plant cells. Trends Plant Sci. 4:287-291. [DOI] [PubMed]
- 16.Gadella, T. W. J., Jr., G. Vereb, Jr., A. E. Hadri, H. Rohrig, J. Schmidt, M. John, J. Schell, and T. Bisseling. 1997. Microspectroscopic imaging of nodulation factor-binding sites on living Vicia sativa roots using a novel bioactive fluorescent nodulation factor. Biophys. J. 72:1986-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadella, T. W. J., Jr., R. M. Clegg, and T. M. Jovin. 1994. Fluorescence lifetime imaging microscopy: pixel-by-pixel analysis of phase modulation data. Bioimaging 2:139-159. [Google Scholar]
- 18.Gadella, T. W. J., Jr., A. van Hoek, and A. J. W. G. Visser. 1997. Construction and characterization of a frequency domain fluorescence lifetime imaging microscopy system. J. Fluoresc. 7:35-43. [Google Scholar]
- 19.Goedhart, J., and T. W. J. Gadella, Jr. 2000. Advanced microspectroscopic methods for monitoring fluorescent molecules in living plant root hairs, p. 65-93. In R. W. Ridge and A. M. C. Emons (ed.), Cell and molecular biology of living plant root hairs. Springer-Verlag, Tokyo, Japan.
- 20.Immink, R. G. H., T. W. J. Gadella, Jr., S. Ferrario, M. Busscher, and G. C. Angenent. 2002. Analysis of MADS box protein-protein interactions in living plant cells. Proc. Natl. Acad. Sci. USA 99:2416-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, K. L., and P. Sarnow. 1991. Three poliovirus 2B mutants exhibit noncomplementable defects in viral RNA amplification and display dosage-dependent dominance over wild-type poliovirus. J. Virol. 65:4341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyawaki, A., O. Griesbeck, R. Heim, and R. Y. Tsien. 1999. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc. Natl. Acad. Sci. USA 96:2135-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollock, B. A., and R. Heim. 1999. Using GFP in FRET-based applications. Trends Cell Biol. 9:57-60. [DOI] [PubMed]
- 24.Sandoval, I., and L. Carrasco. 1997. Poliovirus infection and expression of the poliovirus 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J. Virol. 71:4679-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segrest, J. P., H. de Loof, J. G. Dohlman, C. G. Brouillette, and G. M. Anantharamaiah. 1990. Amphipathic helix motif: classes and properties. Struct. Funct. Genet. 8:103-117. [DOI] [PubMed] [Google Scholar]
- 26.Selvin, P. R. 2000. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 7:730-734. [DOI] [PubMed] [Google Scholar]
- 27.Shah, K., T. W. Gadella, Jr., H. van Erp, V. Hecht, and S. C. de Vries. 2001. Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J. Mol. Biol. 309:641-655. [DOI] [PubMed] [Google Scholar]
- 28.Shai, Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1462:55-70. [DOI] [PubMed] [Google Scholar]
- 29.Squire, A., and P. I. H. Bastiaens. 1999. Three dimensional image restoration in fluorescence lifetime imaging microscopy. J. Microsc. 193:36-49. [DOI] [PubMed] [Google Scholar]
- 30.Tertoolen, C. Blanchetot, G. Jiang, J. Overvoorde, T. W. J. Gadella, Jr., T. Hunter, and J. den Hertog. 2001. Dimerization of receptor protein-tyrosine phosphatase alpha in living cells. BMC Cell Biol. 2:8.. [DOI] [PMC free article] [PubMed]
- 31.Tsien, R. Y. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 32.van Kuppeveld, F. J. M., J. M. D. Galama, J. Zoll, and W. J. G. Melchers. 1995. Genetic analysis of a hydrophobic domain of coxsackie B3 virus protein 2B; a moderate degree of hydrophobicity is required for a cis-acting function in viral RNA synthesis. J. Virol. 69:7782-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Kuppeveld, F. J. M., J. M. D. Galama, J. Zoll, P. J. J. C. van den Hurk, and W. J. G. Melchers. 1996. Coxsackie B3 virus protein 2B contains a cationic amphipathic helix that is required for viral RNA replication. J. Virol. 70:3876-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Kuppeveld, F. J. M., J. G. J. Hoenderop, R. L. L. M. Smeets, P. H. G. M. Willems, H. B. P. M. Dijkman, J. M. D. Galama, and W. J. G. Melchers. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16:3519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Kuppeveld, F. J. M., W. J. G. Melchers, K. Kirkegaard, and J. R. Doedens. 1997. Structure-function analysis of coxsackie B3 virus protein 2B. Virology 227:111-118. [DOI] [PubMed] [Google Scholar]
- 36.van Kuppeveld, F. J. M., P. J. J. C. van den Hurk, W. van der Vliet, J. M. D. Galama, and W. J. G. Melchers. 1997. Chimeric coxsackie B3 virus genomes that express hybrid coxsackievirus-poliovirus 2B proteins: functional dissection of structural domains involved in RNA replication. J. Gen. Virol. 78:1833-1840. [DOI] [PubMed] [Google Scholar]
- 37.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913-916. [DOI] [PubMed] [Google Scholar]