Abstract
The influenza A virus RNA-dependent RNA polymerase consists of three subunits—PB1, PB2, and PA. The PB1 subunit is the catalytically active polymerase, catalyzing the sequential addition of nucleotides to the growing RNA chain. The PB2 subunit is a cap-binding protein that plays a role in initiation of viral mRNA synthesis by recruiting capped RNA primers. The function of PA is unknown, but previous studies of temperature-sensitive viruses with mutations in PA have implied a role in viral RNA replication. In this report we demonstrate that the PA subunit is required not only for replication but also for transcription of viral RNA. We mutated evolutionarily conserved amino acids to alanines in the C-terminal region of the PA protein, since the C-terminal region shows the highest degree of conservation between PA proteins of influenza A, B, and C viruses. We tested the effects of these mutations on the ability of RNA polymerase to transcribe and replicate viral RNA. We also tested the compatibility of these mutations with viral viability by using reverse-genetics techniques. A mutant with a histidine-to-alanine change at position 510 (H510A) in the PA protein of influenza A/WSN/33 virus showed a differential effect on transcription and replication. This mutant was able to perform replication (vRNA→cRNA→vRNA), but its transcriptional activity (vRNA→mRNA) was negligible. In vitro analyses of the H510A recombinant polymerase, by using transcription initiation, vRNA-binding, capped-RNA-binding, and endonuclease assays, suggest that the primary defect of this mutant polymerase is in its endonuclease activity.
Influenza A virus is a negative-strand RNA virus containing eight segments of single-stranded RNA as its genome (39). The RNA genome is transcribed and replicated by the viral RNA-dependent RNA polymerase in the cell nucleus (21). The viral RNAs (vRNA) are transcribed into mRNAs and replicated through a cRNA intermediate to produce more vRNA molecules. Synthesis of these three RNA species requires different modes of initiation and termination (reviewed in references 23 and 34). Synthesis of mRNAs is primed by short capped RNA fragments that are generated from cellular pre-mRNAs by endonucleolytic cleavage. Consequently, viral mRNA molecules contain a 9- to 17-nucleotide (nt) capped host-derived RNA sequence at their 5′ ends. On the other hand, the synthesis of cRNA and vRNA molecules is initiated in a primer-independent manner, resulting in triphosphorylated 5′ ends. Synthesis of mRNAs is prematurely terminated 16 to 17 nucleotides from the 5′ end of the vRNA template at a sequence of 5 to 7 uridines that acts as a polyadenylation signal (30, 47, 49). The poly(A) tail is synthesized by the viral RNA polymerase by repeated copying of the U sequence (47). During the synthesis of cRNA molecules, the polyadenylation signal is ignored, resulting in full-length copies of vRNA.
All three reactions, i.e., vRNA→mRNA (transcription), vRNA→cRNA (first step of replication), and cRNA→vRNA (second step of replication) are catalyzed by the viral RNA polymerase complex. The polymerase complex consists of three subunits: polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), and polymerase acidic protein (PA). The PB1 component functions as the polymerase by catalyzing the sequential addition of nucleotides to RNA transcripts. It contains the conserved motifs characteristic of RNA-dependent RNA polymerases (1, 2, 46). The sequence S-D-D at amino acids 444 to 446 is the most likely candidate for the active site of RNA polymerization (29). The PB2 subunit of the viral RNA polymerase binds to cap-1 structures of host pre-mRNA molecules. The cap-binding site has recently been localized to amino acid residues between 533 and 564 (29). Honda et al. (19), however, proposed that two separate regions of PB2, amino acids 242 to 282 and 538 to 577, constitute the cap-binding site. Early studies indicated that PB2 is also responsible for endonucleolytic cleavage of cellular pre-mRNAs (3, 53), but a recent report suggests that the endonuclease domain resides in the PB1 subunit (29). It has been proposed that amino acids 508 to 522, a region containing three essential acidic amino acids, form the endonuclease active site of PB1.
The role of the PA subunit in the replication cycle of the virus is essentially unknown, although early work with temperature-sensitive (ts) virus mutants suggested that it was involved in vRNA replication (31). PA can induce a generalized proteolysis of both viral and host proteins (50). It is not clear, however, whether this protease activity is a property of PA or of a host protease induced by PA. Deletion analysis of PA showed that the N-terminal one-third of the molecule (amino acids 1 to 247) is sufficient for this activity (52). The role of the proteolysis-inducing activity of PA in viral transcription and replication is controversial. Perales et al. (42) reported that the replication activity of the polymerase was linked to the capacity of PA to induce proteolysis. In particular, point mutations at amino acid positions 157 and 162 of PA, potential sites for phosphorylation by casein kinase II, resulted in decreased proteolytic induction. This decreased activity correlated with a reduced ability to synthesize cRNA from the vRNA template, but the ability of these mutants to transcribe vRNA into mRNA was unaffected. The observed replication-defective phenotype of polymerases with mutant PA proteins was in line with the phenotype of viral ts mutants in which the PA gene was affected (31). In contrast, Naffakh et al. (35) found that proteolysis induced by the PA subunit is not correlated with the transcription-replication activity of the polymerase. Mutations at position 241 of PA reduced proteolytic activity, but the transcription-replication activity of the polymerase was not affected. More recently, Hara et al. (18) reported that PA was a chymotrypsin-like serine protease with S624 at the active site, but the role of this activity in the replication cycle of the virus remains to be elucidated.
The exact nature of the polymerase complexes involved in the synthesis of the three different RNA species—mRNA, cRNA, and vRNA—is not known. The distinct functions—endonuclease cleavage to generate primers, transcription, and replication—must presumably be performed by different polymerase-promoter structures. PB1 alone has been reported to transcribe RNA templates in vitro and catalyze cRNA synthesis in vivo (22, 37). PB1 together with PA (in the presence of NP but without PB2) appears to replicate vRNA and synthesize uncapped poly(A)+ transcripts (36). Others, however, have reported that all three polymerase subunits are required for efficient synthesis of all three influenza virus RNA transcripts (41). The PB1 subunit forms the core of the polymerase complex (6). Interactions between PB1 and PB2, as well as between PB1 and PA, have been demonstrated. PB1 interacts through its N-terminal region with the C-terminal region of PA, while the C-terminal region of PB1 is involved in interaction with the N-terminal region of PB2 (16, 40, 43, 44, 55, 56). A low-resolution three-dimensional structural model of a recombinant influenza virus RNP particle generated by electron microscopy suggests a rather compact structure for the polymerase complex (32).
Interactions of the RNA polymerase with the vRNA and cRNA promoters are crucial in determining the catalytic activities of the polymerase. The 5′- and 3′-terminal sequences of vRNA, which together form the vRNA promoter (11, 12), act as essential cofactors for several activities of the polymerase (17, 28). Binding of the polymerase to the 5′-terminal vRNA sequence is proposed to induce an allosteric change that results in activation of the cap-binding activity of the PB2 subunit. To induce the capped RNA-specific endonuclease activity of the polymerase, required for generating capped RNA primers for transcription initiation, the polymerase has to bind to both the 5′- and 3′-terminal vRNA sequences (17, 28). Detailed mutagenic analyses of the 5′ and 3′ vRNA sequences defined the sequence requirements for inducing endonuclease activity (24, 25, 26). Hairpin loop structures in both the 5′ and 3′ ends of vRNA, as suggested by the corkscrew model for the vRNA promoter (8), are required for endonuclease activity.
The PB1 subunit of the polymerase contains the binding sites for the 5′- and 3′-terminal sequences of vRNA. González and Ortín (14) have proposed that two separate regions of PB1, one localized in the N-terminal 83 amino acids and the other in the C-terminal 264 amino acids, contribute to cooperative binding of the 5′- and 3′-terminal vRNA sequences. According to Li et al. (28), however, the 5′-terminal vRNA sequence binds to an amino acid sequence centered around two arginine residues at positions 571 and 572, while the 3′-terminal vRNA binds to a region at amino acids 249 to 256 containing two phenylalanine residues. The latter sequence was previously identified as a ribonucleoprotein 1 (RNP1)-like RNA binding motif in a computer search of PB1 sequences (13). Although PB1 has been shown to contain the binding sites for the terminal sequences of vRNA, PB2 and PA might also play a role in vRNA recognition, because they can be cross-linked to vRNA sequences (11, 13). Efficient binding of the 5′ end of vRNA has been shown to be dependent on the formation of a complex between PB1 and PA, suggesting that PA might play a role in enhancement of the 5′-end vRNA-binding activity of PB1 (27).
In this report we address the function of the PA subunit of the RNA polymerase complex in the processes of vRNA transcription and replication. We present the construction of a set of PA mutants, their phenotypic characterization, and their activity in transcription and replication of vRNA.
MATERIALS AND METHODS
Plasmids.
The pcDNA-PB1, pcDNA-PB2, pcDNA-PA, and pcDNA-NP protein expression plasmids for the three polymerase subunits and NP of influenza A/WSN/33 virus were generated by subcloning the relevant open reading frames from the pPOLI-PB1-RT, pPOLI-PB2-RT, pPOLI-PA-RT, and pPOLI-NP-RT plasmids (9), respectively, into pcDNA3 (Invitrogen). Briefly, the pcDNA3 vector was modified by inserting an AgeI restriction enzyme site into its polylinker, generating pcDNA3A as follows. pcDNA3 was linearized with HindIII restriction enzyme and ligated to a self-annealed 5′-AGCTTACCGGTA-3′ linker, containing an AgeI site. pPOLI-PB1-RT, pPOLI-PB2-RT, pPOLI-PA-RT, and pPOLI-NP-RT (9) were digested with NgoMIV, and the PB1-, PB2-, PA-, and NP-containing cDNAs were inserted into the AgeI site of pcDNA3A. pPOLI-CAT-RT is a pUC19-based plasmid encoding a vRNA-like RNA containing a chloramphenicol acetyltransferase (CAT) open reading frame in negative sense, flanked by the 5′ and 3′ noncoding regions of the NS vRNA segment of influenza A/WSN/33 virus (45). pPOLI-cCAT-RT is a pUC18-based plasmid encoding a cRNA-like RNA containing a positive-sense CAT open reading frame, flanked by the 5′ and 3′ noncoding regions of the NS cRNA of influenza A/WSN/33 virus. Expression of both RNAs is driven by a truncated human RNA polymerase I promoter (nt −250 to −1). The 3′ ends of both RNAs are generated by ribozyme cleavage (45). The pcDNA-PA-His6 plasmid, based on pcDNA3, encodes the PA protein of influenza A/WSN/33 virus tagged at its C terminus with a sequence of six histidines through an Ala-Ala-Ala-Gly-Ser linker. The open reading frame of PA-His6 lies between the EcoRI and XbaI sites of the pcDNA3 polylinker. Modifications of the plasmids were prepared by site-directed mutagenesis. Sequences of the mutagenic primers are available from us upon request.
Preparation of capped RNA for binding studies and endonuclease assay.
A 2′-O-methylated oligoribonucleotide (5′-AmAAUACUCAAG-3′) was chemically synthesized and 5′ diphosphorylated as described previously (4). The 5′-diphosphorylated oligoribonucleotide was converted into a capped (m7Gppp) oligoribonucleotide by using guanylyl transferase (Gibco BRL). Capping was performed in a reaction volume of 7.5 μl containing 100 pmol of RNA, 50 mM Tris-HCl (pH 8.0), 1.25 mM MgCl2, 60 mM KCl, 2.5 mM dithiothreitol (DTT), 0.1 mM S-adenosylmethionine (New England Biolabs), 30 U of RNasin (Promega), 1 μM [α-32P]GTP (3,000 Ci/mmol) (Amersham), and 2.5 U of guanylyl transferase for 1 h at 30°C. The reaction was terminated by the addition of 7.5 μl of 80% formamide containing 1 mM EDTA, bromophenol blue, and xylene cyanol dyes. The reaction mixture was heated to 95°C for 2 min and loaded onto a 16% acrylamide gel containing 7 M urea in Tris-borate-EDTA (TBE) buffer. The 32P-labeled capped RNA was detected by autoradiography and eluted by soaking the crushed gel piece in 0.5 ml of 0.5 M ammonium acetate (pH 7.5) at 4°C for 16 h. The eluted RNA was precipitated with ethanol in the presence of 20 μg of glycogen carrier and washed with 70% ethanol. Total counts per minute were determined, and the RNA pellet was dissolved in H2O to 2 × 105 cpm/μl. The poly(A)+-capped RNA substrate for the endonuclease assay was prepared by adding a poly(A) sequence to the 3′ end of the 32P-labeled capped RNA. Polyadenylation was performed in a reaction volume of 20 μl containing 32P-labeled capped RNA (2 × 106 cpm), 600 U of yeast poly(A) polymerase (U.S. Biochemicals), 1 mM ATP, 10 mM Tris-HCl (pH 7.0), 50 mM KCl, 0.7 mM MnCl2, 0.2 mM DTT, 2 μg of acetylated bovine serum albumin, and 10% glycerol at 37°C for 1 h. Poly(A)+ RNA was isolated by oligo(dT) chromatography using the Micro-FastTrack 2.0 mRNA isolation kit (Invitrogen) and was eluted in 200 μl of 10 mM Tris-HCl (pH 7.5).
Transfections and CAT assay.
DNA transfections were performed in human kidney 293T cells in suspension in 35-mm dishes (about 106 cells) by using 6 μl of LipofectAMINE 2000 (Gibco BRL) and 1 μg of each of the pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and pPOLI-CAT-RT plasmids in 2 ml of minimal essential medium (MEM) containing 10% fetal calf serum. Cells were harvested 24 h posttransfection, and cell lysates were prepared by resuspension of the cells in 100 μl of 250 mM Tris-HCl (pH 7.5) and by three cycles of freeze-thawing. Fifty microliters of undiluted or diluted cell extracts was incubated in 75-μl reaction mixtures containing 1 mM acetyl coenzyme A (Sigma), 0.1 mM [14C]chloramphenicol (50 mCi/mmol) (Amersham), and 250 mM Tris-HCl (pH 7.5) for 1 h at 37°C. Reaction products were extracted with ethyl acetate, separated by thin-layer chromatography, and detected by autoradiography. Quantitation was performed visually by comparing activities of serially diluted samples.
Western blot analysis of recombinant PA proteins.
DNA transfections were performed as described for the CAT assays (see above). About 48 h posttransfection, cells were harvested and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Cells were lysed by heating to 98°C for 5 min, and cell lysates were analyzed by SDS-8% PAGE. Proteins were electrotransferred to Hybond-C nitrocellulose membranes (Amersham) and probed with a polyclonal anti-PA antibody (6). Bound antibodies were detected by using a horseradish peroxidase conjugated anti-rabbit immunoglobulin G (IgG) secondary antibody (Sigma-Aldrich) and ECL development (Amersham) according to the manufacturer's protocol.
RNA isolation and analysis of vRNA, mRNA, and cRNA by primer extension assay.
Human kidney 293T cells in 35-mm dishes were transfected as for the CAT assays (see above) and harvested 48 posttransfection, and total RNA was isolated by using TRIzol reagent (Gibco BRL). Primer extension assays were performed as described elsewhere (10). Briefly, 5 μg of total RNA was mixed with an excess of DNA primer (about 105 cpm), labeled at its 5′ end with [γ-32P]ATP and T4 polynucleotide kinase, in 6 μl of water and denatured by heating at 95°C for 3 min. The mixture was cooled on ice and transferred to 42°C, and primer extensions were performed after addition of 100 U of SuperScript reverse transcriptase (Gibco BRL) in the reaction buffer provided with the enzyme for 1 h at 42°C. Two CAT-specific primers were used in the same reverse transcription reaction: 5′-CGCAAGGCGACAAGGTGCTGA-3′ (to detect vRNA) and 5′-ATGTTCTTTACGATGCGATTGGG-3′ (to detect mRNA and cRNA). Transcription products were analyzed on 6% polyacrylamide gels containing 7 M urea in TBE buffer and were detected by autoradiography. The expected sizes of the products are 158 nt (vRNA), 98 to 106 nt depending on the length of the capped primer (mRNA), and 89 nt (cRNA). Transcription products were quantitated by phosphorimage analysis.
Viral rescue.
Rescue of recombinant viruses was performed as described elsewhere (9, 38) with modifications. Briefly, human kidney 293T cells (about 1 106 cells) in suspension in 35-mm dishes were transfected with 6 μl of LipofectAMINE 2000 and 1 μg of each of 12 plasmids. Eight pPOLI plasmids encode the eight vRNA segments of influenza A/WSN/33 virus, as described elsewhere (9). Four pcDNA plasmids encode mRNAs for PB1, PB2, PA, and NP, the minimal set of viral proteins required for transcription-replication of vRNA, as described above. Transfection was performed in MEM containing 10% fetal calf serum. Eighteen to 24 h posttransfection, the medium was replaced with MEM containing 0.5% fetal calf serum, penicillin, and streptomycin, and the cells were incubated for another 24 to 48 h. Rescued viruses were amplified and plaqued on Madin-Darby bovine kidney (MDBK) cells.
Preparation of nuclear extracts containing recombinant influenza virus RNA polymerase.
Nuclear extracts were prepared by using a modification of a recently developed method (4a). Human kidney 293T cells in suspension in 8.5-cm dishes (about 3 × 106 cells) were transfected with 30 μl of LipofectAMINE 2000 transfection reagent and 7 μg of each of pcDNA-PB1, pcDNA-PB2, and pcDNA-PA in 10 ml of MEM containing 10% fetal calf serum. About 48 h posttransfection, cells were harvested and washed in 10 ml of ice-cold phosphate-buffered saline (PBS). The cell pellet was resuspended in 1 ml of cell lysis buffer A (10 mM Tris-HCl [pH 7.8], 10 mM KCl, 1 mM EDTA, 1 mM DTT, 0.1% Nonidet P-40, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and incubated on ice for 10 min. Nuclei were pelleted and resuspended in 250 μl of nuclear lysis buffer B (50 mM Tris-HCl [pH 7.8], 200 mM KCl, 0.2 mM EDTA, 1 mM DTT, 0.5% Nonidet P-40, 25% glycerol, and 1 mM PMSF) and incubated at 4°C for 30 min. Nuclear extracts were clarified by centrifugation at 1,000 × g and precipitated by adding an equal volume of 3 M ammonium sulfate (pH 7.5). The protein pellet was dissolved in 25 μl of nuclear lysis buffer B per transfected dish and stored at −20°C. The protein concentration of nuclear extracts was about 2 μg/μl as determined by a Bradford assay (Pierce).
Transcription assays in vitro.
Transcription reactions in vitro were performed (based on reference 4a) in a total reaction volume of 2.5 μl containing 1.25 μl of nuclear extracts (see above), 25 mM Tris-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 0.25% Nonidet P-40, 12.5% glycerol, 4 U of RNasin (Promega), 0.5 mM PMSF, 1 mM ATP, 0.5 mM UTP, 0.5 mM CTP, 0.1 μM GTP, 0.15 μM [α-32P]GTP (3,000 Ci/mmol) (Amersham), 4 pmol of 3′-end vRNA (5′-GGCCUGCUUUUGCU-3′) (Dharmacon), 4 pmol of 5′-end vRNA (5′-AGUAGAAACAAGGCC-3′) (Dharmacon), and 1 mM ApG. In the globin mRNA-primed in vitro transcription assays, the ApG was replaced with 0.03 μg of globin mRNA (Gibco BRL). In the capped RNA-primed transcription assays, the ApG was replaced with an 11-nt 32P-labeled capped RNA (about 5,000 cpm; see above) and 0.5 mM GTP was used instead of [α-32P]GTP. Transcription reactions were performed at 30°C for 1 h and terminated by addition of 7.5 μl of 80% formamide containing 1 mM EDTA, bromophenol blue, and xylene cyanol dyes. The reaction mixture was heated to 95°C for 3 min and loaded onto a 16% acrylamide gel containing 7 M urea in TBE buffer. Transcription products were detected by autoradiography and quantitated by phosphorimage analysis.
Endonuclease assay.
Endonuclease assays were performed in a total reaction volume of 4 μl containing 2 μl of nuclear extracts (see above), poly(A)+-capped RNA substrate (about 2,500 cpm; see above), 25 mM Tris-HCl (pH 7.8), 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 40 U of RNasin (Promega), 0.05 mM EDTA, 0.2% Nonidet P-40, 10% glycerol, 0.3 mM PMSF, 2 pmol of 5′-end vRNA (5′-AGUAGAAACAAGGCC-3′), and 2 pmol of 3′-end vRNA (5′-GGCCUGCUUUUGCU-3′). Endonuclease reactions were performed at 30°C for 30 min and terminated by addition of 7 μl of 80% formamide containing 1 mM EDTA, bromophenol blue, and xylene cyanol dyes. The reaction mixture was heated to 95°C for 3 min and loaded onto an 18% acrylamide gel containing 7 M urea in TBE buffer. Cleavage products were detected by autoradiography and quantitated by phosphorimage analysis.
RNA-binding assay.
RNA probes corresponding to 5′-end vRNA (5′-AGUAGAAACAAGGCC-3′) and 3′-end vRNA (5′-GGCCUGCUUUUGCU-3′) were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase for 1 h at 37°C. Reactions were terminated by addition of an equal volume of 80% formamide containing 1 mM EDTA, bromophenol blue, and xylene cyanol dyes. The reaction mixture was heated to 95°C for 2 min and loaded onto a 16% acrylamide gel containing 7 M urea in TBE buffer. The 32P-labeled RNAs were detected by autoradiography and eluted by soaking the crushed gel piece in 1 ml of H2O for 16 h at 4°C. The eluted RNA was desalted on NAP-10 columns (Pharmacia) and lyophilized by freeze-drying. The RNA pellet was dissolved in H2O to a final concentration of approximately 0.1 pmol/μl. Gel shifts were performed according to the work of Tiley et al. (54) with modifications. Briefly, binding reactions were performed in a total volume of 10 μl containing 5 μl of nuclear extracts (see above), 5,000 cpm of RNA probe, 25 mM HEPES (pH 7.5), 125 mM KCl, 2 mM MgCl2, 1 mM DTT, 0.5 mM EGTA, 0.05 mM EDTA, 1 mM PMSF, 8 U of RNasin (Promega), and 15% glycerol. Reaction mixtures were incubated at room temperature for 10 min, and reactions were terminated by addition of heparin (Sigma) to a final concentration of 5 μg/μl. Complexes were resolved by native gel electrophoresis (4% acrylamide-bisacrylamide [37.5:1], 3% glycerol, 0.5× TBE buffer; 12 V/cm; 4°C) and visualized by autoradiography.
Isolation of His-tagged recombinant influenza virus RNA polymerase.
Human kidney 293T cells in suspension in 8.5-cm dishes (about 3 × 106 cells) were transfected with 7 μg of each of pcDNA-PB1, pcDNA-PB2, and pcDNA-PA-His6 by using 30 μl of LipofectAMINE 2000 transfection reagent in 10 ml of MEM containing 10% fetal calf serum. Three dishes were used per preparation. About 48 h posttransfection, cells were pooled, washed twice in 10 ml of ice-cold PBS, resuspended in 0.9 ml of lysis buffer (50 mM sodium phosphate [pH 8.0], 200 mM NaCl, 25% glycerol, 0.5% Nonidet P-40, 1 mM β-mercaptoethanol, 0.1 mM PMSF, and 1 Complete Mini EDTA-free protease inhibitor cocktail tablet [Roche] per 10 ml), and incubated on ice for 15 min. All subsequent steps were performed at 4°C. The cell lysate was centrifuged at 13,400 × g for 15 min, and the supernatant was transferred into a fresh 1.5-ml tube. A 100-μl volume of a nickel-nitriloacetic acid (NTA) agarose (Qiagen) suspension, washed in lysis buffer, and imidazole to a final concentration of 5 mM were added to the cell lysate. After gentle mixing for 2 h, the Ni-NTA agarose was collected by centrifugation at 13,400 × g for 2 min and washed twice in lysis buffer containing 10 mM imidazole. Proteins were eluted from Ni-NTA agarose in 100 μl of lysis buffer containing 100 mM imidazole.
Cap-binding assay.
A UV-cross-linking assay (13) was used to test the cap-binding activity of recombinant RNA polymerases. Initially, 5 μl of partially purified His-tagged polymerase (see above) was mixed with 5 pmol of 5′-end vRNA (5′-AGUAGAAACAAGGCC-3′) in a total reaction volume of 8 μl containing 10 mM HEPES (pH 7.5), 100 mM KCl, 2 mM MgCl2, 0.5 mM EGTA, 1 mM DTT, 8 U of RNasin (Promega), and 10% glycerol in order to allow binding of the polymerase to the 5′-end vRNA. After 15 min at 30°C, 2 μl of capped RNA (40,000 cpm), labeled with 32P in the cap-1 structure (see above), was added to the reaction mixture. The mixture was incubated for another 15 min at 30°C, then transferred into a U-bottom 96-well plate, and irradiated on ice for 10 min in a UV Stratalinker (Stratagene) equipped with G8T5 bulbs (254 nm). The cross-linked products were denatured at 95°C for 5 min, analyzed by SDS-8% PAGE, and detected by autoradiography. To determine which polymerase subunits were cross-linked to the capped RNA, cross-linked complexes were disrupted by 0.1% SDS treatment and immunoprecipitations were performed as described previously (13).
RESULTS
Strategy of mutagenesis.
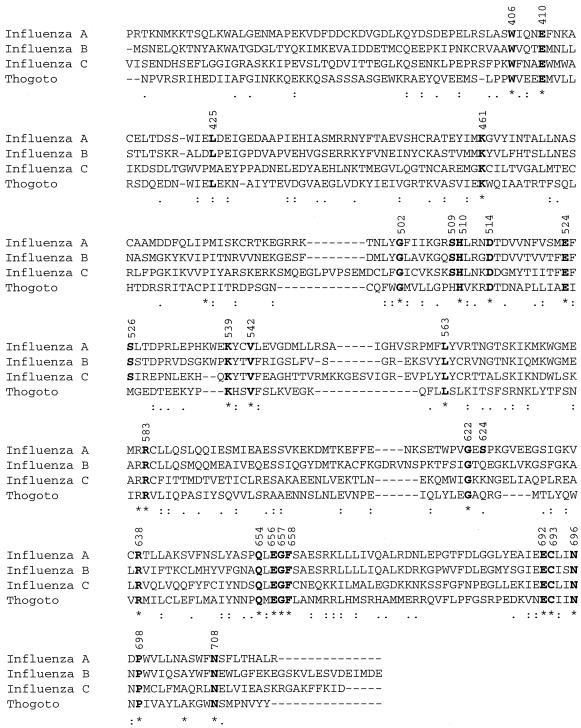
In order to investigate the function of the PA subunit of the influenza virus RNA polymerase in the processes of transcription and replication of vRNA, we decided to perform a mutational analysis of the protein. Similarity between the PA proteins of influenza A, B, and C viruses and Thogoto virus is limited, revealing no clear indication of the possible roles of the different regions of the protein. The PA proteins of influenza A and B viruses are the most closely related, exhibiting about 36% amino acid identity (Table 1). Influenza C virus PA proteins show about 25% identity with PA proteins of both influenza A and B viruses. The PA protein of Thogoto virus, a virus related to influenza viruses and therefore classified in the Orthomyxoviridae family, is the most distant, showing only about 15% amino acid identity with any of the influenza virus PA proteins. A similarity plot of influenza A, B, and C virus PA proteins revealed that the C-terminal one-third exhibited the highest degree of similarity among the three PA proteins (Fig. 1). When only the C-terminal one-third of the protein (amino acids 500 to 716 for influenza A virus) is considered, the identity of amino acids between influenza A and B virus proteins rises from 36 to 48% and the influenza C virus protein also shows a higher degree of identity to both influenza A and B virus proteins (about 32%) (Table 1). Therefore, we decided to focus on the C-terminal region of the protein, arguing that the higher degree of similarity suggests the presence of an evolutionarily conserved domain(s) crucial for PA function(s). We introduced alanine mutations at those amino acid positions that were conserved in an alignment of the PA proteins of influenza A, B, and C viruses and Thogoto virus (Fig. 2). In addition, we included several amino acids that were not absolutely conserved, because they were implicated in various functions, e.g., nucleotide binding (S509) (5), phosphorylation (S526) (51), or serine protease activity (S624) (18), or were present in a region of PA originally implicated in interaction with a cellular protein (L425) (M. Huarte, J. Sanz-Ezquerro, J. Ortín, and A. Nieto, Abstr. 11th Int. Conf. Negative Strand Viruses, abstr. 94, 2000). Moreover, during the cloning procedure, two mutants with deletions were isolated. One of the deletion mutants, Q654A(Δ655-658), contained a glutamine-to-alanine mutation at position 654 and a deletion of four highly conserved amino acids, LEGF, between positions 655 and 658. The other deletion mutant, C693A(Δ694-716), contained a cysteine-to-alanine mutation at position 693 and lacked the C-terminal 23 amino acids. Mutations were introduced into the pcDNA-PA protein expression plasmid by site-directed mutagenesis.
TABLE 1.
Amino acid sequence identities between the PA proteins of influenza A, B, and C viruses and Thogoto virus
| Proteina | % Identity between virusesb
|
|||||
|---|---|---|---|---|---|---|
| A vs B | A vs C | A vs D | B vs C | B vs D | C vs D | |
| PA (aa1-716) | 36 | 25 | 15 | 24 | 15 | 14 |
| PA (aa500-716) | 48 | 31 | 19 | 32 | 18 | 23 |
Numbering refers to amino acid (aa) positions in A/WSN/33.
Influenza A/WSN/33 virus; B, influenza B/Panama/45/90 virus; C, influenza C/JJ/50 virus; D, Thogoto virus.
FIG. 1.
Amino acid sequence similarity plot of influenza A, B, and C virus PA proteins. Influenza A/WSN/33, B/Panama/45/90, and C/JJ/50 virus PA sequences (GenBank accession numbers J02152, AF005738, and M28062, respectively) were aligned, and similarity was calculated and plotted for a window of 10 amino acids with programs in the Genetics Computer Group software package. Horizontal line, average level of overall similarity in the plot.
FIG. 2.
Alignment of amino acid sequences of PA proteins of influenza A, B, and C viruses and Thogoto virus. PA sequences of influenza A/WSN/33, B/Panama/45/90, and C/JJ/50 viruses and Thogoto virus (GenBank accession numbers J02152, AF005738, M28062, and 2253293, respectively) were aligned with CLUSTAL W from the Genetics Computer Group software package by using the default parameters (details available from us on request). Inclusion of the Thogoto virus PA sequence, the most divergent of the four PA sequences used in the alignment, could result in alternative alignments of some amino acid regions if different parameters of CLUSTAL were used. Amino acids selected for mutagenesis are boldfaced. Numbers refer to amino acid positions in the A/WSN/33 sequence. Only the C-terminal halves of the PA sequences are shown (amino acids 355 to 716 for A/WSN/33).
Functionality of the mutant PA proteins.
To test the functionality of the mutant PA proteins, we used an in vivo transcription-replication assay based on in vivo reconstitution of viral RNP complexes and expression of a CAT reporter gene (45). Mutant PA proteins were coexpressed in 293T cells with influenza virus PB1, PB2, and NP proteins (the minimal set of influenza virus proteins required for transcription and replication of vRNA) and a vRNA-like CAT-RNA, encoding a negative-sense CAT reporter gene. Cell extracts were tested for CAT activity 24 h posttransfection. Surprisingly, we found that most of the mutations had little if any effect on CAT activity (Table 2). Only three single-amino-acid mutants (G502A, H510A, and E524A) resulted in <1% or undetectable CAT activity relative to that of the wild-type control. The two deletion mutants [Q654A(Δ655-658) and C693A(Δ694-716)] also showed CAT activities below the level of detection. All the other mutants showed significant CAT activity levels (more than 1% relative to that of the wild type). It is unlikely that the proteolytic activity of the mutant PA proteins showing <1% or undetectable CAT activities was affected, since (i) all the mutations tested are located in a region distinct from that implicated in the induction of proteolysis (52) and (ii) all mutants with low or undetectable CAT activity were expressed at levels similar to those of the wild type, as determined by Western blot analysis with an anti-PA antibody (Table 2)—a result inconsistent with autoproteolysis (52).
TABLE 2.
Phenotypic analyses of mutant PA proteins
| Mutation (amino acid) | CAT activitya | Proteinb | Viral rescuec |
|---|---|---|---|
| WT | +++ | √ | + |
| W406A | ++ | √ | + |
| E410A | ++ | √ | + |
| L425A | +++ | NT | + |
| K461A | +++ | NT | + |
| G502A | — | √ | — |
| S509A | +++ | NT | + |
| H510A | + | √ | — |
| D514A | +++ | NT | + |
| E524A | — | √ | — |
| S526A | +++ | NT | + |
| K539A | +++ | √ | — |
| V542A | +++ | NT | + |
| L563A | +++ | NT | + |
| R583A | +++ | NT | + |
| G622A | +++ | √ | + |
| S624A | +++ | √ | + |
| R638A | +++ | √ | + |
| Q654A | +++ | √ | + |
| E656A | +++ | √ | + |
| G657A | ++ | √ | + |
| F568A | +++ | NT | + |
| E692A | ++ | NT | + |
| C693A | +++ | NT | + |
| N696A | +++ | NT | + |
| P698A | +++ | NT | + |
| N708A | +++ | NT | + |
| Q654A(Δ655-658) | — | √ | — |
| C693A(Δ694-716) | — | √ | — |
+++, 10 to 100%; ++, 1 to 10%; +, <1%; —, not detectable.
√, expressed to wild-type levels; NT, not tested.
+, yes; —, no viral rescue was achieved.
Generation of recombinant influenza viruses with mutations in PA.
Since most of the single-amino-acid mutations had only a marginal or no effect on CAT activity, we proceeded to perform viral rescue. We used the plasmid-based rescue method for generating recombinant influenza viruses from cDNA (9). The mutations were subcloned into the pPOLI-PA-RT plasmid to express mutant PA vRNA. These were cotransfected into 293T cells with seven pPOLI plasmids, encoding the remaining seven vRNA segments, and four protein expression plasmids, encoding PB1, PB2, PA (wild-type), and NP proteins. Only six of the PA mutants could not be rescued into infectious virus (Table 2). These included the three single-amino-acid mutants showing <1% or undetectable CAT activities and the two deletion mutants. In addition, the K539A mutant could not be rescued, although it showed wild-type activity in the CAT reporter assay. All the recombinant viruses, with the exception of the W406A, E410A, R638A, and E656A mutants, produced stocks of at least 106 PFU/ml in MDBK cells, indicating that their growth in cell culture was not severely compromised (data not shown). The W406A, E410A, R638A, and E656A mutants grew poorly, producing pinhead-sized plaques on MDBK cells, which made any further characterization difficult.
Synthesis of mRNA, cRNA, and vRNA by polymerases with mutant PA subunits.
Since early work with viruses carrying ts mutations in their PA genes has indicated that PA is involved in replication, but not necessarily in the transcriptional activity of the polymerase (31), we were particularly interested in whether any of the mutations generated affected viral transcription and replication differentially. Therefore, we set up a “single-tube” primer extension assay to compare mRNA, cRNA, and vRNA levels in cells expressing the three influenza virus RNA polymerase subunits, NP, and a vRNA-like CAT-RNA. Two primers were used in the same primer extension reaction, one specific for the negative-sense RNA (vRNA) and the other specific for the positive-sense RNAs (mRNA and cRNA). Because mRNA and cRNA differ at their 5′ ends, it was possible to distinguish the signals originating from the two RNA species, based on their different lengths. Analysis of total RNA isolated from cells expressing wild-type PB1, PB2, PA, NP, and a vRNA-like CAT-RNA resulted in three radiolabeled bands of the expected sizes for vRNA, mRNA, and cRNA (Fig. 3A, lane 5). While the vRNA and cRNA signals appear as discrete bands, the mRNA signal is a “wide” band representing RNA species of various lengths, due to the heterogeneity in the size of the capped RNA primers used by the influenza virus polymerase to initiate mRNA synthesis. Primer extension performed on poly(A)+ and poly(A)− fractions of total RNA—isolated by oligo(dT) chromatography—showed that the poly(A)+ fraction was enriched in, while the poly(A)− fraction was depleted of, RNA species producing mRNA signals. On the other hand, the poly(A)+ fraction was depleted of, and the poly(A)− fraction was enriched in, RNA species producing the cRNA and vRNA signals (data not shown). Analysis of total RNA from nontransfected cells produced no radiolabeled bands at the expected positions for vRNA, mRNA, or cRNA signals (Fig. 3A, lane 2). If the pPOLI-CAT-RT plasmid was transfected alone or the PA protein expression plasmid was omitted from the transfection mixture, a faint vRNA signal was observed (Fig. 3A, lanes 3 and 4, respectively). This signal represents vRNA transcribed by cellular RNA polymerase I from the pPOLI-CAT-RT plasmid. These results demonstrate that in the presence of PB1, PB2, and NP, but in the absence of PA, no significant vRNA-dependent transcription can occur in vivo (Fig. 3A; compare lanes 4 and 5).
FIG. 3.
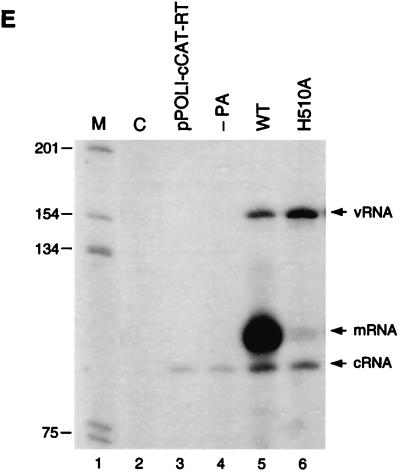
In vivo RNA synthesis mediated by mutant PA proteins. (A through D) Primer extension assays of vRNA, mRNA, and cRNA isolated from cells expressing influenza virus polymerase proteins and vRNA-like CAT-RNA. 293T cells were transfected, as indicated above the lanes, either with pcDNA3 (indicated by “C”), with pPOLI-CAT-RT, with pPOLI-CAT-RT, pcDNA-PB1, and pcDNA-PB2 (−PA), or with pPOLI-CAT-RT, pcDNA-PB1, pcDNA-PB2, and wild-type (WT) or mutant pcDNA-PA. (E) Primer extension assays of vRNA, mRNA, and cRNA isolated from cells expressing influenza virus polymerase proteins and cRNA-like CAT-RNA. 293T cells were transfected, as indicated above the lanes, either with pcDNA3 (indicated by “C”), with pPOLI-cCAT-RT, with pPOLI-cCAT-RT, pcDNA-PB1, and pcDNA-PB2 (−PA), or with pPOLI-cCAT-RT, pcDNA-PB1, pcDNA-PB2, and wild-type (WT) or mutant (H510A) pcDNA-PA. pcDNA-NP was included in all transfections except the negative controls C, pPOLI-CAT-RT, and pPOLI-cCAT-RT. Isolation of RNA and primer extension assays were performed as described in Materials and Methods. Positions of vRNA, mRNA, and cRNA signals are indicated on the right. Stars indicate nonspecific priming on cellular RNAs. M, 32P-labeled 1-kb DNA ladder (Gibco BRL); sizes are indicated on the left.
Having established a simple method for analyzing vRNA, mRNA, and cRNA levels in transfected cells, we proceeded to test the effects of PA mutations on viral transcription and replication (Fig. 3A through C). We found that most of the single-amino-acid mutations had no effect or only a subtle effect on RNA levels (L425A, K461A, S509A, D514A, S526A, V542A, L563A, R583A, G622A, S624A, Q654A, E656A, G657A, F658A, E692A, C693A, N696A, P698A, and N708A), indicating that none of these positions is crucial for transcription and replication. Mutants W406A and E410A resulted in a general decrease in levels of all three RNA species, while mutants G502A and E524A and the two deletion mutants [Q654A(Δ655-658) and C693A(Δ694-716)] showed no detectable levels of RNAs. We observed good correlation between CAT activities (Table 2) and mRNA levels expressed by the PA mutants (Fig. 3), with the exception of the G657A and E692A mutants, which showed 1 to 10% CAT activity but only a subtle effect on mRNA levels. It should be noted that CAT activities were analyzed 24 h posttransfection, while mRNA levels were determined 48 h posttransfection, possibly accounting for the discrepancies.
More interestingly, the remaining three mutants, H510A, K539A, and R638A, showed differential effects on transcription and replication (Fig. 3A, lanes 12 and 16, and Fig. 3C, lane 6). The H510A mutant, which showed less than 1% CAT activity and could not be rescued into infectious virus (Table 2), expressed only background levels of mRNA (<1% compared to the wild type) (Fig. 3A, lane 12). Surprisingly, however, we observed significant levels of both cRNA and vRNA. Although, cRNA levels produced by this mutant were reduced about fivefold, vRNA levels were close to that of the wild type (Fig. 3A; compare lanes 5 and 12). Thus, the H510A mutation specifically inhibits the vRNA-templated transcriptional activity (mRNA and cRNA synthesis) of the RNA polymerase, while its cRNA-templated transcriptional activity (vRNA synthesis) is apparently not affected. Although both mRNA synthesis and cRNA synthesis were inhibited by the mutation, mRNA synthesis was inhibited 20 times more than cRNA synthesis, indicating that the H510A mutation primarily affects the transcriptional (mRNA synthesis) activity of the RNA polymerase.
The K539A mutant showed wild-type CAT activity levels but, surprisingly, did not produce infectious virus (Table 2). Analysis of RNA levels revealed that this mutant produced wild-type levels of mRNA, in agreement with the CAT assay results, but produced about 10-fold less cRNA and vRNA than the wild type (Fig. 3A; compare lanes 5 and 16). Thus, the K539A mutation selectively inhibits replication. The low vRNA levels produced by this mutant might explain why no infectious virus carrying the K539A mutation could be rescued (Table 2).
The R638A mutant, which showed 10 to 100% CAT activity but produced pinhead-sized plaques when rescued into a virus (Table 2), exhibited about a fivefold increase in vRNA levels over that of the wild type. In contrast, the mRNA levels produced by this mutant were reduced fivefold (Fig. 3C; compare lanes 4 and 6). Thus, the R638A mutation in the PA subunit results in upregulation of the replicative and downregulation of the transcriptional activity of the RNA polymerase.
Previous studies of PA implied a role for PA only in cRNA and vRNA synthesis (31, 42), and therefore we decided to study the H510A mutant, which showed a defect in mRNA synthesis, in more detail. First, we confirmed that the observed differences in the ratios of vRNA, mRNA, and cRNA synthesized by this mutant are reproducible with a cRNA-like CAT-RNA template. Analysis of total RNA isolated from cells expressing wild-type PB1, PB2, PA, NP, and cRNA-like CAT-RNA resulted in a pattern of vRNA, mRNA, and cRNA signals comparable to that obtained with the vRNA template (compare lane 5 in Fig. 3E to lane 5 in Fig. 3A). The H510A mutant produced significant amounts of cRNA and vRNA, but only background levels of mRNA, in agreement with the previous results with the vRNA template (compare lane 6 in Fig. 3E to lane 12 in Fig. 3A). This supports the idea that the cRNA→vRNA synthesizing activity of the H510A polymerase is not affected.
If the pPOLI-cCAT-RT plasmid was transfected alone or the PA protein expression plasmid was omitted from the transfection mixture, a faint cRNA signal was observed (Fig. 3E, lanes 3 and 4, respectively). This signal represents cRNA transcribed by cellular RNA polymerase I from the pPOLI-cCAT-RT plasmid. These results demonstrate that in the presence of PB1, PB2, and NP, but in the absence of PA, no significant cRNA-dependent transcription can occur in vivo (Fig. 3E; compare lanes 3 and 4). Thus, in the absence of PA, neither vRNA- nor cRNA-dependent RNA synthesis can occur at significant levels in vivo.
We also tested the effects of two other amino acid mutations at position 510 of PA (Fig. 3D). In contrast to the H510A mutation, a conservative amino acid change (H510K) had no effect on the transcriptional activity of the polymerase, resulting in wild-type levels of mRNA (Fig. 3D, lane 6). A basic-to-acidic amino acid mutation (H510E), however, abolished not only mRNA but also cRNA and vRNA synthesis (Fig. 3D, lane 5). Neither of these mutations could be rescued into infectious virus (data not shown).
In vitro transcriptional activity of the H510A polymerase.
The transfection-based in vivo experiments showed that the H510A mutant could synthesize significant amounts of cRNA and vRNA but undetectable amounts of mRNA. Because the cRNA→vRNA synthesizing activity of the polymerase was not affected, it was unlikely that the H510A mutation in the PA affected the processivity of the polymerase. It was more likely that this mutant was deficient in some specific function required for vRNA-dependent RNA synthesis, in particular mRNA synthesis. The synthesis of viral mRNA molecules is dependent on the ability of the RNA polymerase to generate capped RNA primers for transcription initiation, while replication is independent of capped RNA primers. Therefore, we decided to compare the activities of recombinant RNA polymerases containing wild-type or H510A mutant PA subunits in a transcription initiation assay using either ApG dinucleotide or capped RNA primers in vitro.
Crude nuclear extracts containing recombinant polymerase from cells expressing wild-type PB1, PB2, and PA were able to transcribe a 14-nt 3′-end vRNA template in both ApG- and globin mRNA-primed transcription reactions in vitro (Fig. 4A, lanes 2 and 5). The ApG-primed product appeared as a triplet, the slowest-moving band of which represented the full-length (14-nt) transcription product (4a). We speculate that the faster-moving bands (12 and 13 nt) are the results of nuclease digestion of the RNA template or transcription product, or both. The globin mRNA-primed transcription product ran more slowly than the ApG-primed product due to the presence of the capped RNA sequence at its 5′ end. It also appeared as a triplet, either due to cleavage at multiple sites of the capped primer or due to nuclease digestion, or both. Nuclear extracts from nontransfected cells were inactive (Fig. 4A, lanes 1 and 4). Nuclear extracts from cells expressing PB1 and PB2, but not PA, produced no detectable activity, indicating that PA is required for efficient transcription initiation in vitro with both ApG and capped RNA primers in agreement with previous findings (4a) (data not shown). The activity of the recombinant RNA polymerase was also dependent on the presence of a 15-nt RNA containing the 13 conserved 5′-terminal vRNA residues, as observed previously (4a). If the 5′-end RNA was omitted from the reaction, only low levels of activity were observed with the ApG primer (estimated at <10%) (data not shown). No transcription products could be detected in the absence of the 5′-end vRNA if globin mRNA was used as a primer (data not shown). Although NP is essential for transcription and replication of influenza virus RNA in vivo, the presence of NP is known not to be required for endonuclease cleavage of capped RNA primers or for synthesis of short 14-nt RNA transcripts (4a, 17, 27), and therefore it was not included in our in vitro assays.
FIG. 4.
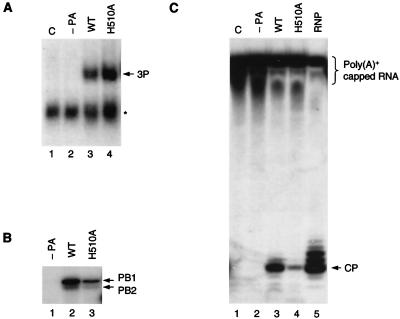
In vitro RNA synthesis mediated by the H510A polymerase. (A) Transcription initiation primed with ApG and globin mRNA primers. Transcription reactions were carried out in vitro with nuclear extracts from cells transfected either with pcDNA3 (C) (lanes 1 and 4) or with pcDNA-PB1, pcDNA-PB2, and wild-type (WT) (lanes 2 and 5) or mutant (H510A) (lanes 3 and 6) pcDNA-PA in the presence of ApG dinucleotide (lanes 1 to 3) or globin mRNA (lanes 4 to 6) primers. The positions of transcription products (TP) are indicated. The star indicates the position of a nonspecific band. (B) Transcription initiation primed with a capped RNA primer. Transcription reactions were carried out in vitro with nuclear extracts from cells transfected either with pcDNA3 (C) (lane 1) or with pcDNA-PB1 and pcDNA-PB2 either without pcDNA-PA (−PA) (lane 2) or with wild-type (WT) (lane3) or mutant (H510A) (lane4) pcDNA-PA in the presence of an 11-nt 32P-labeled capped RNA. The positions of transcription products (TP) and the primer are indicated.
Having established the conditions of the in vitro transcription reaction using recombinant RNA polymerase, we proceeded to test the activity of the H510A mutant. We found that the H510A mutant was able to initiate transcription with the ApG primer as efficiently as the wild type (Fig. 4A; compare lane 3 to lane 2). In contrast, the H510A mutant showed dramatically reduced activity with the globin mRNA primer (Fig. 4A; compare lane 6 to lane 5). Although the activities of the individual recombinant polymerase preparations varied, we consistently observed a greater degree of reduction in the activity of the H510A mutant with capped RNA primers than with the ApG primer. On average, the H510A mutant was about 10-fold less efficient in priming with capped RNA than with ApG. These results suggested that the defect of the H510A polymerase could, at least in part, be directly related to its reduced ability to initiate with capped RNA primers.
Endonuclease and RNA-binding activities of the H510A polymerase.
In order to initiate transcription with capped RNA primers, the RNA polymerase must be able to perform several activities. It must bind to both ends of vRNA, because the 5′ vRNA sequence acts as an activator of the cap-binding activity of the polymerase and, in addition, binding to 3′-end vRNA is required for the endonuclease activity. Since the H510A mutant is active in ApG-primed transcription initiation in vitro and this activity is dependent on both 5′-end and 3′-end RNAs (see above), it is unlikely that the H510A polymerase is deficient in template RNA binding. To test this further, we analyzed the vRNA-binding activity of the H510A polymerase in a gel shift assay. We found that the ability of the mutant polymerase to bind 5′- or 3′-end vRNA was not significantly affected (Fig. 5A, compare lanes 3 and 4; and data not shown). Next we tested the ability of the H510A mutant polymerase to bind capped RNA in a UV-cross-linking assay. However, because of the presence of a cellular protein in the crude recombinant RNA polymerase preparations that cross-linked to the radiolabeled capped RNA and produced a strong signal at around 90 kDa on protein gels—at the expected position of the influenza virus polymerase proteins—it was not possible to study cap-binding functions with the polymerase preparations that we used in the in vitro transcription assay (see above). Therefore, we prepared partially purified recombinant polymerases with wild-type or H510A mutant PA subunits. To this end, we fused a short histidine tag to the C terminus of the PA protein to enable purification on a Ni affinity column. Partially purified wild-type RNA polymerase, cross-linked to capped RNA radiolabeled in its cap structure, produced two distinct bands on protein gels (Fig. 5B, lane 2). Immunoprecipitation of the cross-linked RNA polymerase subunits, after disruption of the complex with 0.1% SDS treatment, confirmed that the band with lower mobility corresponded to PB1 and the band with higher mobility corresponded to PB2 (data not shown). These results are in agreement with a previous report (29) showing that a capped RNA could be cross-linked to PB1, in addition to the cap-binding PB2 subunit, possibly because PB1 contains the endonuclease domain involved in cleaving capped RNAs.
FIG. 5.
RNA-binding and endonuclease activities of the H510A polymerase. (A) 5′-end vRNA-binding activity of the H510A polymerase. Nuclear extracts from cells transfected either with pcDNA3 (lane 1), with pcDNA-PB1 and pcDNA-PB2 (−PA) (lane 2), or with pcDNA-PB1, pcDNA-PB2, and either wild-type (WT) or H510A mutant pcDNA-PA (lanes 3 and 4, respectively) were used in gel shifts with 32P-labeled 15-nt 5′-end vRNA as described in Materials and Methods. The position of the polymerase complex (3P) is indicated. The star indicates a nonspecific band. (B) Capped RNA-binding activity of the H510A polymerase. His-tagged polymerases, partially purified from cells transfected either with pcDNA-PB1 and pcDNA-PB2 (−PA) (lane1) or with pcDNA-PB1, pcDNA-PB2, and either pcDNA-PA-His6 wild-type (WT) or H510A mutant pcDNA-PA-His6 (lanes 2 and 3, respectively), were cross-linked to 32P-labeled capped RNA as described in Materials and Methods. The positions of the PB1 and PB2 bands are indicated. (C) Endonuclease activity of the H510A polymerase. Nuclear extracts from cells transfected either with pcDNA3 (lane 1), with pcDNA-PB1 and pcDNA-PB2 (−PA) (lane 2), or with pcDNA-PB1, pcDNA-PB2, and either wild-type (WT) or H510A mutant pcDNA-PA (lanes 3 and 4, respectively) were incubated with poly(A)+-capped RNA, and cleavage products were analyzed as described in Materials and Methods. As a positive control, endonuclease cleavage was carried out with viral cores isolated from purified virus (RNP) (lane 5). The position of the specific cleavage product (CP) is indicated. The poly(A)+-capped RNA (indicated) runs as a smear due to the variety in the length of the poly(A) tail.
Recombinant partially purified His-tagged H510A polymerase produced a similar pattern of cross-linked PB1 and PB2 bands, although the intensity of the bands was weaker than with the wild type, suggesting that the cap-binding activity of the H510A mutant was affected (Fig. 5B, lane3). We cannot, however, exclude the possibility that the C-terminal His tag perturbs the function of PA to some extent, and it might do so to a greater extent in the case of the H510A mutant than in the case of the wild type. Indeed, preliminary studies suggest that a recombinant polymerase containing a His-tagged H510A mutant PA subunit results in slightly reduced CAT reporter expression compared to that for a polymerase containing non-His-tagged H510A mutant PA (data not shown). Nevertheless, it is clear that the H510A mutant polymerase retains a significant level of capped RNA-binding activity.
It was unlikely that the observed reduction in capped RNA binding could fully explain the severe inhibition of the ability of the H510A mutant to prime transcription with capped RNA. Therefore, we proceeded to test whether recombinant RNA polymerases containing wild-type or H510A mutant PA subunits were active in endonucleolytic cleavage of capped RNAs. As a substrate, we used a synthetic 11-nt capped RNA, radiolabeled with 32P in its cap structure, that was extended by a poly(A) polymerase in vitro, resulting in capped and poly(A)-tailed products. Because the substrate contained RNA molecules with poly(A) tails of various lengths, it ran as a “smear” rather than a discrete band near the top of the gel (Fig. 5C, lane 1). Incubation of wild-type recombinant polymerase with the substrate RNA resulted in the appearance of a discrete band (Fig. 5C, lane 3) that comigrated with an 11-nt capped RNA marker (data not shown) and a cleavage product generated by influenza virus cores (lane 5). This band was absent if the RNA substrate was incubated with nuclear extracts from cells expressing no influenza virus polymerase proteins (Fig. 5C, lane 1) or expressing only PB1 and PB2, but no PA (lane 2). These results showed that the wild-type recombinant RNA polymerase possessed the specific endonuclease activity characteristic of influenza virus RNA polymerases. In contrast, we observed only low levels of specific cleavage products with the H510A mutant polymerase (Fig. 5C, lane 4).
We speculated that if the major defect of the H510A polymerase was in its endonuclease activity, as suggested by the results described above, it might be able to initiate transcription with short capped RNA primers not requiring endonuclease cleavage. To address this question, we tested the activity of the recombinant RNA polymerase preparations with an 11-nt capped RNA primer which has been previously shown to act as a primer in transcription initiation reactions in vitro without the need for endonuclease cleavage (4). We found that the H510A mutant polymerase could extend the capped RNA primer almost as efficiently as the wild type (Fig. 4B; compare lanes 3 and 4). We detected no specific extension products if we used nuclear extracts from nontransfected cells or nuclear extracts containing only PB1 and PB2 (Fig. 4B, lanes 1 and 2, respectively).
Taken together, these results suggest that the severe inhibition of the capped RNA-priming activity of the H510A mutant could be primarily due to a defect in its endonuclease cleavage activity.
DISCUSSION
We have identified an amino acid position in the PA subunit of the influenza A virus RNA polymerase complex that is required for the endonuclease activity of the polymerase. Mutation of a histidine at amino acid position 510 to an alanine resulted in inhibition of the transcriptional activity of the polymerase complex in vivo. In agreement with this, the H510A mutant showed severely reduced activity (<1%) in a CAT reporter gene expression assay. In addition, no mutant virus could be constructed with an H510A mutation in the PA protein, most likely due to the low transcriptional activity and, consequently, the low protein levels expressed in cells. Surprisingly, however, this mutant RNA polymerase was active in RNA replication, as determined by analysis of RNA levels in transfected cells.
Detailed analyses of the activities of the H510A mutant in vitro revealed that the endonuclease activity of this polymerase was severely affected. Although cap-binding activity appeared to be reduced, this reduction could not fully account for the reduction in endonuclease activity. In support of this, the H510A mutant was able to initiate transcription efficiently when provided with a short capped RNA primer that did not require cleavage prior to transcription initiation. To our knowledge, this is the first report suggesting a specific role of PA in transcription initiation and in endonuclease activity in particular.
Early work with ts virus mutants suggested that PA is involved in RNA replication (31), but its role in RNA transcription was not appreciated. Our results clearly indicate that PA is required for transcription initiation, possibly by directly participating in the endonuclease cleavage of cellular pre-mRNAs. Early work suggested that the endonuclease domain resides in the PB2 subunit (3, 53); however, a more recent study proposed that the PB1 subunit of the polymerase is responsible for the cleavage of capped pre-mRNAs (29). Although the PB2 subunit contains the cap-binding motif, the actual cleavage is performed by the PB1 subunit. In support of this, a capped RNA can be cross-linked to both PB1 and PB2, and amino acid sequences involved in binding and cleaving capped RNA have been identified. Three acidic amino acids (E508, E519, and D522) of PB1 have been shown to be crucial for the cleavage reaction (29). Our results suggest that PA forms an active part of the influenza virus cap-dependent endonuclease, although we have found no evidence as yet that a capped RNA could be directly cross-linked to PA (data not shown).
PA is known to interact with PB1, and the histidine is present in the region of PA which is proposed to interact with PB1 (55, 56). Although the proposed endonuclease active site in PB1 is outside the region that is known to interact with PA (44), we speculate that in the three-dimensional structure, H510 might be close to the endonuclease active site of the PB1 subunit. It is possible that the active site is formed on the interface of the PB1 and PA subunits with amino acids from both subunits participating. Whether H510 participates directly in the catalytic mechanism (proposed to be a two-metal ion catalysis [7]), however, would need further investigation. Our result that another basic amino acid residue can replace H510 without loss of transcriptional activity (H510K) suggests that H510 is not directly involved in catalysis. It should be noted, however, that no infectious virus could be generated with an H510K mutation in the PA protein.
Masunaga et al. (33) reported that the cap-dependent endonuclease was sensitive to inhibition by monoclonal antibodies targeted against amino acids 401 to 517 of PA and the N-terminal region of PB1 (amino acids 1 to 222). The anti-PB1 antibody could interfere with the binding of PA to PB1, since the N-terminal region is implicated in binding PA (44), while the anti-PA antibody could directly target the region of PA forming part of the endonuclease active site, as suggested by this report.
Amino acid position H510 is located just downstream of a region which has been identified as a putative nucleotide-binding motif based on a homology search (5). Zürcher et al. (56) identified the putative nucleotide-binding domain as a potential active site of PA, based on a mutagenesis study. In particular, a mutant that had a serine insertion at position 509, located just upstream of the H510 position, did not show any polymerase activity, as measured by the expression of a CAT reporter. However, this mutant bound to PB1, induced proteolytic activity like the wild type, and acted as a dominant-negative mutant, suggesting that the S509 insertion did not affect folding. The inhibition of CAT reporter activity indicates that mRNA synthesis was affected but provides no information on RNA replication. It would be interesting to see whether this insertion affected the replicative activity of the polymerase or whether the inhibitory effect was limited to mRNA synthesis.
The close proximity of H510 to the putative nucleotide-binding domain might suggest that the H510A mutation interferes with nucleotide binding, but it is difficult to speculate what role nucleotide binding could play in nuclease activity. As part of our mutagenesis strategy, we mutated two amino acid residues, G502 and S509, which form part of the putative nucleotide-binding motif (G502FIIKG507R508S509) (5). The G502A mutant showed no detectable activity in CAT reporter gene expression or primer extension analysis of RNA in transfected cells. In contrast, the S509A was fully active in both assays. A direct nucleotide-binding assay would be required to evaluate whether this region of PA is involved in nucleotide binding.
Although the finding that the H510A polymerase retains many functions, e.g., primer-independent transcription initiation and RNA elongation, would suggest that the PA is properly folded and the H510A mutation has a direct effect on endonuclease activity, we cannot exclude an indirect effect. In fact, the observation that the H510A mutation in PA also affects cRNA synthesis, although less dramatically than mRNA synthesis, but not vRNA synthesis indicates that the H510A polymerase has a template-specific defect. While the vRNA-dependent activities of the polymerase were affected, the cRNA-dependent activity was unaffected. PB1 has been shown to contain the binding sites for vRNA and cRNA, although there is some controversy as to exactly which amino acid regions of PB1 form these binding sites (14, 15, 28). Recently, Lee et al. (27) suggested that PA might act as an enhancer of the RNA-binding activities of PB1, since they observed binding of the 5′-end vRNA only when PA was present. In addition, it was shown that PA can be cross-linked to vRNA and cRNA terminal sequences, suggesting a role for PA in promoter RNA binding (11, 13, 48). Although we have not been able to demonstrate significant differences in vRNA-binding properties between the H510A mutant and the wild type, we cannot exclude the possibility that the H510A mutation might specifically interfere with vRNA promoter recognition. Subtle differences in binding, not detectable in gel shifts, might lead to significant effects in vivo, resulting in inhibition of some vRNA-dependent polymerase activities. On the other hand, cRNA binding could be performed by a different domain of PB1 (15), which might be independent of PA or involve a different region of PA. Therefore, the H510A mutation would not affect cRNA-dependent polymerase activities. However, the observations that, in the presence of an ApG primer or a short capped RNA primer (not requiring cleavage), the H510A polymerase can initiate transcription in vitro on a 14-nt vRNA-like template as efficiently as the wild-type polymerase argue against such a scenario and suggest a more direct role. Further studies with highly purified recombinant polymerase preparations would be required to address the question of how the H510A mutation in PA might affect the binding of individual polymerase subunits to vRNA and cRNA promoter sequences in cross-linking assays.
We tested mutations at several other positions of PA that were implicated in various functions. Most importantly, we tested the effect of a serine-to-alanine mutation at position 624 that has been shown by Hara et al. (18) to be at the active site of a serine protease motif. The S624A mutation had no measurable effect on transcription and replication by the RNA polymerase, as determined by CAT reporter gene expression and primer extension analysis of RNA in transfected cells. In addition, a recombinant virus with an S624A mutation in the PA protein had growth properties in cell culture comparable to those of the wild type, indicating that the serine protease activity, as determined by Hara et al. (18), is not required for viral growth in cell culture. The role of the serine protease activity in the viral life cycle remains to be determined.
Recently, it was reported that PA interacts with a cellular protein (hCLE) with homology to a family of transcriptional activators, and it was proposed that this interaction could be the basis of the mechanism by which PA regulates the transcription and replication activities of the polymerase (20). Several of the mutations tested in this report are in regions of PA that were implicated as important for this interaction. Among these, H510A showed a differential effect on transcription and replication, while G502A abolished both transcription and replication. It would be interesting to test whether these mutations interfere with the binding of PA to hCLE. None of the other mutations (L425A, S509A, L563A, and R583A) in the regions of PA implied in hCLE-binding had a significant effect on transcription and replication.
It is interesting that all three amino acid mutations that resulted in differential effects on transcription and replication (H510A, K539A, and R638A) involve basic amino acid residues. The H510A mutation severely inhibited transcription but not replication, the K539A mutation inhibited replication but not transcription, and the R638A mutation inhibited transcription and upregulated replication. Although we have not investigated how the K539A and R638A mutations induce the observed phenotypes, our observations are consistent with the view that PA plays a regulatory role in transcription and replication. Our results, however, also suggest that the hypothesis that the mere presence or absence of PA determines whether PB1 acts as a transcriptase or a replicase is too simplistic. In fact, our data support the view that PA is absolutely required for both transcription and replication.
In addition, our results with the E656A mutant suggest that PA might have unrecognized functions that are not directly related to transcription and replication of viral RNA. This mutant showed wild-type activity in the CAT reporter gene assay (Table 2) and expressed wild-type or near-wild-type levels of all viral RNA species (Fig. 3C), yet produced a virus with pinhead-sized plaques which was severely compromised in its growth in cell culture.
Perhaps one of the most intriguing findings described in this paper is that how many evolutionarily conserved amino acids in the C-terminal region of PA could be mutated without affecting RNA polymerase activity. In particular, individual amino acid mutations in two clusters of amino acids, at positions 654 to 658 and 692 to 698, that are conserved not only among the three influenza virus types A, B, and C but also in the homologous protein of Thogoto virus had very little if any effect on polymerase activity. The question arises whether these clusters have a role in a function of PA which is not related to RNA polymerase activity or, in fact, to viral growth in cell culture, since viable recombinant viruses with individual mutations in these clusters could be generated. Experiments in animal models may be required to answer the question of what role these highly conserved regions of PA might play.
Acknowledgments
E.F. and M.C. contributed equally to this work.
We thank P. Palese for plasmids, S. Inglis and A. Portela for antibodies, and A. Taylor and D. Peiris for DNA sequencing. We also thank L. Tiley and D. Elton for helpful discussions.
This work was supported by the MRC (program grant G9523972 and cooperative component grant G9901312 to G.G.B.).
REFERENCES
- 1.Argos, P. 1988. A sequence motif in many polymerases.Nucleic Acids Res. 16:9909-9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas, S. K., and D. P. Nayak. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blok, V., C. Cianci, K. W. Tibbles, S. C. Inglis, M. Krystal, and P. Digard. 1996. Inhibition of the influenza virus RNA-dependent RNA polymerase by antisera directed against the carboxy-terminal region of the PB2 subunit. J. Gen. Virol. 77:1025-1033. [DOI] [PubMed] [Google Scholar]
- 4.Brownlee, G. G., E. Fodor, D. C. Pritlove, K. G. Gould, and J. J. Dalluge. 1995. Solid phase synthesis of 5′-diphosphorylated oligoribonucleotides and their conversion to capped m7Gppp-oligoribonucleotides for use as primers for influenza A virus RNA polymerase in vitro. Nucleic Acids Res. 23:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Brownlee, G. G., and J. L. Sharps. 2002. The RNA polymerase of influenza A virus is stabilized by interaction with its viral RNA promoter. J. Virol. 76:7103-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Luna, S., C. Martinez, and J. Ortín. 1989. Molecular cloning and sequencing of influenza virus A/Victoria/3/75 polymerase genes: sequence evolution and prediction of possible functional domains. Virus Res. 13:143-155. [DOI] [PubMed] [Google Scholar]
- 6.Digard, P., V. C. Blok, and S. C. Inglis. 1989. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology 171:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doan, L., B. Handa, N. A. Roberts, and K. Klumpp. 1999. Metal ion catalysis of RNA cleavage by the influenza virus endonuclease. Biochemistry 38:5612-5619. [DOI] [PubMed] [Google Scholar]
- 8.Flick, R., G. Neumann, E. Hoffmann, E. Neumeier, and G. Hobom. 1996. Promoter elements in the influenza vRNA terminal structure. RNA 2:1046-1057. [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor, E., P. Palese, G. G. Brownlee, and A. García-Sastre. 1998. Attenuation of influenza A virus mRNA levels by promoter mutations. J. Virol. 72:6283-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1995. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 69:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor, E., B. L. Seong, and G. G. Brownlee. 1993. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J. Gen. Virol. 74:1327-1333. [DOI] [PubMed] [Google Scholar]
- 14.González, S., and J. Ortín. 1999. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 73:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González, S., and J. Ortín. 1999. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J. 18:3767-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González, S., T. Zürcher, and J. Ortín. 1996. Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 24:4456-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagen, M., T. D. Y. Chung, J. A. Butcher, and M. Krystal. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 68:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara, K., M. Shiota, H. Kido, Y. Ohtsu, T. Kashiwagi, J. Iwahashi, N. Hamada, K. Mizoue, N. Tsumura, H. Kato, and T. Toyoda. 2001. Influenza virus RNA polymerase PA subunit is a novel serine protease with Ser624 at the active site. Genes Cells 6:87-97. [DOI] [PubMed] [Google Scholar]
- 19.Honda, A., K. Mizumoto, and A. Ishihama. 1999. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 4:475-485. [DOI] [PubMed] [Google Scholar]
- 20.Huarte, M., J. J. Sanz-Ezquerro, F. Roncal, J. Ortín, and A. Nieto. 2001. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 75:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, D. A., A. J. Caton, S. J. McCready, and P. R. Cook. 1982. Influenza virus RNA is synthesized at a fixed site in the nucleus. Nature 296:366-368. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, M., T. Toyoda, and A. Ishihama. 1996. Influenza virus PB1 protein is the minimal and essential subunit of RNA polymerase. Arch. Virol. 141:525-539. [DOI] [PubMed] [Google Scholar]
- 23.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1530. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. R. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Leahy, M. B., H. C. Dobbyn, and G. G. Brownlee. 2001. A hairpin loop structure in the 3′ arm of the influenza A virus virion RNA promoter is required for endonuclease activity. J. Virol. 75:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leahy, M. B., D. C. Pritlove, L. M. Poon, and G. G. Brownlee. 2001. Mutagenic analysis of the 5′ arm of the influenza A virus virion RNA promoter defines the sequence requirements for endonuclease activity. J. Virol. 75:134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leahy, M. B., G. Zecchin, and G. G. Brownlee. 2002. Differential activation of influenza A virus endonuclease activity is dependent on multiple sequence differences between the virion RNA and cRNA promoters. J. Virol. 76:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, M. T. M., K. Bishop, L. Medcalf, D. Elton, P. Digard, and L. Tiley. 2002. Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res. 30:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, M. L., B. C. Ramirez, and R. M. Krug. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17:5844-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, G., W. Luytjes, M. Enami, and P. Palese. 1991. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J. Virol. 65:2861-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahy, B. W. J., T. Barrett, S. T. Nichol, C. R. Penn, and A. J. Wolsteinholme. 1981. Analysis of the functions of influenza virus genome RNA segments by use of temperature-sensitive mutants of fowl plague virus, p. 379-387. In D. H. L. Bishop and R. W. Compans (ed.), The replication of negative stranded viruses. Elsevier, New York, N.Y.
- 32.Martín-Benito, J., E. Area, J. Ortega, O. Llorca, J. M. Valpuesta, J. L. Carrascosa, and J. Ortín. 2001. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2:313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masunaga, K., K. Mizumoto, H. Kato, A. Ishihama, and T. Toyoda. 1999. Molecular mapping of influenza virus RNA polymerase by site-specific antibodies. Virology 256:130-141. [DOI] [PubMed] [Google Scholar]
- 34.Mikulasova, A., E. Vareckova, and E. Fodor. 2000. Transcription and replication of the influenza A virus genome. Acta Virol. 44:273-282. [PubMed] [Google Scholar]
- 35.Naffakh, N., P. Massin, and S. van der Werf. 2001. The transcription/replication activity of the polymerase of influenza A viruses is not correlated with the level of proteolysis induced by the PA subunit. Virology 285:244-252. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa, Y., N. Kimura, T. Toyoda, K. Mizumoto, A. Ishihama, K. Oda, and S. Nakada. 1995. The RNA polymerase PB2 subunit is not required for replication of the influenza virus genome but is involved in capped mRNA synthesis. J. Virol. 69:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa, Y., K. Oda, and S. Nakada. 1996. The PB1 subunit alone can catalyze cRNA synthesis, and the PA subunit in addition to the PB1 subunit is required for viral RNA synthesis in replication of the influenza virus genome. J. Virol. 70:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palese, P. 1977. The genes of influenza virus. Cell 10:1-10. [DOI] [PubMed] [Google Scholar]
- 40.Perales, B., S. de la Luna, I. Palacios, and J. Ortín. 1996. Mutational analysis identifies functional domains in the influenza A virus PB2 polymerase subunit. J. Virol. 70:1678-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perales, B., and J. Ortín. 1997. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J. Virol. 71:1381-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perales, B., J. J. Sanz-Ezquerro, P. Gastaminza, J. Ortega, J. F. Santarén, J. Ortín, and A. Nieto. 2000. The replication activity of influenza virus polymerase is linked to the capacity of the PA subunit to induce proteolysis. J. Virol. 74:1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez, D. R., and R. O. Donis. 1995. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J. Virol. 69:6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez, D. R., and R. O. Donis. 2001. Functional analysis of PA binding by influenza A virus PB1: effects on polymerase activity and viral infectivity. J. Virol. 75:8127-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pleschka, S., S. R. Jaskunas, O. G. Engelhardt, T. Zürcher, P. Palese, and A. García-Sastre. 1996. A plasmid-based reverse genetics system for influenza A virus. J. Virol. 70:4188-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poon, L. L., D. C. Pritlove, E. Fodor, and G. G. Brownlee. 1999. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73:3473-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pritlove, D. C., E. Fodor, B. L. Seong, and G. G. Brownlee. 1995. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J. Gen. Virol. 76:2205-2213. [DOI] [PubMed] [Google Scholar]
- 49.Robertson, J. S., M. Schubert, and R. A. Lazzarini. 1981. Polyadenylation sites for influenza virus mRNA. J. Virol. 38:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanz-Ezquerro, J. J., S. de la Luna, J. Ortín, and A. Nieto. 1995. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J. Virol. 69:2420-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanz-Ezquerro, J. J., J. Fernández-Santarén, T. Sierra, T. Aragón, J. Ortega, J. Ortín, G. L. Smith, and A. Nieto. 1998. The PA influenza polymerase subunit is a phosphorylated protein. J. Gen. Virol. 79:471-478. [DOI] [PubMed] [Google Scholar]
- 52.Sanz-Ezquerro, J. J., T. Zürcher, S. de la Luna, J. Ortín, and A. Nieto. 1996. The amino-terminal one-third of the influenza virus PA protein is responsible for the induction of proteolysis. J. Virol. 70:1905-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi, L., D. F. Summers, Q. Peng, and J. M. Galarza. 1995. Influenza A virus RNA polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology 208:38-47. [DOI] [PubMed] [Google Scholar]
- 54.Tiley, L. S., M. Hagen, J. T. Matthews, and M. Krystal. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68:5108-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toyoda, T., D. M. Adyshev, M. Kobayashi, A. Iwata, and A. Ishihama. 1996. Molecular assembly of the influenza virus RNA polymerase: determination of the subunit-subunit contact sites. J. Gen. Virol. 77:2149-2157. [DOI] [PubMed] [Google Scholar]
- 56.Zürcher, T., S. de la Luna, J. J. Sanz-Ezquerro, A. Nieto, and J. Ortín. 1996. Mutational analysis of the influenza virus A/Victoria/3/75 PA protein: studies of interaction with PB1 protein and identification of a dominant negative mutant. J. Gen. Virol. 77:1745-1749. [DOI] [PubMed] [Google Scholar]