Abstract
CpG methylation of the human T-cell leukemia virus type 1 (HTLV-1) long terminal repeat (LTR) has been implicated in proviral latency, but there is presently little information available regarding the pattern of LTR methylation and its effect on viral gene expression. To gain insight into the mechanisms of HTLV-1 latency, we have studied methylation of individual CpG sites in the U3-R region of the integrated proviral LTR by using bisulfite genomic sequencing methods. Surprisingly, our results reveal selective hypermethylation of the 5′ LTR and accompanying hypomethylation of the 3′ LTR in both latently infected cell lines and adult T-cell leukemia (ATL) cells having a complete provirus. Moreover, we observed a lack of CpG methylation in the LTRs of 5′-defective proviruses recovered from ATL samples, which is consistent with the selective hypomethylation of the 3′ LTR. Thus, the integrated HTLV-1 provirus in these carriers appears to be hypermethylated in the 5′ LTR and hypomethylated in the 3′ LTR. These results, together with the observation that proviral gene expression is reactivated by 5-azacytidine in latently infected cell lines, indicate that selective hypermethylation of the HTLV-1 5′ LTR is common both in vivo and in vitro. Thus, hypermethylation of the 5′ LTR appears to be an important mechanism by which HTLV-1 gene expression is repressed during viral latency.
Human T-cell leukemia virus type 1 (HTLV-1), the first human pathogenic retrovirus isolated, is the etiologic agent of adult T-cell leukemia (ATL), tropical spastic paraparesis/HTLV-1-associated myelopathy, and HTLV-1 uveitis. HTLV-1 is mainly transmitted through breastfeeding, and a long latency period precedes development of these diseases, which occur in carriers during middle or older age (12, 18, 28, 29, 33, 34, 42, 44).
HTLV-1-infected cells in the peripheral blood rarely express viral genes. Detection of HTLV-1 transcripts in peripheral blood mononuclear cells (PBMC) by reverse transcriptase PCR (RT-PCR) showed low levels of virus expression that are independent of the number of circulating HTLV-1-infected cells of asymptomatic carriers and tropical spastic paraparesis/HTLV-1-associated myelopathy patients (11, 13). Furthermore, fresh ATL cells do not express viral antigens until they are cultured in the presence of fetal calf serum, when in some cases ATL cells become positive for viral antigens (9, 17, 43) or viral transcripts (11, 22). However, the possibility that viral transcripts are derived from contaminating untransformed HTLV-1-infected cells was not ruled out. Thus, it appears that most HTLV-1-infected cells in vivo harbor a provirus that is transcriptionally silent. A recent study of an infection model using squirrel monkeys (Saimiri sciureus) revealed transient expression of tax/rex mRNA and early induction of latency, suggesting that primary HTLV-1 infection consists of a first transient step of reverse transcription and viral expression, followed by a latency of HTLV-1-bearing T cells (21). These observations collectively suggest that latent infection may be the norm for HTLV-1 in vivo. However, the mechanisms for HTLV-1 latency remain to be characterized.
CpG methylation has been implicated in the silencing of integrated proviral genomes (15, 20) as well as in the regulation of many imprinted genes (27). Moreover, demethylation induced by a potent inhibitor of DNA methyltransferase, 5-azacytidine (5-AzaC), reactivates proviral gene expression in a model of retroviral latency (31). For HTLV-1, methylation of proviral DNA has been studied by Southern blotting with methylation-sensitive restriction enzymes. CpG methylation was demonstrated in ATL cells (5, 23) and in an infected T-cell line, MT-4 (37). Importantly, treatment with 5-AzaC induced HTLV-1 gene expression in MT-4 cells (7, 37), while in vitro DNA methylation by HpaII or SssI methylase was shown to suppress the basal promoter activity of the HTLV-1 long terminal repeat (LTR) as well as responses to activating stimuli (3, 36). Thus, suppression of LTR promoter activity by CpG methylation appears to be involved in HTLV-1 latency. However, due to technical limitations, the pattern of CpG methylation within an integrated proviral LTR has not been explored. Consequently, it is not yet known whether LTR promoter activity is determined by methylation of specific CpG sites or, alternatively, by the density of methylation.
To gain insight into the mechanisms of HTLV-1 latency, we characterized the methylation status of each CpG site in the U3-R region of the LTR by the bisulfite genomic sequencing method (4). We differentially profiled methylation of CpG sites in the 5′ and 3′ LTRs of the integrated proviruses, using latently infected T-cell lines, leukemic ATL cells, and PBMC of asymptomatic carriers. The results revealed for the first time a selective methylation pattern of the 5′ and 3′ LTRs in the infected cells in vitro and in vivo: hypermethylation of the 5′ LTR versus hypomethylation of 3′ LTR. Since 5-AzaC treatment reactivated proviral gene expression in latently infected cell lines, the 5′-LTR-selective hypermethylation appears to be involved in silencing HTLV-1 gene expression. Our results raise interesting questions regarding the mechanisms that control differential methylation of the two LTRs of integrated HTLV-1 proviruses and also regarding the possible promoter activity of the hypomethylated 3′ LTR that require reevaluation of the promoter insertion model in HTLV-1 leukemogenesis.
MATERIALS AND METHODS
Cells.
HTLV-1-infected cell lines include MT-1 and TL-Om1, which were kindly provided by M. Miyoshi (Kochi Medical School) and by K. Sugamura (Tohoku University School of Medicine), respectively. Sez, ATL-16, and HUT102 are HTLV-1-infected cell lines that constitutively express virus genes. The first two of these cell lines were kindly provided by M. Maeda (Kyoto University), and the third was provided by R. Gallo (University of Maryland). These cell lines were cultured in RPMI1640 supplemented with 10% fetal calf serum and antibiotics. PBMC samples were obtained from ATL patients and asymptomatic carriers. The research followed the tenets of the Helsinki Declaration. Written informed consent for the study was obtained from each participant.
Nucleotide sequence analysis of the 5′ flanking region by the inverse PCR technique.
The nucleotide sequence of the 5′ flanking region of the integrated provirus was determined by the inverse PCR method (40). Briefly, genomic DNA was first digested with Sau3AI and self-ligated with T4 DNA ligase. PCR amplification of self-ligated DNA was achieved using the sense primer 5′-CTGAAGACAAATCATAAGCTCAGACC-3′ (nucleotide positions 191 to 216) and the antisense primer 5′-GAAAAGATTTGGCCCATTGCCTAGG-3′ (nucleotide positions 50 to 26). PCR was performed according to the LA-PCR (Takara, Kyoto, Japan) protocol of denaturation at 95°C for 30 s followed by annealing and extension at 68°C for 8 min. After 35 cycles, products were analyzed by 1% agarose gel electrophoresis. PCR products were subcloned into plasmid pGEM-T (Promega, Madison, Wis.), and the sequences were determined by automated DNA sequencing (SQ-5500; Hitachi, Tokyo, Japan). The determined sequences were confirmed by successful PCR amplification with a sense primer located in the determined sequence and an antisense primer located in the LTR.
CpG methylation analysis.
Methylation of the cytosine residue of the CpG site was analyzed by the bisulfite genomic sequencing method (4) with slight modifications. Briefly, 5 μg of genomic DNA was used for bisulfite treatment. The DNA sample in 0.3 N NaOH was heat denatured at 75°C for 20 min, followed by incubation at 55°C for 5 h in 4.2 M Na2S2O5 and 0.5 M hydroquinone. The sample DNA was purified using the Wizard DNA Clean-Up system (Promega) and treated with 0.3 N NaOH. DNA was precipitated with ethanol and dissolved in 50 μl of H2O, and 1/20 of this solution was subjected to PCR amplification with primers designed to amplify the sense strand of the bisulfite-modified LTR sequence. PCR was performed according to the touchdown protocol as follows: three cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min; three cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min; and then 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. The primer pair for nonselective analysis was as follows, where the nucleotide sequences are described as unmodified ones to facilitate identification of their localization: sense primer, 5′-CAATGACCATGAGCCCCAAAT-3′ (nucleotide positions 4 to 24); antisense primer, 5′-CAGTTCAGGAGGCACCACAGG-3′ (nucleotide positions 456 to 436). For the analysis of PBMC obtained from asymptomatic carriers, the following antisense primer was used to amplify a shorter fragment: 5′-TTTTTGAGGTGAGGGGTTGT-3′ (nucleotide positions 280 to 261). For the differential analysis of the 5′ LTR, the antisense primer was the same as was used for amplification of the larger fragment, and the sense primers were prepared from the nucleotide sequences of the 5′ flanking DNA as indicated in Table 1. For selective analysis of the 3′ LTR sequence, the sense primer was located in pX region: 5′-ACACCAACATCCCCATTTCT-3′ (nucleotide positions 8252 to 8233). Amplified PCR products were subcloned into pGEM-T, and the nucleotide sequences of at least 10 clones were determined.
TABLE 1.
Nucleotide sequences of 5′ flanking DNAs
| Sample | 5′ flanking sequencea | Repetitive sequence |
|---|---|---|
| TL-Om1 | ---ACTCATTGTTTTTTATGGCATAGTATCCCATGGTATATATGT-LTR | L1 |
| MT-1 | ---CCCATTCTGAAGTACTGGGTGTTACACCTTCAACGTATCTTTTTTAGGGGACAAAATT CAACCCATAACAGATGGCCTTCACATTTAGGAAGAATATCTT-LTR | MLT |
| ---TGGTTTCACATGCCTGGTAGGCCCCGGTTATCAATGAAATACTTCTTTTCTCAAGAT ATGAATTCTTAATTTTACTTTGAACTTAAATTAACATTAACAGATAATGTACGTGTAG TACAGAAAGAGAATGCAGGCAAGCAAAAACAAAATGAACATAAC-LTR | L1 | |
| ATL case 1 | ---CAGACAGTAAATCTTGAGATGTAGAGATGAAGAAGGGTCGGGGGAATGTTTCTTGG TATAATTTAGACTAAAGGCAGGTATAAACTTTGTTTTGGAGATTTTGGATTGACTGTC TCCATGGTCCTGAATCCCAGGGCCAATCAGAGGCAAAACTGAAA-LTR | AluSx |
Underlining indicates the sequences used for primers in the bisulfite genomic sequencing analysis.
LA-PCR.
The LA-PCR (Takara) system was used to amplify the integrated provirus sequence according to the manufacturer's instructions. Using 100 ng of genomic DNA sample, 35 cycles of a two-step PCR consisting of 94°C for 30 s and 68°C for 8 min were done. The primers used were as follows: sense primers 5′-CTTTGCTGACCCTGCTTGCTCAACTCAC-3′ (nucleotide positions 4 to 28) and 5′-GAGAGGCCTTACAAACTGGAATCACCCTTG-3′ (nucleotide positions 6499 to 6528) and antisense primers 5′-CAAGGGTGATTCCAGTTTGTAAGGCCTCTC-3′ (nucleotide positions 6528 to 5499) and 5′-GTAGAGTTGAGCAAGCAGGGTCAGGCAAAG-3′ (nucleotide positions 8831 to 8860). Amplified products were analyzed by 1% agarose gel electrophoresis.
Confocal immunofluorescence microscopy.
Expression of the Gag protein was detected by using a laser confocal microscope (Radiance 2000; Bio-Rad). The primary antibody was anti-p19/27 antibody (Fujirebio), and the secondary antibody was fluorescein isothiocyanate-labeled anti-mouse immunoglobulin G antibody.
ELISA.
The concentration of p19 Gag protein in the culture supernatant was measured with the RETRO-TEK HTLV-I/II p19 antigen enzyme-linked immunosorbent assay (ELISA) kit (ZeptoMetrix Corporation, Buffalo, N.Y.) according to the manufacturer's instructions. Triplicate samples were assayed, and the mean and standard deviation were calculated.
RESULTS
Viral gene expression and CpG methylation of the provirus LTR in cell lines.
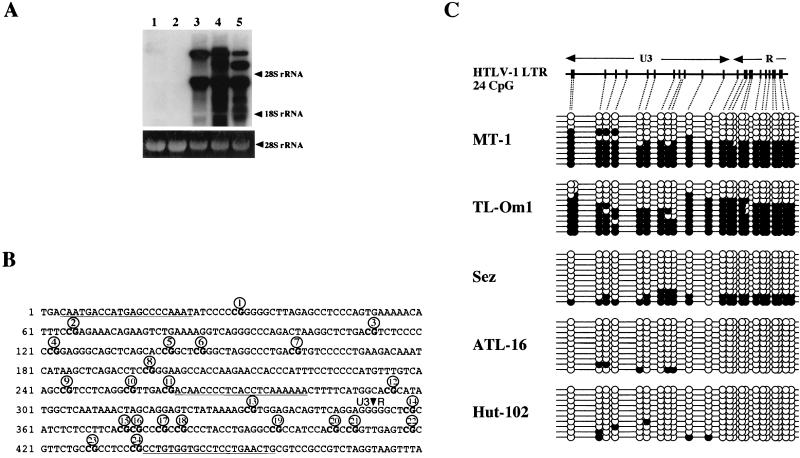
To study the relationship between viral gene expression and CpG methylation of the integrated provirus LTR, we characterized CpG methylation by the bisulfite genomic sequencing method (4) and detected viral gene expression by Northern blot analysis. Viral gene expression was not detected in MT-1 and TL-Om1 cells, whereas viral transcripts were abundantly expressed in Sez, ATL16, and HUT102 cells (Fig. 1A). We then analyzed the methylation status of CpG sites in the U3-R region of the HTLV-1 provirus shown in Fig. 1B. The results showed relatively heavy methylation in the latently infected cell lines, MT-1 and TL-Om1, whereas a low level of methylation or almost a total lack of methylation was found in virus-expressing cell lines (Fig. 1C). In MT-1 and TL-Om1 cells, 128 and 130 out of 240 CpG sites, respectively, were methylated. On the other hand, in Sez, ATL-16, and HUT102 cells, only 48, 4, and 6 out of 240 CpG sites, respectively, were methylated. Other characteristics were also noted. First, all CpG sites in a single PCR-amplified copy tended to show a uniform methylation or hypomethylation pattern. Second, almost uniformly methylated copies constituted about half or more of the analyzed ones in TL-Om1 cells and MT-1 cells. Specifically, in both cell lines, 6 out of 10 copies had fully methylated CpG sites or fewer than three unmethylated sites, whereas in Sez cells 8 out of 10 copies and in ATL16 and HUT102 cells all copies had fewer than three methylated CpG sites (Fig. 1B). These results collectively show an inverse correlation between the levels of CpG methylation and virus gene expression and provide support for the idea that CpG methylation of the LTR is involved in suppression of HTLV-1 expression, as reported previously (36, 37).
FIG. 1.
HTLV-1 gene expression and CpG methylation of LTR in cell lines. (A) Upper panel, Northern blot analysis of HTLV-1 transcripts in HTLV-1-infected T-cell lines. Samples of 10 μg of total RNA were subjected to formalin-agarose gel electrophoresis and blotted onto a nylon membrane (Biodyne A; Pall, Ann Arbor, Mich.). HTLV-1 transcripts were detected with a 32P-labeled LTR probe. Positions of 18S and 28S rRNAs are indicated on the right. Lower panel, photograph of the ethidium bromide-stained agarose gel, showing that equal amounts of RNA were applied. Lane 1, MT-1; lane 2, TL-Om1; lane 3, Sez; lane 4, ATL-16; lane 5, HUT102. (B) CpG sites in the U3-R region of the HTLV-1 provirus. CpG sites are in boldface and numbered from the 5′ end of the LTR. Sequences used for primers are indicated by underlining. (C) Levels of CpG methylation of the HTLV-1 LTR in HTLV-1-infected T-cell lines. Results of bisulfite genomic sequencing coupled with TA cloning are shown. The methylation status of 10 clones for each sample is presented; methylation of each CpG site is expressed as a filled circle, and unmethylated sites are shown as open circles. Top, schematic description of CpG sites in the U3-R region studied in this experiment.
Selective CpG methylation of the 5′ LTR of the integrated HTLV-1 provirus in latently infected cell lines.
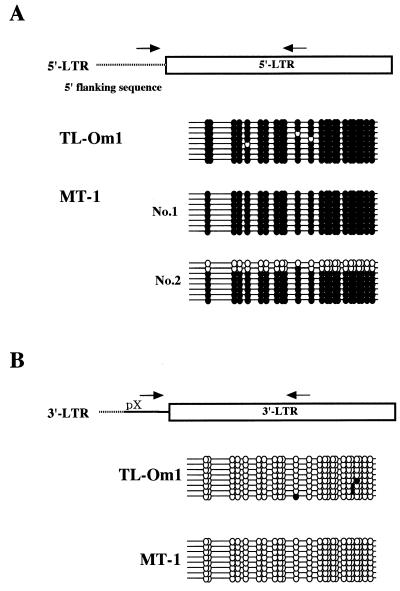
The results described above raised the possibility that each LTR may be differentially methylated. Therefore, we tried to characterize CpG methylation of the two LTRs independently. To enable the differential analysis of the 5′ LTR, we determined the 5′ flanking DNA sequences of the integrated provirus by using the inverse PCR technique (40). TL-Om1 cells have 1 copy of integrated provirus, whereas MT-1 cells have 10 copies, based on the Southern blot analysis. MT-1 cells were also shown to have only two copies of apparently complete provirus (data not shown). Inverse PCR using an LTR primer pair amplified two and five fragments from DNA samples of TL-Om1and MT-1 cells, respectively (data not shown). Nucleotide sequence analysis of these fragments clearly demonstrated that one of the fragments was derived from the pX 3′-LTR region and revealed one DNA sequence flanking the 5′ LTR from TL-Om1 and four from MT-1. Sequences that flank defective proviruses were discriminated from those flanking complete ones by PCR analysis with sense primers located in the flanking sequence and antisense primers in the LTR or gag region. Two sequences from MT-1 were thus confirmed to flank the 5′ LTR of the complete provirus (Table 1). We then analyzed these nucleotide sequences by using the BLAST program and found that all of the sequences are human repetitive sequences. The sequence of TL-Om1 and one of MT-1 were homologous to LINE1, and the other of MT-1 was homologous to the MLT (39) (Table 1). These results are in line with a previous report indicating that HTLV-1 frequently integrates into repetitive sequences (45).
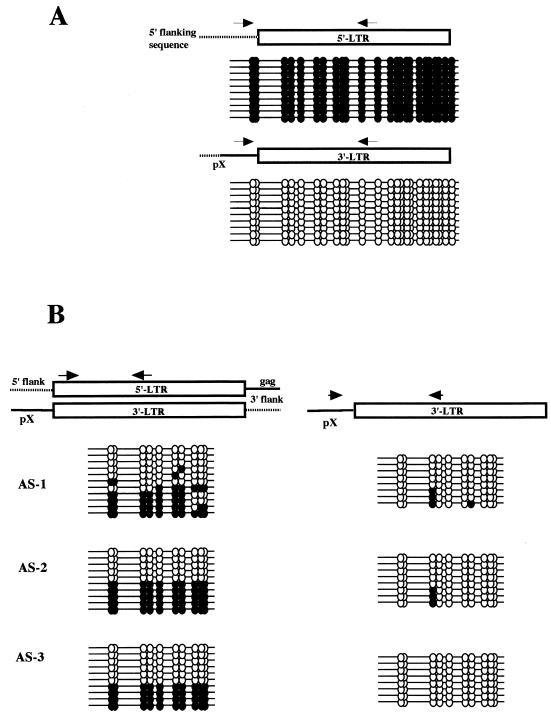
We then characterized CpG methylation of the 5′ LTR U3-R region with sense primers located in the 5′ flanking DNA. The results demonstrated almost complete CpG methylation of the 5′ LTR U3-R region in TL-Om1 and MT-1 cells, with the exception of two copies from the latter (Fig. 2A). We next analyzed CpG methylation of the 3′ LTR by using a sense primer located in the pX region. In clear contrast to the results described above, the 3′ LTRs of TL-Om1 and MT-1 cells were completely unmethylated, with exceptional copies having a single methylated CpG site in TL-Om1 cells (Fig. 2B). These results demonstrate for the first time that in latently infected cell lines the integrated HTLV-1 provirus exhibits a differential pattern of LTR methylation. Specifically, the 5′ LTR is hypermethylated, while the 3′ LTR is unmethylated.
FIG. 2.
Selective hypermethylation of the 5′ LTR in latently infected cell lines. (A) Results of 5′-LTR-selective bisulfite genomic sequencing analysis. After bisulfite modification, sample DNA was amplified with a sense primer located in the 5′ flanking sequence of the HTLV-1 provirus. Results for the single provirus in TL-Om1 cells and for two complete proviruses in MT-1 cells are shown. Methylated and unmethylated CpG sites are shown as filled and open circles, respectively. (B) Results of 3′-LTR-selective bisulfite genomic sequencing analysis. After bisulfite modification, sample DNA was amplified with a sense primer located in the pX region. Methylated CpG sites are shown as filled circles, and unmethylated CpG sites are shown as open circles.
HTLV-1 with a methylated 5′ LTR is reactivated by 5-AzaC.
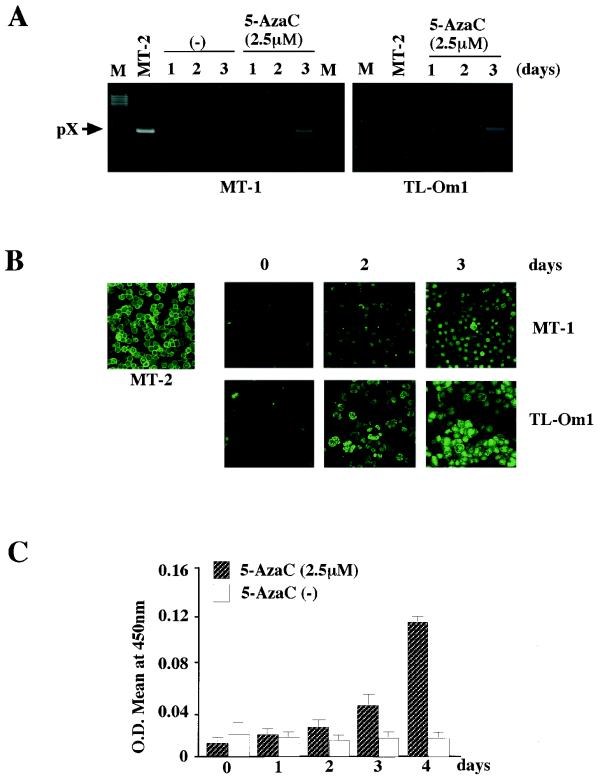
To examine whether hypermethylation of the 5′ LTR suppresses proviral gene expression in latently infected cell lines, we tested reactivation of viral gene expression in these cell lines by 5-AzaC treatment. Cells were treated with 2.5 μM 5-AzaC or vehicle alone for the indicated periods, and viral gene expression was examined by RT-PCR, confocal immunofluorescence microscopy, and ELISA. On the second day of 5-AzaC treatment and thereafter, pX transcripts and Gag antigen were detected by RT-PCR and confocal immunofluorescence microscopy, respectively (Fig. 3A and B). Production of p19 Gag protein in the culture supernatants became evident after 2 days of 5-AzaC treatment when measured by ELISA and showed a dramatic increase by the fourth day following treatment (Fig. 3C). Taken collectively, the results suggest that heavy CpG methylation of the 5′ LTR plays a critical role in repression of viral gene expression in latently infected cell lines.
FIG. 3.
Reactivation of HTLV-1 gene expression by the methylation inhibitor 5-AzaC. (A) Detection of viral gene expression by RT-PCR. HTLV-1 pX mRNA was detected by RT-PCR with a primer pair located in the second exon and pX region. After 30 cycles of amplification, one-fifth of the products was analyzed by agarose gel electrophoresis. (B) Induction of Gag antigen expression detected by confocal immunofluorescence microscopy with anti-p19/27 antibody. Magnification, ×80. (C) Induction of p19 antigen production by MT-1 cells. Levels of p19 antigen in the culture supernatants were measured with a RETRO-TEK p19 ELISA kit. O.D., optical density. Error bars indicate standard deviations.
CpG methylation of integrated HTLV-1 provirus LTR in ATL cells.
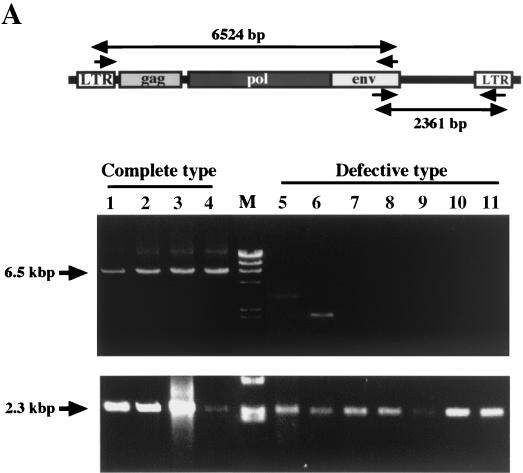
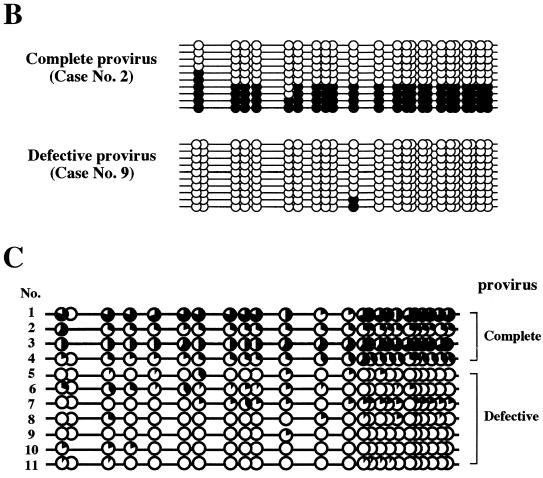
Selective hypermethylation of the integrated provirus 5′ LTR was next examined with ATL samples to determine whether hypermethylation of the 5′ LTR also occurs in vivo. Since the HTLV-1 provirus is defective in the 5′ region covering the 5′ LTR and gag-pol region in more than half of ATL cases (41), defects of the provirus in ATL samples were first examined with the LA-PCR system (Takara). A primer pair of a sense LTR primer and an antisense env primer can detect defects in the 5′ region of the provirus, while a primer pair of a sense env primer and an antisense LTR primer can detect the 3′ region that remains even if the provirus is defective (Fig. 4A, top). Of 11 ATL samples, only 4 were shown to have a complete provirus, whereas the other 7 had defects in the 5′ region (Fig. 4A). Analysis of LTR methylation with primers located in the LTR was carried out, which is indiscriminate of 5′ and 3′ LTRs. Representative results for a sample with a complete provirus and one with a defective provirus are shown in Fig. 4B, which shows methylation of CpG sites of 10 amplified copies. In a sample with a complete provirus (sample 2 in Fig. 4C), four copies had no or one unmethylated CpG site and four copies were totally unmethylated. In contrast, a sample with a defective provirus (sample 9 in Fig. 4C) had 8 copies with totally unmethylated CpG sites, and only 2 CpG sites in 10 copies each having 24 CpG sites were methylated. These results are summarized in Fig. 4C, where levels of methylation at each CpG site in 10 copies are shown. Although the results are by no means absolute, significantly heavy CpG methylation was observed in four cases with a complete provirus, whereas very low levels of methylation were observed in seven cases with a defective provirus. These results are consistent with, and appear to reflect, a lack of CpG methylation at the 3′ LTR.
FIG. 4.
CpG methylation of the LTR of the HTLV-1 provirus in ATL cells. (A) Long-PCR analysis of the integrated provirus in the ATL cells. One-fifth of the amplified products was analyzed by agarose gel electrophoresis. Top panel, schematic presentation of the primer locations and the expected sizes of PCR products. Middle panel, results of LA-PCR of the 5′ region. Bottom panel, results of LA-PCR of the 3′ region. (B) Representative results of bisulfite genomic sequencing analysis of ATL samples. After bisulfite modification, PCR amplification was done with a nonselective primer pair located in the LTR U3-R region. Results of one amplification with a complete provirus (case 1) and one amplification with a defective provirus (case 9) are shown. (C) Summary of results. The level of CpG methylation at each site is expressed as the amount of filled area of the circle.
Selective CpG methylation of the 5′ LTR of the integrated HTLV-1 provirus in ATL cells and PBMC of asymptomatic carriers.
Since the results described above suggested to us that 5′-LTR-selective methylation takes place in ATL cells, we next tried to directly confirm this by using an ATL sample having a single complete provirus and infected cells in the PBMC of asymptomatic carriers. As described for the analysis of latently infected cell lines, we determined the nucleotide sequence of the 5′ flanking region of the ATL sample (case 1), which also belonged to a human repetitive sequence, AluSx (Table 1). The sense primer was prepared by using the determined sequence, and 5′-specific CpG analysis was done. The results revealed complete methylation of the amplified copies, showing methylation of all 240 sites (Fig. 5A, upper panel). On the other hand, analysis of the 3′ LTR with a sense pX primer showed a complete lack of methylated CpG sites out of 240 sites (Fig. 5A, lower panel). These results provide support for the notion that 5′-LTR-selective CpG methylation also takes place in HTLV-1-infected cells in vivo.
FIG. 5.
5′-LTR-selective hypermethylation of the provirus LTR in HTLV-1-infected cells in vivo. (A) Results of differential analysis of an ATL sample (case 1). Upper panel, 5′ LTR; lower panel, 3′ LTR. (B) Results of differential analysis of PBMC samples of asymptomatic carriers. Left panel, results determined by nonselective analysis; right panel, results determined by 3′-LTR-selective analysis. The antisense primer used in this study is located upstream from that in the analysis shown in Fig. 2 and amplifies a shorter fragment containing 11 CpG sites.
To eliminate the possibility that the 5′-selective hypermethylation may be found only in transformed cells, we next analyzed proviruses in the PBMC of asymptomatic carriers. Because fewer than a few percent of the PBMC are infected by HTLV-1 in asymptomatic carriers and PCR amplification of bisulfite-modified DNA is very inefficient, we used a primer pair that amplifies smaller fragments. In these samples, 5′-LTR-selective analysis was impossible, since HTLV-1-infected cells are polyclonal in asymptomatic carriers. Thus, in this study we compared the results of nonselective CpG methylation analysis and those of 3′-LTR-specific analysis with a sense primer located in the pX sequence as described above. The nonselective primer pair will yield results that reflect the CpG methylation status of both LTRs, whereas the latter will yield those specific for the 3′ LTR. The results with the nonselective primer pair showed relatively high levels of methylation at each CpG site, with about half of the copies being methylated at almost all CpG sites (Fig. 5B, left column). On the other hand, the results of 3′-LTR-selective analysis demonstrated almost complete demethylation of every CpG site in all copies analyzed, except for a few with one or two methylated sites (Fig. 5B, right column). These results indicate that the 5′ LTR is heavily methylated in HTLV-1-infected cells in the asymptomatic carriers. Taken collectively the results indicate that selective hypermethylation of the 5′ LTR appears to be the norm in HTLV-1-infected cells in vivo irrespective of the state of transformation.
DISCUSSION
In the present study, we demonstrate by selective bisulfite genomic sequence analysis that selective hypermethylation of the 5′ LTR and hypomethylation of the 3′ LTR are the norm for the integrated HTLV-1 provirus in vivo. Reactivation of viral gene expression by 5-AzaC treatment in latently infected cell lines with a heavily methylated 5′ LTR suggests that CpG methylation of the 5′ LTR plays a major role in suppression of provirus expression. Furthermore, hypermethylation of the 5′ LTR in ATL cells with a complete provirus, but not in those with a 5′-defective provirus, indicates that HTLV-1 is inactivated in transformed cells by CpG methylation or defects in the 5′ region of the proviral genome.
Suppression of HTLV-1 LTR promoter activity by methylation in vitro was reported previously with transient-transfection assays (3, 36, 37). Reactivation of viral gene expression by 5-AzaC treatment was also reported with MT-4 cells (6, 37). These data suggested the involvement of CpG methylation in HTLV-1 latency, as observed for other viruses (8, 15, 19, 20, 31, 32). We have confirmed that in vitro methylation of an LTR-driven luciferase construct results in profound suppression of basal activities as well as loss of its response to stimulating signals such as Tax (data not shown). Here we showed that in two latently infected cell lines the 5′ LTR is selectively and highly methylated and that viral gene expression was induced after 2 days by 5-AzaC treatment (Fig. 2A and 3). The induction kinetics of viral gene expression appeared to conform to those of passive demethylation through inhibition of maintenance methylase Dnmt1, which requires DNA replication for inducing unmethylated cytosine at the CpG sites (2). These data, together with previously reported results (3, 5, 7, 36, 37), provide further support for the involvement of CpG methylation in HTLV-1 latency.
In the present study, we showed for the first time the 5′-LTR-selective hypermethylation of integrated HTLV-1 provirus in vivo and in vitro, which was based on the differential characterization of the methylation status of the two LTRs. In striking contrast, the 3′ LTR was almost free from CpG methylation in all samples studied. To our knowledge, this is the first report of differential characterization of CpG methylation of the integrated proviral LTR. Our results are consistent with those of a previous study in which methylation-sensitive restriction enzyme analysis revealed hypomethylation of pX and the LTR region; however, detailed characterization was not performed in that work (23).
In the present study, differential analysis of LTR methylation showed that ATL cells and HTLV-1-infected cells in the PBMC of asymptomatic carriers have proviral integrants carrying hypermethylated 5′ LTRs. Thus, we suggest that this selective methylation is the norm for the HTLV-1 provirus in vivo and that suppression of proviral gene expression by CpG methylation is one important mechanism for achieving HTLV-1 latency. The results of the present study provide support for the notion that the HTLV-1 provirus in ATL cells is inactivated by hypermethylation of the 5′ LTR or by deletions within the 5′ end of the provirus; such deletions are observed in more than half of the ATL cases examined (41). Consequently, it appears that the role of Tax in the leukemogenesis of HTLV-1-infected T cells is limited to those early stages of disease when the provirus is intact and Tax expression induces clonal T-cell proliferation. Thus, full transformation of virus-infected cells is achieved by accumulation of genetic events, and at least some of these events are Tax independent. This multistep leukemogenesis model is also supported by recent reports that describe frequent mutations and/or deletions of the integrated HTLV-1 provirus in ATL cells and untransformed infected cells in vivo (10, 30, 35).
Earlier studies showed that the integration sites of the HTLV-1 provirus are located in repetitive sequences such as LINE and SINE (25, 26). In line with those reports, the 5′ flanking sequences determined in the present study with the latently infected cell lines MT-1 and TL-Om1 and also with an ATL sample were shown to belong to repetitive sequences (Table 1). It is interesting that copies of the human Alu repetitive element are methylated at high levels in human somatic cells (16, 24), and spreading of methylation from Alu elements has been suggested to play a causal role in epigenetic inactivation of some tumor suppressor genes (1, 14). Thus, the heavy methylation of the 5′ LTR could result from de novo methylation initiated by the flanking repetitive sequence. However, this mechanism cannot explain the sparing 3′ LTR CpG methylation. Our data indicate that two identical sequences located within 10 kbp are differentially methylated, and earlier studies showed that a sequence that is repeated, even if it is not the repetitive sequence, is a good target of do novo methylation (38). Therefore, it is reasonable to suppose that there are some specific mechanisms that make a boundary between the two LTRs and protect the 3′ LTR from do novo methylation. Studies of the mechanisms underlying differential methylation of two LTRs are under way in our laboratory.
In conclusion, we have demonstrated a novel pattern of CpG methylation for the HTLV-1 provirus, namely, 5′-LTR hypermethylation and 3′-LTR hypomethylation, in latently infected cell lines and HTLV-1-infected cells in vivo. Delineation of the mechanisms for this differential methylation will provide insights into HTLV-1 latency and the molecular mechanisms of pathogenicity.
Acknowledgments
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Japan Society of Promotion of Science.
REFERENCES
- 1.Baylin, S. B., J. G. Herman, J. R. Graff, P. M. Vertino, and J. P. Issa. 1998. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 72:141-196. [PubMed] [Google Scholar]
- 2.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. [DOI] [PubMed] [Google Scholar]
- 3.Cassens, S., U. Ulrich, P. Beimling, and D. Simon. 1994. Inhibition of human T cell leukaemia virus type I long terminal repeat expression by DNA methylation: implications for latency. J. Gen. Virol. 75:3255-3259. [DOI] [PubMed] [Google Scholar]
- 4.Clark, S. J., J. Harrison, C. L. Paul, and M. Frommer. 1994. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22:2990-2997. [DOI] [PMC free article] [PubMed]
- 5.Clarke, M. F., C. D. Trainor, D. L. Mann, R. C. Gallo, and M. S. Reitz. 1984. Methylation of human T-cell leukemia virus proviral DNA and viral RNA expression in short- and long-term cultures of infected cells. Virology 135:97-104. [DOI] [PubMed] [Google Scholar]
- 6.Datta, S., N. H. Kothari, and H. Fan. 2000. In vivo genomic footprinting of the human T-cell leukemia virus type 1 (HTLV-1) long terminal repeat enhancer sequences in HTLV-1-infected human T-cell lines with different levels of Tax I activity. J. Virol. 74:8277-8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta, S., N. H. Kothari, and H. Fan. 2001. Induction of Tax I expression in MT-4 cells by 5-azacytidine leads to protein binding in the HTLV-1 LTR in vivo. Virology 283:207-214. [DOI] [PubMed] [Google Scholar]
- 8.Falk, K., and I. Ernberg. 1993. An origin of DNA replication (oriP) in highly methylated episomal Epstein-Barr virus DNA localizes to a 4.5-kb unmethylated region. Virology 195:608-615. [DOI] [PubMed] [Google Scholar]
- 9.Franchini, G., F. Wong-Staal, and R. C. Gallo. 1984. Human T-cell leukemia virus (HTLV-I) transcripts in fresh and cultured cells of patients with adult T-cell leukemia. Proc. Natl. Acad. Sci. USA 81:6207-6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa, Y., R. Kubota, M. Tara, S. Izumo, and M. Osame. 2001. Existence of escape mutant in HTLV-I tax during the development of adult T-cell leukemia. Blood 97:987-993. [DOI] [PubMed] [Google Scholar]
- 11.Furukawa, Y., M. Osame, R. Kubota, M. Tara, and M. Yoshida. 1995. Human T-cell leukemia virus type-1 (HTLV-1) Tax is expressed at the same level in infected cells of HTLV-1-associated myelopathy or tropical spastic paraparesis patients as in asymptomatic carriers but at a lower level in adult T-cell leukemia cells. Blood 85:1865-1870. [PubMed] [Google Scholar]
- 12.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407-410. [DOI] [PubMed]
- 13.Gessain, A., F. Saal, O. Gout, M. T. Daniel, G. Flandrin, G. de The, J. Peries, and F. Sigaux. 1990. High human T-cell lymphotropic virus type I proviral DNA load with polyclonal integration in peripheral blood mononuclear cells of French West Indian, Guianese, and African patients with tropical spastic paraparesis. Blood 75:428-433. [PubMed] [Google Scholar]
- 14.Graff, J. R., J. G. Herman, S. Myohanen, S. B. Baylin, and P. M. Vertino. 1997. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J. Biol. Chem. 272:22322-22329. [DOI] [PubMed] [Google Scholar]
- 15.Harbers, K., A. Schnieke, H. Stuhlmann, D. Jahner, and R. Jaenisch. 1981. DNA methylation and gene expression: endogenous retroviral genome becomes infectious after molecular cloning. Proc. Natl. Acad. Sci. USA 78:7609-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellmann-Blumberg, U., M. F. Hintz, J. M. Gatewood, and C. W. Schmid. 1993. Developmental differences in methylation of human Alu repeats. Mol. Cell. Biol. 13:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinuma, Y., H. Komoda, T. Chosa, T. Kondo, M. Kohakura, T. Takenaka, M. Kikuchi, M. Ichimaru, K. Yunoki, I. Sato, R. Matsuo, Y. Takiuchi, H. Uchino, and M. Hanaoka. 1982. Antibodies to adult T-cell leukemia-virus-associated antigen (ATLA) in sera from patients with ATL and controls in Japan: a nation-wide sero-epidemiologic study. Int. J. Cancer 29:631-635. [DOI] [PubMed] [Google Scholar]
- 18.Hinuma, Y., K. Nagata, M. Hanaoka, M. Nakai, T. Matsumoto, K. I. Kinoshita, S. Shirakawa, and I. Miyoshi. 1981. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. USA 78:6476-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, L. F., J. Minarovits, S. L. Cao, B. Contreras-Salazar, L. Rymo, K. Falk, G. Klein, and I. Ernberg. 1991. Variable expression of latent membrane protein in nasopharyngeal carcinoma can be related to methylation status of the Epstein-Barr virus BNLF-1 5′-flanking region. J. Virol. 65:1558-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, W. S., T. G. Fanning, and R. D. Cardiff. 1984. Mouse mammary tumor virus: specific methylation patterns of proviral DNA in normal mouse tissues. J. Virol. 49:66-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kazanji, M., A. Ureta-Vidal, S. Ozden, F. Tangy, B. de Thoisy, L. Fiette, A. Talarmin, A. Gessain, and G. de The. 2000. Lymphoid organs as a major reservoir for human T-cell leukemia virus type 1 in experimentally infected squirrel monkeys (Saimiri sciureus): provirus expression, persistence, and humoral and cellular immune responses. J. Virol. 74:4860-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinoshita, T., M. Shimoyama, K. Tobinai, M. Ito, S. Ito, S. Ikeda, K. Tajima, K. Shimotohno, and T. Sugimura. 1989. Detection of mRNA for the tax1/rex1 gene of human T-cell leukemia virus type I in fresh peripheral blood mononuclear cells of adult T-cell leukemia patients and viral carriers by using the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86:5620-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitamura, T., M. Takano, H. Hoshino, K. Shimotohno, M. Shimoyama, M. Miwa, F. Takaku, and T. Sugimura. 1985. Methylation pattern of human T-cell leukemia virus in vivo and in vitro: pX and LTR regions are hypomethylated in vivo. Int. J. Cancer 35:629-635. [DOI] [PubMed] [Google Scholar]
- 24.Kochanek, S., D. Renz, and W. Doerfler. 1993. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 12:1141-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leclercq, I., F. Mortreux, M. Cavrois, A. Leroy, A. Gessain, S. Wain-Hobson, and E. Wattel. 2000. Host sequences flanking the human T-cell leukemia virus type 1 provirus in vivo. J. Virol. 74:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leclercq, I., F. Mortreux, A. S. Gabet, C. B. Jonsson, and E. Wattel. 2000. Basis of HTLV type 1 target site selection. AIDS Res. Hum. Retroviruses 16:1653-1659. [DOI] [PubMed] [Google Scholar]
- 27.Li, E., C. Beard, and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature 366:362-365. [DOI] [PubMed] [Google Scholar]
- 28.Mochizuki, M., K. Tajima, T. Watanabe, and K. Yamaguchi. 1994. Human T lymphotropic virus type 1 uveitis. Br. J. Ophthalmol. 78:149-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mochizuki, M., T. Watanabe, K. Yamaguchi, K. Takatsuki, K. Yoshimura, M. Shirao, S. Nakashima, S. Mori, S. Araki, and N. Miyata. 1992. HTLV-I uveitis: a distinct clinical entity caused by HTLV-I. Jpn. J. Cancer Res. 83:236-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortreux, F., I. Leclercq, A. S. Gabet, A. Leroy, E. Westhof, A. Gessain, S. Wain-Hobson, and E. Wattel. 2001. Somatic mutation in human T-cell leukemia virus type 1 provirus and flanking cellular sequences during clonal expansion in vivo. J. Natl. Cancer Inst. 93:367-377. [DOI] [PubMed] [Google Scholar]
- 31.Niwa, O., and T. Sugahara. 1981. 5-Azacytidine induction of mouse endogenous type C virus and suppression of DNA methylation. Proc. Natl. Acad. Sci. USA 78:6290-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa, Y., N. Sagata, J. Tsuzuku-Kawamura, M. Onuma, H. Izawa, and Y. Ikawa. 1985. Methylation pattern of the bovine leukemia provirus genome in bovine leukemic cells. Jpn. J. Cancer Res. 76:5-8. [PubMed] [Google Scholar]
- 33.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i:1031-1032. [DOI] [PubMed]
- 34.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose, N. J., J. H. Richardson, U. Desselberger, and A. M. Lever. 2000. Virus inactivation in a proportion of human T-cell leukaemia virus type I-infected T-cell clones arises through naturally occurring mutations. J. Gen. Virol. 81:97-104. [DOI] [PubMed] [Google Scholar]
- 36.Saggioro, D., M. Forino, and L. Chieco-Bianchi. 1991. Transcriptional block of HTLV-I LTR by sequence-specific methylation. Virology 182:68-75. [DOI] [PubMed] [Google Scholar]
- 37.Saggioro, D., M. Panozzo, and L. Chieco-Bianchi. 1990. Human T-lymphotropic virus type I transcriptional regulation by methylation. Cancer Res. 50:4968-4973. [PubMed] [Google Scholar]
- 38.Selker, E. U. 1999. Gene silencing: repeats that count. Cell 97:157-160. [DOI] [PubMed] [Google Scholar]
- 39.Smit, A. F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9:657-663. [DOI] [PubMed] [Google Scholar]
- 40.Takemoto, S., M. Matsuoka, K. Yamaguchi, and K. Takatsuki. 1994. A novel diagnostic method of adult T-cell leukemia: monoclonal integration of human T-cell lymphotropic virus type I provirus DNA detected by inverse polymerase chain reaction. Blood 84:3080-3085. [PubMed] [Google Scholar]
- 41.Tamiya, S., M. Matsuoka, K. Etoh, T. Watanabe, S. Kamihira, K. Yamaguchi, and K. Takatsuki. 1996. Two types of defective human T-lymphotropic virus type I provirus in adult T-cell leukemia. Blood 88:3065-3073. [PubMed] [Google Scholar]
- 42.Uchiyama, T., J. Yodoi, K. Sagawa, K. Takatsuki, and H. Uchino. 1977. Adult T-cell leukemia: clinical and hematologic features of 16 cases. Blood 50:481-492. [PubMed] [Google Scholar]
- 43.Vyth-Dreese, F. A., P. Rumke, M. Robert-Guroff, G. de Lange, and R. C. Gallo. 1983. Antibodies against human T-cell leukemia/lymphoma virus (HTLV) and expression of HTLV p19 antigen in relatives of a T-cell leukemia patient originating from Surinam. Int. J. Cancer 32:337-342. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe, T. 1997. HTLV-1-associated diseases. Int. J. Hematol. 66:257-278. [DOI] [PubMed] [Google Scholar]
- 45.Wattel, E., J. P. Vartanian, C. Pannetier, and S. Wain-Hobson. 1995. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J. Virol. 69:2863-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]