Abstract
To facilitate identification of rhesus cytomegalovirus (RhCMV)-infected cells, a recombinant virus expressing enhanced green fluorescent protein (EGFP), designated RhCMV-EGFP, was constructed. An expression cassette for EGFP under the control of the simian virus 40 (SV40) early promoter was inserted into the intergenic region between unique short 1 (US1) and US2 of the RhCMV genome by homologous recombination. RhCMV-EGFP exhibited comparable growth kinetics to that of wild-type virus in rhesus fibroblast cultures and retained its pathogenicity in monkey fetuses. Typical neurologic syndromes caused by CMV infection were observed in all fetuses experimentally inoculated with RhCMV-EGFP, as evidenced by sonographic and gross examinations. Systemic RhCMV infections were established in all fetuses, as viral antigen was detected in multiple organs and virus was isolated from fetal blood samples. The engineered viral genome was stable following rapid serial passages in vitro and multiple rounds of replication in vivo. Infected cells could be readily distinguished by green fluorescence both in tissue cultures and in the fetuses. In addition, EGFP expression was detected in various cell types that were permissive to RhCMV infection, consistent with a broad tissue tropism of the SV40 promoter. These results demonstrate that RhCMV can be successfully engineered without loss of wild-type replication and pathogenic potential. Further, the spectrum of cortical anomalies and the distribution of infected cells in the brain tissues indicated that RhCMV may have preferentially targeted immature neuronal cells. The pattern of RhCMV infection in the central nervous system may offer an explanation for the severe developmental outcomes associated with congenital human CMV infection early in gestation.
Human cytomegalovirus (HCMV) is the most prevalent congenital viral infection in the world, with an average incidence of 1% of all live births (44). It is also a significant cause of central nervous system (CNS) and other developmental anomalies in congenitally infected infants. The risk of congenital HCMV infection and its associated fetal sequelae have been correlated with the immune status of the mother (14). Fetuses of women undergoing a primary infection are at a significantly greater risk for both infection and HCMV-associated disease than those of women with nonprimary HCMV infections. Up to 8,000 congenitally infected infants in the United States suffer various degrees of neurological impairment, including diminished intellectual capacity, sensorineural hearing loss, visual disorder, seizures, and epilepsy (11, 31). However, the true rate of congenital HCMV infection in seropositive women remains uncertain and may be greater than generally perceived (5).
The mechanism of HCMV neuropathogenesis in the developing CNS has not been fully elucidated. It has been suggested that the severity of neuropathological changes and clinical outcome may be associated with the stage of CNS development when fetal infection is established (4). Microcephaly and polymicrogyria are the most prominent features of brain abnormalities in congenital HCMV infection (32). These may result from the disturbance of cellular events within the neuroepithelium after viral infection, including proliferation of neuronal stem cells, differentiation of progenitor cells for neurons or glia, migration of differentiating cells, and cell loss during migration. This hypothesis is supported by the fact that ventricular and subventricular zones are the most susceptible regions to HCMV infection (32). Multiple cell types (neuroepithelial stem cells, differentiating neuronal cells, and neuroglia) in this region are permissive for HCMV infection (29).
The rhesus CMV (RhCMV) nonhuman primate model of HCMV infection represents an excellent opportunity to address viral mechanisms of pathogenesis (50). HCMV and RhCMV exhibit parallel natural histories, and the hosts share strong developmental, physiological, and evolutionary similarities. Since HCMV exhibits a strict species specificity for replication and studies of human subjects have obvious limitations, utilizing the rhesus monkey model for studying the neuropathogenesis of congenital RhCMV infection can provide key insights into the mechanisms for CMV-related diseases in the developing brain. Since we have previously reported that all current breeding-age female macaques at the California National Primate Research Center (CNPRC) are RhCMV seropositive, simulating the condition of congenital HCMV infection that parallels primary infection in gravid women is not possible (54). However, previous studies have demonstrated that RhCMV can cause neuropathogenic outcomes in infected fetal monkeys (Macaca mulatta) similar to those observed in congenitally infected humans (25, 50).
In this study, the construction and characterization of a recombinant RhCMV that expresses enhanced green fluorescent protein (EGFP) under the control of simian virus 40 (SV40) early promoter (designated RhCMV-EGFP) in infected cells is described. We show that (i) RhCMV-EGFP is fully pathogenic in experimentally inoculated fetuses, (ii) a high level of EGFP expression was achieved in virus-infected cells, and (iii) the SV40 promoter was constitutively active in multiple cell types that were permissive to RhCMV infection. Importantly, the results also establish that insertion of an exogenous gene expression cassette into the RhCMV genome does not perturb its pathogenic potential.
MATERIALS AND METHODS
Cells and viruses.
RhCMV strain 68-1 (3) was used for recombinant virus construction and was originally obtained from the American Type Culture Collection (ATCC VR-677). The culture conditions for primary rhesus fibroblasts (RF) and Telo-RF (19) have been previously described (7). For the preparation of virus stocks, Telo-RF were infected at a multiplicity of infection (MOI) of 0.01 and cultured until 100% of the cells exhibited cytopathic effect (CPE). Supernatant was collected, filtered through a 0.45-μm filter, and stored at −70°C in a 1:1 mixture with 9% sterile skim milk. The virus stock used for fetus inoculation was prepared by high-speed centrifugation. In brief, virion particles in filtered supernatant were pelleted by centrifugation at 15,000 × g at 4°C for 2 h. Pellets were washed twice with phosphate-buffered saline (PBS) containing 5% fetal calf serum. Aliquots of resuspended virions were stored in liquid nitrogen. Virus titers were determined by plaque assays on Telo-RF according to previously described methods (7).
Plasmid and recombinant virus construction.
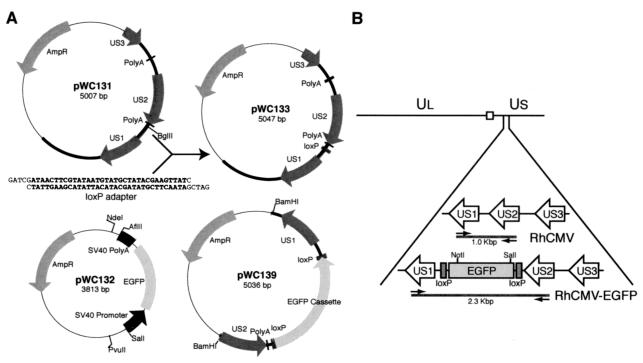
To construct the recombination vector, an oligonucleotide adapter (PAB464/PAB465) (Table 1) containing a 34-bp loxP site was inserted into the BglII site located in the intergenic region between unique short 1 (US1) and US2 of the RhCMV genome fragment of pWC131 (Fig. 1A). The resulting plasmid, pWC133, was sequenced to confirm the orientation of the loxP site and then used as a template for the PCR. Two regions of pWC133, corresponding to loxP-US1 and US2-loxP, were amplified with PCR primer pairs PAB466/PAB467 and PAB468/PAB469 (Table 1), respectively. PCR products were cloned, sequenced, and subcloned into the AflII/NdeI and PvuII/SalI sites of pWC132 to generate the recombination vector pWC139 (Fig. 1A). pWC132 was constructed in a two-step procedure to generate an EGFP expression cassette. The SV40 promoter was amplified with PSV40R/PSV40F by using pEGFP-1 vector (Clontech Laboratories) as a template. The PCR amplicon and EGFP open reading frame-SV40 polyadenylation signal from pEGFP-1 was sequentially cloned into pUC19. The BamHI site between the SV40 promoter and the EGFP coding region was deleted by treatment with T4 DNA polymerase. The DNA fragment for homologous recombination was cleaved from pWC139 by BamHI digestion (Fig. 1A), and 10 μg of digested DNA was transfected into 2.5 × 106 primary RF by electroporation with a 0.4-cm Gene Pulser cuvette (Bio-Rad Laboratories) under the conditions of 0.31 kV and 960 μF (0.3°C). Cells were subsequently infected with RhCMV at an MOI of 1 at 48 h posttransfection. Supernatant was collected when the cells exhibited 100% CPE and was used to infect fresh Telo-RF cultures. Fluorescent plaques were visualized with an Axiovert inverted fluorescence microscope (Carl Zeiss Microimaging) and individually collected. Recombinant clones were obtained by six rounds of serial plaque purification.
TABLE 1.
Sequences of primers used for PCR and 3′ RACE
| Primer | Sequence (5′-3′) |
|---|---|
| PAB201 | CCCTTCCTGACTACTAATGTAC |
| PAB431 | AACGTGACGAGTCGGAGTCCAGAGTC |
| PAB435 | GCAAGCTGTTCATGACGGGATAC |
| PAB464 | GATCGATAACTTCGTATAATGTATGCTATACGAAGTTATC |
| PAB465 | GATCGATAACTTCGTATAGCATACATTATACGAAGTTATC |
| PAB466 | CCCCTTAAGCTTATAACTTCGTATAATGTATGC |
| PAB467 | CGACATATGGATCCCCGCATGGTTTCCATTGAG |
| PAB468 | TTTCAGCTGGATCCAGCTCGGACTGACATTCGG |
| PAB469 | GGGTCGACAAGCTTATAACTTCGTATAGCATACAT |
| PAB489 | AACATCCTGGGGCACAAGCTGGAGTAC |
| PAB534 | GCATCCTGGGCTACACTGAG |
| PAB548 | GCAGTGGAGACTGCAAATCAAGG |
| PAB549 | GGTATGCCGAGAGAGTATCTCGC |
| PSV40F | ACGCGTCGACAGTTAGGGTGTGGAAAGTCC |
| PSV40R | CGCGGATCCTCACTACTTCTGGAATAGC |
| SFV119 | GATGGAACCGGATCCAAGTC |
FIG. 1.
(A) Configuration of the plasmids used in this study. The pUC19 sequences in each plasmid are shown as a thin line, and the viral sequences are presented as a thick line. For the construction of pWC133, a loxP adapter was inserted into the BglII site of pWC131. (B) Schematic diagram of viral genome structure with expansion of the region from US1 to US3 of RhCMV and RhCMV-EGFP. The locations of the EGFP expression cassette flanked by two loxP sites and the additional NotI/SalI sites in the viral genome introduced by homologous recombination are shown. The square represents the junction region between UL and US. Arrows indicate the locations of diagnostic PCR primers PAB431 and PAB435 specific to US1 and US2, respectively.
Viral DNA isolation and analysis.
Viral nucleocapsid DNA was isolated by the methods described by Sinzger et al. (39). In brief, infected cells were harvested when cultures reached 100% CPE, collected by low-speed centrifugation, and washed two times with cold PBS. Cells were first treated with a membrane permeabilization buffer and then incubated with micrococcal nuclease (1,500 U/ml; U.S. Biochemicals) at 37°C for 60 min. After nuclease treatment, cells were digested with proteinase K (100 μg/ml; Invitrogen) at 56°C overnight. Total DNA was extracted with phenol-chloroform and precipitated with isopropanol. Restriction endonuclease-digested DNA was separated by electrophoresis on 0.8% agarose gels for 18 h at 40 V and visualized by ethidium bromide staining.
PCR, RFLP, and 3′ RACE.
The diagnostic primer set PAB431 and PAB435 (Table 1) was used to amplify the region between the US1 and US2 of wild-type and recombinant RhCMV. Supernatants collected from infected cultures were heated at 100°C for 10 min and used as templates for PCR with Takara EX Taq polymerase (Takara Shuzo). An amplification program of 94°C for 3 min, 55°C for 2 min, and 72°C for 5 min for 40 cycles was used for diagnostic PCR. For restriction fragment length polymorphism (RFLP) analyses, amplicons of diagnostic PCR were purified with a QIAquick PCR purification kit (Qiagen) and digested with HaeIII, HinfI, RsaI, or TaqI for 2 h at 37°C. Electrophoresis of digested DNA was carried out on 2% Metaphor agarose gels (FMC BioProducts) at 70 V for 2 h. Extraction of cytoplasmic RNA from infected cells and the synthesis of cDNA for 3′ rapid amplification of cDNA ends (3′ RACE) were performed as previously described (7). 3′ RACE reactions were performed with reverse primer SFV119 paired with one of the following gene-specific primers: PAB201 (IE2), PAB548 (US1), PAB435 (US2), PAB549 (US3), PAB489 (EGFP), and PAB534 (rhesus glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) (Table 1). A touch-down PCR program was used for 3′ RACE reactions as follows: 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min for 5 cycles, followed by 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min for 25 cycles. For 3′ RACE of US1 and GAPDH, the reaction mixtures were adjusted to 10% dimethyl sulfoxide, and the program was modified by either a 2-min extension at 72°C for US1 or a 30-s annealing at 58°C for GAPDH.
Virus replication kinetics and FACS analyses.
Viral replication kinetics were determined by multiple-step growth curve analyses on Telo-RF cells according to previously described methods (7). Triplicate cultures of cells were infected with either RhCMV or RhCMV-EGFP at an MOI of 0.01. Supernatants from infected cultures were collected daily for plaque assays, and the fluorescence intensity within infected cells was monitored by fluorescence microscopy and fluorescence-activated cell sorter (FACS) assay. Fluorescent cell scanning was carried out on a FACSCalibur cytometer (BD Immunocytometry Systems), and data were analyzed and illustrated with FlowJo software (Tree Star).
Animals.
All animal procedures conformed to the requirements of the Animal Welfare Act, and protocols were approved prior to implementation by the Institutional Animal Use and Care Administrative Advisory Committee at the University of California at Davis. Normally cycling, adult female rhesus macaques (M. mulatta) seropositive for RhCMV, ranging in age from 7 to 13 years and with body weights of 5 to 10 kg, with a history of prior pregnancies were bred and identified as pregnant using established methods (48). Pregnancy in the rhesus monkey is divided into trimesters by 55-day increments, with 0 to 55 days gestation representing the first trimester, 56 to 110 days gestation representing the second trimester, and 111 to 165 days gestation representing the third trimester (term ≈165 ± 10 days) (46). Activities related to animal care (diet, housing) and screening animals for endogenous retroviruses (simian retrovirus and simian T-lymphotropic virus) prior to assignment to the study were performed as per standard CNPRC operating procedures. Maternal health was monitored daily and body weights were assessed monthly.
Virus inoculation and fetal monitoring.
All pregnancies were sonographically assessed to confirm normal growth and development prior to fetal inoculation with RhCMV-EGFP (47). The dams were administered ketamine hydrochloride (10 mg/kg) for these and subsequent ultrasound examinations. Immobilized dams were aseptically prepared for transabdominal ultrasound-guided fetal intracranial inoculation on day 50 of gestation (first trimester). A total volume of 150 μl with 5 × 104 or 1 × 105 PFU of RhCMV-EGFP was injected into the lateral ventricle, using established techniques (45) (see Table 2). Postinoculation, sonographic measurements of the fetal head (biparietal and occipitofrontal diameters, area and circumference), abdomen (area and circumference), and limbs (humerus and femur lengths), in addition to gross anatomical evaluations (axial and appendicular skeleton, viscera, membranes, placenta, amniotic fluid), were assessed weekly, as previously described, and all measures were compared to normative growth curves for rhesus fetuses (47).
TABLE 2.
Outcome for fetal rhesus monkeys (M. mulatta) intracranially inoculated with RhCMV-EGFP in utero at day 50 of gestationa
| Fetus (i.d. no.) | Inoculum titer (PFU) | Necropsy (dpi) | Sonographic findings | Gross findings at necropsy | Viral IE1 antigen found by IHC | Virus isolation
|
|
|---|---|---|---|---|---|---|---|
| 10 dpi | 22 ± 1 dpi | ||||||
| 1 (001-0645) | 5 × 104 | NDb | Microcephalyc | NDb | NDb | ND | ND |
| 2 (001-0801) | 1 × 105 | 10 | Flocculent AF | Mild ventricular dilatation | Brain | ND | ND |
| Ventricular dilatation | Lower limb deformity | ||||||
| Severe hydrops | Ileocolic stricture, dilated colon | ||||||
| Microcephaly | |||||||
| Echogenic bowel | |||||||
| 3 (001-0879) | 1 × 105 | 21 | Severe hydrops | Ventricular dilatation | Brain | −(FB) | +(FB) |
| Severe microcephaly | Lower limb deformity | Adrenals | −(AF) | −(P) | |||
| Abdominal distension | Ileocolic stricture, dilated colon | Kidney | −(AF) | ||||
| Lower limb deformity | SD (cranium and neck) | Liver | |||||
| IUGR | Gonad | ||||||
| Pancreas | |||||||
| 4 (001-0888) | 1 × 105 | 23 | Severe microcephaly | Ventricular dilatation | Brain | +(FB) | +(FB) |
| Severe hydrops | Lower limb deformity | Adrenal | −(AF) | +(P) | |||
| Abdominal distension | Ileocolic stricture, dilated colon | Kidney | −(AF) | ||||
| Lower limb deformity | SD (cranium, face, and neck) | Liver | |||||
| IUGR | Gonad | ||||||
| Pancreas | |||||||
| Lung | |||||||
| Duodenum | |||||||
| Colon | |||||||
i.d., identification; ND, not done; AF, amniotic fluid; IUGR, intrauterine growth restriction; SD, subcutaneous discoloration; IHC, immunohistochemistry; FB, fetal blood; P, plasma.
Fetus death at ≈22 dpi, which was attributed to virus pathogenicity.
Evident within 10 dpi, severe at death.
Fetal sample collection and necropsy.
Fetal blood (0.5 ml) and amniotic fluid (0.5 ml) samples were collected via ultrasound guidance using standard techniques (45) from fetuses 3 and 4 at 10 days postinoculation (dpi) (60 days gestation) for virus isolation and complete blood counts. Tissues were harvested by hysterotomy at defined ages of gestation (62 to 75 days gestation [second trimester]). Fetal blood and amniotic fluid were collected for virus isolation, and then complete tissue harvests were performed using standard techniques (49). Total body weights and measures (hand, foot, humerus, and femur lengths; biparietal and occipitofrontal diameters; head, arm, and chest circumferences; crown-rump lengths) were assessed and tissues were grossly evaluated, and then all organs were removed, including the brain, thymus, spleen, liver, lymph nodes (axillary, inguinal, and mesenteric), pancreas, right and left adrenals, right and left kidneys, right and left gonads, stomach, duodenum, jejunum, ileum, colon, heart, skin, muscle, and bone. Select tissues were weighed, including the brain, thymus, spleen, liver, adrenals, and kidneys. The placenta, membranes, umbilical cord, and decidua were also collected and assessed, and the placenta was weighed. The brain was divided into the right and left cerebral hemispheres, and each hemisphere was then divided by three coronal sections into four specimens which were alternately placed in optimal cutting temperature compound (OCT) or formalin. The first and third sections of the right and left hemispheres (with midbrain and cerebellum) were placed in OCT and then quick-frozen over liquid nitrogen, and the second and fourth sections were placed into cassettes and then immersed in 10% buffered formalin. Samples of all of the above were collected and then snap-frozen in liquid nitrogen. Representative sections of all tissues were preserved in formalin, embedded and sectioned at 5 to 6 μm, and then stained with hematoxylin and eosin for routine histopathology. Specimens from animals of comparable age without any interventions (controls) were similarly processed and analyzed in parallel.
Fluorescence microscopy and immunohistochemistry.
Each of the OCT-embedded cranial specimens (four total sections) were sectioned at 9 μm on the day of hysterotomy on a cryostat, air dried, and stored at −20°C for 24 h. Sections were fixed in 10% buffered formalin at room temperature for 2 h, equilibrated briefly with PBS, and counterstained with 300 nM 4′,6′-diamidino-2-phenylindole (DAPI) using the protocols provided by the manufacturer (Molecular Probes). Slides were mounted with Fluoromount G (Electron Microscopy Sciences) and visualized under fluorescent light using single band-pass filters (Omega Optical). Formalin-fixed fetal tissues, including brain sections, were embedded in paraffin, sectioned, and immunoperoxidase stained with polyclonal antibodies for RhCMV IE1 (1:3,200 dilution) or GFP (1:50 dilution) (Clontech Laboratories) and counterstained with hematoxylin according to published procedures (24). Photographs were taken on an Axioskop fluorescence microscope equipped with an Axiocam digital camera (Carl Zeiss Microimaging) interfaced with a PC computer. The contrast of images was adjusted with Adobe Photoshop (Adobe Systems).
Nucleotide sequence accession number.
US1, US2, and US3 3′ RACE products were cloned into the TOPO TA cloning vector (Invitrogen) and sequenced. The sequences of the US1, US2, and US3 open reading frames, as well as the internal UL-US junction region of RhCMV have been submitted to GenBank (accession number AF474179).
RESULTS
Construction of recombinant EGFP-expressing RhCMV.
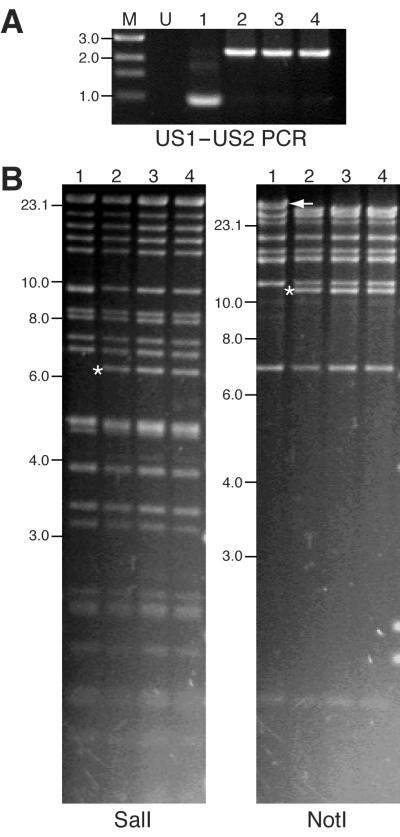
An EGFP expression cassette, under the transcriptional control of the SV40 promoter and polyadenylation signal, was inserted between US1 and US2 of the RhCMV genome by homologous recombination (Fig. 1B). This location was chosen because there is a 102-bp intergenic region between the US2 polyadenylation signal and the US1 transcription start site. A DNA fragment containing US1-loxP-EGFP-loxP-US2 sequences was transfected into primary RF cells by electroporation. Transfected cultures were subsequently infected with RhCMV (strain 68-1) at 48 h posttransfection. A pure population of EGFP-expressing virus was obtained after six rounds of plaque purification. The purity of the recombinant clone was confirmed by diagnostic PCR designed to amplify the region between US1 and US2 of the viral genome (Fig. 1B). Amplification of DNA purified from EGFP-expressing virus yielded the predicted 2,332-bp fragment instead of the 964-bp amplicon of wild-type RhCMV (Fig. 2A). The precise location of the EGFP gene cassette was further confirmed by sequencing (data not shown). To confirm that additional genomic rearrangements did not occur during homologous recombination, viral nucleocapsid DNA was analyzed by restriction endonuclease digestion. Novel SalI and NotI restriction fragments were detected by digestion and gel electrophoresis (Fig. 2B), consistent with the insertion of the EGFP cassette between US1 and US2 (Fig. 1B). The loss of restriction fragments resulting from insertion of the EGFP cassette was obscured by either comigration of identically sized SalI restriction fragments (9.3 kb) or the large size of the NotI fragment (48 kb) (Fig. 2B).
FIG. 2.
Analyses of RhCMV and RhCMV-EGFP genome organizations. (A) Diagnostic PCR for RhCMV and RhCMV-EGFP isolates using supernatants collected from virus-infected cultures as templates. (B) Gel electrophoresis of SalI- and NotI-digested viral nucleocapsid DNA. Novel restriction fragments generated following recombination are marked with asterisks. The loss of the NotI restriction fragment is marked with an arrow. Size standards are displayed on the left of the gel pictures and are indicated in kilobases. Lane M, DNA marker; lane U, uninfected control; lane 1, wild-type RhCMV; lane 2, plaque-purified RhCMV-EGFP; lane 3, virus recovered from fetus 3; lane 4, virus recovered from fetus 4.
RhCMV-EGFP exhibited comparable replication properties as wild-type virus in vitro.
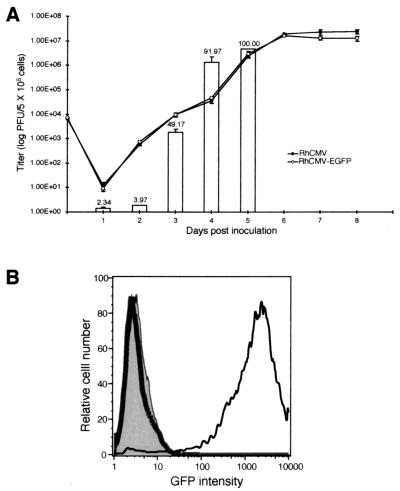
The replication kinetics of the recombinant virus were compared to those of wild-type RhCMV to determine whether the insertion of the EGFP cassette into the RhCMV genome perturbed viral replication in vitro. Multiple-step growth curve analysis confirmed that insertion and expression of the EGFP cassette did not significantly alter the replication properties of RhCMV. RhCMV-EGFP exhibited near-identical kinetics to that of wild-type RhCMV in Telo-RF cultures (Fig. 3A). During the course of these studies, it was noted that the peak titers of RhCMV-EGFP were lower than those of wild-type RhCMV. Compared to wild-type virus, the reduction of progeny RhCMV-EGFP titer occurred after all of the cells in the culture exhibited CPE and expressed a high level of EGFP. It is believed that accumulation of high levels of EGFP may be somewhat cytotoxic, resulting in the reduced production of progeny virions. It is unlikely that the decrease in titer resulted from the increased genome size (1.5 kb) of RhCMV-EGFP. Both viruses showed comparable replication kinetics at the first two rounds of replication cycle in the curves (Fig. 3A).
FIG. 3.
Replication kinetics of RhCMV-EGFP and EGFP expression in infected cells. (A) The multiple-step growth curve of RhCMV-EGFP is compared to that of its parental RhCMV strain. Telo-RF cells were infected in triplicate with each virus at an MOI of 0.01. Supernatants and cells were collected longitudinally from the infected cultures for standard plaque assays and FACS analyses, respectively. The titers of infectious virions in the samples are shown as a solid line. The percentages of GFP-positive cells in total gated populations are presented as bars with values shown. Data points represent the mean of three independent cultures, with the standard deviations indicated by error bars. (B) Overlaid histogram of fluorescence cytometry profiles of cells infected with either RhCMV (thick line) or RhCMV-EGFP (thin line). Mock-infected cells are also shown (shaded). Telo-RF cells were infected at an MOI of 0.01, collected on 4 dpi, fixed with 1% paraformaldehyde, and analyzed by FACS.
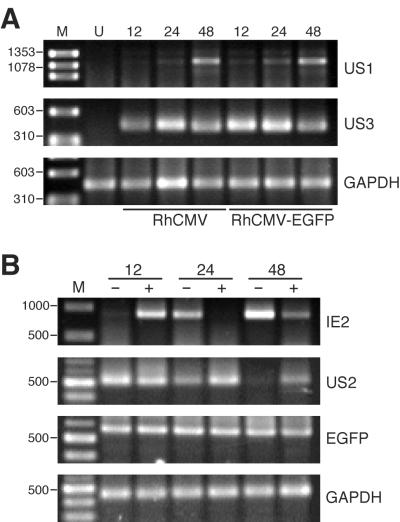
The gene loci from US1 to US11 of HCMV have been shown to be dispensable in cell cultures (15, 16). It has not been determined whether they are essential for viral replication in vivo. US2, -3, -6, and -11 may be important for HCMV pathogenicity due to their potential modulation of antigen presentation (1, 17, 18, 26, 51). To evaluate the impact of the EGFP cassette on the expression of neighboring viral genes, the steady-state levels of US1, US2, and US3 transcripts in RhCMV-EGFP-infected cells were examined by 3′ RACE. Cytoplasmic RNA from Telo-RF infected with RhCMV or RhCMV-EGFP (MOI, 1) was isolated at different time points postinoculation. The temporal expression profiles of US1, US2, and US3 in RhCMV-EGFP-infected cells were comparable to those in wild-type RhCMV-infected cells (Fig. 4; for 3′ RACE of US2, only RhCMV-EGFP is shown). Although 3′ RACE analysis does not discriminate subtle changes in expression levels, the results demonstrate that the EGFP cassette did not significantly interfere with the expression and regulation of these viral genes.
FIG. 4.
3′ RACE analyses of RhCMV-EGFP gene expression profiles. (A) US1 and US3 expression levels in RhCMV- or RhCMV-EGFP-infected Telo-RF at different time points (indicated in hours) after virus inoculation. (B) Constitutive expression of EGFP open reading frame and the temporal regulation of viral IE2 and US2 genes in RhCMV-EGFP-infected cells. Cytoplasmic RNA was isolated from infected cultures at different time points in the presence (+) or absence (−) of either 200 μg of cycloheximide/ml (12 hpi) or 400 μg of phosphonoformic acid/ml (24 and 48 hpi). 3′ RACE for GAPDH was performed as an internal control. Size standards are displayed on the left of the gel pictures and are indicated in base pairs. Lane M, DNA marker; lane U, uninfected control.
Although RhCMV-EGFP exhibited comparable replication properties to those of wild-type virus in vitro (Fig. 3A), viral populations with deletion of nonessential genome sequences (e.g., EGFP cassette) may have arisen due to selection pressure. The stability of the recombinant viral genome was assessed by serial passages in vitro. Telo-RF cultures were inoculated with RhCMV-EGFP at a low MOI, and supernatants were collected for the next passage as soon as the majority of cells exhibited CPE. The exogenous EGFP cassette remained stable in the viral genome after 20 passages, as all infected cells expressed bright fluorescence. PCR and RFLP analyses of US1-US2 regions with RhCMV-EGFP after the sixth round of plaque purification (passage 6) and passage 18 were consistent with the predicted amplification and restriction fragment sizes of the recombinant viral genome (Fig. 5). To investigate whether deletions occurred at other sites in the viral genome, full-length nucleocapsid DNA was analyzed by endonuclease digestion. The NotI and SalI restriction patterns of RhCMV-EGFP DNA remained unchanged with prolonged serial passage (data not shown).
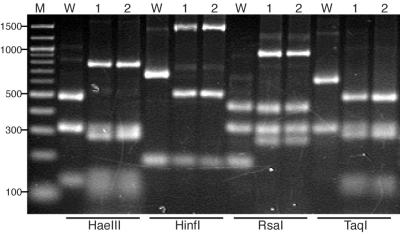
FIG. 5.
PCR-RFLP analyses of RhCMV and RhCMV-EGFP of different passages. PCR amplicons derived from primers within US1 and US2 were digested with four- or five-base cutters. Digested fragments were separated by electrophoresis on 2% Metaphor agarose gels and visualized by ethidium bromide staining. Size standards are displayed on the left of the gel pictures and are indicated in base pairs. Lane M, DNA marker; lane W, wild-type RhCMV; lane 1, RhCMV-EGFP passage 6; lane 2, RhCMV-EGFP passage 18.
Constitutive expression of EGFP in infected cells.
Fluorescence in RhCMV-EGFP-infected cells was readily detected as early as 1 dpi by flow cytometry (Fig. 3A). There was a large increase in the ratio of GFP-positive cells from 2 to 3 dpi, which occurred 1 day subsequent to the initial release of progeny virions on 2 dpi. The overlaid histogram of GFP profiles of cells collected on 4 dpi from the growth curve study showed that RhCMV-EGFP-infected cells displayed an intense fluorescence and could be readily distinguished from the mock-infected or wild-type RhCMV-infected cells (Fig. 3B). Compared with FACS, fluorescence microscopy was less sensitive. Infected cells were barely discernible at 36 h postinoculation (hpi). The intensity of fluorescence in the infected cells steadily increased during the course of infection. To evaluate the effects of RhCMV gene products at different stages of viral replication on the SV40 early promoter, 3′ RACE for EGFP transcripts was performed using cytoplasmic RNA isolated from infected cell cultures in the presence or absence of cycloheximide or phosphonoformic acid. Unlike the viral IE2 and US2 genes that were regulated during RhCMV replication, EGFP RNA expression was constitutive throughout the RhCMV replication cycle (Fig. 4B). The results from EGFP RNA steady-state levels and the accumulation of fluorescence in the infected cells indicated that the SV40 early promoter functioned independently of RhCMV temporal regulation.
Infection of monkey fetuses in utero by intracranial inoculation with RhCMV-EGFP.
RhCMV-EGFP exhibited wild-type parameters of replication and gene expression in vitro. A fundamental question was whether the process of genome manipulation attenuated the pathogenicity of the virus. To assess this possibility, fetal rhesus macaques were experimentally inoculated in utero with purified RhCMV-EGFP on day 50 of gestation (late first trimester) by the intracranial route. Fetuses were evaluated by ultrasound for developmental abnormalities. All four fetuses exhibited evidence of severe RhCMV-caused brain anomalies, including microcephaly and ventriculomegaly, and other non-CNS sequelae, consistent with those observed following inoculation with wild-type RhCMV (50; P. A. Barry and A. F. Tarantal, unpublished data). One fetus (fetus 1) died from RhCMV infection at approximately 25 dpi, and tissues were too autolyzed for analysis. The other three fetuses were terminated at select time points based on the severity of disease (Table 2), and extensive RhCMV infection in the CNS and other tissues was observed. Gross cerebral and cerebellar abnormalities were detected in fetuses 2 to 4. The extent of microcephaly and ventricular dilatation was greater in fetuses 3 and 4 (terminated at 21 and 23 dpi, respectively) than in fetus 2 (terminated at 10 dpi).
Neuropathological changes caused by RhCMV-EGFP infection.
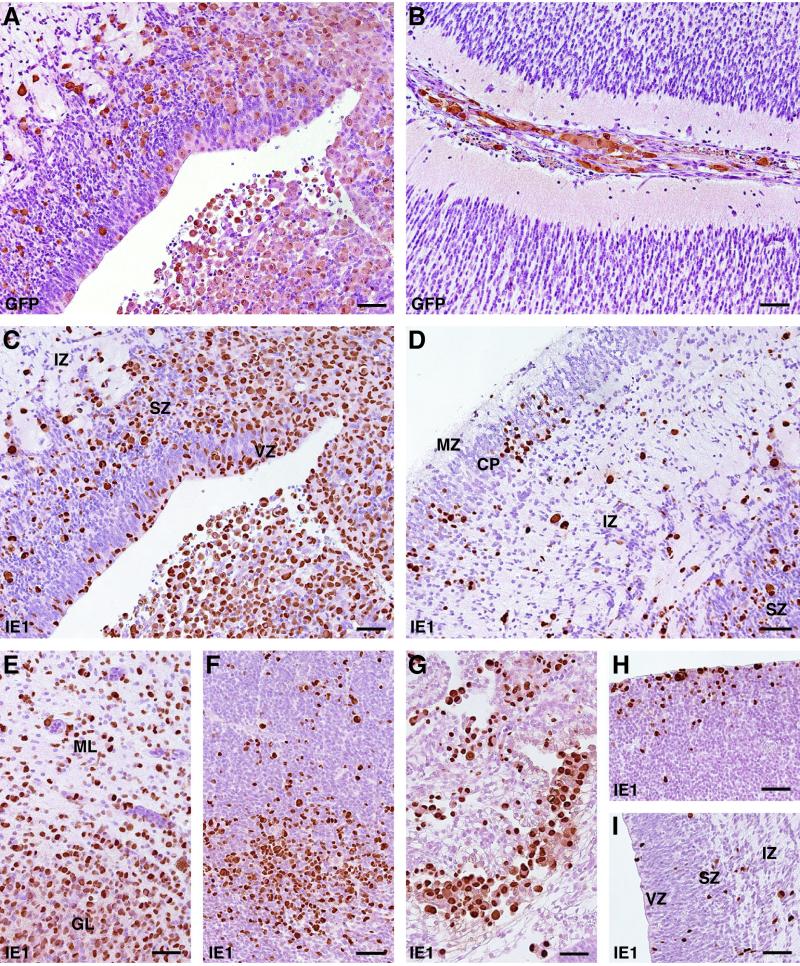
The neuropathological findings revealed both inflammatory changes and neuronal migration defects, similar to congenital CMV infection in humans (32). Periventricular necrosis associated with low-level calcification was observed in cerebral hemispheres of fetuses 3 and 4. Numerous enlarged cells, immunoperoxidase stained with antibody for either EGFP or IE1, were observed in various areas of the cerebral and cerebellar tissues from fetuses 3 and 4 (Fig. 6A to E; only brain sections of fetus 4 are shown). Scattered inclusion-bearing neuronal cytomegaly was detected throughout the brain but with a distinct pattern of infected cell localization. Aggregates and clusters of RhCMV antigen-positive cells were most dense in the granular layer of neuroepithelium surrounding the lateral ventricles (Fig. 6C, F, H, and I). This was seen consistently in all animals studied. The ventricular and subventricular zones of the cerebrum, where the neuronal stem cells proliferate and from which the immature neuronal and glial cells migrate outward (35), have been thought to be the most susceptible site for CMV infection in the embryonic stage of humans and mice (22, 32). Infected cells were also found scattered in the intermediate zone and cortical plate of cerebral cortices (Fig. 6D) and meninges (Fig. 6B). Cytomegaly and infected epithelial cells were noted in the choroid plexus of fetus 2 (Fig. 6G) after 10 dpi. This finding suggests that epithelial cells of the choroid plexus are highly susceptible to CMV infection and may play an important role in the establishment of CMV infection in the fetal CNS.
FIG. 6.
Distribution of RhCMV-EGFP-infected cells in fetal brain. Tissues were collected from fetus 2 (F to I) and fetus 4 (A to E) at 10 and 23 dpi, respectively. Immunostaining was carried out using the polyclonal antibody for GFP (A and B) or RhCMV IE1 (C to I). (A and C) Serial brain sections with periventricular foci stained with different antibodies; (B) cells in the meninges are labeled after immunoperoxidase staining of GFP; (D) cerebral neocortex; (E) cerebellum; (F) tectal neuroepithelium; (G) choroid plexus; (H) striatal neuroepithelium; (I) cerebral neocortex. CP, cortical plate; GL, germinal layer; IZ, intermediate zone; ML, marginal layer; MZ, marginal zone; SZ, subventricular zone; VZ, ventricular zone. Bars, 50 μm.
RhCMV-EGFP established systemic infection in inoculated fetuses.
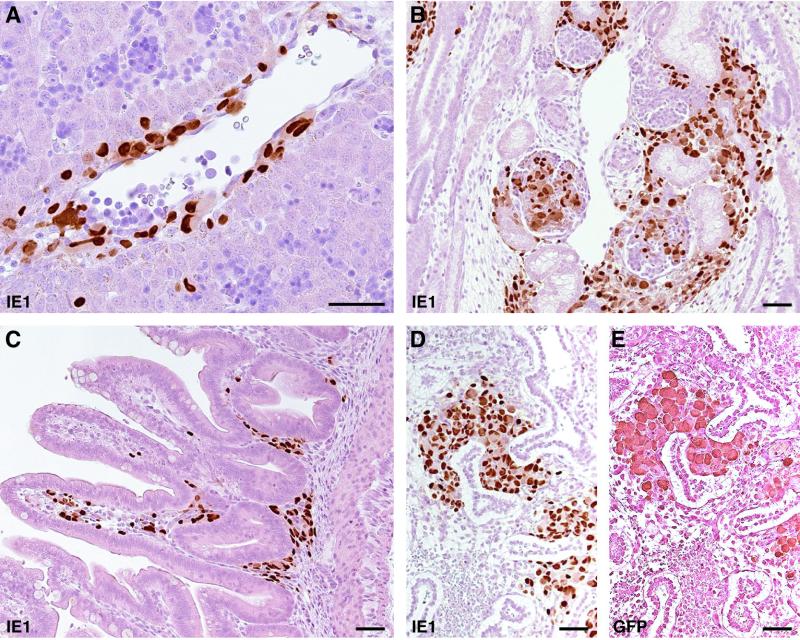
Intracranially inoculated RhCMV-EGFP also established systemic infection in multiple fetal organs. RhCMV-EGFP was isolated from fetal blood, and viral IE1 antigen was detected in multiple organs by immunohistochemistry (Table 2). Blood samples were collected from fetuses 3 and 4 for virus isolation at 10 dpi and at necropsy. Although both fetuses received identical titers of virus on the same day of gestation, the development of viremia in fetus 4 occurred prior to that of fetus 3 (Table 2). In addition, infected cells were detected in more organs than in fetus 3. Viral antigen was detected within cells in the perivascular region of portal veins and some hepatocytes (Fig. 7A), suggesting that the virus disseminated through the bloodstream to multiple organs. It was also noted that the virus showed preferential replication in the interstitial regions of multiple fetal tissues (Fig. 7B, C, and D). Since inoculations were performed during organogenesis, these developing mesenchymal cells may have exhibited higher susceptibility to RhCMV. Although RhCMV-EGFP established systemic infection in both fetuses, no viral antigen was detected in the placenta, and attempts to isolate virus from amniotic fluid were not successful (Table 2). The genome integrity and stability of the EGFP expression cassette of RhCMV-EGFP isolates from both fetuses were examined by diagnostic PCR (Fig. 2A), restriction digestion (Fig. 2B), and plaque assays. Deletion variants were not detected in the isolated RhCMV-EGFP populations.
FIG. 7.
Immunoperoxidase staining of tissues collected from fetus 4, using polyclonal antibody for RhCMV IE1 (A to D) or GFP (E). (A) Infected cells adjacent to the endothelial cells around the portal vein were frequently found in the liver. (B) Infected cells in developing glomeruli and mesenchyme of the kidney. (C) In the duodenum, stained cells were observed in the lamina propria. (D and E) Mesenchymal cells in the lung were highly infected. Bars, 50 μm.
Expression of EGFP in virus-infected tissues.
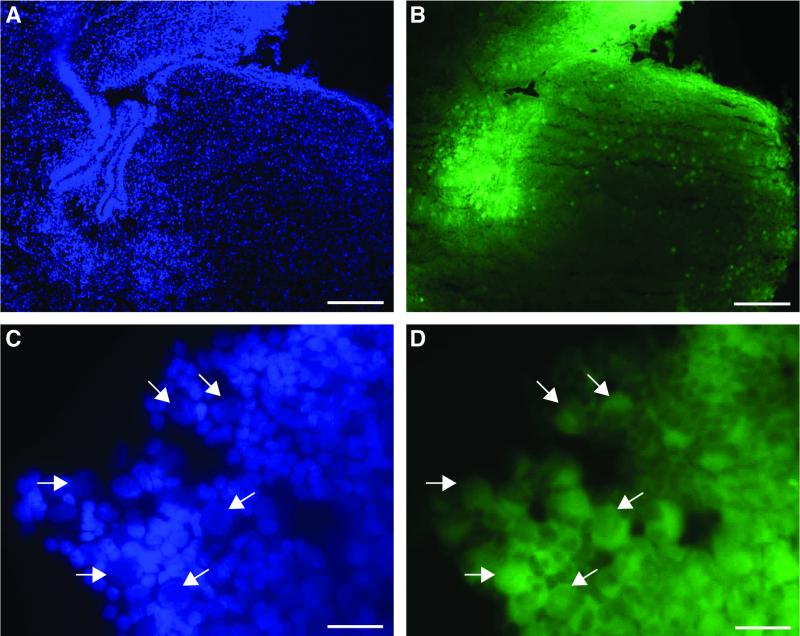
Subcutaneous discoloration was observed in the cranial base and subcutaneous region around the head and neck of fetuses 3 and 4 and was attributed to the accumulation of EGFP in these infected tissues. At the time of necropsy, it was noted that the lateral ventricles were filled with a thick, yellow-green fluid. RhCMV-EGFP-infected cells in frozen brain sections from fetuses 2 to 4 were readily observed by fluorescence microscopy. Focal areas of fluorescence were confined within the parenchyma of brain sections. Comparably processed tissues from uninfected fetuses do not exhibit detectable fluorescence (A. F. Tarantal, unpublished data). A large number of GFP-positive cells were located in the periventricular regions of the brain from fetuses 3 (Fig. 8A and B) and 4 (data not shown). A high-magnification view (Fig. 8C and D) demonstrates the enlarged and pale DAPI-stained nuclei in GFP-positive cells. Serial sections, stained with either anti-RhCMV IE1 or anti-GFP polyclonal antibody, confirmed that there was colocalization of EGFP expression and RhCMV infection. The distributions of IE1-positive and GFP-positive cells in the infected fetal tissues were essentially identical (Fig. 6A and C and 7D and E). Unlike the nuclear localization of IE1 in infected cells, EGFP accumulated in the cytoplasm. The pathognomonic “owl's eye” inclusions were readily observed within the RhCMV-infected cells in sections stained by immunoperoxidase for GFP (Fig. 6A and 7E).
FIG. 8.
Fluorometric detection of EGFP expression under the control of SV40 early promoter from brain sections from fetus 3 (B and D) and nuclei of cells counterstained with DAPI (A and C). Strong RhCMV-infected regions were found in the periventricular area. Some infected cells scattered in the cerebral cortex were also noted. Arrows indicate individual infected cells that had weaker DAPI staining but enlarged nuclei. GFP-positive cells were frequently surrounded by green halos, presumably from diffusing EGFP of adjacent cells caused by cell lysis or sectioning. Bars, 200 μm (A and B) and 25 μm (C and D).
DISCUSSION
This is the first reported demonstration of a genetically manipulated nonhuman primate CMV genome that retained a wild-type phenotype in vitro and in vivo. Recovery of the EGFP-expressing recombinant RhCMV from the experimentally inoculated macaque fetuses and the absence of deletion or reversion are evidence that insertion of the EGFP cassette can be readily tolerated during viral replication. Typical CMV neuropathological changes in the CNS and dissemination of inoculated virus to multiple organs indicate that the expression of EGFP did not attenuate its pathogenic potential.
Previous reports have demonstrated that herpesviruses represent stable, replication-competent vectors for exogenous gene expression in vitro and in vivo. These include herpes simplex virus 1 (12, 13), pseudorabies virus (41), Epstein-Barr virus (43), murine CMV (MCMV) (52, 53), and HCMV (6, 28). There are several advantages of using CMV as a vector for foreign gene expression due to its broad cell tropism (24, 38, 40). CMV has the largest genome among the herpesviruses (30), ranging in size from 190 kb for equine CMV (10) to 220 to 230 kb for HCMV (8), MCMV (36), and RhCMV (D. G. Anders and S. Wong, personal communication). Although the quantity of viral DNA that can be efficiently packaged into the progeny capsids is not known, inserting extra sequences into the CMV genome may impede viral replication at the step of viral DNA packaging to a greater extent than for other herpesviruses. The results of serial passage in vitro and in vivo show that inclusion of an additional 1.5 kb of sequence does not interfere with RhCMV replication.
Despite the large size of the HCMV genome, there are relatively few noncoding regions of significant length within the viral genome (8). The results of this report demonstrate that as little as 210 bp of intergenic sequence between US1 and US2 enable expression of the EGFP cassette without altering the temporal expression kinetics of the neighboring viral genes. In addition, the SV40 early promoter is refractory to RhCMV temporal regulation and is constitutively active throughout the viral replication cycle. The HCMV IE promoter has been reported to be transcriptionally more active than other commonly used viral promoters in cultures of sensory neurons (42) and muscle cells (21). However, transcription of the HCMV IE promoter is negatively regulated by HCMV IE2 via a short target sequence, the cis repression signal (crs), overlapping the transcription start site (9, 23, 33). Since the putative RhCMV crs differs only by a single base pair from the HCMV crs (2), the SV40 early promoter was used for construction of the EGFP cassette to preclude potential repression by RhCMV IE2. More importantly, the SV40 early promoter was active in various types of cells in multiple organs that are susceptible to RhCMV infection (Fig. 6A and C and 7D and E). Constitutive expression of the SV40 promoter, together with the wide tissue tropism exhibited by RhCMV-EGFP, may facilitate systemic and local immune responses to exogenously expressed antigens.
Expression of an easily detectable reporter gene, such as EGFP, provides a sensitive system to address mechanisms of viral pathogenesis. The pattern of EGFP expression and RhCMV histopathology offer insights into intrauterine neuropathogenesis in fetal primates. The developing brain is highly susceptible to CMV infection. Preferential brain infection in fetuses has also been shown in both the monkey and mouse models after intraperitoneal inoculation with RhCMV and placental infection with MCMV (22, 50). In the study described here, the periventricular zone containing a high density of RhCMV-infected cells was observed early after intracranial inoculation (Fig. 6H and I). The results are consistent with findings in humans and rodents, where the cerebral periventricular zone has been shown to be one of the most susceptible sites for CMV infection during embryogenesis (22, 32). There are different possibilities to account for the distinct localization of CMV infection in the CNS. These include (i) that neuronal stem cells and neuronal precursor cells may be more susceptible to CMV infection than other cells in the developing brain, and/or (ii) that CMV may readily reach the CNS through the blood-cerebrospinal fluid route during early gestation. It has been reported that CNS stem cells and undifferentiated neurons are permissive to HCMV and MCMV replication (20, 29, 34). Identification of the choroid plexus as another major target for RhCMV replication early after inoculation (Fig. 6G) supports the hypothesis that CMV may be able to readily cross the blood-cerebrospinal fluid barrier to the developing CNS.
Microcephaly was found in all inoculated fetuses (Table 2), consistent with an early insult to CNS development. The pattern of neuropathological outcomes was identical to those reported for HCMV. The most common pathological changes associated with early HCMV congenital infection include cerebral malformations, such as microcephaly, lissencephaly, or polymicrogyria. The degree of developmental anomalies is dependent on the stage of development when infection occurs (4, 32). According to the radial unit hypothesis of brain development (35), the size of individual cytoarchitectonic areas depends on the number of contributing proliferative units, which are established prior to day 40 of gestation in both humans and nonhuman primates. In turn, the thickness of the cortex is dependent on the number of neurons produced within each unit. In this study, viral inoculations were performed on day 50 of gestation, and the infections were terminated before day 73 of gestation, a period of active neurogenesis. The cortical malformation in these fetuses may have resulted from early defects in proliferation and migration of neuronal cells in the individual proliferative unit, similar to findings in mice after intraventricular infection with MCMV (37).
In addition to infection of protoneuronal cells in the periventricular regions, migration and organization of normal cells may also be perturbed by RhCMV infection. Infected radial glia were observed scattered within the intermediate zone (Fig. 6D). Radial glia play an important role in guiding immature neuronal cells during migration from the proliferative units in the periventricular zone to the corresponding ontogenetic columns within expanding cortex (35). Compromised radial glial cells may have contributed to cortical malformation by failing to guide differentiating neuronal precursor cells to migrate to the appropriate venue. It has been shown in the mouse model that small numbers of infected neuronal cells can still migrate during brain development (37). Small numbers of IE1-positive cells were observed in a laminar pattern in the cerebral cortex of inoculated fetal macaques (Fig. 6D). It is unknown whether those virally infected cells were infected prior to or subsequent to migration from the ventricular zone.
Intrauterine CMV sequelae may be the result of disruption of any of multiple events during CNS development. These include proliferation of neuronal stem cells, determination of neuronal or glial lineages, differentiation and migration of these cell types, interaction and connection between these cells, and selective removal of the neuronal cells by apoptosis. Disruption of vascular integrity may have also contributed to the extent of neuropathological outcomes. It has been suggested that CMV-induced microgyri result from insufficient blood supply possibly related to endothelial cell infection rather than direct damage of neurons (27). Further studies in the rhesus monkey model will be essential for understanding the mechanisms of neuropathogenesis. These investigations will be enhanced with the use of RhCMV-EGFP, since infected cells can be detected by FACS as early as 1 dpi. This will enable isolation of living infected cells by sorting or laser capture microdissection early after infection, as well as the identification of infected cells prior to development of overt CPE. In summary, experimental infection of rhesus macaques with RhCMV-EGFP will facilitate the study of pathogenesis in clinical settings especially relevant to HCMV, including intrauterine infection, immunodeficiency, and transplant-associated immunosuppression.
Acknowledgments
We thank Robert Cardiff and Stephen Barthold for comments and suggestions and Jason Ekert for assistance in providing brain sections. We also acknowledge the expert technical assistance of Abigail Spinner with flow cytometry.
This work was supported by NIH grants NS-36859, AI49342, and RR-00169.
REFERENCES
- 1.Ahn, K., A. Angulo, P. Ghazal, P. A. Peterson, Y. Yang, and K. Fruh. 1996. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. USA 93:10990-10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcendor, D. J., P. A. Barry, E. Pratt-Lowe, and P. A. Luciw. 1993. Analysis of the rhesus cytomegalovirus immediate-early gene promoter. Virology 194:815-821. [DOI] [PubMed] [Google Scholar]
- 3.Asher, D. M., C. J. Gibbs, Jr., D. J. Lang, D. C. Gajdusek, and R. M. Chanock. 1974. Persistent shedding of cytomegalovirus in the urine of healthy rhesus monkeys. Proc. Soc. Exp. Biol. Med. 145:794-801. [DOI] [PubMed] [Google Scholar]
- 4.Barkovich, A. J., and C. E. Lindan. 1994. Congenital cytomegalovirus infection of the brain: imaging analysis and embryologic considerations. Am. J. Neuroradiol. 15:703-715. [PMC free article] [PubMed] [Google Scholar]
- 5.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 6.Borst, E., and M. Messerle. 2000. Development of a cytomegalovirus vector for somatic gene therapy. Bone Marrow Transplant. 25:S80-S82. [DOI] [PubMed] [Google Scholar]
- 7.Chang, W. L., V. Kirchoff, G. S. Pari, and P. A. Barry. 2002. Replication of rhesus cytomegalovirus in life-expanded rhesus fibroblasts expressing human telomerase. J. Virol. Methods 104:135-146. [DOI] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus ie2 negatively regulates α gene expression via a short target sequence near the transcription start site. J. Virol. 65:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colacino, J. M., C. C. Flowers, J. Menna, D. J. O'Callaghan, and J. Staczek. 1989. Physical structure and molecular cloning of equine cytomegalovirus DNA. Virology 173:566-580. [DOI] [PubMed] [Google Scholar]
- 11.Conboy, T. J., R. F. Pass, S. Stagno, W. J. Britt, C. A. Alford, C. E. McFarland, and T. J. Boll. 1986. Intellectual development in school-aged children with asymptomatic congenital cytomegalovirus infection. Pediatrics 77:801-806. [PubMed] [Google Scholar]
- 12.Elliott, G., and P. O'Hare. 1999. Intercellular trafficking of VP22-GFP fusion proteins. Gene Ther. 6:149-151. [DOI] [PubMed] [Google Scholar]
- 13.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 1998. Expression of the enhanced green fluorescent protein by herpes simplex virus type 1 (HSV-1) as an in vitro or in vivo marker for virus entry and replication. J. Virol. Methods 75:151-160. [DOI] [PubMed] [Google Scholar]
- 14.Fowler, K. B., S. Stagno, R. F. Pass, W. J. Britt, T. J. Boll, and C. A. Alford. 1992. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N. Engl. J. Med. 326:663-667. [DOI] [PubMed] [Google Scholar]
- 15.Jones, T. R., and V. P. Muzithras. 1992. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J. Virol. 66:2541-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, T. R., V. P. Muzithras, and Y. Gluzman. 1991. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J. Virol. 65:5860-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, T. R., and L. Sun. 1997. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J. Virol. 71:2970-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, T. R., E. J. Wiertz, L. Sun, K. N. Fish, J. A. Nelson, and H. L. Ploegh. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchoff, V., S. Wong, S. St. Jeor, and G. S. Pari. 2002. Generation of a life-expanded rhesus monkey fibroblast cell line for the growth of rhesus rhadinovirus (RRV). Arch. Virol. 147:321-333. [DOI] [PubMed] [Google Scholar]
- 20.Kosugi, I., Y. Shinmura, H. Kawasaki, Y. Arai, R. Y. Li, S. Baba, and Y. Tsutsui. 2000. Cytomegalovirus infection of the central nervous system stem cells from mouse embryo: a model for developmental brain disorders induced by cytomegalovirus. Lab. Investig. 80:1373-1383. [DOI] [PubMed] [Google Scholar]
- 21.Lee, A. H., Y. S. Suh, J. H. Sung, S. H. Yang, and Y. C. Sung. 1997. Comparison of various expression plasmids for the induction of immune response by DNA immunization. Mol. Cells 7:495-501. [PubMed] [Google Scholar]
- 22.Li, R. Y., and Y. Tsutsui. 2000. Growth retardation and microcephaly induced in mice by placental infection with murine cytomegalovirus. Teratology 62:79-85. [DOI] [PubMed] [Google Scholar]
- 23.Liu, B., T. W. Hermiston, and M. F. Stinski. 1991. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 65:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockridge, K. M., G. Sequar, S. S. Zhou, Y. Yue, C. P. Mandell, and P. A. Barry. 1999. Pathogenesis of experimental rhesus cytomegalovirus infection. J. Virol. 73:9576-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.London, W. T., A. J. Martinez, S. A. Houff, W. C. Wallen, B. L. Curfman, R. G. Traub, and J. L. Sever. 1986. Experimental congenital disease with simian cytomegalovirus in rhesus monkeys. Teratology 33:323-331. [DOI] [PubMed] [Google Scholar]
- 26.Machold, R. P., E. J. Wiertz, T. R. Jones, and H. L. Ploegh. 1997. The HCMV gene products US11 and US2 differ in their ability to attack allelic forms of murine major histocompatibility complex (MHC) class I heavy chains. J. Exp. Med. 185:363-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marques Dias, M. J., G. Harmant-van Rijckevorsel, P. Landrieu, and G. Lyon.1984. Prenatal cytomegalovirus disease and cerebral microgyria: evidence for perfusion failure, not disturbance of histogenesis, as the major cause of fetal cytomegalovirus encephalopathy. Neuropediatrics 15:18-24. [DOI] [PubMed] [Google Scholar]
- 28.Marschall, M., M. Freitag, S. Weiler, G. Sorg, and T. Stamminger. 2000. Recombinant green fluorescent protein-expressing human cytomegalovirus as a tool for screening antiviral agents. Antimicrob. Agents Chemother. 44:1588-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy, M., D. Auger, and S. R. Whittemore. 2000. Human cytomegalovirus causes productive infection and neuronal injury in differentiating fetal human central nervous system neuroepithelial precursor cells. J. Hum. Virol. 3:215-228. [PubMed] [Google Scholar]
- 30.Mocarski, E. S. 1996. Cytomegaloviruses and their replication, p. 2447-2492. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 31.Pass, R. F., S. Stagno, G. J. Myers, and C. A. Alford. 1980. Outcome of symptomatic congenital cytomegalovirus infection: results of long-term longitudinal follow-up. Pediatrics 66:758-762. [PubMed] [Google Scholar]
- 32.Perlman, J. M., and C. Argyle. 1992. Lethal cytomegalovirus infection in preterm infants: clinical, radiological, and neuropathological findings. Ann. Neurol. 31:64-68. [DOI] [PubMed] [Google Scholar]
- 33.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poland, S. D., L. L. Bambrick, G. A. Dekaban, and G. P. Rice. 1994. The extent of human cytomegalovirus replication in primary neurons is dependent on host cell differentiation. J. Infect. Dis. 170:1267-1271. [DOI] [PubMed] [Google Scholar]
- 35.Rakic, P. 1988. Specification of cerebral cortical areas. Science 241:170-176. [DOI] [PubMed] [Google Scholar]
- 36.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinmura, Y., I. Kosugi, S. Aiba-Masago, S. Baba, L. R. Yong, and Y. Tsutsui. 1997. Disordered migration and loss of virus-infected neuronal cells in developing mouse brains infected with murine cytomegalovirus. Acta Neuropathol. 93:551-557. [DOI] [PubMed] [Google Scholar]
- 38.Sinzger, C., A. Grefte, B. Plachter, A. S. Gouw, T. H. The, and G. Jahn. 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J. Gen. Virol. 76:741-750. [DOI] [PubMed] [Google Scholar]
- 39.Sinzger, C., J. Knapp, K. Schmidt, M. Kahl, and G. Jahn. 1999. A simple and rapid method for preparation of viral DNA from cell associated cytomegalovirus. J. Virol. Methods 81:115-122. [DOI] [PubMed] [Google Scholar]
- 40.Sinzger, C., B. Plachter, A. Grefte, T. H. The, and G. Jahn. 1996. Tissue macrophages are infected by human cytomegalovirus in vivo. J. Infect. Dis. 173:240-245. [DOI] [PubMed] [Google Scholar]
- 41.Smith, B. N., B. W. Banfield, C. A. Smeraski, C. L. Wilcox, F. E. Dudek, L. W. Enquist, and G. E. Pickard. 2000. Pseudorabies virus expressing enhanced green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. USA 97:9264-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, R. L., D. L. Traul, J. Schaack, G. H. Clayton, K. J. Staley, and C. L. Wilcox. 2000. Characterization of promoter function and cell-type-specific expression from viral vectors in the nervous system. J. Virol. 74:11254-11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speck, P., and R. Longnecker. 1999. Epstein-Barr virus (EBV) infection visualized by EGFP expression demonstrates dependence on known mediators of EBV entry. Arch. Virol. 144:1123-1137. [DOI] [PubMed] [Google Scholar]
- 44.Stagno, S., R. F. Pass, G. Cloud, W. J. Britt, R. E. Henderson, P. D. Walton, D. A. Veren, F. Page, and C. A. Alford. 1986. Primary cytomegalovirus infection in pregnancy. Incidence, transmission to fetus, and clinical outcome. JAMA 256:1904-1908. [PubMed] [Google Scholar]
- 45.Tarantal, A. F. 1990. Interventional ultrasound in pregnant macaques: embryonic/fetal applications. J. Med. Primatol. 19:47-58. [PubMed] [Google Scholar]
- 46.Tarantal, A. F., and S. E. Gargosky. 1995. Characterization of the insulin-like growth factor (IGF) axis in the serum of maternal and fetal macaques (Macaca mulatta and Macaca fascicularis). Growth Regul. 5:190-198. [PubMed] [Google Scholar]
- 47.Tarantal, A. F., and A. G. Hendrickx. 1988. Characterization of prenatal growth and development in the crab-eating macaque (Macaca fascicularis) by ultrasound. Anat. Rec. 222:177-184. [DOI] [PubMed] [Google Scholar]
- 48.Tarantal, A. F., and A. G. Hendrickx. 1988. Use of ultrasound for early pregnancy detection in the rhesus and cynomolgus macaque (Macaca mulatta and Macaca fascicularis). J. Med. Primatol. 17:105-112. [PubMed] [Google Scholar]
- 49.Tarantal, A. F., M. L. Marthas, S. E. Gargosky, M. Otysula, M. B. McChesney, C. J. Miller, and A. G. Hendrickx. 1995. Effects of viral virulence on intrauterine growth in SIV-infected fetal rhesus macaques (Macaca mulatta). J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10:129-138. [DOI] [PubMed] [Google Scholar]
- 50.Tarantal, A. F., M. S. Salamat, W. J. Britt, P. A. Luciw, A. G. Hendrickx, and P. A. Barry. 1998. Neuropathogenesis induced by rhesus cytomegalovirus in fetal rhesus monkeys (Macaca mulatta). J. Infect. Dis. 177:446-450. [DOI] [PubMed] [Google Scholar]
- 51.Tomazin, R., J. Boname, N. R. Hegde, D. M. Lewinsohn, Y. Altschuler, T. R. Jones, P. Cresswell, J. A. Nelson, S. R. Riddell, and D. C. Johnson. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 5:1039-1043. [DOI] [PubMed] [Google Scholar]
- 52.van den Pol, A. N., E. Mocarski, N. Saederup, J. Vieira, and T. J. Meier. 1999. Cytomegalovirus cell tropism, replication, and gene transfer in brain. J. Neurosci. 19:10948-10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van den Pol, A. N., J. Vieira, D. D. Spencer, and J. G. Santarelli. 2000. Mouse cytomegalovirus in developing brain tissue: analysis of 11 species with GFP-expressing recombinant virus. J. Comp. Neurol. 427:559-580. [DOI] [PubMed] [Google Scholar]
- 54.Vogel, P., B. J. Weigler, H. Kerr, A. G. Hendrickx, and P. A. Barry. 1994. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab. Anim. Sci. 44:25-30. [PubMed] [Google Scholar]